Co-crystal structures of P-glycoprotein with a series of engineered ligands reveal multiple ligand-binding modes, a ligand-binding site on the outer surface of the transporter and a conformational change that may couple to ATP hydrolysis.
Keywords: P-glycoprotein
Abstract
P-glycoprotein (P-gp) is a transporter of great clinical and pharmacological significance. Several structural studies of P-gp and its homologs have provided insights into its transport cycle, but questions remain regarding how P-gp recognizes diverse substrates and how substrate binding is coupled to ATP hydrolysis. Here, four new P-gp co-crystal structures with a series of rationally designed ligands are presented. It is observed that the binding of certain ligands, including an ATP-hydrolysis stimulator, produces a large conformational change in the fourth transmembrane helix, which is positioned to potentially transmit a signal to the nucleotide-binding domains. A new ligand-binding site on the surface of P-gp facing the inner leaflet of the membrane is also described, providing vital insights regarding the entry mechanism of hydrophobic drugs and lipids into P-gp. These results represent significant advances in the understanding of how P-gp and related transporters bind and export a plethora of metabolites, antibiotics and clinically approved and pipeline drugs.
1. Introduction
Membrane-embedded transporter proteins mediate the passage of metabolites and toxins across cellular membranes. The transporter P-glycoprotein (P-gp; ABCB1; EC 3.6.3.44) is expressed in the intestines, liver, kidney and blood–brain barrier, and affects the bioavailability, pharmacokinetics and efficacy of drugs (Cascorbi, 2006 ▶). P-gp also causes cellular multidrug resistance, hindering the treatment of several diseases, including cancers and HIV (Eckford & Sharom, 2009 ▶; Gottesman & Ling, 2006 ▶; Falasca & Linton, 2012 ▶). As such, the US Food and Drug Administration (FDA) now mandates the documentation of P-gp–drug interactions for the approval of any new drug (Giacomini et al., 2010 ▶; US Food and Drug Administration, 2012 ▶). Thus, the inhibition or evasion of P-gp without compromising therapeutic efficacy is a major goal of the pharmaceutical industry.
P-gp belongs to a superfamily of ATP-binding cassette (ABC) transporters found throughout all kingdoms of life. The transporter consists of two pseudosymmetric halves encoded into a single polypeptide. Each half is formed by six transmembrane helices (TMs) and one cytosolic nucleotide-binding domain (NBD) along with interconnecting loops and short helices. The two structural halves enclose a central pocket/cavity which contains multiple discrete binding sites for ligands of P-gp, drugs and transport substrates. P-gp and related ABC transporters drive substrate translocation using ATP binding and hydrolysis at the cytosolic NBDs. Because P-gp can transport a wide array of structurally diverse compounds, a molecular framework for understanding how different ligands enter and bind is crucial to overcoming P-gp-mediated drug efflux.
The crystallography of protein–ligand complexes can be a challenge at low to moderate diffraction resolutions. For co-crystals comprising transporters in complex with ligands of relatively low binding affinity (in this case the transport substrates), the level of difficulty in resolving their positions increases. Our previous structures of P-gp bound to two enantiomeric cyclopeptides provided the first structural view detailing how P-gp recognizes and binds ligands (Aller et al., 2009 ▶). In that study, we confirmed the presence of selenium-labelled ligands using anomalous diffraction methods. Unfortunately, the lower resolution of those structures (4.4 and 4.35 Å) limited the interpretation of ligand–P-gp interactions. Here, we report higher resolution (3.4–3.8 Å) structures of P-gp bound to a series of rationally engineered selenium-labelled cyclopeptide compounds that were designed to probe the ligand-binding pocket of P-gp. In doing so, we reveal how drug substrates (i) may enter the transporter, (ii) bind at multiple, shared yet discrete sites in the transmembrane domain (TMD) and (iii) might transmit this information to the NBDs to stimulate ATP hydrolysis. Together, these new structures provide snapshots of how structurally similar ligands, differing only in side-chain R-group size, can bind prior to drug efflux. These higher resolution P-gp structures also resolve, to an extent, the issue of TM registry (Jin et al., 2012 ▶; Li et al., 2014 ▶).
2. Materials and methods
2.1. Synthesis of QZ-homotrimers
N-Boc-(S)-amino-acid selenazole esters were prepared from the corresponding N-Boc-(S)-amino acids according to a previously published procedure (Tao et al., 2011 ▶). The selenazole ester derived from N-Boc-(S)-Ala (500 mg, 1.44 mmol) was subjected to hydrolysis by a solution of NaOH in mixed solvents (5:1:2 THF:MeOH:H2O) followed by Boc deprotection by 50% trifluoroacetic acid in dichloromethane. The resulting amino acid (∼1.4 mmol) was dissolved in anhydrous acetonitrile (14 ml) and treated with diisopropylethylamine (DIEA; 1.0 ml, 6.1 mmol) and pentafluorophenol diphenylphosphinate (FDPP; 1.1 g, 2.9 mmol). The reaction was stirred at room temperature for 24 h before being concentrated under vacuum. The residue was dissolved in dichloromethane and washed successively with NaHCO3, 5% HCl aqueous solution and NaCl solution. Organic phases were concentrated and the residue was purified by silica-gel chromatography. The major product was collected and identified as the desired product QZ-Ala (92 mg, 32%).
