Abstract
Acute muscle injury and physiological stress from chronic muscle diseases and aging lead to impairment of skeletal muscle function. This raises the question of whether p53, a cellular stress sensor, regulates muscle tissue repair under stress conditions. By investigating muscle differentiation in the presence of genotoxic stress, we discovered that p53 binds directly to the myogenin promoter and represses transcription of myogenin, a member of the MyoD family of transcription factors that plays a critical role in driving terminal muscle differentiation. This reduction of myogenin protein is observed in G1-arrested cells and leads to decreased expression of late but not early differentiation markers. In response to acute genotoxic stress, p53-mediated repression of myogenin reduces post-mitotic nuclear abnormalities in terminally differentiated cells. This study reveals a mechanistic link previously unknown between p53 and muscle differentiation, and suggests new avenues for managing p53-mediated stress responses in chronic muscle diseases or during muscle aging.
The tumor suppressor p53 promotes cell cycle arrest or apoptosis in response to diverse stress signals such as DNA damage, thus preventing propagation of genetically compromised cells.1, 2, 3 Among the diverse functions attributed to p53, a growing body of evidence supports its role in regulation of differentiation and maintenance of cellular function and integrity.1, 4, 5, 6, 7 For example, p53 represses Nanog to maintain genetic stability of the stem cell pool by promoting differentiation of mouse embryonic stem cells (mESCs) after DNA damage.6
Skeletal muscle differentiation, a key step during muscle tissue formation, is orchestrated by the MyoD family of myogenic regulatory factors (MRFs). MyoD determines the myogenic lineage, whereas myogenin, a member of the MRF family, functions downstream of MyoD and plays a critical role in driving terminal differentiation as myogenin-null mice show a lethal deficiency of differentiated skeletal muscle.8, 9, 10, 11, 12, 13 The dynamic differentiation program of skeletal muscle is characterized by the orderly expression of genes and structural changes that can be recapitulated in vitro, as myogenic cells undergo cell cycle withdrawal and express early and then late differentiation genes with the formation of mononucleated myocytes to elongated multinucleated myotubes.8, 9, 14 Under normal unstressed conditions, p53 has been shown to promote muscle differentiation in vitro.15, 16, 17, 18, 19, 20 In contrast, under stress-associated conditions such as inflammation, chronic exposure to double-strand DNA breaks, and aging, enhanced p53 activity has been shown to correlate with skeletal muscle atrophy in vivo.21, 22, 23, 24, 25 In addition, it has been shown that p53 and its downstream effectors are required for an inflammatory cytokine-mediated inhibition of myogenic differentiation in vitro.22 Intriguingly, cell cycle arrest induced by acute genotoxic stress is associated with decreased myogenic differentiation in contrast to the cell cycle arrest that promotes differentiation under non-stressed conditions.26 This seminal observation suggests the existence of alternative mechanisms regulating differentiation in the presence of stress.
Despite evidence pointing to a mechanistic link between p53-mediated stress responses and impaired myogenic differentiation,21, 22, 23 there is little biochemical evidence associating p53 directly with regulation of key MRFs, particularly the MyoD family of transcription factors. Here we report a mechanistic link previously unknown between p53 regulation and muscle differentiation by showing that p53 directly represses myogenin and regulates muscle differentiation in response to genotoxic stress.
Results
p53 transcriptionally represses myogenin
We first used human embryonal rhabdomyosarcoma cells (RD cells)27, 28 as a model system to study whether p53 has a direct role in regulating myogenic differentiation. RD cells carry a homozygous p53 Arg248Trp mutation,29, 30 which renders the protein defective in DNA binding.3 Wild-type p53 function was restored in RD cells by expressing a tamoxifen-inducible p53-estrogen receptor (ER) fusion (Figure 1a). By tuning the translation level31 of the p53-ER (Supplementary Figures 1 and 2a–c), we showed that restoration of wild-type p53 over a range of expression levels did not result in apoptotic and/or necrotic cells32 in RD cells (Supplementary Figures 2d–g). Transcriptional activity of the p53-ER was validated by measuring activation of p53 target genes, p21 and Mdm2, (Figure 1b, Supplementary Table 1, and Supplementary Figure 3a) and cell cycle arrest in RD cells (Supplementary Figures 3b and c).
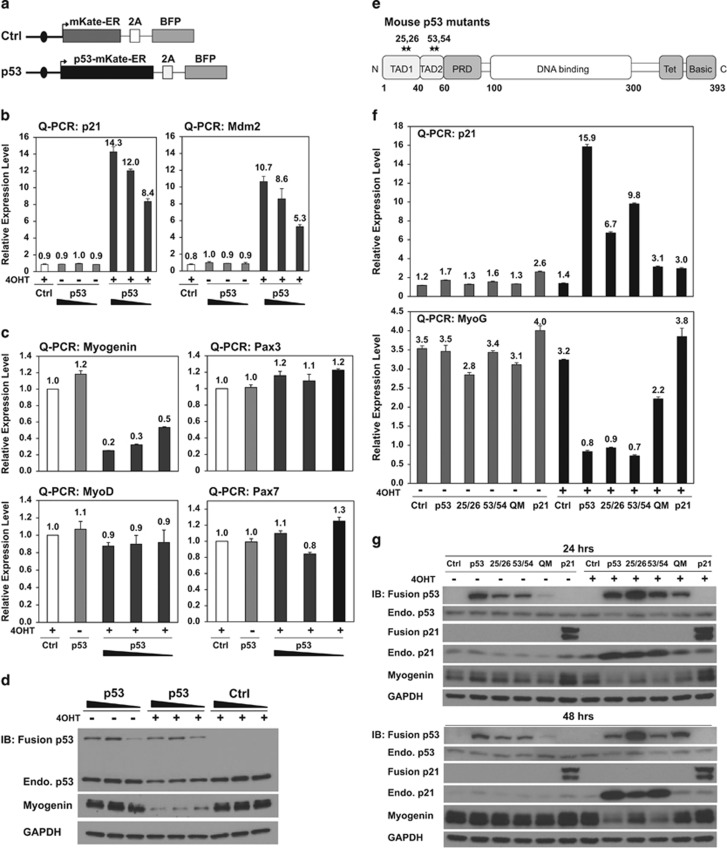
Figure 1.
p53 transcriptionally represses myogenin. (a) Schematics of the control- and p53-ER fusion constructs. Protein translation levels are modulated by synthetic upstream open reading frames as shown by black ovals (details in Supplementary Figure 1). Arrow: transcription start site (TSS); mKate: red fluorescent protein; 2A: ribosomal slippage sites; BFP: blue fluorescent protein. (b) Transcriptional activity of p53-ER fusion constructs was determined by measuring expression of p53-activated target genes. Nuclear localization of the ER fusion proteins was induced by 4-hydroxytamoxifen (4OHT). Ethanol treatment served as the uninduced control. Quantitative PCR (Q-PCR) graph represents transcription level relative to that of the control in the absence of 4OHT induction. The fusion proteins were expressed from high to low levels as represented by the black gradient bars here and in all following figures. (c) Q-PCR analysis shows dose-dependent repression of myogenin in response to ectopic p53 in RD cells. (d) Immunoblotting (IB) shows corresponding reduction of myogenin protein upon p53 activation. (e) Schematic of p53 mutants, p532526, p535354, and p5325265354. TAD: transcriptional activation domain; PRD: proline-rich domain; DNA binding: DNA-binding domain; Tet: tetramerization domain; Basic: basic domain. (f) Q-PCR analyses show p21 activation and myogenin repression in response to ectopic p53 and its mutants at 48 h of 4OHT induction. Ectopic expression of mouse p21 served as a negative control for myogenin mRNA level in RD cells. Quadruple mutation (QM): p5325265354. (g) IBs show p21 activation and myogenin repression in response to ectopic p53 and its mutants at 48 h of 4OHT induction. Ectopic p21 in RD cells led to accumulation of myogenin at the protein level and served as a positive control for IB. QM: p5325265354
Next we examined the gene expression of the following factors in RD cells upon p53 activation: MRFs8, 9, 14 (MyoD and myogenin), muscle stem cell markers33, 34 (Pax3, Pax7, and cMet), and cancer stem cell-like markers35 (Oct4, Nanog, Sox2, and CD133). We did not observe significant changes in expression of MyoD, Pax genes, and most of the cancer stem cell-like markers (Figure 1c and Supplementary Figure 4a). However, p53 repressed myogenin transcription in a dosage-dependent manner (Figure 1c), which correlated with a corresponding reduction in protein levels (Figure 1d and Supplementary Figure 4b).
To further confirm that p53 transcriptionally represses myogenin, we generated three previously characterized mutants in the N-terminal transcriptional activation domains (TAD): p532526, p535354, and p5325265354 (Figure 1e). Previous microarray analysis of mouse embryonic fibroblasts (MEFs) derived from the knockin mutant mouse strains revealed that p532526 shows severely impaired transactivation activity, p535354 retains the wild-type transactivation function, whereas p5325,26,53,54 is a transactivation-dead mutant.36 The extent of myogenin repression by p53 mutants echoed the transactivation activity observed in MEFs: p535354 repressed myogenin as strongly as wild-type p53, whereas p5325265354 demonstrated compromised repression of myogenin. p532526 exhibited a relatively intermediate level of repression (Figures 1f and g). Therefore, p53 transcriptionally represses myogenin, as demonstrated by the repression following restoration of wild-type p53 in RD cells and by the attenuated repression following expression of p53 N-terminal mutants.
