A variant of the Aurora A kinase domain in which both surface cysteines are mutated to alanine generates crystals that diffract to higher resolution than the equivalent crystals produced using wild-type protein. The structure confirms that single phosphorylation on Thr288 is insufficient to stabilize a conformation of the activation loop in a ‘fully active’ state.
Keywords: protein kinase, phosphorylation, activation
Abstract
Aurora A is a Ser/Thr protein kinase that functions in cell-cycle regulation and is implicated in cancer development. During mitosis, Aurora A is activated by autophosphorylation on its activation loop at Thr288. The Aurora A catalytic domain (amino acids 122–403) expressed in Escherichia coli autophosphorylates on two activation-loop threonine residues (Thr288 and Thr287), whereas a C290A,C393A double point mutant of the Aurora A catalytic domain autophosphorylates only on Thr288. The structure of the complex of this mutant with ADP and magnesium was determined to 2.1 Å resolution using molecular replacement. This is an improvement on the existing 2.75 Å resolution structure of the equivalent wild-type complex. The structure confirms that single phosphorylation of the activation loop on Thr288 is insufficient to stabilize a ‘fully active’ conformation of the activation loop in the absence of binding to TPX2.
1. Introduction
During mitosis, there is a peak in protein phosphorylation that regulates changes in the appearance of cells in readiness for chromosome segregation (Olsen et al., 2010 ▶). Ser/Thr protein kinases of the Aurora family contribute to mitotic protein phosphorylation, particularly of proteins associated with microtubules such as TPX2, TACC3 and NUSAP (Sardon et al., 2010 ▶). Aurora A localizes to centrosomes and spindle microtubules and regulates mitotic entry, centrosome maturation and bipolar spindle assembly (Carmena & Earnshaw, 2003 ▶; Barr & Gergely, 2007 ▶; Nikonova et al., 2012 ▶). Aurora A is frequently dysregulated in cancer and the gene is located at 20q13, a region of the genome that is amplified in breast cancer (Asteriti et al., 2010 ▶). The potential of Aurora A inhibition as a cancer-treatment strategy is currently under investigation in clinical trials (Manchado et al., 2012 ▶).
Aurora A consists of an N-terminal region of unknown structure and a C-terminal kinase catalytic domain. The catalytic activity of Aurora A is regulated by phosphorylation of its activation loop within the kinase domain at Thr288/Thr287 and through the binding of protein partners such as TPX2 (Bayliss et al., 2003 ▶). In mitosis, Aurora A activation is promoted by TPX2, which stimulates Aurora A autophosphorylation on Thr288 (Eyers et al., 2003 ▶; Dodson et al., 2013 ▶). Phosphorylation of Thr287 can also stimulate kinase activity, and is implicated in a nonmitotic role of Aurora A (Mori et al., 2009 ▶; Rowan et al., 2013 ▶). Previous structures of the catalytic domain have captured the activation loop in several conformations dependent on the phosphorylation state, the presence or absence of TPX2 and the nature of the ligand bound to the active site (Cheetham et al., 2002 ▶; Bayliss et al., 2003 ▶; Dodson et al., 2010 ▶; Gustafson et al., 2014 ▶).
We have explored the site-specific labelling of Aurora A through chemical modification of cysteines (Rowan et al., 2013 ▶). To facilitate this process, we made a variant of the Aurora A catalytic domain that lacks surface-exposed cysteines. This involved the mutation of Cys290 and Cys393 to alanine, producing a protein that we call C290A:C393A Aurora A. This protein has a slightly reduced catalytic activity relative to the wild-type catalytic domain, quantified as a factor of two decrease in k cat/K M(ATP) (Rowan et al., 2013 ▶). Residue 393 of Aurora A resides in a disordered region at the very C-terminus of the protein and is unlikely to affect the activity. However, residue 290 resides in the activation loop and is therefore most likely to have an impact on activity. In order to characterize the structural consequences of these mutations on the active kinase, we expressed and purified C290A:C393A Aurora A, co-crystallized the protein with ADP/MgCl2 and determined its structure.
