Abstract
Despite the sizable number of components that comprise Mapk cascades, Map3k1 is the only element that contains both a kinase domain and a plant homeodomain (PHD) motif, allowing Map3k1 to regulate the protein phosphorylation and ubiquitin proteasome systems. As such, Map3k1 has complex roles in the regulation of cell death, survival, migration and differentiation. Numerous mouse and human genetic analyses have demonstrated that Map3k1 is of critical importance for the immune system, cardiac tissue, testis, wound healing, tumorigenesis and cancer. Recent gene knockin of Map3k1 to mutate the E2 binding site within the Map3k1 PHD motif and high throughput ubiquitin protein array screening for Map3k1 PHD motif substrates provide critical novel insights into Map3k1 PHD motif signal transduction and bring a brand-new understanding to Map3k1 signaling in mammalian biology.
Facts
Of the 19 Map3ks only Map3k1 contains a plant homeodomain (PHD) motif, and is an E3 ubiquitin (Ub) ligase.
The Map3k1 PHD motif regulates both Mapk cascade protein stability following hyperosmotic stress and Mapk pathway activation from transforming growth factor-β (Tgf-β) and epidermal growth factor (Egf) cytokine receptors by the Ub-proteasome system.
The Map3k1 PHD motif is critical for stem cell differentiation, tumorigenesis, B-cell development, T-cell signaling, protecting cardiac tissue from damage and testis development.
Open Questions
Are divergent roles for the Map3k1 kinase domain and PHD motifs present within human breast cancers?
Does the Map3k1 PHD motif regulate Tabs by non-canonical ubiquitination following CD40, and other Tnfrs, signal transduction?
Are novel Map3k1 PHD motif substrates targeted by the Ub-proteasome system during apoptosis?
Discovery and Early Characterization
Mitogen-activated protein kinase (Mapk) kinase (Map2k) kinases (Map3ks) activate Mapks by the binding and phosphorylation of Map2ks (Figure 1).1, 2, 3, 4 Map3k1 (encoded by Map3k1, and also known as MEKK1) was initially partially cloned as a complimentary DNA (cDNA) that encoded the C-terminal 672 amino acid residues of the Map3k and contains the kinase domain.5 Notably, the Map3k1 kinase domain shows a significant sequence homology to the Schizosaccharomyces pombe kinase Byr2 and the Saccharomyces cerevisiae kinase Ste11, both Map3ks of the yeast pheromone response pathway.5, 6 But, despite its relatively high sequence similarity, Map3k1 cannot replace the function of Ste11 in yeast.6 Map3k1 is a serine and threonine kinase and a phospho-protein that was the second mammalian Map3k, after c-Raf, demonstrated to phosphorylate Map2k1 (also known as MEK1) within its activation domain.7, 8, 9 Subsequently, Map3k1 was shown to bind and activate Map2k4 (also known as MKK4 or JNKK1) that, in turn, phosphorylates the c-Jun N-terminal kinase (JNK) Mapk8 (also known as JNK1), Mapk9 (also known as JNK2) and Mapk14 (also known as p38-α).10, 11, 12, 13 Early work showed that both Tumor necrosis factor (Tnf) -α14 and crosslinking of the Tnf receptor (Tnfr) family member CD40 with antibodies activate Map3k1 in cell lines.15
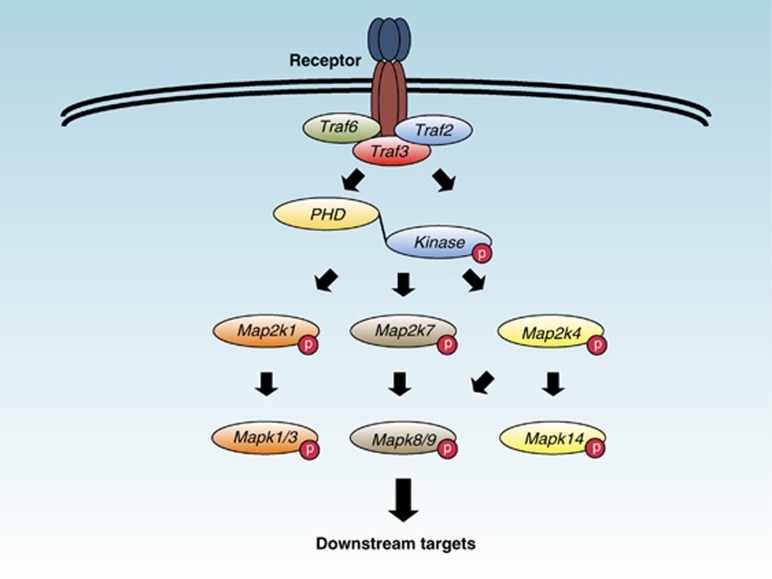
Figure 1.
Schematic illustration of our early understanding of Map3k1 signal transduction. The Map3k1 kinase domain can phosphorylate and activate Map2ks that in turn phosphorylate and activate Mapks. Activated Mapk then phosphorylates downstream signaling targets in cells.
The later cloning of the full-length rat Map3k1 cDNA revealed that Map3k1 possessed, in addition to its Ste11-like C-terminal kinase domain, a large N-terminal regulatory region.16 Overexpression of full-length Map3k1 activates Mapk1/3, Mapk8/9 and Mapk14 pathways in fibroblast cell lines.1, 16 Transfection of Map3k1 encoding cDNA into NIH3T3 cells leads to the activation of the nuclear factor κ-light-chain-enhancer of activated B-cell (NF-κB) pathway.17 Under these circumstances Map3k1 can form a complex with and induce the phosphorylation of IκBα kinases (Ikks) to activate the NF-κB pathway in cell lines.18 By similar bioinformatics methods that identified the Map3k1 kinase domain, other functional protein motifs have been identified within Map3k1 through computer sequence alignment techniques, including the PHD,19 Ub interacting motifs19 and SWI2/SNF2 and MuDR domain.20 A multitude of Map3k1 binding partners have been found to date by a wide variety of molecular approaches (Table 1).
Table 1. Listing of Map3k1 binding partners.
| Protein | Function | Reference |
|---|---|---|
| Nck interacting kinase | Kinase | 85 |
| 14-3-3 | Scaffold | 86 |
| Mapk9 | Kinase | 87 |
| α-actinin | Microfilament protein | 41 |
| c-Raf | Kinase | 88 |
| Mapk1 | Kinase | 88 |
| RhoA | Small GTPase | 89 |
| Cdc42 | Small GTPase | 90 |
| Ras | Small GTPase | 91 |
| Rac | Small GTPase | 90 |
| p115 RhoGAP | GTPase-activating protein | 92 |
| Map2k4 | Kinase | 12 |
| Map2k7 | Kinase | 12 |
| Map2k1 | Kinase | 5 |
| c-Jun | Transcription factor | 32 |
| Itch | HECT E3 Ub ligase | 51 |
| Traf2 | Scaffold | 93 |
| Grb2 | Adapter | 94 |
| Axin | Scaffold | 95 |
| Fak | Kinase | 96 |
| Deltex 1 | E3 Ub ligase | 73 |
| Ikkα | Kinase | 18 |
| Ikkβ | Kinase | 18 |
| Ikkγ | Scaffold | 53 |
| Tax | Nuclear factor | 97 |
| Han11 | Scaffold | 98 |
| MarvelD3 | Tight junction protein | 37 |
| Tab1 | Scaffold | 72 |
Is Map3k1 a Mediator of Cell Death, Survival or Both?