The reduction of myogenin protein on p53 activation is independent of cell cycle phases
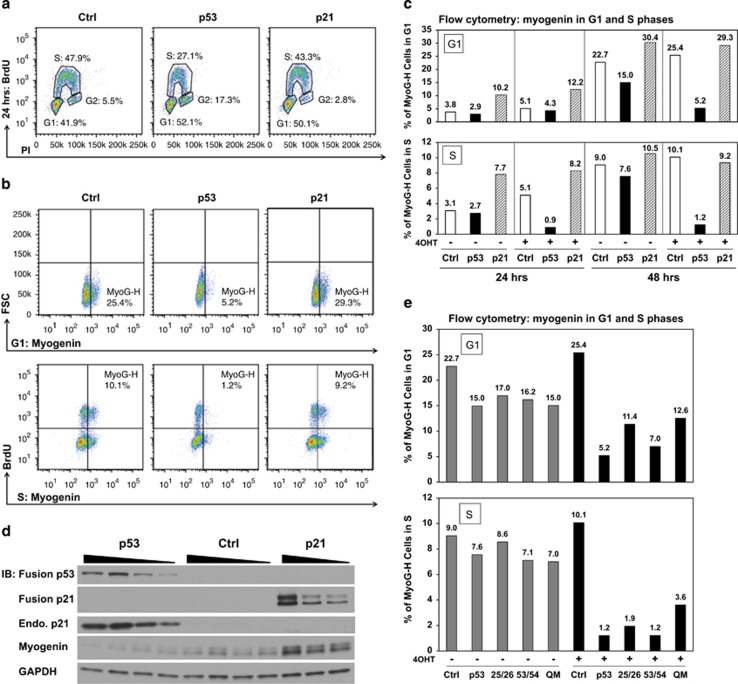
It is believed that proliferating myoblasts exit the G1 phase of the cell cycle in order to terminally differentiate.37, 38 Because of its critical role in driving terminal differentiation, we quantified myogenin expression and bromodeoxyuridine (BrdU) incorporation by flow cytometry analysis to determine whether p53-mediated repression of myogenin occurs at a specific phase of the cell cycle (Figure 2a). Owing to the heterogeneous expression of myogenin observed in RD muscle cancer cells,39 we defined quadrants to denote a myogenin-high (MyoG-H) population in either G1 or S phase (Figure 2b). We observed a prominent reduction of MyoG-H cells in both the G1 and S phases in response to p53 expression (Figures 2b and c). It has been demonstrated that upregulation of both p21 and myogenin is required for establishing the state of irreversible cell cycle withdrawal and terminal differentiation.11, 16, 40 In line with this notion, ectopic expression of p21 in RD cells induced G1 arrest (Figures 2a and c), accumulation of myogenin protein (Figure 2d), and enhanced late-stage differentiation (shown in Figure 5d). In contrast, ectopic p53 in RD cells repressed myogenin protein in G1-arrested cells (G1: Myogenin in Figure 2c) despite a strong activation of endogenous p21 (Figure 2d). This result indicates that repression of myogenin is p53-specific and independent of the G1 cell cycle arrest that promotes differentiation. This is further supported by the observation that p5325265354 exhibited a severe impairment in repression of myogenin in both G1 and S phases, even though this mutant did not affect either cell cycle phase distribution (Figure 2e and Supplementary Figures 5a and b).
Figure 2.
The reduction of myogenin protein in response to p53 activation is independent of cell cycle phases. (a) Distribution of cell cycle phases was quantified in RD cells ectopically expressing control, p53, or p21. The flow cytometry plot shows G1, S, and G2 phases based on BrdU and PI staining. (b) Reduction of myogenin protein in each cell cycle phase was evaluated by co-immunostaining of myogenin and BrdU, followed by flow cytometry analysis. Quadrants were drawn to denote a MyoG-H population in either G1 or S phase at 48 h of 4OHT induction. Ectopic expression of p21 served as a positive control. FSC: forward scatter. (c) Summary of the percentage of cells with MyoG-H expression in either G1 or S phase in response to ectopic p53 or p21 expression. (d) Immunoblotting (IB) shows that p53 activation induced expression of endogenous p21 and repressed myogenin expression in contrast to ectopic expression of p21, which led to accumulation of myogenin at 24 h of 4OHT induction. (e) Expression of myogenin protein in response to ectopic p53 and its mutants is summarized as the percentage of MyoG-H cells in either G1 or S phase at 48 h of 4OHT induction
p53 binds to the myogenin promoter
To further investigate p53-mediated transcriptional repression of myogenin, we examined previously published ChIP-seq data (chromatin immunoprecipitation followed by high-throughput DNA sequencing) to determine whether p53 binds to the myogenin promoter. A putative p53 response element (p53RE) at position −2560 was identified when MEFs were treated with a DNA damage-inducing drug, doxorubicin41 (Figure 3a, the upper panel). Corresponding RNA sequencing analysis showed reduced myogenin mRNA41 (Supplementary Figure 6a). The same binding region was also identified when mESCs were exposed to doxorubicin42 (Supplementary Figure 6b). This p53RE shares 95% sequence similarity with the canonical element43 (Figure 4a).
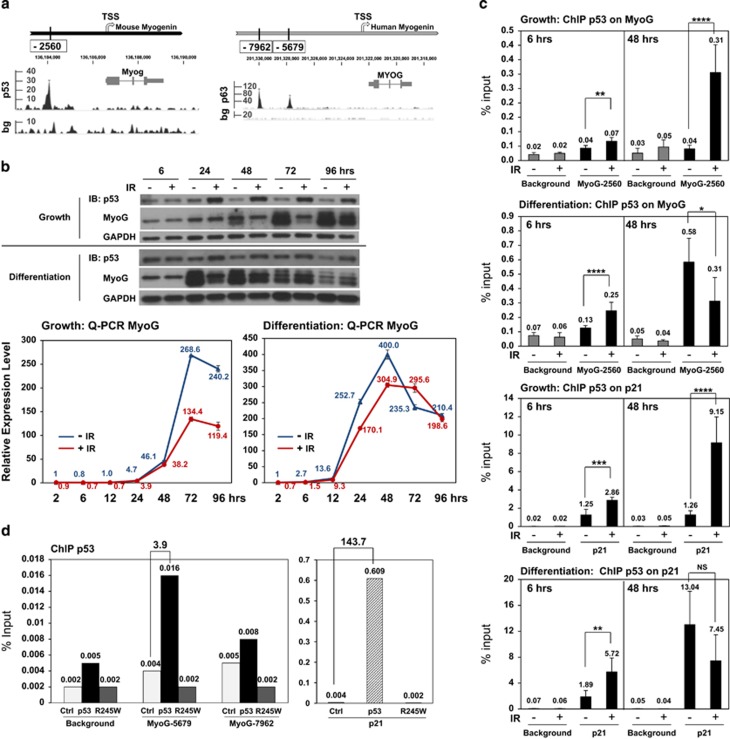
Figure 3.