2. Materials and methods
2.1. Macromolecule production
C290A:C393A Aurora A was generated in a previous study (Rowan et al., 2013 ▶). The protein was expressed and purified as described previously (Bayliss et al., 2003 ▶). Chelating Sepharose was used instead of TALON resin according to the manufacturer’s instructions (GE Healthcare). The protein was subjected to size-exclusion chromatography using a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) equilibrated with 20 mM Tris pH 7.0, 200 mM NaCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 10% glycerol as a final purification step. Macromolecule-production information is summarized in Table 1 ▶.
Table 1. Macromolecule-production information.
| Source organism | Homo sapiens |
| Expression vector | pET-30-TEV |
| Expression host | Escherichia coli |
| Complete amino-acid sequence of the construct produced† | MHHHHHHSSGLVPRGSGMKETAAAKFEENLYFNGAMESKKRQWALEDFEIGRPLGKGKFGNVYLAREKQSKFILALKVLFKAQLEKAGVEHQLRREVEIQSHLRHPNILRLYGYFHDATRVYLILEYAPLGTVYRELQKLSKFDEQRTATYITELANALSYCHSKRVIHRDIKPENLLLGSAGELKIADFGWSVHAPSSRRTTLAGTLDYLPPEMIEGRMHDEKVDLWSLGVLCYEFLVGKPPFEANTYQETYKRISRVEFTFPDFVTEGARDLISRLLKHNPSQRPMLREVLEHPWITANSSKPSNAQNKESASKQS |
The TEV cleavage site is underlined.
2.2. Crystallization
C290A:C393A Aurora A was concentrated to 16.5 mg ml−1 and incubated with 5 mM ADP/MgCl2 on ice for 1 h. Crystallization screens were set up in 96-well sitting-drop MRC plates using a Mosquito LCP crystallization robot (TTP Labtech) and incubated at 295 K. Crystals were obtained after 1–2 d in a variety of conditions. Crystals produced using 0.1 M Tris–HCl pH 8.5, 0.2 M MgCl2, 0.5 M NaCl, 32.5% PEG 3350 as a precipitant were cryoprotected in the crystallization solution with the addition of 30% glycerol and were flash-cooled to completely eliminate ice-crystal formation. Crystallization information is summarized in Table 2 ▶.
Table 2. Crystallization.
| Method | Vapour diffusion |
| Plate type | Sitting-drop MRC 96-well |
| Temperature (K) | 295 |
| Protein concentration (mgml1) | 16.5 |
| Buffer composition of protein solution | 20mM Tris pH 7.0, 200mM NaCl, 5mM MgCl2, 5mM -mercaptoethanol, 10% glycerol |
| Composition of reservoir solution | 0.1M TrisHCl pH 8.5, 0.2M MgCl2, 0.5M NaCl, 32.5% PEG 3350 |
2.3. Data collection and processing
X-ray diffraction data were collected from a single crystal on beamline I04-1 at Diamond Light Source (DLS), Oxford, England. Data processing was carried out using the xia2 automated data-reduction platform (Winter, 2010 ▶) available at DLS. Data-collection and processing statistics are summarized in Table 3 ▶.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | I04-1, DLS |
| Wavelength () | 0.92 |
| Temperature (K) | 100 |
| Space group | P6122 |
| a, b, c () | 80.95, 80.95, 174.53 |
| , , () | 90.00, 90.00, 120.00 |
| Resolution range () | 70.102.10 (2.152.10) |
| Total No. of reflections | 20539 |
| Completeness (%) | 99.9 (99.6) |
| Multiplicity | 19.2 (17.3) |
| I/(I) | 36.8 (4.60) |
| R merge (%) | 5.0 (67.0) |
| Overall B factor from Wilson plot (2) | 42.09 |
2.4. Structure solution and refinement
The structure of C290A:C393A Aurora A was solved by molecular replacement using Phaser-MR (McCoy et al., 2007 ▶) with the structure of Aurora A 122–403 (PDB entry 1ol7; Bayliss et al., 2003 ▶) as a model. The structure was solved and rigid-body refined with phenix.refine (Adams et al., 2010 ▶; Afonine et al., 2012 ▶). Model building was carried out with Coot (Emsley et al., 2010 ▶). MolProbity (Chen et al., 2010 ▶) was used for structure validation, including Ramachandran plot analysis. Structure-solution and refinement statistics are summarized in Table 4 ▶.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range () | 70.102.10 |
| No. of reflections, working set | 20525 |
| Final R cryst (%) | 19.98 |
| Final R free (%) | 23.75 |
| No. of non-H atoms | |
| Protein | 2141 |
| Ligand + ion | 45 |
| Water | 69 |
| Total | 2255 |
| R.m.s. deviations | |
| Bonds () | 0.008 |
| Angles () | 1.223 |
| Average B factors (2) | |
| Protein | 37.59 |
| Ligand + ion | 36.62 |
| Water | 37.65 |
| MolProbity analysis | |
| All-atom clashscore | 4.64 |
| Rotamer outliers (%) | 5.36 |
| Favoured regions (%) | 96.96 |
| Outliers (%) | 0 |
| MolProbity score | 1.98 |
2.5. Mass spectrometry
Samples of wild-type and C290A:C393A Aurora A were excised from Instant Blue-stained gels and subjected to mass spectrometry.