Map3k1 was initially suggested to be a pro-apoptotic kinase after several failed attempts to create stable cell lines that overexpress this Map3k.21 Indeed, inducible expression of the Map3k1 kinase domain can sensitize Swiss 3T3 cells to UV-irradiation-induced cell death.21 Similarly, inducible expression of the Map3k1 kinase domain in L929 fibrosarcoma cells can increase their susceptibility to Tnf-α-induced apoptosis.22 Sequence analysis of full-length Map3k1 identified a short Cysteine-aspartic acid protease (Caspase) -3 cleavage site (871DTVD874), and its mutagenesis can prevent Map3k1-induced apoptosis caused by the overexpression of full-length Map3k1 encoding cDNA in cells.22 Caspase-3 cleavage at this site generates two Map3k1 fragments, the N-terminal fragment containing the PHD motif and the C-terminal fragment containing the kinase domain.5, 19 Expression of Map3k1 with a mutant Capase-3 cleavage site can also prevent Map3k1 cleavage in response to genotoxic stress in fibroblast cell lines and following CD95 (also known as Fas) -mediated apoptosis in the Jurkat T-cell line.23, 24 Anoikis, apoptosis induced by cellular detachment from extracellular matrices, can also initiate Map3k1 cleavage by Caspase-3, and an inactive kinase domain form of Map3k1 can inhibit anoikis-induced programmed cell death upon overexpression in Madin-Darby canine kidney cells.25 By contrast, Map3k1 is not susceptible to Caspase-3 cleavage following Madin-Darby canine kidney cell death induced by microtubule disruption drug treatment,26 demonstrating that Caspase-3 cleavage is not the only mechanism of Map3k1-mediated cell death. Expression of the anti-apoptotic protein B-cell lymphoma 2 can block Map3k1-mediated apoptosis, though B-cell lymphoma 2 overexpression by itself is insufficient to prevent the Map3k1 cleavage.26 Overexpression of Map3k1 initiates a substantial Mapk8/9 activation in many cell types and is the likely mechanism for the pro-apoptotic role of the Map3k1 kinase domain.10, 13 However, the induction of apoptosis by Mapk8/9 is a complicated process in cells and numerous mechanisms have been proposed to account for it, including activation of Itch by Mapk8 phosphorylation and Lys48-linked poly-Ub transfer onto cellular FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein (c-Flip) leading to c-Flip proteasomal degradation and the promotion of apoptosis,27 the control of p53 stability and cell death by Mapk8/9 phosphorylation of p53 at Ser6,28 and Mapk8/9 phosphorylation and regulation of B-cell lymphoma 2 family members to promote cell death.4
While the early analysis of Map3k1 by transfection and overexpression of its cDNA into cell lines suggested a pro-apoptotic role, these initial results in cell lines were then complicated by the contradictory finding that null mutant Map3k1 (Map3k1−/−) mouse embryonic stem (ES) cells display enhanced cell death in response to hyperosmotic stress, low temperature shock and microtubule disruption drug treatment.29, 30 These important results demonstrated that Map3k1 has a more critical role in protecting mammalian cells from many types of cell death, and that the role of Map3k1 in promoting apoptosis may well be a labyrinthine one. Similarly, Map3k1−/− ES cell-derived cardiac myocytes show enhanced cell death in response to oxidative stress.31 Most likely Map3k1-dependent Mapk activation reduces cell death by the activation of pro-survival targets.1, 3
In addition to the Map3k1 kinase domain, roles for the Map3k1 PHD motif in cell death have been described.19, 32, 33 The Map3k1 PHD can mediate the transfer of Lys48-linked poly-Ub onto Mapk1, leading to the subsequent proteasomal degradation of Mapk1 in cell lines undergoing hyperosmotic stress-induced apoptosis.19, 34 Similarly, the Map3k1 PHD motif has been reported to mediate the transfer of Lys48-linked poly-Ub onto the c-Jun transcription factor to promote its degradation by the proteasome in Map3k1−/− mouse embryonic fibroblast (MEF) cells undergoing hyperosmotic stress-induced apoptosis.32 The Map3k1 PHD may also act as E3 Ub ligase for c-Jun in cells undergoing cisplatin-induced apoptosis.35 Both the Map3k1 PHD and kinase domains are essential for microtubule disruption drug-induced Mapk8/9 activation and apoptosis in Map3k1−/− DT40 cells.36
Recently, MarvelD3, a transmembrane component of tight junctions that is required for epithelial monolayer integrity during hyperosmotic stress, has been identified as a protein that forms a complex with Map3k1 in cells.37 MarvelD3 can relocalize Map3k1 in response to hyperosmotic stress and by this means can regulate Mapk8/9 activation.37 MarvelD3-mediated attenuation of Map3k1 signaling is critical for epithelial cell survival while undergoing hyperosmotic stress.37
Cell Migration and Wound Healing
The generation of kinase-deficient Map3k1 (encoded by Map3k1ΔKD) expressing ES cells revealed that Map3k1 is critical for both serum- and lysophosphatidic acid (LPA) -induced Mapk8/9 phosphorylation.38 Map3k1ΔKD ES cells also display reduced serum-induced migration in the Boyden chamber chemotaxis assays.38 Epidermal keratinocytes extracted from Map3k1ΔKD mice have defective Tgf-β, Activin A- and Activin B-induced migration in cell culture plate-based wound healing assays.2, 39 Map3k1ΔKD keratinocytes display reduced Mapk8/9 phosphorylation following treatment with Tgf-β, Activin A or Activin B.39 The molecular basis underpinning defective Map3k1-dependent migration during wound healing may be the reduced formation of actin stress fibers in Activin B-treated Map3k1ΔKD keratinocytes.39 The formation of Activin B-induced actin stress fibers in keratinocytes is dependent upon Mapk8/9 activity because they can be ablated by the pre-treatment of keratinocytes with the SP600125 inhibitor compound.2, 39
Map3k1−/− MEF cells are defective in their adherence to cell culture plates when centrifuged at low speed.40 Like Map3k1ΔKD ES cells, Map3k1−/− MEF cells display significantly reduced migration toward serum in the transwell migration assays.38, 40 Similarly, migration toward fibronectin or fibronectin and Egf is reduced in Map3k1−/− MEF cells.40 Map3k1 has been shown by two groups to localizes to focal adhesions in fibroblasts,40, 41 and less Vinculin, a critical cytoskeletal protein found in focal adhesions, is detected at the focal adhesions of Map3k1−/− MEF cells.40 Egf treatment of MEF cells leads to the formation of a complex between focal adhesion kinase (Fak) and Map3k1.40 Map3k1−/− MEF cells also display both reduced Mapk1/3 phosphorylation in response to Egf or Fibroblast growth factor-2 treatment and decreased Calpain activation, a calcium-dependent cysteine protease that is activated by Mapk1/3 phosphorylation.40, 42
Lymphocyte Differentiation and Effector Responses