p53 binds to the myogenin promoter and regulates its transcription in response to genotoxic stress. (a) Schematics of putative p53 or p63 response elements (REs) on the mouse and human myogenin promoter with p53 and p63 ChIP-seq profiles,41, 46 respectively. Arrow: transcription start site (TSS); Myog or MYOG: myogenin gene; bg: background signal. (b) Protein or RNA expression kinetics of p53 and myogenin from 2 to 96 h post IR in C2C12 myoblasts maintained under either growth or differentiation condition. (c) ChIP analysis of p53 binding to mouse myogenin 2560 site at 6 and 48 h post IR in C2C12 myoblasts maintained under either growth or differentiation condition. ChIP background was measured by binding of the p53 antibody to a gene desert region. p53 enrichment on the p21 promoter, p21, served as a positive control. ns, not significant; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. (d) ChIP analysis of p53 binding to the human myogenin promoter. ChIP was performed with an anti-ER antibody of RD cells ectopically expressing ER fusion of the control, p53, or p53R245W. p53R245W is the mouse equivalent of human p53R248W expressed in RD cells and is defective in DNA binding. Fold enrichment relative to the control is shown as bold numbers on the top of each bracket. ChIP background was measured by binding of the ER-fusion proteins to a gene desert region. p53 enrichment on the p21 promoter, p21, served as a positive control
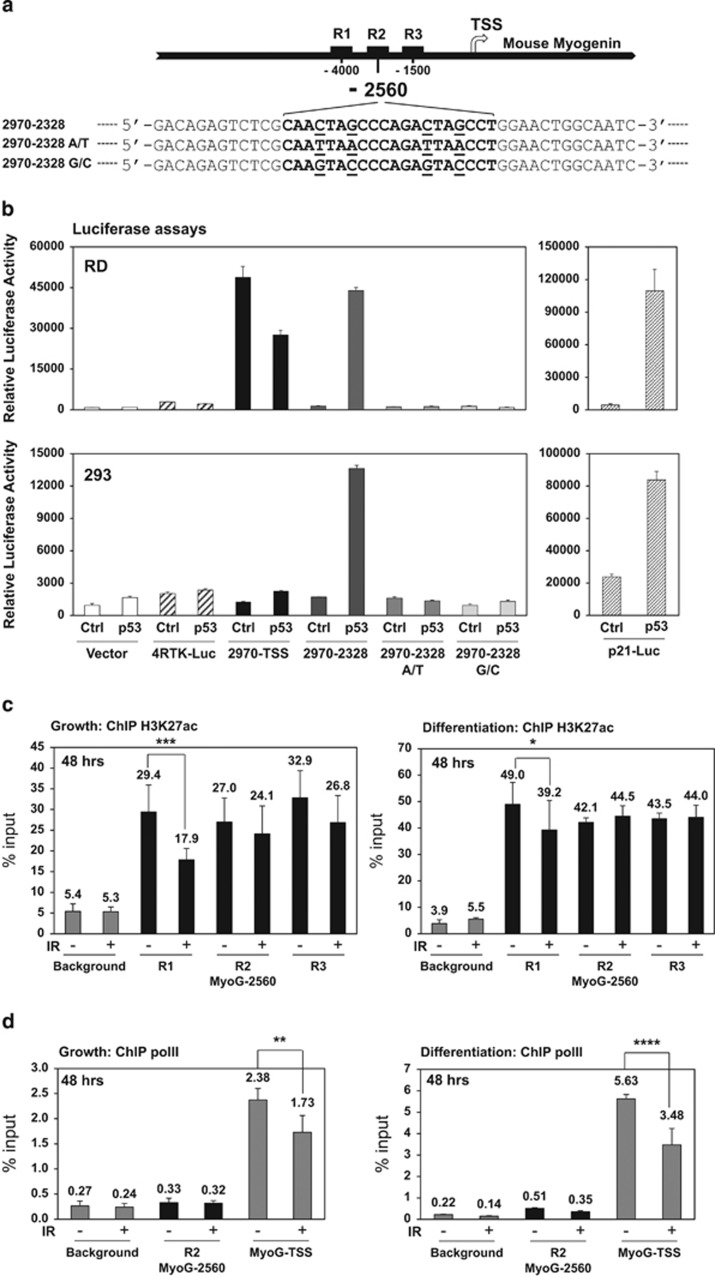
Figure 4.
Repression of myogenin by p53 is partially mediated through a distal enhancer region upstream of the mouse myogenin gene. (a) Schematics of the distal enhancer region upstream of the mouse myogenin gene. Black squares: R1, R2, and R3 enhancer elements; arrow: transcription start site (TSS). The p53 consensus binding sequence is characterized as two half sites of RRRCWWGYYY (R=purine, W=adenine or thymine, and Y=pyrimidine) with a spacer of 0–13 base pairs between each half site.43 Mouse myogenin p53 RE at−2560 site is shown in the bracket and mutations at the consensus sites are underlined as 2970–2328 A/T or G/C. (b) p53-ER transcriptional activity at 24 h of 4OHT induction was validated by luciferase reporter assays in RD or 293 cells. The reporter 2970-TSS includes the fragment from −2970 to TSS. The reporter 2970-2328 includes the fragment from −2970 to −2328. pGL3 vector and 4RTK-Luc served as negative controls. p21-Luc served as a positive control. (c) ChIP analysis of histone H3K27ac enrichment to mouse myogenin enhancer elements at 48 h post IR in C2C12 myoblasts maintained under either growth or differentiation condition. ChIP background was measured by binding of the H3K27ac antibody to a gene desert region. *P<0.05; ***P<0.001. (d) ChIP analysis of polymerase II enrichment to mouse myogenin R2 (MyoG-2560) and TSS at 48 h post IR in C2C12 myoblasts maintained under either growth or differentiation condition. ChIP background was measured by binding of the polymerase II antibody to a gene desert region. **P<0.01; ****P<0.0001
Before we could validate p53 binding to the mouse myogenin promoter in response to genotoxic stress, we first characterized expression kinetics of p53 and myogenin in C2C12 myoblasts at both RNA and protein levels under two culture conditions, growth versus differentiation, over a period of 96 h post ionizing radiation (IR) (Figure 3b and Supplementary Figure 6c). p53 can be rapidly activated in C2C12 cells within 2 to 3 h upon exposure to IR.44, 45 Based on the results of our time-course experiments, we chose to examine both early and late promoter occupancy of p53 at 6 and 48 h post IR, respectively, since myogenin showed distinct mRNA expression between the growth and differentiation condition after 48 h post IR (Figure 3b, Q-PCR MyoG).
Through quantitative ChIP analysis, we observed p53 enrichment at the myogenin p53RE, −2560 site, at 6 h post IR under both culture conditions (Figure 3c). A robust enrichment of p53 at 48 h under the growth condition (Figure 3c, Growth) was correlated with strong repression of myogenin until 96 h (Figure 3b, Growth MyoG). In contrast, under the differentiation condition, p53 enrichment at 48 h was decreased post IR (Figure 3c, Differentiation), with a corresponding recovery of myogenin mRNA and protein at the late time points, 72 and 96 h (Figure 3b, Differentiation MyoG). As a positive control, p53 binding to the p21 promoter showed similar patterns as compared with those binding to myogenin p53RE (Figure 3c, the lower half). Our results suggest that p53 binds to the myogenin p53RE at early time points and represses myogenin in response to genotoxic stress under both growth and differentiation conditions.
To our knowledge, the binding of p53 to the human myogenin promoter has not been reported. Instead, we analyzed a published human p63 ChIP-seq data set based on the observation that p63, a p53 family member, is estimated to bind 61.8 to 82.3% of p53 target genes.41 We found two p63-binding sites at positions −7962 and −5679 on the human myogenin promoter based on a genome-wide profiling of p63-binding sites using human primary keratinocytes cultured under the non-stressed growth condition46 (Figure 3a, the lower panel, and Supplementary Figure 6d). ChIP analyses validated p53 binding at position −5679 but not −7962 in RD cells (Figure 3d). The DNA-binding defective mutant, p53R245W, showed no enrichment at the position −5679.
Repression of myogenin by p53 is partially mediated through a distal enhancer region upstream of the mouse myogenin gene
Global ChIP sequencing analysis has shown that p53-repressed genes tend to associate with p53 peak enrichment at the distal enhancers in mESC exposed to doxorubicin.42 A recent study on mapping the genome-wide histone marks during myogenic differentiation identified three upstream enhancers, R1, R2, and R3, in the distal region upstream of the mouse myogenin gene47 (Figure 4a). These three enhancers are proposed to function as a switch control that regulates myogenin expression from proliferation to differentiation.47 We noted that the p53RE is located in the R2 enhancer and asked whether repression of myogenin by p53 could be mediated through an enhancer-associated mechanism.
We first used luciferase reporter assays to determine whether the p53RE could mediate transcriptional repression of the reporter. A 2970-bp region upstream of the TSS, 2970-TSS, showed repression by ectopic p53 in RD cells but not in 293 cells, suggesting cell-type-specific regulation by p53 (Figure 4b). Intriguingly, fragment, 2970–2328, containing ~300 bp upstream and downstream from position −2560, was transcriptionally activated by p53. Furthermore, mutation of the conserved CWWG motif to either TWWA or GWWC completely abolished the activation by p53, indicating the specificity of p53 binding on this p53RE (Figure 4b). Interestingly, we did not observe release of repression by p53 when TWWA or GWWC mutation was introduced to 2970-TSS reporter, suggesting this enhancer-mediated repression of myogenin may require a native chromatin state.
It is known that active enhancers are associated with histone marks, H3K27Ac and H3K4me1, and polymerase II.47, 48 Using quantitative ChIP analyses, we found a significant decline of histone H3K27Ac enrichment at the R1 and subtle yet consistent reduction at the R2 and R3 enhancers at the growth condition post IR (Figure 4c). In agreement with our early observation, diminished p53 enrichment at 48 h of differentiation post IR was correlated with a subtle reduction of H3K27Ac mark at the R1 but not at the R2 and R3 enhancers (Figure 4c). We did not observe any significant change of H3K4m1 enrichment with or without IR (data not shown). In addition, ChIP analyses revealed positive enrichment of Ser5-phosphorylated polymerase II49 at the R2 enhancer and confirmed the previous observation that only under the differentiation condition, polymerase II is recruited to the R2 enhancer47 (Figure 4d). Although reduced enrichment of polymerase II was observed at R2 under the differentiation condition post IR, we could not draw any conclusions on its biological significance due to the background variation (Figure 4d, Differentiation, R2 versus background). Nevertheless, we observed less polymerase II enrichment at TSS under both growth and differentiation conditions (Figure 4d), indicating reduced transcriptional initiation of the myogenin gene in response to genotoxic stress. Therefore, p53-mediated repression of myogenin is partially regulated through a distal enhancer region where loss of active histone marks negatively modulates myogenin expression in the presence of genotoxic stress.