LC-MS/MS was carried out using an RSLCnano HPLC system (Thermo Scientific) and an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). Samples were loaded at 0.25 ml min−1 onto a Widepore C4 (butyl) 4 × 2 mm internal diameter Security Guard cartridge (Phenomenx, UK). The protein was desalted for 10 min on-column in the loading buffer (0.1% formic acid) before elution using a 10 min linear gradient from 3 to 96% buffer B (80% acetonitrile/0.1% formic acid). The output of the column was sprayed directly into the H-ESI2 electrospray ion source of the mass spectrometer maintained at 6 kV. The LTQ Orbitrap Velos was set to acquire a ten-microscan FTMS scan event at 100 000 resolution over the m/z range 700–2000 in positive-ion mode. The maximum injection time for MS was 50 ms and the AGC target setting was 1 × 105. Accurate calibration of the FTMS scan was achieved using a background ion lock mass for C6H10O14S3 (401.922718 Da). Protein charge-state distributions were deconvoluted using the Xtract software within the Xcalibur program (v.2.1.0.1139; Thermo Scientific).
3. Results and discussion
Crystals of phosphorylated C290A:C393A Aurora A diffracted to 2.1 Å resolution, a marked improvement on the usual diffraction limit that we obtain for phosphorylated Aurora A (∼3.0 Å and at best 2.75 Å; PDB entry 1ol7; Bayliss et al., 2003 ▶), with both exhibiting the same crystal form and space group (P6122). Structures of phosphorylated Aurora A at resolutions better than 2.75 Å have been obtained in the presence of TPX2 (2.5 Å; PDB entry 1ol5; space group P212121; Bayliss et al., 2003 ▶), potent Aurora A inhibitors such as AT9283 (2.71 Å; PDB entry 2w1g; space group P6122; Howard et al., 2009 ▶) and MLN8054 (2.4 Å; PDB entry 2wtv; space group P31; Dodson et al., 2010 ▶), or a combination of TPX2 and an inhibitor (2.3 Å; PDB entry 3e5a; space group C2221; Zhao et al., 2008 ▶). However, the phosphorylated C290A:C393A Aurora A crystals that we obtained routinely diffracted to higher resolution than was obtained for these structures, despite the absence of the stabilizing factors TPX2 or inhibitors.