Naïve CD4+ T cells purified from the secondary lymphoid tissues of Map3k1ΔKD mice and cultured under T helper (Th) 2 polarizing conditions secrete enhanced levels of Interleukins 4, 5, 10 and 13.43 By contrast, Th1 differentiation proceeds normally for CD4+ T cells isolated from Map3k1ΔKD mice.43 The aberrant Th2 phenotype identified in CD4+ T cells derived from Map3k1ΔKD mice resembles the overproduction of Th2 cytokines found in Itchy mice, that harbor a promoter rearrangement mutation that ablates the expression of homologous to the E6-AP carboxyl terminus (HECT) E3 Ub ligase Itch.44, 45 Overproduction of Th2 cytokines is also a phenotype of CD4+ T cells isolated from Mapk8−/− mice and transgenic mice engineered to overproduce JunB.46, 47 Map3k1ΔKD, Mapk8−/− and Itchy Th2 cells all produce similarly deficient responses in a mouse T-cell allergy model.48 As well as having an important role in CD4+ T-cell differentiation the Map3k1 kinase domain also has a negative regulatory role in the proliferative-expansion of CD8+ T cells.49
Either Map3k1 or Mapk8-Map2k7 fusion protein can enhance Itch E3 Ub ligase activity toward its substrate JunB in HEK 293 cells.43, 45 After T-cell receptor engagement Map3k1-dependent Mapk8 signaling is activated in T cells and Itch undergoes Mapk8-mediated phosphorylation of Ser199, Ser232 and Thr222 within the Itch Pro-rich region.50 Direct Itch phosphorylation by Mapk8 disrupts an inhibitory interaction that occurs between the Itch WW domains, that mediate protein–protein interactions, and the HECT domain, the E3 Ub ligase, and this change in Itch conformation leads to the significantly increased activity of the HECT domain.50 Map3k1 and Itch also form a complex within T cells as Itch is activated by Mapk8 phosphorylation.51
Despite Map3k1ΔKD T cells displaying skewed Th2 cytokine production, Map3k1ΔKD mice show both significantly reduced germinal center formation within their secondary lymphoid tissues and production of antibodies in response to thymus-dependent, but not thymus-independent, antigens.52, 53, 54 As suggested by early work measuring Map3k1 activation in B-cell lines following CD40 engagement with antibodies, Map3k1 was found to be necessary for CD40 ligand (CD40L, also known as CD154) -mediated activation of Mapk8/9 and Mapk14 in B cells.15, 52, 54 Map3k1ΔKD B cells have significantly reduced c-Jun phosphorylation and defective expression of both Cyclin D2, a regulator of cyclin-dependent kinases, and Activation-induced deaminase, a protein important in antibody diversity, that likely explains the poor humoral immune responses seen in Map3k1ΔKD mice.52, 54
Following the engagement of CD40 by CD40L, and also many other Tnfrs by their ligands, Tnf receptor-associated factor (Traf) 2 (Traf2), Traf3, Ub-conjugating enzyme E2 N (Ube2N, also known as Ubc13), cellular inhibitor of apoptosis proteins 1 and 2 (c-Iap1/2), Ikkγ and Map3k1 are recruited rapidly to the CD40 receptor.52, 53 Traf2, Ube2N and Ikkγ are all necessary components for both the assembly of the signal transduction complex at the receptor and the activation of Map3k1 and its downstream Mapks.52, 53 The CD40 Mapk signaling complex is inactive at the receptor, but the complex then translocates from the CD40 receptor into the cytosol to become active after the transfer of Lys48-linked poly-Ub by c-Iap1/2 onto Traf3 and the subsequent degradation of Traf3 by the proteasome.52, 53
Role in Stem Cells and Cardiac Myocytes
Initial analyses using Map3k1−/− ES cells showed they are deficient in Mapk activation in response to a wide variety of stimuli, including microtubule disrupting drugs, low temperature shock, hyperosmotic stress and growth factors present within the serum.29, 30 As stated above, Map3k1−/− ES cells have a greater propensity to enter apoptosis following hyperosmotic stress or treatment with microtubule disrupting drugs than wild-type (WT) ES cells.29, 30 Map3k1ΔKD ES cells display reduced Mapk8/9 activation in response to pro-inflammatory agonists, serum and LPA.38 Map3k1−/− ES cell-derived cardiac myocytes display increased sensitivity to apoptosis following hydrogen peroxide-induced stress. In response to hydrogen peroxide-induced stress Map3k1−/− ES cell-derived cardiac myocytes show reduced Mapk8/9 phosphorylation.31
Map3k1 can be activated by heart-restricted expression of Gαq, a heterotrimeric G protein subunit, in transgenic mice.55 Map3k1−/− ES cell-derived cardiac myocytes have reduced Mapk8/9 phosphorylation following treatment with the α1-Adrenergic receptor agonist phenylephrine.55 Map3k1−/− mice display increased cardiac mass, larger cardiac myocytes, elevated Atrial natriuretic factor (Anf) expression, impaired Mapk8/9 phosphorylation by Gαq and improved ventricular function.55 Map3k1−/− mice also show reduced Mapk8/9 phosphorylation following transverse aortic constriction, an indicator that Map3k1 may regulate Mapk8/9 activation following cardiac pressure overload, where cardiac muscle is forced to contract while undergoing excessive afterload.56 Pressure overload causes significant cardiac hypertrophy and increased expression of Anf in Map3k1−/− mice, which also show higher mortality and a greater ratio of lung to body mass.56 Map3k1 may be required for pressure overload-induced Mapk8/9 activation and enhanced production of Tnf-α and Tgf-β cytokines.56 Map3k1 can function in the cardiovascular system to promote cardiac myocyte cell survival, reduce inflammation and protect against cardiac failure.56
Roles in Cancer
Early analyses identified a potential role for MAP3K1 in Androgen receptor signaling in prostate cancer cell lines.57 The Androgen receptor-positive LNCaP cell line, derived from Androgen-sensitive human prostate adenocarcinoma cells, undergoes apoptosis when transduced with a retroviral vector that overexpresses the Map3k1 kinase domain.57 By contrast, Androgen receptor-negative DU145 cells and PC3 cells do not enter apoptosis when transduced with retrovirus expressing the Map3k1 kinase domain.57 Co-transduction of the Androgen receptor and Map3k1 kinase domain into DU145 cells leads to apoptosis.57 Conversely, transfection of kinase-inactive Map3k1 expressing cDNA into BxPC-3, PANC-1, MIAPaCa-2 and AsPC-1 pancreatic cancer cell lines promoted cell death in pancreatic cancer cell lines, suggesting a role for Map3k1 kinase domain signaling in tumor cell survival.58
Small interfering RNA knockdown of MAP3K1 expression in the invasive human adenocarcinoma cell line MDA-MB-231, that contains an aberrant Wnt7b oncogene, causes a significant reduction in urokinase-type plasminogen activator (uPA), a serine protease whose expression can correlate with tumor malignancy, activity.59, 60 Knockdown of MAP3K1 in MDA-MB-231 cells causes reduced migration towards serum growth factors in transwell migration assays when compared with MDA-MB-231 cells transfected with a control small interfering RNA.60 These results suggest a potential role for MAP3K1 in regulating both tumor malignancy and cancer cell invasion into normal tissues.