Repression of myogenin by p53 leads to reduction of late but not early differentiation markers
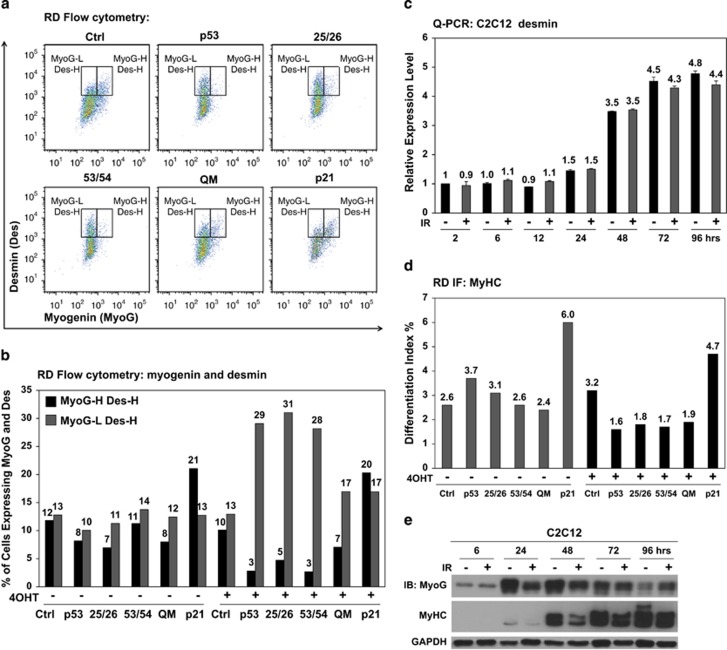
Next, we investigated the phenotypic outcome of p53-mediated repression of myogenin in RD and C2C12 cells by examining expression of differentiation markers. We did not observe any significant change in desmin expression, an early differentiation marker, in RD cells with ectopic p53 (Supplementary Figures 7a and b). Flow cytometry analysis confirmed that p53-mediated repression of myogenin led to a marked reduction in the percentage of MyoG-H cells, yet did not significantly change the overall percentage of desmin-high (Des-H) cells in RD cells (Figure 5a and quantification in Figure 5b). Similarly, no significant change was observed in desmin expression in C2C12 cells with or without IR (Figure 5c).
Figure 5.
Repression of myogenin by p53 leads to reduction of late but not early differentiation markers. (a) In response to ectopic p53, expression of an early differentiation marker, desmin, was quantitatively analyzed by co-immunostaining myogenin in RD cells and followed by flow cytometry analysis at 48 h of 4OHT induction. Two gates were drawn as myogenin-low and desmin-high (MyoG-L Des-H) on the left and myogenin-high and desmin-high (MyoG-H Des-H) on the right. (b) Summary of the percentage of RD cells in either MyoG-H Des-H or MyoG-L Des-H. QM: p5325265354. (c) Q-PCR analysis of desmin expression from 2 to 96 h post IR in C2C12 myoblasts maintained under differentiation condition. (d) Expression of MyHC in RD cells in response to ectopic p53 and mutants was analyzed by immunofluorescence (IF) and quantified by the Differentiation Index, which is the percentage of MyHC-positive cells above the total number of nuclei. Ectopic p21 served as a positive control. (e) Immunoblotting shows expression of myogenin and MyHC from 6 to 96 h post IR in C2C12 myoblasts maintained under differentiation condition
Next, we examined the expression of a late differentiation marker, myosin heavy chain (MyHC), in response to myogenin repression by p53. RD cells showed reduction of MyHC expression in response to ectopic p53 (Figure 5d and Supplementary Figure 7c). Reduction of MyHC expression was further confirmed in differentiated C2C12 myoblasts exposed to IR, in agreement with published results26, 50 (Figure 5e and Supplementary Figure 7d). Our ChIP results indicated decreased p53 binding at 48 h (Figure 3c, Differentiation) with a corresponding recovery of myogenin at 72 and 96 h of differentiation in C2C12 cells exposed to IR (Figure 3b, Differentiation). In line with this result, we observed recovery of MyHC expression at late-stage differentiation in C2C12 cells. Therefore, p53-mediated repression of myogenin leads to decreased and delayed expression of late but not early myogenic differentiation genes. Altogether, our findings suggest that p53 regulates late-stage differentiation at the myogenin step in response to genotoxic stress.
p53-mediated repression of myogenin reduces post-mitotic nuclear abnormality in terminally differentiated cells in response to acute DNA damage
We have discovered that p53 transcriptionally represses myogenin in response to genotoxic stress. To further investigate the broader biological significance behind this new mechanism, we asked whether repression of myogenin by p53 imposes a quality control mechanism that ensures cellular function and integrity by either delaying or terminating a myogenic differentiation program. To answer this question, we used IR to introduce DNA damage and compromised genomic integrity in differentiating myogenic cells. To simplify the complex cross-regulation between members of MRFs in response to DNA damage,51 our approach was to assay the myogenic conversion properties of ectopic MyoD and myogenin in the presence of DNA damage in MEFs. Both MyoD and myogenin can program non-muscle cells to myogenic fate and induce differentiation in vitro through a process called myogenic conversion.52 As discussed earlier, MyoD acts upstream of myogenin and transcriptionally activates myogenin.8, 9, 13, 14 We anticipated that p53 would repress endogenous myogenin transcription activated by ectopic MyoD and would not repress ectopic myogenin driven by a constitutive promoter. This experimental design allowed us to characterize myogenin function in either a p53-dependent or p53-independent manner.
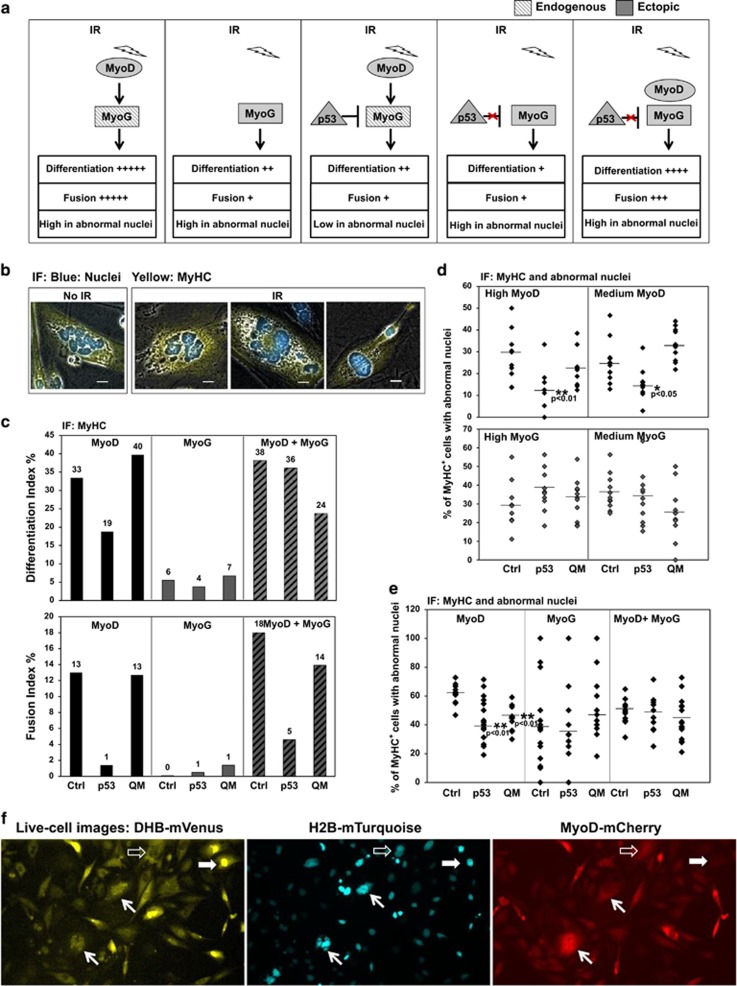
We first confirmed myogenic conversion of p53-null MEFs induced by ectopic MyoD or myogenin (Supplementary Figure 8a) and induction of myogenin expression by ectopic MyoD (Supplementary Figure 8b). We then examined the level of myogenic conversion on p53-null MEFs by MyoD and/or myogenin with or without p53 (protocol summarized in Supplementary Figure 8c; experimental design and results are summarized in Figure 6a). In myogenically converted MEFs exposed to IR, we discovered that a high percentage of MyHC-positive cells carried abnormal nuclei in the absence of p53 (Figure 6b). In the presence of p53, MyoD-mediated differentiation was strongly inhibited as measured by both Differentiation Index and Fusion Index (MyoD in Figure 6c and Supplementary Figure 8d). We also observed a p53-dependent inhibition of myogenic conversion mediated by myogenin as measured by the Differentiation Index (MyoG in Figure 6c). This inhibition of myogenin-mediated conversion possibly reflects indirect detrimental effects on differentiation as the result of ectopic p53. Nevertheless, inhibition of MyoD-mediated differentiation in the presence of p53 can be partially rescued by ectopic expression of myogenin (MyoD+MyoG in Figure 6c). Most importantly, in the presence of p53, the percentage of MyHC-positive cells carrying abnormal nuclei was significantly reduced upon MyoD-mediated conversion. This reduction was compromised in the presence of the p5325265354 mutant, a transactivation-null mutant. In contrast, myogenin-converted cells did not show a p53-dependent reduction in the number of abnormal nuclei (Figure 6d). Furthermore, ectopic expression of myogenin rescued and restored post-mitotic nuclear defects with an increased number of abnormal nuclei in MyHC-positive cells even in the presence of p53 (MyoD+MyoG in Figure 6e), suggesting that myogenin suppression by p53 is a mechanism for maintaining cellular integrity during the differentiation process.