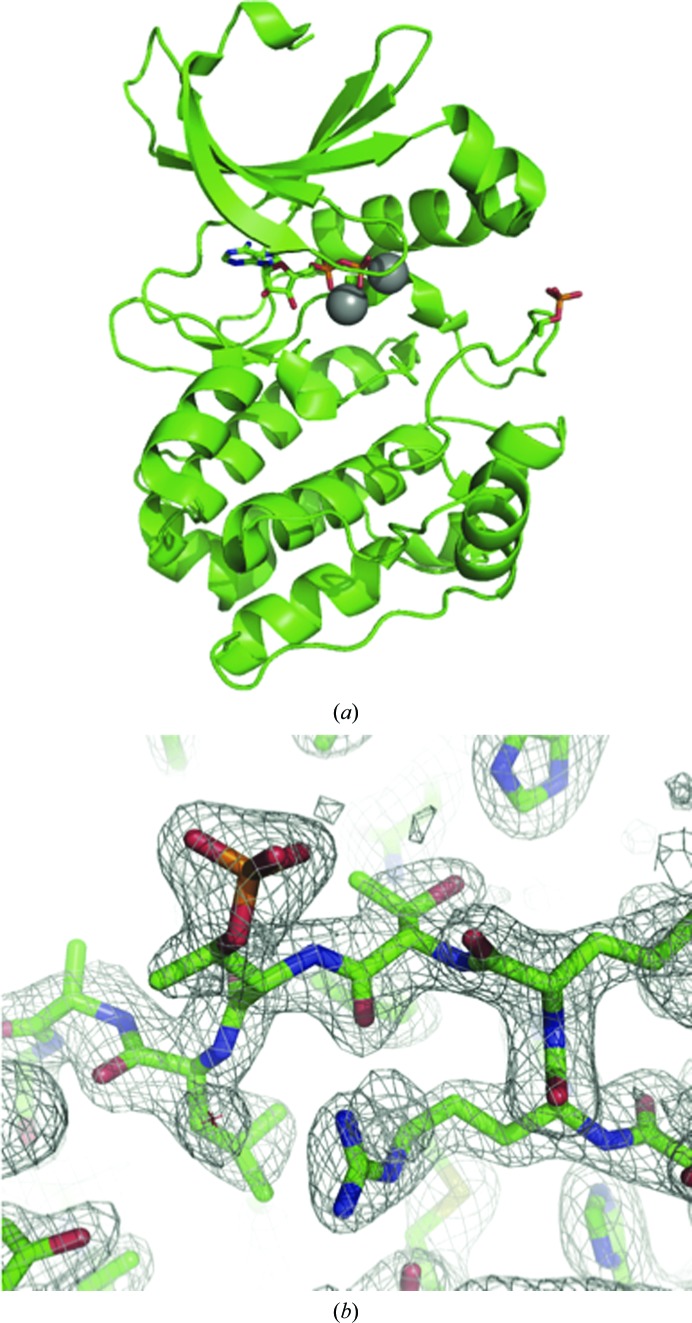
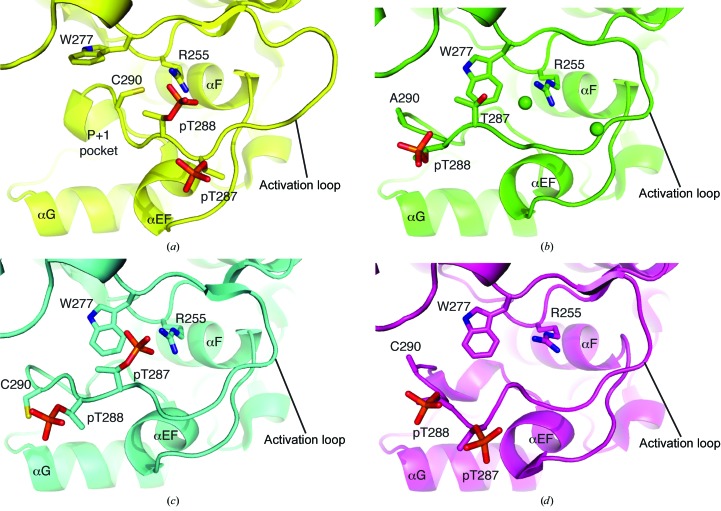
The final C290A:C393A Aurora A model comprises residues 127–391, ADP and two magnesium ions modelled in the active site (Fig. 1 ▶ a). The activation loop was highly ordered and could be modelled unambiguously (Fig. 1 ▶ b). The side chain of residue 290 was solvent-exposed and made no contacts. In the structure of Aurora A in a fully active conformation bound to TPX2 and ADP, the side chain of Cys290 is buried, at least in part, by the side chain of Trp277 (Fig. 2 ▶ a). This interaction would be decreased by the mutation of Cys290 to alanine, which might destabilize the active conformation of C290A:C393A Aurora A relative to the wild type, resulting in the reduced activity of the mutant protein. We examined the activation loop more carefully to investigate the basis of the observed conformation.
Figure 1.
Structure of C290A:C393A Aurora A. (a) The main chain of C290A:C393A Aurora A is shown as a cartoon, with ADP and the side chain of phospho-Thr288 as sticks and magnesium ions as grey spheres. (b) The activation loop of C290A:C393A Aurora A is ordered and the tracing of the chain is unambiguous. The wire mesh shows a 2mF o − DF c electron-density map contoured at 1.0σ in the vicinity of Thr287 and phospho-Thr288.
Figure 2.