Map3k1−/− mice crossed to a polyoma virus middle T antigen (PyMT) transgene under control of the mouse mammary tumor virus long terminal repeat (MMTV LTR) show significantly delayed development of lung metastases.60 The dissemination of cancerous cells from Map3k1−/− mammary tumors is reduced, though the eventual formation of lung metastases still occurs in mice.60 The delay in the development of lung metastasis observed in Map3k1−/− mice is perhaps caused by the reduced integrity of the basement membranes surrounding the Map3k1−/− mammary tumors.60 Map3k1 may regulate both proteolytic degradation and migration of tumor cells in mice.60
Screening for breast cancer susceptibility alleles has identified, amongst several genes, MAP3K1 as a causative gene for breast cancer.61 In fact, ~12% of all luminal A breast cancer tumors contain mutations within MAP3K1 or MAP2K4, kinases that can regulate MAPK8/9 and MAPK14 signaling.62 Almost all breast cancer MAP3K1 and MAP2K4 mutations are found within luminal A tumors, and MAP3K1 and MAP2K4 mutations are largely mutually exclusive of each other.62 Although the role for MAP3K1 in some forms of breast cancer is substantiated by numerous reports,61, 63, 64, 65, 66, 67, 68 the importance of MAP3K1 in other cancers is far less certain. A recent low-copy transposon mutagenesis screening methodology in mice identified Map3k1 as a potentiator of melanoma.69 Point mutations within introns 9 and 10 of Map3k1 produce truncated forms of Map3k1 that lack the N-terminal regulatory region.69 N-terminal truncation of Map3k1 leads to enhanced Mapk1/3 phosphorylation in melanoma tumors, and provides a possible mechanism to explain how Map3k1 may drive malignancy in melanoma.69 It is also notable that host immunodeficiency may also contribute to the prognosis of Map3k1-dependent cancers.70
New Insights into Map3k1 PHD Motif Signaling by Gene Knockin of Map3k1
To better understand the functions of the PHD motif in mammalian biology we first modeled the Map3k1 PHD upon the known structure of the Deltex 2 Really Interesting New Gene (RING), which has homology with the Map3k1 PHD motif, and mutated conserved residues within the E2 binding region of the PHD structure (Cys438Ala and Ile440Ala, the Map3k1 mPHD mutation) to inactivate the E3 Ub ligase, and the Map3k1 mPHD mutation does not significantly reduce kinase domain activity.34, 71, 72 Transfection of Map3k1 mPHD expressing cDNA into HEK 293 cells revealed that the Map3k1 mPHD protein displays significantly impaired Map3k1 auto-ubiquitination.19, 33, 72 Analysis of the E2 conjugating enzymes that can act in concert with Ube1 and the Map3k1 PHD revealed that Ub-conjugating enzyme E2D (Ube2D) 2 (Ube2D2), Ube2D3 and Ube2N:Ub-conjugating enzyme E2 variant 1 (Ube2V1) can all mediate Map3k1 auto-ubiquitination.72 Previously it was known that the Map3k1 PHD may utilize Ube2D2 to transfer poly-Ub onto Mapk1.19, 33 Immunoblotting with anti-Lys63-linked Ub monoclonal antibodies demonstrated that the Map3k1 PHD motif largely forms Lys63-linked poly-Ub chains as opposed to linear poly-Ub chains.72 Lys63-linked poly-Ub is efficiently removed from Map3k1 by the deubiquitinating enzymes Ub-specific protease 2, 7 and 8.72
Mutation of Map3k1 alleles to express the Map3k1 mPHD (encoded by Map3k1mPHD) revealed that Map3k1mPHD ES cells do not exhibit defective Mapk1 expression following long-term hyperosmotic stress as has previously been suggested by the overexpression analysis in cell lines.19, 33, 72 This finding suggested that while Mapk1 is a substrate for Map3k1 PHD ubiquitination in cells, Map3k1 may not have a critical role in regulating Mapk1 by poly-Ub and there may also be other Mapk1 E3 Ub ligases that perform this function in cells undergoing hyperosmotic stress.19, 33 However, the kinetics of the full-length Map3k1 degradation in response to hyperosmotic stress are altered in Map3k1mPHD ES cells.72 Our findings suggested that the Map3k1 PHD can critically regulate the rapidity of its own degradation while undergoing hyperosmotic stress by auto-ubiquitination, but that other E3 Ub ligases (e.g. Deltex family E3 Ub ligases) may also act as E3 Ub ligases toward Map3k1 following hyperosmotic stress in the absence of a functional PHD motif.72, 73 Surprisingly, we identified a defective Mapk activation in Map3k1mPHD ES cells following their treatment with microtubule disrupting drugs and the cytokines Tgf-β or Egf, suggesting that the Map3k1 PHD, in fact, has an unexpected and critical role in regulating Mapk activation.72
Neither of the known Map3k1 PHD substrates (Mapk1 and c-Jun) provide an obvious explanation for the new critical role found for the PHD motif in cytokine-induced Mapk activation,19, 32, 33, 61 so we searched for novel substrates using a high throughput screening approach that analyzed a library of over 9400 human full-length proteins.74, 75 Our Ub protein array screening methodology identified 82 new proteins as potential substrates for a ubiquitination reaction containing Ube1, Ube2N:Ube2V1 and the Map3k1 PHD motif.72 Many of the PHD motif substrates are molecular scaffold proteins involved in signal transduction, and bioinformatics analysis suggested that Tgf-β activated kinase 1-binding protein (Tab) 1 (Tab1) was critical for Tgf-β-induced Mapk activation.72, 76
Ubiquitination assays by orthogonal approaches confirmed that the Map3k1 PHD can transfer Lys63-linked poly-Ub onto recombinant Tab1, and also other scaffold proteins, namely Traf2, TNFAIP3 interacting protein (Tnip) 1 (Tnip1), Tnip2 and Signal transducing adapter molecule 1 (Stam1).72 Of these Map3k1 PHD motif substrates, and after immunoprecipitation from the WT and Map3k1mPHD ES cells stimulated by Tgf-β, only Tab1 was found to be significantly ubiquitinated by Lys63-linked poly-Ub in WT and not Map3k1mPHD ES cells.72 Generation of Tab1−/− ES cells revealed that they, like Map3k1mPHD ES cells, are deficient in Egf- and Tgf-β-induced Mapk and Map3k7 (also known as Tak1) activation.72 Mapping of the Tab1 ubiquitination sites mediated by the Map3k1 PHD and Ube2N:Ube2V1 identified Lys294, Lys319, Lys335 and Lys350 as being important for Tab1 ubiquitination by the PHD motif. Map3k1 can interact with and transfer Lys63-linked poly-Ub onto Tab1 by its PHD motif to potentiate the protein–protein interaction between Tab1 and Map3k7.72 Tab2, though not itself a Map3k1 PHD substrate, can be recruited into the Tab1:Map3k1 Ub complex to form a ternary complex that is dependent upon the Tab2 zinc finger (ZnF), a motif that can interact with proteins that possess Lys63-linked poly-Ub chains.72, 77 Recruitment of Tab2 into the Map3k1:Tab1 signaling complex may facilitate a further downstream signaling from Tgf-β receptors (Tgfβrs) and Egf receptors (Egfrs) (Figure 2). The formation of a Ub signaling complex between Tab1:Map3k1:Map3k7 offers a plausible explanation for why Map3k1mPHD ES cells or WT ES cells treated with the Map3k7 chemical inhibitor (5Z)-7-oxozeaenol both lose Mapk activation following stimulation by Tgf-β or Egf cytokines.72 Chemical inhibition using the Ube2N inhibitor NSC697923 demonstrates that Ube2N is also critical for Map3k7 and Mapk activation in ES cells following treatment with Tgf-β or Egf, and Ube2N is also important for Tgf-β-induced Mapk activation in breast cancer cells.72, 78 However, unlike CD40 signaling in B cells, where Traf2 is critical for Mapk8/9 and Mapk14 activation,52, 53 instead for Tgfβrs Traf6 is critical for Mapk activation.79 The identification of a new PHD motif substrate that forms the lynchpin between Tgf-β-dependent Mapk pathway activation and stem cell differentiation further complicates the role for the Map3k1 PHD motif in apoptosis, and suggests that under conditions of cell death induced by hyperosmotic stress Map3k1 may silence Mapk signaling, while when stimulated by cytokines the Map3k1 PHD plays a role in promoting cell survival and differentiation by activating Mapks (Figure 3).
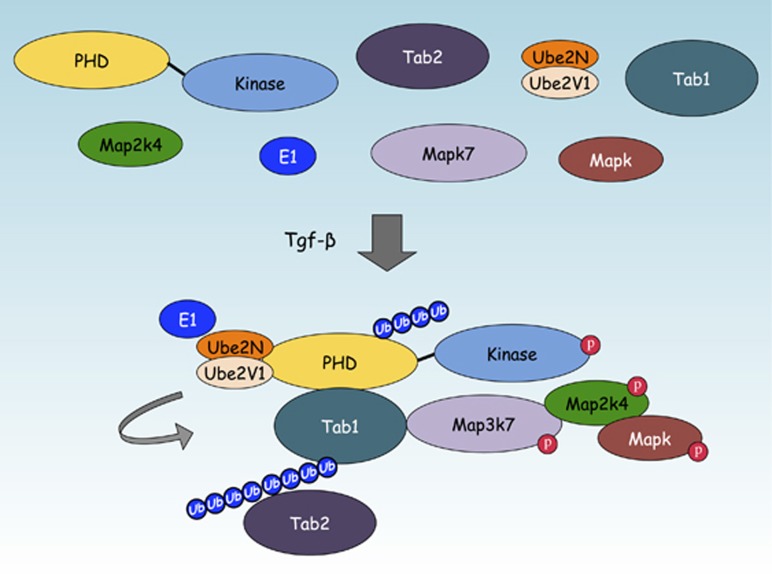
Figure 2.