Figure 6.
p53-mediated repression of myogenin reduces post-mitotic nuclear abnormality in terminally differentiated cells in response to acute DNA damage. (a) Experimental design and summary of results on p53-null MEFs for assays of myogenin function in either a p53-dependent or p53-independent manner. IR: ionizing radiation. Red-cross sign: p53 is unable to repress ectopic myogenin. Endogenous expression is shown as hashed grey and ectopic expression as filled grey. Myogenic differentiation was quantified using both Differentiation Index (the percentage of MyHC+ cells above the total number of nuclei) and Fusion Index (the percentage of the number of nuclei in MyHC+ cells containing at least three nuclei above the total number of nuclei). The percentage of abnormal nuclei in differentiated muscle cells (MyHC+) is summarized as either high or low in abnormal nuclei. (b) Examples of nuclear abnormality observed in irradiated cells versus non-irradiated cells. Using immunofluorescence (IF), nuclei were scored as abnormal if they were micronuclei, macronuclei, lobulated nuclei, or fragmented nuclei. Scale bar: 20 μm. (c) Myogenic differentiation induced by MyoD, myogenin, or both in the presence of ectopic p53 or the QM mutant, p5325265354. (d) The percentage of abnormal nuclei in MyHC+ cells expressing a high or medium level of MyoD or myogenin in the presence of ectopic p53 or the QM mutant. Statistical significance of reduction in numbers of abnormal nuclei relative to that of the control was determined by a two-tailed Student's t-test. (e) The percentage of abnormal nuclei in MyHC+ cells induced by MyoD, myogenin, or both in the presence of ectopic p53 or the QM mutant. (f) Live-cell images show cells undergoing myogenic conversion and acquiring post-mitotic nuclear defects. Images are shown in three channels from a single frame at 36 h post IR. DHB-mVenus: a fragment of DHB fused to a yellow fluorescent protein. H2B-mTurquoise: a histone H2B fused to a cyan fluorescent protein. MyoD-mCherry: MyoD fused to a red fluorescent protein. Solid white block arrows indicate a cell in G1 and open white block arrows indicate a cell in G2. White slender arrows indicate two MyoD-positive cells showing severely fragmented nuclei arrested in G1 phase at 36 h post IR after one or two rounds of mitosis
Next, we used p53-null MEFs to visualize live cells undergoing myogenic conversion and acquiring post-mitotic nuclear abnormalities in the absence of p53-mediated suppression of myogenin. To monitor cell cycle exit to a post-mitotic state, we used a recently developed live-cell sensor for cyclin-dependent kinase 2 (CDK2) activity, human DNA helicase B (DHB)-Venus.53 DHB-Venus is located in the nucleus during G1 phase and progressively translocates to the cytoplasm during cell cycle progression (Supplementary Figure 8e). To monitor the integrity of nuclei, we used histone H2B-Turquoise.53 Slender white arrows highlight two cells expressing MyoD-mCherry (Figure 6f) that went through one or two rounds of mitosis and subsequently developed highly fragmented nuclei (Supplementary Video 1). As indicated by nuclear-localized DHB-Venus, these two myogenic cells became arrested at 36 h post IR and remained arrested in G1 until conclusion of the time-lapse microscopy at 72 h post IR. This live-imaging approach enabled us to confirm that nuclear abnormality occurs in DNA-damaged myogenic cells that undergo cell cycle withdrawal and terminal differentiation.
We have demonstrated that p53 represses myogenin to prevent nuclear abnormality and maintain cellular integrity in terminally differentiated muscle cells. This was supported by the following observations: (1) in the absence of p53, myogenic cells can terminally differentiate with severe nuclear abnormality after exposure to IR; (2) in the presence of p53, this nuclear abnormality is significantly reduced in MyoD-mediated but not in myogenin-mediated differentiation; (3) in bypassing p53-mediated myogenin repression, a high level of myogenin permits terminal differentiation and is accompanied by the accumulation of post-mitotic nuclear abnormalities, suggesting a carryover of compromised genomic integrity and cellular function to terminally differentiated cells.
Discussion
It has been shown that genotoxic stress inactivates MyoD function through inhibitory mechanisms that is p53-independent.26, 50, 51, 54 Here we have discovered a previously unknown mechanism by which p53 directly represses myogenin and regulates myogenic differentiation in response to genotoxic stress (Figure 7). We also found that p53 mediates a quality-control program of reducing the incidence of nuclear abnormality by delaying or terminating a myogenic differentiation program. Based on our findings, we propose that regulation of myogenin plays a critical role not only in driving terminal differentiation but also in ensuring cellular function and integrity in terminally differentiated muscle cells by virtue of its conditional repression by p53.
Figure 7.

p53-mediated repression of myogenin regulates muscle differentiation in response to genotoxic stress. Muscle differentiation is represented in black lines and p53-mediated regulation in heavy red lines. Skeletal muscle differentiation is orchestrated by members of the MyoD family of transcription factors. MyoD determines the myogenic lineage and acts upstream of p21 and myogenin.8, 9, 13, 56 Under normal conditions, expression of both p21 and myogenin is critical to establish the state of irreversible cell cycle arrest and terminal differentiation.11, 16, 40 In contrast to the cell cycle arrest that promotes terminal differentiation under non-stressed conditions, it is known that cell cycle arrest induced by DNA damage is associated with decreased myogenic differentiation.26 In addition to the previously characterized mechanism that genotoxic stress inactivates MyoD function through a p53-independent mechanism,26, 50, 51, 54 we describe a direct regulatory mechanism by which p53 represses myogenin in response to genotoxic stress. This repression of myogenin leads to reduction of late-stage differentiation and prevents formation of post-mitotic nuclear abnormality in terminally differentiated muscle cells. We propose that the regulation of myogenin plays a critical role not only in driving terminal differentiation but also in maintaining cellular integrity in terminally differentiated muscle cells by virtue of its conditional repression by p53.
A recent study reports a novel function of p53 in ensuring accurate chromosome segregation and postulates a new mechanism of tumor suppression by preventing formation of aneuploidy.55 It is reasonable to hypothesize that p53 represses myogenin to ensure DNA damage repair and proper chromosome segregation when the differentiation program intersects with the cell cycle checkpoint control. Post-mitotic nuclear abnormalities that we observed in highly differentiated myotubes may result in poor fitness and viability as a consequence of the accumulation of unrepaired DNA lesions, aberrant formation of DNA and chromosomal structures, and aneuploidy. Our study supports a previously proposed ‘myogenic differentiation checkpoint' that ensures genetic stability in differentiated cells.26
Molecular mechanisms of p53-mediated transcriptional repression are less understood in comparison with transcriptional activation mechanisms.7, 42, 43 Here we report for the first time that p53 binds to a highly conserved p53RE in a distal enhancer region of the myogenin gene and transcriptionally represses myogenin in response to genotoxic stress. Using luciferase reporter assays, we found that this p53RE can be activated by p53, suggesting that genomic context and native chromatin states are required for repression by p53. Mechanisms behind this context-dependent repression are further investigated by examining the level of histone modification, H3K27ac, in the distal enhancer region. Under both growth and differentiation conditions in response to genotoxic stress, we found a decline in enrichment of H3K27ac and a corresponding repression of myogenin as indicated by reduced recruitment of polymerase II to the transcriptional starting site. Although there are several previously proposed modes of repression,6, 7, 42, 43, 57 the exact mechanism of myogenin repression by p53 is still unclear to us. We hypothesize that p53 interferes with the activity of enhancer elements by competing with histone acetylation complex, MyoD, or polymerase II binding.
The long-term repression of myogenin by p53 under the growth condition may prevent aberrant or sporadic differentiation of proliferating myogenic cells after exposure to genotoxic stress (Figure 3b). We speculate that p53 or p63 enrichment on myogenin regulatory elements previously observed in non-myogenic cells, such as MEFs,41 mESC,42 or primary keratinocytes,46 may reflect a poised global surveillance system that maintains cell identity and prevents, at least, a myogenic fate transition in response to stress signals. This notion is supported by the observation that elimination of p53 enhances cellular plasticity and efficiency of reprogramming somatic cells to pluripotency58 and differentiated hepatocytes to malignant cell fates.59 Intriguingly, under the differentiation condition we observed short-term p53 binding, loss of myogenin repression, and recovery of late-stage differentiation post IR. This observation is in line with the previous report that myoblasts can eventually overcome a block on differentiation if damaged DNA is repaired and/or genotoxic stress is removed.26 We unexpectedly found that p53 is significantly enriched at the p53RE of the myogenin gene under the normal differentiation condition (Figure 3c). We propose two phases of conditional regulation by p53: under the non-stressed condition, p53 is poised at the myogenin promoter but exerts low or minimal repression; under stressed conditions, p53 switches to a negative regulator of myogenin and prevents aberrant muscle differentiation.