Comparison of activation-loop conformations of phosphorylated Aurora A. (a) Aurora A–TPX2 complex (PDB entry 1ol5; Bayliss et al., 2003 ▶). The P+1 pocket is a key feature of the recognition site for protein substrates, the formation of which requires TPX2 binding and phosphorylation of Thr288. (b) C290A:C393A Aurora A (PDB entry 4ceg). Chloride ions are shown as green spheres. (c) V174M Aurora A (PDB entry 4bn1; Rowan et al., 2013 ▶). (d) Aurora A (PDB entry 1ol7; Bayliss et al., 2003 ▶). Three α-helices are marked as reference points to aid in the visualization of conformational changes in the activation loop and Trp277.
Phosphorylation of Thr288 in the activation loop was clear, but there was no evidence for phosphorylation of Thr287 (Fig. 1 ▶ b). We confirmed the phosphorylation state of C290A:C393A Aurora A by mass spectrometry (Supplementary Fig. S1). This contrasted with preparations of wild-type Aurora A, which uniformally have full phosphorylation on Thr287 and Thr288 (Dodson & Bayliss, 2012 ▶). Although Thr288 was the only phosphorylated activation-loop residue, the phospho-group was exposed to solvent and did not form the expected interaction with Arg255 found in active kinase structures such as the Aurora A–TPX2 complex (Fig. 2 ▶ b). The side chain of Thr287 is positioned such that if it were phosphorylated the phospho-group would interact with Arg255. This arrangement has previously been observed in the structure of the V174M mutant of Aurora A (Fig. 2 ▶ c; Rowan et al., 2013 ▶). However, in the C290A:C393A Aurora A structure a chloride ion sits in the pocket close to Arg255, neutralizing the charge on the arginine side chain.
Phosphorylation of the activation loop of a subset of protein kinases is required for them to adopt a fully active conformation capable of binding substrate (Bayliss et al., 2012 ▶; Endicott et al., 2012 ▶; Johnson & Lewis, 2001 ▶). To date, this conformation of Aurora A has only been observed in complexes of phosphorylated Aurora A with TPX2 (Bayliss et al., 2003 ▶; Zhao et al., 2008 ▶). Previous structures had phosphorylation on both Thr287 and Thr288, which might affect the conformation of the activation loop (Figs. 2 ▶ c and 2 ▶ d). The structure of C290A:C393A Aurora A provides further evidence that phosphorylation on Thr288 alone is insufficient for the kinase to adopt a fully active conformation. This is fully consistent with kinetic data showing that optimal substrate binding requires binding of TPX2 (Anderson et al., 2007 ▶). Our data suggest that caution should be applied when using a monophosphospecific (pThr288) antibody as a marker for Aurora A activity. We are currently investigating the conformation of the activation loop of Aurora A in solution, aided by the availability of the stable C290A:C393A Aurora A protein. The crystal structure of this mutant will be a useful reference point for these studies. The C290A:C393A Aurora A protein yielded crystals that reproducibly diffracted to ∼2.1 Å resolution. This improved crystallization system will be valuable in the ongoing development of Aurora A inhibitors for the treatment of cancer (Bebbington et al., 2009 ▶; Dodson et al., 2010 ▶; Bavetsias et al., 2013 ▶).