The Map3k1 PHD motif regulates Tabs in response to cytokine stimulation. Following Tgf-β treatment, the Map3k1 PHD motif binds and transfers Lys63-linked poly-Ub onto Tab1 to enhance Map3k7 activation. Tab2, although not a Map3k1 PHD motif substrate, can be recruited to the Map3k1:Map3k7 Ub signaling complex by the Ub binding ZnF motif of Tab2.
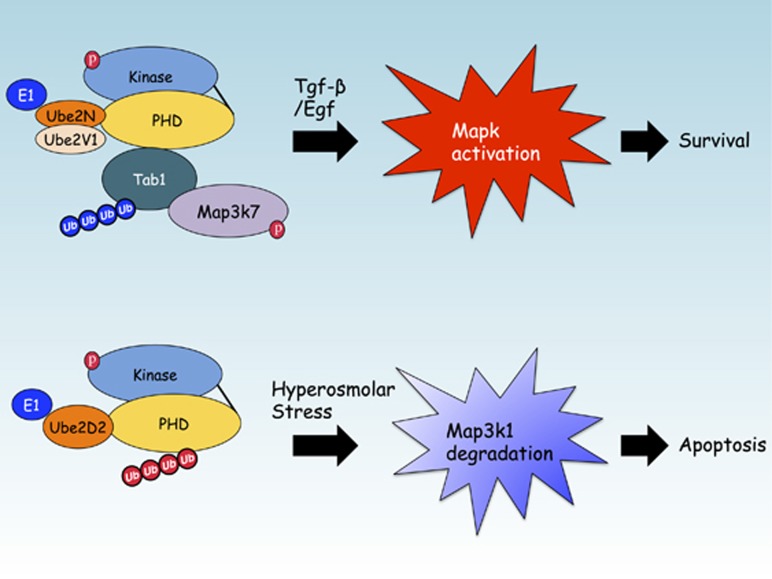
Figure 3.
The signal transduction role for the Map3k1 PHD motif during cell survival or apoptosis. In response to Tgf-β or Egf cytokines Map3k1 ubiquitinates Tab1 with Lys63-linked poly-Ub to activate Map3k7 and Mapk. This promotes both the differentiation and survival of stem cells. In response to hyperosmotic stress, stem cells enter apoptosis and the Map3k1 PHD motif transfers Lys48-linked poly-Ub onto itself, in conjunction with other E3 Ub ligases, to potentiate the degradation of the Mapk signaling cascade by the proteasome.
Both Tab1 and the Map3k1 PHD can negatively regulate neuroectoderm genes and enhance long-term expression of mesoderm genes in ES cells as they differentiate from pluripotent stem cells into embryoid bodies in cell culture.72 Mapk8−/−/Mapk9−/− double deficiency in ES cells has demonstrated that Mapk8/9 are important for driving ES cell differentiation.80 Mapk14 is also known to have an important role in ES cell differentiation by regulating neuroectoderm and mesoderm formation.81, 82 Despite these results no compound mutant Mapk8−/−/Mapk9−/−- and Mapk14−/−-deficient ES cells have been generated and analyzed to date that would produce the similar differentiation defects observed in Map3k1mPHD or Tab1−/− ES cells. Transplantation of immunodeficient host mice with either Tab1−/− or Map3k1mPHD ES cells causes aberrant tumors to form, and these have altered tissue composition and are of smaller mass and size.72 Add-back of Tab1, but not lysine-mutated (Lys294Ala, Lys319Ala, Lys335Ala and Lys350Ala) Tab1, into Tab1−/− ES cells restores normal ES cell differentiation and tumorigenesis, demonstrating that the lysines in Tab1 ubiquitinated by the Map3k1 PHD motif are critically important for ES cell differentiation and tumor formation in mice.72
Analysis of Map3k1mPHD knockin mice is complicated by their early lethality during embryogenesis, a more severe phenotype than the partial lethality observed in Map3k1ΔKD mice.72, 83 Indeed, the combination of aberrant regulation of the Ub-proteasome system and defects in Mapk signaling provide a plausible explanation for the more severe phenotypes of Map3k1mPHD mice.19, 33 However, mature Map3k1mPHD/+ mice are viable for phenotypic analysis and have demonstrated that signal transduction by the Map3k1 PHD motif is critical for B-cell development beyond the Pro-B-cell stage, T-cell receptor signal transduction and Itch phosphorylation within the pro-rich region , protecting cardiac tissue and maintaining the Leydig cell population within testis.72
Summary
From its initial discovery as the second Map2k1 kinase to defining the role of Map3k1 signal transduction in tumorigenesis and breast cancer, the road to understanding the intricate mechanisms of Map3k1 signaling has seemed to go on forever (Figure 4). Our new insights into the role of the Map3k1 PHD motif provide a fresh perspective into how Map3k1 signaling can regulate both the Ub-proteasome and protein phosphorylation systems. Analysis of Map3k1 domain-specific signaling has now revealed several important brand-new insights. At cytokine receptors and in response to microtubule disruption the Map3k1 PHD motif and kinase domain are both required for Mapk activation. But, while the Map3k1 kinase domain is required for hyperosmotic stress-dependent Mapk activation, the PHD motif is dispensable for Mapk activation under this circumstance, and instead enhances the kinetics of full-length Map3k1 degradation. Analysis of the Map3k1 PHD by mouse genetics has also demonstrated that, like the Map3k1 kinase domain, the PHD motif is important for lymphocyte T-cell receptor signaling, cardiac tissue damage and stem cells. However, it is notable that disruption of the Map3k1 PHD motif has a more dramatic effect upon B-cell development than kinase domain ablation, the degree of embryonic lethality encountered is more severe in Map3k1mPHD than Map3k1ΔKD mice and there are more severe mesoderm and neuroectoderm differentiation defects in Map3k1mPHD than Map3k1ΔKD stem cells. As such, modulation of the Map3k1 PHD motif may eventually provide an attractive alternative target to the kinase domain for future drug discovery.84
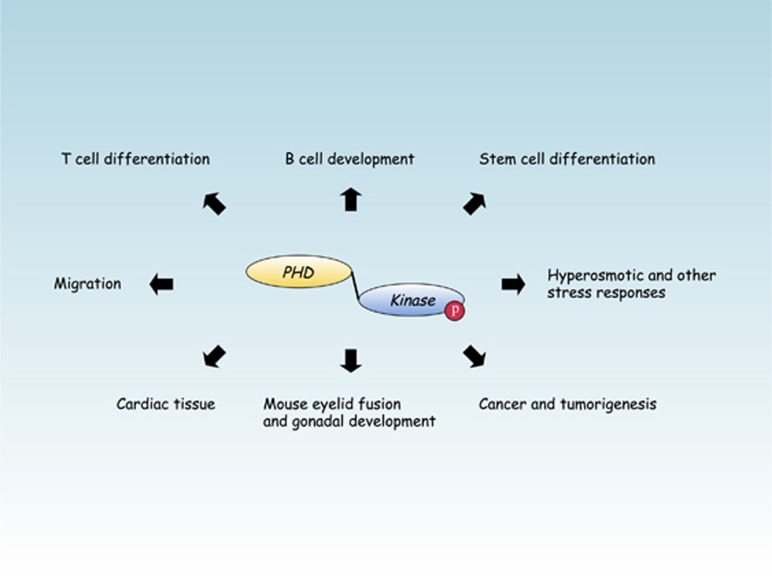
Figure 4.
Tissues, cells and diseases where mouse or human genetics have demonstrated Map3k1 signaling to be of importance.2, 30, 45, 52, 99
Acknowledgments
Our research was supported by grants from the Wellcome Trust (WT090939MA) and Cancer Research UK (C26616/A12679).