Our findings have led us to consider whether p53 activation in response to chronic physiological stress occurring in myopathic diseases21, 22, 23, 60, 61 could persistently repress myogenin and in part contribute to development of muscle atrophy at the stage of myogenic differentiation.21, 22, 23, 24, 25, 62, 63, 64 Moreover, we are intrigued by a possible connection between repression of myogenin by p53 and muscle aging. Skeletal muscle aging is characterized by muscle atrophy and a decline in muscle function and regenerative capacity.24, 25, 65, 66, 67 Many studies have shown a close connection between p53 function and muscle aging, as well as perturbation of the muscle stem cell pool.21, 22, 23, 24, 25 Our work suggests new avenues for investigating muscle aging and muscle regeneration, where p53-mediated repression of myogenin in response to stress signals may have a critical role in mediating quality control of differentiation and in maintaining muscle tissue homeostasis.
In this study, we have discovered a mechanism by which p53 represses myogenin, thus directly regulating late-stage myogenic differentiation in response to genotoxic stress. In a broader context, our study illustrates how p53 can regulate tissue-specific differentiation to maintain cellular integrity and function under stress conditions.
Materials and Methods
Tuning translational level of the ectopic p53-ER expression in RD cells
Using upstream open reading frames (ORFs) and different translation initiation sites to modulate p53 translation levels31 (Supplementary Figure 1), we generated a series of p53-ER constructs that could express a range of protein comparable to endogenous p53 expression activated when MEFs were exposed to acute DNA damage (Supplementary Figures 2a–c). p53-ER expression in RD cells showed a degree of cell viability similar to that of the control-ER fusion, indicating that restoration of wild-type p53 within this range did not result in apoptotic and/or necrotic cells32 (Supplementary Figures 2d–g). Transcriptional activity of the p53 ER fusion was first evaluated in Mdm2/p53 double-knockout MEFs (Supplementary Table 1) and then validated by measuring activation of p53 target genes and cell cycle arrest in RD cells. Wild-type ectopic p53 strongly activated p21 and Mdm2 (Figure 1b and Supplementary Figure 3a), and induced cell cycle arrest (Supplementary Figure 3b), which was further confirmed by induction of hypophosphorylated pRb (Supplementary Figure 3c). Thus, the p53-ER fusion is capable of activating downstream target genes and inducing cell cycle arrest in a tunable manner.
Retroviral expression constructs using translational initiation library
Mouse p53-mKate-ER fusion and the control fusion were cloned into a pCru5 puromycin-selectable retroviral expression library.31 The control has a nuclear localization signal fused to mKate. The red-fluorescent protein mKate enabled us to monitor protein expression in live cells when protein expression level was modulated by upstream ORFs and different translation initiation sites31 (Supplementary Figure 1). The gene encoding the blue fluorescent protein (BFP) mTagBFP was cloned downstream of the control- or p53-mKate-ER and 2A ribosomal slippage sites (TaV2A and ERAV 2A), which results in expression of separate mKate-ER fusion and BFP proteins from a single ORF. Detection of mTagBPF fluorescence aided in both live-cell detection and determination of viral titers. Human p21 gene was PCR-amplified from human cDNA. p21-BFP-ER fusion constructs were cloned into the pCru5 expression library.
Mutations in p532526 are L25Q and W26S in transactivation domain 1 (TAD1). Mutations in p535354 are F53Q and F54S in TAD2. p5325265354 are quadruple mutations (QM) at positions 25, 26, 53, and 54. Mouse p53R245W, an equivalent human p53R248W mutant expressed in RD cells, renders the protein defective in DNA binding. All mutations were introduced by site-directed PCR mutagenesis using Pfu Ultra (Agilent Technologies, Santa Clara, CA, USA) to pCru5 3.14A p53-mKate-ER construct (#5 construct, Supplementary Figure 1).
MyoD- and myogenin-mCherry fusion constructs with a blasticidin selection marker were cloned into the pCru5 expression library as described above. NFP (non-fluorescent protein) served as a negative control with the same selection marker. The mouse MyoD gene was PCR amplified from pclBabe-MyoD.27 Myogenin gene was PCR amplified from human cDNA prepared from RD cells. Lentiviral expression constructs, DHB-mVenus and H2B-mTurquoise, were kindly provided by Dr. Tobias Meyer.53
Virus production, infection, and titer determination
All retroviral expression plasmids were packaged in either amphotropic or ecotropic 293T cells using the calcium precipitation method according to the protocol described by the manufacturer (CalPhos, Clontech, Mountain View, CA, USA). Viral titer for each batch of viruses was determined by flow cytometry analysis of BFP expression to ensure infection consistency among p53, p53 mutants, and the control. RD cells (1 × 105) or 0.5 × 105 of p53-null MEFs were infected by 50–100 μl of viruses in the presence of 8 μg/ml of polybrene the day before. After 24 h, cells were washed and fed with fresh medium in the presence of 100 nM of 4-hydroxytamoxifen (4OHT). After 24 h, cells were trypsinized and analyzed on an LSRII flow cytometry (BD Biosciences, San Jose, CA, USA). RD or MEFs cells were infected with viruses expressing p53- or the control-ER fusion protein to generate a 30% initial infection frequency to minimize multiple integrations per cell. After 48 h of puromycin selection at 0.5 μg/ml, we normally observed 50 to 60% of BFP+ cells. Lentiviral expression constructs were packaged using the CalPhos method in 293T cells in the presence of three packaging plasmids: pMDLg, pRSV-Rev, and HCMV-VSVG, kindly provided by Dr Tobias Meyer.
Cell culture
Low passage-number cell lines, RD, C2C12, 293, and p53-null MEFs, were maintained in Dulbecco's modified eagle medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 10% FBS (HyClone, ThermoFisher Scientific, Waltham, MA, USA) in the presence of penicillin and streptomycin. Differentiation medium is serum-free DMEM supplemented with 2% horse serum (GemCell, Gemini Bio-Products, West Sacramento, CA, USA). RD cells (2 × 105) were infected by viruses expressing p53- or the control-ER fusion protein at a 30% infection frequency the day before in each well per six-well tissue culture plates. After overnight infection, cell were trypsinized and re-seeded to 12-well tissue culture plates. To reach a higher cell density, one fourth of the total cells from each well of the 6-well plate were seeded to each well of the 12-well plate. To reach a lower cell density, one fifth of the total cells were seeded to each well of the 12-well plates. Cells were allowed to expand for 24 h and then washed 3 times with serum-free DMEM before switching to differentiation medium. Cells ectopically expressing ER fusion constructs were treated with 100 nM of 4OHT to induce nuclear translocation. Ethanol was used as the uninduced control. Puromycin selection was carried out at 0.5 μg/ml for all ectopic ER constructs expressed in either RD or MEFs. Cells were either collected for quantitative reverse transcriptase-PCR (qRT-PCR) and immunoblotting, or fixed for immunostaining at 24, 48, or 72 h of 4OHT induction.
Before IR, C2C12 were maintained at a sub-confluent density. C2C12 (2.5–3 × 104) were seeded onto each 35-mm tissue culture dish or each well per 6-well plate the day before. On the next day, the no-IR cells were either changed to fresh growth medium or washed four times with serum-free DMEM before switching to differentiation medium and were harvested at 2, 6, 12, 24, 48, 72, and 96 h after switching to fresh growth medium or differentiation medium. Medium was changed every 48 h. At the same time, another set of cells in either growth or differentiation medium were 5-Gy γ-irradiated and were collected at 2, 6, 12, 24, 48, 72, and 96 h post IR. We also tested cells that were allowed recovery in growth medium for 24 h post IR before switching to differentiation medium and collected at 24, 48, 72 and 96 h of differentiation time.
qRT-PCR, immunoblotting, and immunofluorescence microscopy
RD RNA was isolated using Trizol (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed using high-capacity cDNA RT kit (Applied Biosystems, Foster City, CA, USA). Q-PCR was performed in triplicates using Fast SYBR Green mixture (Applied Biosystems) on a StepOnePlus real-time PCR machine (Applied Biosystems).
Immunoblotting was performed according to standard methods. Protein lysate was prepared in Triton X-100 lysis buffer in the presence of protease inhibitor cocktail (Roche, Indianapolis, IN, USA) and phosphatase inhibitors (20 mM Tris-HCl pH7.6, 1% Triton X-100, 10% glycerol, 2 mM EDTA, 150 mM NaCl, 0.25 mM Na Orthovanadate, 25 mM β-glycerophosphate, 25 mM NaF). Proteins were separated and blotted using Bio-Rad Mini Trans-Blot system (Hercules, CA, USA). p53 was detected by antibodies 1C12 (#2524, Cell Signaling, Beverly, MA, USA) or CM5 (VP-P956, Vector Laboratories, Burlingame, CA, USA) for the N-terminal mutants. Note that CM5 antibody has weak reactivity to the endogenous human p53 as compared with the 1C12 antibody (Figure 1d versus Figure 1g). p21 was detected by antibody DCS60 (#2946, Cell Signaling), myogenin by M-225 (sc-576, Santa Cruz Biotechnology, Santa Cruz, CA, USA), pRb by G3–245 (554136, BD Pharmingen, San Jose, CA, USA), MyHC by H-300 (sc-20641, Santa Cruz Biotechnology), desmin by D33 (M0760, Dako, Glostrup, Denmark), and GAPDH by HRP-conjugated 14C10 (#3683, Cell Signaling). HRP activity was detected with the WesternBright ECL reagents (Advansta, Menlo Park, CA, USA).