Supplementary Material
PDB reference: C290A:C393A Aurora A, 4ceg
Mass spectrometry of Aurora A.. DOI: 10.1107/S2053230X15002290/no5070sup1.pdf
Acknowledgments
We thank the beamline staff at Diamond Light Source beamline I04-1. We thank Dr Andrew R. Bottrill (University of Leicester, PNACL Proteomics Facility) for his help with mass spectrometry. Research in the Bayliss laboratory is funded by Cancer Research UK (grant No. C24461/A13231).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Anderson, K., Yang, J., Koretke, K., Nurse, K., Calamari, A., Kirkpatrick, R. B., Patrick, D., Silva, D., Tummino, P. J., Copeland, R. A. & Lai, Z. (2007). Biochemistry, 46, 10287–10295. [DOI] [PubMed]
- Asteriti, I. A., Rensen, W. M., Lindon, C., Lavia, P. & Guarguaglini, G. (2010). Biochim. Biophys. Acta, 1806, 230–239. [DOI] [PubMed]
- Barr, A. R. & Gergely, F. (2007). J. Cell Sci. 120, 2987–2996. [DOI] [PubMed]
- Bavetsias, V. et al. (2013). J. Med. Chem. 56, 9122–9135. [DOI] [PMC free article] [PubMed]
- Bayliss, R., Fry, A., Haq, T. & Yeoh, S. (2012). Open Biol. 2, 120136. [DOI] [PMC free article] [PubMed]
- Bayliss, R., Sardon, T., Vernos, I. & Conti, E. (2003). Mol. Cell, 12, 851–862. [DOI] [PubMed]
- Bebbington, D. et al. (2009). Bioorg. Med. Chem. Lett. 19, 3586–3592. [DOI] [PubMed]
- Carmena, M. & Earnshaw, W. C. (2003). Nature Rev. Mol. Cell Biol. 4, 842–854. [DOI] [PubMed]
- Cheetham, G. M. T., Knegtel, R. M. A., Coll, J. T., Renwick, S. B., Swenson, L., Weber, P., Lippke, J. A. & Austen, D. A. (2002). J. Biol. Chem. 277, 42419–42422. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Dodson, C. A. & Bayliss, R. (2012). J. Biol. Chem. 287, 1150–1157. [DOI] [PMC free article] [PubMed]
- Dodson, C. A., Kosmopoulou, M., Richards, M. W., Atrash, B., Bavetsias, V., Blagg, J. & Bayliss, R. (2010). Biochem. J. 427, 19–28. [DOI] [PubMed]
- Dodson, C. A., Yeoh, S., Haq, T. & Bayliss, R. (2013). Sci. Signal. 6, ra54. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Endicott, J. A., Noble, M. E. M. & Johnson, L. N. (2012). Annu. Rev. Biochem. 81, 587–613. [DOI] [PubMed]
- Eyers, P. A., Erikson, E., Chen, L. G. & Maller, J. L. (2003). Curr. Biol. 13, 691–697. [DOI] [PubMed]
- Gustafson, W. C., Meyerowitz, J. G., Nekritz, E. A., Chen, J., Benes, C., Charron, E., Simonds, E. F., Seeger, R., Matthay, K. K., Hertz, N. T., Eilers, M., Shokat, K. M. & Weiss, W. A. (2014). Cancer Cell, 26, 414–427. [DOI] [PMC free article] [PubMed]
- Howard, S. et al. (2009). J. Med. Chem. 52, 379–388.
- Johnson, L. N. & Lewis, R. J. (2001). Chem. Rev. 101, 2209–2242. [DOI] [PubMed]
- Manchado, E., Guillamot, M. & Malumbres, M. (2012). Cell Death Differ. 19, 369–377. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mori, D., Yamada, M., Mimori-Kiyosue, Y., Shirai, Y., Suzuki, A., Ohno, S., Saya, H., Wynshaw-Boris, A. & Hirotsune, S. (2009). Nature Cell Biol. 11, 1057–1068. [DOI] [PubMed]
- Nikonova, A. S., Astsaturov, I., Serebriiskii, I. G., Dunbrack, R. L. Jr & Golemis, E. A. (2012). Cell. Mol. Life Sci. 70, 661–687. [DOI] [PMC free article] [PubMed]
- Olsen, J. V., Vermeulen, M., Santamaria, A., Kumar, C., Miller, M. L., Jensen, L. J., Gnad, F., Cox, J., Jensen, T. S., Nigg, E. A., Brunak, S. & Mann, M. (2010). Sci. Signal. 3, ra3. [DOI] [PubMed]
- Rowan, F. C., Richards, M., Bibby, R. A., Thompson, A., Bayliss, R. & Blagg, J. (2013). ACS Chem. Biol. 8, 2184–2191. [DOI] [PMC free article] [PubMed]
- Sardon, T., Pache, R. A., Stein, A., Molina, H., Vernos, I. & Aloy, P. (2010). EMBO Rep. 11, 977–984. [DOI] [PMC free article] [PubMed]
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Zhao, B., Smallwood, A., Yang, J., Koretke, K., Nurse, K., Calamari, A., Kirkpatrick, R. B. & Lai, Z. (2008). Protein Sci. 17, 1791–1797. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: C290A:C393A Aurora A, 4ceg
Mass spectrometry of Aurora A.. DOI: 10.1107/S2053230X15002290/no5070sup1.pdf