Glossary
- Anf
Atrial natriuretic factor
- Bcl-2
B-cell lymphoma 2
- c-Iap1/2
Cellular inhibitor of apoptosis proteins 1 and 2
- Caspase
Cysteine-aspartic acid protease
- DUBs
deubiquitinating enzymes
- Egf
Epidermal growth factor
- Egfrs
Egf receptors
- ES
embryonic stem
- Fak
Focal adhesion kinase
- c-Flip
FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein
- HECT
Homologous to the E6-AP Carboxyl Terminus
- Ikks
IκBα kinases
- LPA
lysophosphatidic acid
- MDCK
Madin-Darby canine kidney
- MMTV LTR
mammary tumor virus long terminal repeat
- Map2k
Mapk kinase
- Map3k
Map2k kinase
- Mapk
Mitogen-activated protein kinase
- MEF
mouse embryonic fibroblast
- NF-κB
Nuclear factor κ-light-chain-enhancer of activated B cells
- PHD
plant homeodomain
- PyMT
Polyoma Virus middle T antigen
- PRR
Pro-rich region
- RING
Really Interesting New Gene
- Stam1
Signal transducing adapter molecule 1
- SWIM
SWI2/SNF2 and MuDR
- siRNA
Small interfering RNA
- Tab
Tgf-β activated kinase 1-binding protein
- Tgfβrs
Tgf-β receptors
- Th
T helper
- Tnip
TNFAIP3 interacting protein
- Tnfr
Tnf receptor
- Traf
Tnf receptor-associated factor
- Tgf-β
Transforming growth factor-β
- Tnf
Tumor necrosis factor
- Ube2N
Ub-conjugating enzyme E2 N
- UIMs
Ub-interacting motifs
- Ub
Ubiquitin
- Upa
urokinase-type Plasminogen Activator
- Ube2D
Ub-conjugating enzyme E2D
- Ube2V1
Ub-conjugating enzyme E2 variant 1
- Usp
Ub-specific protease
- ZnF
zinc finger
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Xia Y, Karin M. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends in cell biology. 2004;14:94–101. doi: 10.1016/j.tcb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Karin M, Gallagher E, From JNK. to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57:283–295. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Lange-Carter CA, Pleiman CM, Gardner AM, Blumer KJ, Johnson GL. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- Blumer KJ, Johnson GL, Lange-Carter CA. Mammalian mitogen-activated protein kinase kinase kinase (MEKK) can function in a yeast mitogen-activated protein kinase pathway downstream of protein kinase C. Proc Natl Acad Sci USA. 1994;91:4925–4929. doi: 10.1073/pnas.91.11.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AM, Vaillancourt RR, Lange-Carter CA, Johnson GL. MEK-1 phosphorylation by MEK kinase, Raf, and mitogen-activated protein kinase: analysis of phosphopeptides and regulation of activity. Mol Biol Cell. 1994;5:193–201. doi: 10.1091/mbc.5.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Templeton DJ. Identification of 2 serine residues of MEK-1 that are differentially phosphorylated during activation by raf and MEK kinase. J Biol Chem. 1994;269:19067–19073. [PubMed] [Google Scholar]
- Gallagher ED, Xu S, Moomaw C, Slaughter CA, Cobb MH. Binding of JNK/SAPK to MEKK1 is regulated by phosphorylation. J Biol Chem. 2002;277:45785–45792. doi: 10.1074/jbc.M207702200. [DOI] [PubMed] [Google Scholar]
- Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis RJ, et al. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- Yan M, Dai T, Deak JC, Kyriakis JM, Zon LI, Woodgett JR, et al. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- Xia Y, Wu Z, Su B, Murray B, Karin M. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 1998;12:3369–3381. doi: 10.1101/gad.12.21.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Minden A, Martinetto H, Claret FX, Lange-Carter C, Mercurio F, et al. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- Winston BW, Lange-Carter CA, Gardner AM, Johnson GL, Riches DW. Tumor necrosis factor alpha rapidly activates the mitogen-activated protein kinase (MAPK) cascade in a MAPK kinase kinase-dependent, c-Raf-1-independent fashion in mouse macrophages. Proc Natl Acad Sci USA. 1995;92:1614–1618. doi: 10.1073/pnas.92.5.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata N, Patel HR, Terada N, Aruffo A, Johnson GL, Gelfand EW. Selective activation of c-Jun kinase mitogen-activated protein kinase by CD40 on human B cells. J Biol Chem. 1995;270:30823–30828. doi: 10.1074/jbc.270.51.30823. [DOI] [PubMed] [Google Scholar]
- Xu S, Robbins DJ, Christerson LB, English JM, Vanderbilt CA, Cobb MH. Cloning of rat MEK kinase 1 cDNA reveals an endogenous membrane-associated 195-kDa protein with a large regulatory domain. Proc Natl Acad Sci USA. 1996;93:5291–5295. doi: 10.1073/pnas.93.11.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Osada S, Aoki T, Hirai S, Hosaka M, Inoue J, et al. MEK kinase is involved in tumor necrosis factor alpha-induced NF-kappaB activation and degradation of IkappaB-alpha. J Biol Chem. 1996;271:13234–13238. doi: 10.1074/jbc.271.22.13234. [DOI] [PubMed] [Google Scholar]
- Lee FS, Hagler J, Chen ZJ, Maniatis T. Activation of the IkappaB alpha kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell. 2002;9:945–956. doi: 10.1016/s1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- Rieger MA, Duellman T, Hooper C, Ameka M, Bakowska JC, Cuevas BD. The MEKK1 SWIM domain is a novel substrate receptor for c-Jun ubiquitylation. Biochem J. 2012;445:431–439. doi: 10.1042/BJ20120406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NL, Gardner AM, Diener KM, Lange-Carter CA, Gleavy J, Jarpe MB, et al. Signal transduction pathways regulated by mitogen-activated/extracellular response kinase kinase kinase induce cell death. J Biol Chem. 1996;271:3229–3237. doi: 10.1074/jbc.271.6.3229. [DOI] [PubMed] [Google Scholar]
- Widmann C, Johnson NL, Gardner AM, Smith RJ, Johnson GL. Potentiation of apoptosis by low dose stress stimuli in cells expressing activated MEK kinase 1. Oncogene. 1997;15:2439–2447. doi: 10.1038/sj.onc.1201421. [DOI] [PubMed] [Google Scholar]
- Widmann C, Gerwins P, Johnson NL, Jarpe MB, Johnson GL. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol Cell Biol. 1998;18:2416–2429. doi: 10.1128/mcb.18.4.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak JC, Cross JV, Lewis M, Qian Y, Parrott LA, Distelhorst CW, et al. Fas-induced proteolytic activation and intracellular redistribution of the stress-signaling kinase MEKK1. Proc Natl Acad Sci USA. 1998;95:5595–5600. doi: 10.1073/pnas.95.10.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MH, Salvesen GS, Widmann C, Johnson G, Frisch SM. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- Gibson S, Widmann C, Johnson GL. Differential involvement of MEK kinase 1 (MEKK1) in the induction of apoptosis in response to microtubule-targeted drugs versus DNA damaging agents. J Biol Chem. 1999;274:10916–10922. doi: 10.1074/jbc.274.16.10916. [DOI] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Adler V, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yujiri T, Fanger GR, Garrington TP, Schlesinger TK, Gibson S, Johnson GL. MEK kinase 1 (MEKK1) transduces c-Jun NH2-terminal kinase activation in response to changes in the microtubule cytoskeleton. J Biol Chem. 1999;274:12605–12610. doi: 10.1074/jbc.274.18.12605. [DOI] [PubMed] [Google Scholar]
- Yujiri T, Sather S, Fanger GR, Johnson GL. Role of MEKK1 in cell survival and activation of JNK and ERK pathways defined by targeted gene disruption. Science. 1998;282:1911–1914. doi: 10.1126/science.282.5395.1911. [DOI] [PubMed] [Google Scholar]