Immunostaining was performed by fixing cells in 2% formaldehyde (16%, Thermo Scientific, Waltham, MA, USA) at room temperature for 8–10 min followed by permeabilization with PBS and 0.1% Triton X-100. Cells were then blocked in PBS and 1% BSA for 1 h before overnight incubation with the primary antibody. MyHC was detected by antibody H-300 (sc-20641, Santa Cruz Biotechnology), desmin by D33 (M0760 Dako), and myogenin by M-225 (sc-576, Santa Cruz Biotechnology) or F5D (556358, BD Pharmingen). Cells were washed the next day before incubating with Alexa Fluor 488-conjugated anti-mouse (Cell Signaling) or R-PE-conjugated anti-rabbit (Life Technologies) secondary antibody. Nuclei were stained with Hoechst 33342 at 1 μg/ml (1 mg/ml, BD Pharmingen). Images were acquired on Zeiss Observer.Z1 microscope (Cambridge, UK) and analyzed by ImageJ (National Institutes of Health; http://rsb.info.nih.gov/ij/docs/faqs.html). Imaging was performed either directly on tissue culture plates or glass slides pretreated with 0.1 mg/ml of Poly-D-Lysine (P7886, Sigma-Aldrich, St. Louis, MO, USA) for overnight at room temperature.
BrdU incorporation for cell cycle analysis and myogenin co-staining followed by flow cytometry analysis
RD cells were infected and induced by 4OHT as described above. BrdU were added at a final concentration of 10 μM 2.5 h before collecting cells. Cell pellets were re-suspended and fixed in PBS with 4% formaldehyde for 15 min at room temperature. Following the standard procedure, cells were then HCl-treated in PBST (0.2% Tween 20) for 30 min at room temperature and subsequently neutralized by NaB4O7. Cells were then re-suspended in cold PBS, and fixed and permeabilized in 90% ice-cold methanol for 30 min. After washing with PBS, cells were blocked in PBST with 1% BSA for 1 h at room temperature. Meanwhile, cells in each sample were manually counted to ensure that the same number of cells was used for immunostaining. Normally, 0.2 to 1 × 105 cells per experiment were immunostained for BrdU and myogenin. Cells were then suspended in 100 μl of pre-mixed 488-conjugated BrdU antibody (1 μl per 1 × 105 cells, Life Technologies) and anti-myogenin antibody (1 : 1200, either M-225 or F5D). After overnight incubation and washing, cells were then incubated with APC-conjugated anti-mouse or -rabbit secondary antibody for 1 h at room temperature. Right before flow cytometry analysis, cells were stained with propidium iodide (PI) using the standard protocol for 30 min at room temperature and kept on ice before flow cytometry analysis on an LSRII flow cytometer (BD Biosciences).
We first gated retroviral-infected BFP+ cells and then gated BFP+ cells into G1, S, and G2 phases based on BrdU and PI staining. Owing to heterogeneity in myogenin protein levels in RD cells, quadrants were drawn to denote a MyoG-H population in each cell cycle phase. Myogenin expression in MyoG-H population was quantified in each cell cycle phase. This heterogeneity of myogenin expression was also confirmed by immunostaining with either M-225 (Santa Cruz Biotechnology) or F5D (BD Pharmingen) antibody, followed by immunofluorescence (IF) microscopy.
Cell death evaluation by SYTOX Green and Annexin V staining
Cell viability was evaluated by staining RD cells expressing ectopic p53 or the control with both SYTOX Green and APC-conjugated Annexin V (Life Technologies, A35110). RD cells were infected, induced, and selected as described above. We first estimated overall cell viability, including cells killed by puromycin selection (no debris). We then gated retrovirus-infected and BFP+ cells, and estimated cell viability in p53-expressing cells as compared with that of the control-expressing cells. Both SYTOX Green and Annexin V staining were carried out according to the protocol recommended by the manufacturer (Life Technologies).
ChIP assay
A total of six 10-cm tissue culture dishes with 2 × 106 of RD cells per dish were infected with p53-, the control-, and p53R245W-ER viruses in the presence of 8 μg/ml of polybrene the day before. After 24 h, infected RD cells were expanded to seven 15-cm dishes. After 24 h, cells were washed 3 times with serum-free DMEM and then switched to the differentiation medium in the presence of 100 nM 4OHT and 0.5 μg/ml of puromycin. After 24 h of induction and selection, 1 dish of cells was trypsinized and counted. Other dishes were scraped and pooled with the trypsinized cells for fixation. RD cells (1 × 107) or 0.5–1 × 107 of C2C12 cells were used for each ChIP assay. ChIP assay was carried out as described.41 ChIP assay was performed using a biotinylated anti-ER antibody (TE111.5D11, Thermo Scientific, MA5–13062) to pull down p53-ER fusion in order to avoid using anti-p53 antibodies, which recognize endogenous mutant p53R248W expressed in RD cells. Streptavidin Dynabeads (M-280) were used for immunoprecipitation according to the protocol recommended by the manufacturer (Life Technologies). DNA was purified and eluted using MinElute PCR purification kit (Qiagen, Valencia, CA, USA) before Q-PCR analysis. For C2C12 ChIP assays, we used anti-p53 antibody (CM5, Vector Laboratories, VP-P956), anti-Ser5 polymerase II antibody (4H8, Thermo Scientific, MA1–46093), anti-H3K4m1 (ab8895, Abcam, Cambridge, MA, USA), and anti-H3K27ac antibody (ab4729, Abcam). C2C12 cells were collected and directly fixed by formaldehyde at 6 and 48 h post IR under the growth condition. For C2C12 cells collected at 48 h under the differentiation condition, cells were allowed recovery in growth medium for 24 h post IR before switching to differentiation medium and collected 48 h later. At the same time, the no-IR cells were also grown for another 24 h before switching to differentiation medium and collected 48 h later.
Transient transfection and luciferase assays
RD or 293 cells were transiently transfected using Superfect (Qiagen). Cells (5 × 104) were seeded on to each well of the 12-well plate the day before. Cells were transfected with 200 ng of p53- or the control-ER expression plasmid along with 250 ng of luciferase reporter and 125 ng of pCS2-β-galactosidase, which served as an internal control. After overnight incubation, cells were induced with 100 nM 4OHT or the ethanol control and lysed at 24 h post induction in 1 × lysis buffer (Promega, Madison, WI, USA). Luciferase activity was assayed using beetle luciferin (Promega) buffer on Tecan Infinite M1000 (Tecan Group Ltd., Männedorf, Switzerland) and β-galactosidase activity was assayed using β-D-galactoside buffer on a Molecular Devices plate reader (Sunnyvale, CA, USA). pT81-4RTK-luciferase, p21-luciferase, and pCS2-β-galactosidase were kindly provided by Dr Stephen Tapscott. pT81-4RTK-luciferase contains four copies of the MyoD-binding site. Mouse myogenin promoter region from −2970 to ATG was PCR amplified from mouse genomic DNA and inserted into the pGL3 basic vector to generate 2970-TSS luciferase reporter. Fragment containing the region from −2970 to −2328 was then subcloned as the 2970–2328 luciferase reporter. Subsequent site-directed mutagenesis was performed to introduce point mutations at the p53 consensus binding site and to generate luciferase reporters, 2970–2328 A/T and 2970–2328 G/C.
Myogenic conversion of p53-null MEFs and time-lapse microscopy
p53-null MEFs less than two rounds of passages after the initial expansion were used for myogenic conversion. All viruses were titered to achieve 100% infection of p53-null MEFs with MyoD-mCherry or myogenin-mCherry, and 30 to 50% of infection with p53-, p53 QM- or the control-ER fusion. The control virus for MyoD-mCherry and myogenin-mCherry was NFP as described early. Freshly thawed and expanded p53-null MEFs (1 × 105) were infected by viruses in each well of the six-well plate the day before. After overnight infection, one fourth of the infected cells were seeded onto each well of the 12-well plate. On the next day, right before 5 Gy of IR, cells were induced with 100 nM 4OHT. Cells were then recovered overnight before adding 0.5 μg/ml puromycin. Cells were selected and induced for another 24 h before switching to the differentiation medium in the presence of 4OHT and puromycin. After 24 h, cells were fixed for IF analysis. For live-cell tracking using time-lapse microscopy, p53-null MEFs were first infected with lentiviruses expressing DHB-mVenus or H2B-mTurquoise. Double-positive cells were sorted using BD FACSAria flow cytometer (BD Biosciences) and further expanded. One-eighth infected p53-null MEFs described above were seeded in each well per 24-well μ-plates (Ibidi, GmbH, Planegg/Martinsried, Germany). Right before IR, cells were induced with 4OHT and then recovered for 30 min post IR before moving to 37 °C chamber supplied with CO2 and humidity for 72 h of observation on the time-lapse microscopy. Images were acquired every 15 min in three color channels on Zeiss Observer.Z1 microscope and analyzed by ImageJ.
Statistical analysis and repeatability of experiments
Statistical significance and P-values were determined using the two-tailed Student's t-test. Two to four independent experiments were performed on Q-PCR, immunostaining and flow cytometry analysis, and immunoblotting in Figures 1,2,3, and 5; three independent experiments were performed for ChIP analysis in Figures 3 and 4; two to three independent experiments were performed for immunostaining and microscopy in Figure 5; three to five independent experiments were performed on characterizing myogenic conversion and morphology of abnormal nuclei using immunostaining and microscopy in Figure 6. Two independent experiments were performed on live imaging using time-lapse microscopy in Supplementary Video 1.
Acknowledgments
We thank members of the Wang laboratory for technical support and stimulating discussions. We are grateful to Drs Ingrid Lawhorn, Phillip Patten, Stefano Biressi, and Maddalena Adorno for valuable discussions and comments on the manuscript; Teddy Yewdell from the Dr Aaron Straight laboratory for helpful discussions; Emma Chory from the Dr Gerald Crabtree laboratory, and Rose Tran and Brook Hunt from the Dr Paul Khavari laboratory for generously providing histone antibodies for preliminary ChIP validations. We thank Dr Foteini Mourkioti for providing antibodies for initial testing of muscle differentiation markers. We also thank the Stanford FACS facility for technical support. This study was supported by NIH-5R21AG040360-02 to CLW and Swiss National Science Foundation Fellowships to DKB.
Glossary
- MRF
myogenic regulatory factor
- ER
estrogen receptor
- BFP
blue fluorescent protein
- 4OHT
4-hydroxytamoxifen
- MEF
mouse embryonic fibroblast
- IR
ionizing radiation
- PI
propidium iodide
- BrdU
bromodeoxyuridine
- ChIP-seq
chromatin immunoprecipitation followed by high-throughput DNA sequencing
- RE
response element
- TAD
transcriptional activation domain
- DHB
human DNA helicase B
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by B Zhivotovsky
Supplementary Material
References
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31:1501–1508. doi: 10.1093/carcin/bgq101. [DOI] [PubMed] [Google Scholar]
- Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Wang B, Xiao Z, Ko HL, Ren EC. The p53 response element and transcriptional repression. Cell Cycle. 2010;9:870–879. doi: 10.4161/cc.9.5.10825. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Sem Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4:pii:a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, et al. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Rawls A, Valdez MR, Zhang W, Richardson J, Klein WH, Olson EN. Myogenin's functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev Biol. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H, Griesmann H, Beitzinger M, Hofmann L, Beinoraviciute-Kellner R, Sauer M, Hüttinger-Kirchhof N, et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell. 2006;10:281–293. doi: 10.1016/j.ccr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, et al. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- Porrello A, Cerone MA, Coen S, Gurtner A, Fontemaggi G, Cimino L, et al. p53 regulates myogenesis by triggering the differentiation activity of pRb. J Cell Biol. 2000;151:1295–1304. doi: 10.1083/jcb.151.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soddu S, Blandino G, Scardigli R, Coen S, Marchetti A, Rizzo MG, et al. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J Cell Biol. 1996;134:1–12. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir Y, Bengal E. p53 protein is activated during muscle differentiation and participates with MyoD in the transcription of muscle creatine kinase gene. Oncogene. 1998;17:347–356. doi: 10.1038/sj.onc.1201929. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Hauschka S, Tapscott SJ. The MCK enhancer contains a p53 responsive element. Proc Natl Acad Sci USA. 1991;88:4570–4571. doi: 10.1073/pnas.88.11.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier N, Hourdé C, Amthor H, Marazzi G, Sassoon D. Loss of a single allele for Ku80 leads to progenitor dysfunction and accelerated aging in skeletal muscle. EMBO Mol Med. 2012;4:910–923. doi: 10.1002/emmm.201101075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf M, Coletti D, Sassoon D, Marazzi G. Muscle cachexia is regulated by a p53-PW1/Peg3-dependent pathway. Genes Dev. 2006;20:3440–3452. doi: 10.1101/gad.412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf M, Coletti D, Marazzi G, Sassoon D. Chronic p53 activity leads to skeletal muscle atrophy and muscle stem cell perturbation. Basic Appl Myol. 2008;18:131–138. [Google Scholar]
- Rodier F, Campisi J, Bhaumik D. Two faces of p53: aging and tumor suppression. Nucleic Acids Res. 2007;35:7475–7484. doi: 10.1093/nar/gkm744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MG, Anderson RM, Yuan M, Kendziorski CM, Weindruch R, Prolla TA. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics. 2007;8:80. doi: 10.1186/1471-2164-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. A myogenic differentiation checkpoint activated by genotoxic stress. Nat Genet. 2002;32:585–593. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- Yang Z, MacQuarrie KL, Analau E, Tyler AE, Dilworth FJ, Cao Y, et al. MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev. 2009;23:694–707. doi: 10.1101/gad.1765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, Thayer MJ, Weintraub H. Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science. 1993;259:1450–1453. doi: 10.1126/science.8383879. [DOI] [PubMed] [Google Scholar]
- Merlino G, Helman LJ. Rhabdomyosarcoma—working out the pathways. Oncogene. 1999;18:5340–5348. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- Felix CA, Kappel CC, Mitsudomi T, Nau MM, Tsokos M, Crouch GD, et al. Frequency and diversity of p53 mutations in childhood rhabdomyosarcoma. Cancer Res. 1992;52:2243–2247. [PubMed] [Google Scholar]
- Ferreira JP, Overton KW, Wang CL. Tuning gene expression with synthetic upstream open reading frames. Proc Natl Acad Sci USA. 2013;110:11284–11289. doi: 10.1073/pnas.1305590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011;19:177–191. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Walter D, Satheesha S, Albrecht P, Bornhauser BC, D'Alessandro V, Oesch SM, et al. CD133 positive embryonal rhabdomyosarcoma stem-like cell population is enriched in rhabdospheres. PLoS One. 2011;6:e19506. doi: 10.1371/journal.pone.0019506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M, Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell Mol Life Sci. 2001;58:571–579. doi: 10.1007/PL00000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Dilworth FJ. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J. 2013;280:3991–4003. doi: 10.1111/febs.12188. [DOI] [PubMed] [Google Scholar]
- Hostein I, Andraud-Fregeville M, Guillou L, Terrier-Lacombe MJ, Deminière C, Ranchère D, et al. Rhabdomyosarcoma: value of myogenin expression analysis and molecular testing in diagnosing the alveolar subtype: an analysis of 109 paraffin-embedded specimens. Cancer. 2004;101:2817–2824. doi: 10.1002/cncr.20711. [DOI] [PubMed] [Google Scholar]
- Andrés V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Latella L, Lukas J, Simone C, Puri PL, Bartek J. Differentiation-induced radioresistance in muscle cells. Mol Cell Biol. 2004;24:6350–6361. doi: 10.1128/MCB.24.14.6350-6361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kim M, Choi YH, Goemans B, Yeung C, Hu Z, et al. Diminished G1 checkpoint after gamma-irradiation and altered cell cycle regulation by insulin-like growth factor II overexpression. J Biol Chem. 1999;274:13118–13126. doi: 10.1074/jbc.274.19.13118. [DOI] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci USA. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Kurabayashi M, Jeyaseelan R, Kedes L. Antineoplastic agent doxorubicin inhibits myogenic differentiation of C2 myoblasts. J Biol Chem. 1993;268:5524–5529. [PubMed] [Google Scholar]
- Innocenzi A, Latella L, Messina G, Simonatto M, Marullo F, Berghella L, et al. An evolutionarily acquired genotoxic response discriminates MyoD from Myf5, and differentially regulates hypaxial and epaxial myogenesis. EMBO Rep. 2011;12:164–171. doi: 10.1038/embor.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabayashi M, Jeyaseelan R, Kedes L. Doxorubicin represses the function of the myogenic helix-loop-helix transcription factor MyoD. Involvement of Id gene induction. J Biol Chem. 1994;269:6031–6039. [PubMed] [Google Scholar]
- Nam HJ, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat Cell Biol. 2014;16:538–549. doi: 10.1038/ncb2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, et al. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- Ori A, Zauberman A, Doitsh G, Paran N, Oren M, Shaul Y. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17:544–553. doi: 10.1093/emboj/17.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh DF, Xue W, Calvisi DF, Evert M, Michurina TV, Dow LE, et al. p53-dependent nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell. 2014;158:579–592. doi: 10.1016/j.cell.2014.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, et al. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol. 2011;69:540–752. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Skotte NH, Ladha S, Nguyen YT, Qiu X, Deng Y, et al. p53 increases caspase-6 expression and activation in muscle tissue expressing mutant huntingtin. Hum Mol Genet. 2014;23:717–729. doi: 10.1093/hmg/ddt458. [DOI] [PubMed] [Google Scholar]
- Powers SK, Kavazis AN, McClung JM. Oxidative stress and disuse muscle atrophy. J Appl Physiol (1985) 2007;102:2389–2397. doi: 10.1152/japplphysiol.01202.2006. [DOI] [PubMed] [Google Scholar]
- Ebert SM, Dyle MC, Kunkel SD, Bullard SA, Bongers KS, Fox DK, et al. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem. 2012;287:27290–27301. doi: 10.1074/jbc.M112.374777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Brack AS. Cellular mechanisms of somatic stem cell aging. Curr Top Dev Biol. 2014;107:405–438. doi: 10.1016/B978-0-12-416022-4.00014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Piccirillo R, Goldberg AL, Perrimon N. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis Model Mech. 2013;2013;6:1339–1352. doi: 10.1242/dmm.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.