- Minamino T, Yujiri T, Papst PJ, Chan ED, Johnson GL, Terada N. MEKK1 suppresses oxidative stress-induced apoptosis of embryonic stem cell-derived cardiac myocytes. Proc Natl Acad Sci USA. 1999;96:15127–15132. doi: 10.1073/pnas.96.26.15127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Wang J, Xu S, Johnson GL, Hunter T, Lu Z. MEKK1 mediates the ubiquitination and degradation of c-Jun in response to osmotic stress. Mol Cell Biol. 2007;27:510–517. doi: 10.1128/MCB.01355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hunter T. Degradation of activated protein kinases by ubiquitination. Annu Rev Biochem. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Robertson M. Ubiquitin ligases and beyond. BMC biology. 2012;10:22. doi: 10.1186/1741-7007-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yang W, Bu W, Ji H, Zhao X, Zheng Y, et al. Differential regulation of c-Jun protein plays an instrumental role in chemoresistance of cancer cells. J Biol Chem. 2013;288:19321–19329. doi: 10.1074/jbc.M113.475442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker E, Arvand A, Kwan R, Chen GY, Gallagher E, Cheng G. Apoptosis induced by cytoskeletal disruption requires distinct domains of MEKK1. PloS One. 2011;6:e17310. doi: 10.1371/journal.pone.0017310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed E, Elbediwy A, Vacca B, Dupasquier S, Hemkemeyer SA, Suddason T, et al. MarvelD3 couples tight junctions to the MEKK1-JNK pathway to regulate cell behavior and survival. J Cell Biol. 2014;204:821–838. doi: 10.1083/jcb.201304115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Makris C, Su B, Li E, Yang J, Nemerow GR, et al. MEK kinase 1 is critically required for c-Jun N-terminal kinase activation by proinflammatory stimuli and growth factor-induced cell migration. Proc Natl Acad Sci USA. 2000;97:5243–5248. doi: 10.1073/pnas.97.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang W, Hayashi Y, Jester JV, Birk DE, Gao M, et al. A role for MEK kinase 1 in TGF-beta/activin-induced epithelium movement and embryonic eyelid closure. EMBO J. 2003;22:4443–4454. doi: 10.1093/emboj/cdg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas BD, Abell AN, Witowsky JA, Yujiri T, Johnson NL, Kesavan K, et al. MEKK1 regulates calpain-dependent proteolysis of focal adhesion proteins for rear-end detachment of migrating fibroblasts. EMBO J. 2003;22:3346–3355. doi: 10.1093/emboj/cdg322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christerson LB, Vanderbilt CA, Cobb MH. MEKK1 interacts with alpha-actinin and localizes to stress fibers and focal adhesions. Cell Motil Cytoskeleton. 1999;43:186–198. doi: 10.1002/(SICI)1097-0169(1999)43:3<186::AID-CM2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Glading A, Bodnar RJ, Reynolds IJ, Shiraha H, Satish L, Potter DA, et al. Epidermal growth factor activates m-calpain (calpain II), at least in part, by extracellular signal-regulated kinase-mediated phosphorylation. Mol Cell Biol. 2004;24:2499–2512. doi: 10.1128/MCB.24.6.2499-2512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Fang D, Elly C, Gao B, Fang N, Altman Y, Joazeiro C, et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nature Immunol. 2002;3:281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- Gao M, Karin M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol Cell. 2005;19:581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Dong C, Yang DD, Tournier C, Whitmarsh AJ, Xu J, Davis RJ, et al. JNK is required for effector T-cell function but not for T-cell activation. Nature. 2000;405:91–94. doi: 10.1038/35011091. [DOI] [PubMed] [Google Scholar]
- Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad K, Elly C, Gao M, Salek-Ardakani S, Harada Y, Luo JL, et al. Convergence of Itch-induced ubiquitination with MEKK1-JNK signaling in Th2 tolerance and airway inflammation. J Clin Invest. 2006;116:1117–1126. doi: 10.1172/JCI26858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda T, Christensen JP, Rasmussen S, Bonnesen B, Karin M, Thomsen AR, et al. MEK kinase 1 is a negative regulator of virus-specific CD8(+) T cells. Eur J Immunol. 2006;36:2076–2084. doi: 10.1002/eji.200535163. [DOI] [PubMed] [Google Scholar]
- Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzler T, Chang X, Facchinetti V, Melino G, Karin M, Su B, et al. MEKK1 binds HECT E3 ligase Itch by its amino-terminal RING motif to regulate Th2 cytokine gene expression. J Immunol. 2009;183:3831–3838. doi: 10.4049/jimmunol.0803412. [DOI] [PubMed] [Google Scholar]
- Karin M, Gallagher E. TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol Rev. 2009;228:225–240. doi: 10.1111/j.1600-065X.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, et al. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science. 2008;321:663–668. doi: 10.1126/science.1157340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher E, Enzler T, Matsuzawa A, Anzelon-Mills A, Otero D, Holzer R, et al. Kinase MEKK1 is required for CD40-dependent activation of the kinases Jnk and p38, germinal center formation, B cell proliferation and antibody production. Nature Immunol. 2007;8:57–63. doi: 10.1038/ni1421. [DOI] [PubMed] [Google Scholar]
- Minamino T, Yujiri T, Terada N, Taffet GE, Michael LH, Johnson GL, et al. MEKK1 is essential for cardiac hypertrophy and dysfunction induced by Gq. Proc Natl Acad Sci USA. 2002;99:3866–3871. doi: 10.1073/pnas.062453699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoshima J, Montagne O, Wang Q, Yang G, Warden J, Liu J, et al. The MEKK1-JNK pathway plays a protective role in pressure overload but does not mediate cardiac hypertrophy. J Clin Invest. 2002;110:271–279. doi: 10.1172/JCI14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu-Martin MT, Chari A, Palladino AA, Craft NA, Sawyers CL. Mitogen-activated protein kinase kinase kinase 1 activates androgen receptor-dependent transcription and apoptosis in prostate cancer. Mol Cell Biol. 1999;19:5143–5154. doi: 10.1128/mcb.19.7.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Shino Y, Saito T, Komoda F, Okutomi Y, Takeda A, et al. Dominant negative MEKK1 inhibits survival of pancreatic cancer cells. Oncogene. 2002;21:5923–5928. doi: 10.1038/sj.onc.1205643. [DOI] [PubMed] [Google Scholar]
- Blasi F, Vassalli JD, Dano K. Urokinase-type plasminogen activator: proenzyme, receptor, and inhibitors. J Cell Biol. 1987;104:801–804. doi: 10.1083/jcb.104.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas BD, Winter-Vann AM, Johnson NL, Johnson GL. MEKK1 controls matrix degradation and tumor cell dissemination during metastasis of polyoma middle-T driven mammary cancer. Oncogene. 2006;25:4998–5010. doi: 10.1038/sj.onc.1209507. [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanale D, Amodeo V, Corsini LR, Rizzo S, Bazan V, Russo A. Breast cancer genome-wide association studies: there is strength in numbers. Oncogene. 2012;31:2121–2128. doi: 10.1038/onc.2011.408. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif A, Hadfield KD, Roberts SA, Shenton A, Lalloo F, Black GC, et al. Breast cancer susceptibility variants alter risks in familial disease. J Med Genet. 2010;47:126–131. doi: 10.1136/jmg.2009.067256. [DOI] [PubMed] [Google Scholar]
- Gorodnova TV, Kuligina E, Yanus GA, Katanugina AS, Abysheva SN, Togo AV, et al. Distribution of FGFR2, TNRC9, MAP3K1, LSP1, and 8q24 alleles in genetically enriched breast cancer patients versus elderly tumor-free women. Cancer Genet Cytogenet. 2010;199:69–72. doi: 10.1016/j.cancergencyto.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Santisteban M, Reiman JM, Asiedu MK, Behrens MD, Nassar A, Kalli KR, et al. Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Cancer Res. 2009;69:2887–2895. doi: 10.1158/0008-5472.CAN-08-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger T, Gadzicki D, Meindl A, Schlegelberger B. Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur J Human Genet. 2009;17:722–731. doi: 10.1038/ejhg.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni TK, Landrette SF, Bjornson RD, Bosenberg MW, Xu T. Low-copy piggyBac transposon mutagenesis in mice identifies genes driving melanoma. Proc Natl Acad Sci USA. 2013;110:E3640–E3649. doi: 10.1073/pnas.1314435110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol. 2007;19:209–216. doi: 10.1016/j.coi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, Barrick D. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure. 2005;13:1599–1611. doi: 10.1016/j.str.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Charlaftis N, Suddason T, Wu X, Anwar S, Karin M, Gallagher E. The MEKK1 PHD ubiquitinates TAB1 to activate MAPKs in response to cytokines. EMBO J. 2014;33:2581–2596. doi: 10.15252/embj.201488351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WH, Lai MZ. Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol Cell Biol. 2005;25:1367–1378. doi: 10.1128/MCB.25.4.1367-1378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbl Y, Kirschner MW. Protein microarrays for genome-wide posttranslational modification analysis. WIRE Syst Biol Med. 2011;3:347–356. doi: 10.1002/wsbm.120. [DOI] [PubMed] [Google Scholar]
- Merbl Y, Kirschner MW. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci USA. 2009;106:2543–2548. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, et al. TAB1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science. 1996;272:1179–1182. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Wu X, Zhang W, Font-Burgada J, Palmer T, Hamil AS, Biswas SK, et al. Ubiquitin-conjugating enzyme Ubc13 controls breast cancer metastasis through a TAK1-p38 MAP kinase cascade. Proc Natil Acad Sci USA. 2014;111:13870–13875. doi: 10.1073/pnas.1414358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Davis RJ. c-Jun NH2-terminal kinase is required for lineage-specific differentiation but not stem cell self-renewal. Mol Cell Biol. 2010;30:1329–1340. doi: 10.1128/MCB.00795-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barruet E, Hadadeh O, Peiretti F, Renault VM, Hadjal Y, Bernot D, et al. p38 mitogen activated protein kinase controls two successive-steps during the early mesodermal commitment of embryonic stem cells. Stem Cells Dev. 2011;20:1233–1246. doi: 10.1089/scd.2010.0213. [DOI] [PubMed] [Google Scholar]
- Gaur M, Ritner C, Sievers R, Pedersen A, Prasad M, Bernstein HS, et al. Timed inhibition of p38MAPK directs accelerated differentiation of human embryonic stem cells into cardiomyocytes. Cytotherapy. 2010;12:807–817. doi: 10.3109/14653249.2010.491821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnesen B, Orskov C, Rasmussen S, Holst PJ, Christensen JP, Eriksen KW, et al. MEK kinase 1 activity is required for definitive erythropoiesis in the mouse fetal liver. Blood. 2005;106:3396–3404. doi: 10.1182/blood-2005-04-1739. [DOI] [PubMed] [Google Scholar]
- Goto M, Chow J, Muramoto K, Chiba K, Yamamoto S, Fujita M, et al. E6201 [(3S,4R,5Z,8S,9S,11E)-14-(ethylamino)-8, 9,16-trihydroxy-3,4-dimethyl-3,4,9,19-tetrahydro-1H-2-benzoxacyclotetradecine-1,7 (8H)-dione], a novel kinase inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK)-1 and MEK kinase-1: in vitro characterization of its anti-inflammatory and antihyperproliferative activities. J Pharmacol Exp Therapeut. 2009;331:485–495. doi: 10.1124/jpet.109.156554. [DOI] [PubMed] [Google Scholar]
- Su YC, Han J, Xu S, Cobb M, Skolnik EY. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanger GR, Widmann C, Porter AC, Sather S, Johnson GL, Vaillancourt RR. 14-3-3 proteins interact with specific MEK kinases. J Biol Chem. 1998;273:3476–3483. doi: 10.1074/jbc.273.6.3476. [DOI] [PubMed] [Google Scholar]
- Xu S, Cobb MH. MEKK1 binds directly to the c-Jun N-terminal kinases/stress-activated protein kinases. J Biol Chem. 1997;272:32056–32060. doi: 10.1074/jbc.272.51.32056. [DOI] [PubMed] [Google Scholar]
- Karandikar M, Xu S, Cobb MH. MEKK1 binds raf-1 and the ERK2 cascade components. J Biol Chem. 2000;275:40120–40127. doi: 10.1074/jbc.M005926200. [DOI] [PubMed] [Google Scholar]
- Gallagher ED, Gutowski S, Sternweis PC, Cobb MH. RhoA binds to the amino terminus of MEKK1 and regulates its kinase activity. J Biol Chem. 2004;279:1872–1877. doi: 10.1074/jbc.M309525200. [DOI] [PubMed] [Google Scholar]
- Fanger GR, Johnson NL, Johnson GL. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997;16:4961–4972. doi: 10.1093/emboj/16.16.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell M, Lange-Carter CA, Johnson GL. Direct interaction between Ras and the kinase domain of mitogen-activated protein kinase kinase kinase (MEKK1) J Biol Chem. 1995;270:11757–11760. doi: 10.1074/jbc.270.20.11757. [DOI] [PubMed] [Google Scholar]
- Christerson LB, Gallagher E, Vanderbilt CA, Whitehurst AW, Wells C, Kazempour R, et al. p115 Rho GTPase activating protein interacts with MEKK1. J Cell Physiol. 2002;192:200–208. doi: 10.1002/jcp.10125. [DOI] [PubMed] [Google Scholar]
- Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerance M, Multon MC, Parker F, Venot C, Blondeau JP, Tocque B, et al. Grb2 interaction with MEK-kinase 1 is involved in regulation of Jun-kinase activities in response to epidermal growth factor. J Biol Chem. 1998;273:24301–24304. doi: 10.1074/jbc.273.38.24301. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Neo SY, Wang X, Han J, Lin SC. Axin forms a complex with MEKK1 and activates c-Jun NH(2)-terminal kinase/stress-activated protein kinase through domains distinct from Wnt signaling. J Biol Chem. 1999;274:35247–35254. doi: 10.1074/jbc.274.49.35247. [DOI] [PubMed] [Google Scholar]
- Yujiri T, Nawata R, Takahashi T, Sato Y, Tanizawa Y, Kitamura T, et al. MEK kinase 1 interacts with focal adhesion kinase and regulates insulin receptor substrate-1 expression. J Biol Chem. 2003;278:3846–3851. doi: 10.1074/jbc.M206087200. [DOI] [PubMed] [Google Scholar]
- Yin MJ, Christerson LB, Yamamoto Y, Kwak YT, Xu S, Mercurio F, et al. HTLV-I Tax protein binds to MEKK1 to stimulate IkappaB kinase activity and NF-kappaB activation. Cell. 1998;93:875–884. doi: 10.1016/s0092-8674(00)81447-6. [DOI] [PubMed] [Google Scholar]
- Ritterhoff S, Farah CM, Grabitzki J, Lochnit G, Skurat AV, Schmitz ML. The WD40-repeat protein Han11 functions as a scaffold protein to control HIPK2 and MEKK1 kinase functions. EMBO J. 2010;29:3750–3761. doi: 10.1038/emboj.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman A, Loke J, Le Caignec C, White S, Chin L, Friedman A, et al. Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. Am J Human Genet. 2010;87:898–904. doi: 10.1016/j.ajhg.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]