The other cyclopeptides were obtained by following the same procedure. The purity of the individual cyclopeptides was >98% based on NMR spectroscopy (Supplementary Fig. S1) and reverse-phase and chiral high-performance liquid-chromatography (HPLC) analyses.
QZ-Ala was obtained as a white solid with a 32% yield. 13C NMR (75 MHz, CDCl3) δ (p.p.m.): 178.1 (3C), 160.0 (3C), 149.7 (3C), 131.4 (3C), 50.3 (3C), 25.2 (3C). 1H NMR (300 MHz, CDCl3) δ (p.p.m.): 8.86 (s, 3 × 1H), 8.50 (d, J = 7.9 Hz, 3 × 1H), 5.67 (dq, J = 13.7, 6.8 Hz, 3 × 1H), 1.70 (d, J = 6.8 Hz, 3 × 3H). HRMS (ESI-TOF) calculated for C18H19N6O3Se3: (M + H)+, 606.9009; found, 606.9008.
The data for the characterization of QZ-Val have previously been reported (Aller et al., 2009 ▶).
QZ-Leu was obtained as a white solid with a 45% yield. 13C NMR (75 MHz, CDCl3) δ (p.p.m.): 177.3 (3C), 160.1 (3C), 150.1 (3C), 130.9 (3C), 51.7 (3C), 47.9 (3C), 25.2 (3C), 23.0 (3C), 22.5 (3C). 1H NMR (300 MHz, CDCl3) δ (p.p.m.): 8.76 (s, 3 × 1 H), 8.25 (d, J = 9.6 Hz, 3 × 1H), 5.85–5.56 (m, 3 × 1H), 1.98–1.87 (m, 3 × 1H), 1.84–1.68 (m, 3 × 2H), 1.04 (d, J = 5.9 Hz, 3 × 3H), 1.00 (d, J = 6.1 Hz, 3 × 3H). HRMS (ESI-TOF) calculated for C27H37N6O3Se3: (M + H)+, 733.0417; found, 733.0418.
QZ-Phe was obtained as a white solid with a 40% yield. 13C NMR (75 MHz, CDCl3) δ (p.p.m.): 175.0 (3C), 160.1 (3C), 149.5 (3C), 136.3 (3 × 2C), 131.5 (3C), 130.0 (3 × 2C), 128.9 (3C), 127.5 (3C), 55.5 (3C), 44.0 (3C). 1H NMR (300 MHz, CDCl3) δ (p.p.m.): 8.72 (s, 3 × 1H), 8.48 (d, J = 8.5 Hz, 3 × 1H), 7.32–7.21 (m, 3 × 3H), 7.17–7.11 (m, 3 × 2H), 5.76 (td, J = 9.0, 4.8 Hz, 3 × 1H), 3.50 (dd, J = 13.1, 4.7 Hz, 3 × 1H), 3.04 (dd, J = 13.0, 9.4 Hz, 3 × 1H). HRMS (ESI-TOF) calculated for C36H31N6O3Se3: (M + H)+, 834.9948; found, 834.9949.
2.2. Measurement of ATPase activity
The ATPase activity of P-gp was measured at 310 K using the ATP-regenerating system described by Vogel & Steinhart (1976 ▶) as modified by Urbatsch et al. (1995 ▶). Briefly, 1 µg P-gp was added to 100 µl 50 mM Tris–HCl pH 7.5 buffer containing 10 mM ATP, 12 mM MgCl2, 6 mM phosphoenolpyruvate, 1 mM NADH, 10 units of lactate dehydrogenase, 10 units of pyruvate kinase and test compounds over a range of concentrations. ATP hydrolysis was determined by the decrease in NADH absorbance at OD340 using a Filtermax F5 Multiplate Spectrophotometer. The ATPase activity was calculated using the equation ΔOD/(∊ × [protein] × time), where ΔOD is the change in absorbance and ∊ is the extinction coefficient of NADH. The concentration of purified P-gp was estimated by comparing the SDS–PAGE intensity of Coomassie-stained protein bands with known amounts of BSA. EC50 values were calculated using GraphPad Prism (GraphPad Software, San Diego, California, USA) using nonlinear regression (curve fit) from the entire concentration range.
2.3. Calcein-AM transport assay
Calcein-AM (calcein acetoxymethyl ester) is a membrane-permeable P-gp substrate, while its free acid, calcein, hydrolyzed by endogeneous esterases, is trapped in the cytosol and exhibits strong fluorescence. It has commonly been used in fluorescence-based transport assays for P-gp in whole-cell systems (Al-Shawi & Senior, 1993 ▶). Chinese hamster ovary CR1R12 cells were cultured in the presence of 5 µg ml−1 colchicine to maintain P-gp overexpression. In general, ∼5 × 105 CR1R12 cells per well were pretreated with test compounds in serially increasing concentrations at room temperature for 15 min; calcein-AM (0.25 µM) was then added and incubated for an additional 15 min at room temperature while monitoring the fluorescence intensity (excitation at 485 nm, emission at 535 nm). 100% was the maximum fluorescence achieved by full inhibition of P-gp in CR1R12 cells. The means and standard deviations were obtained from quadruplet experiments.
2.4. Sensitization assay
CR1R12 cells were grown in the presence of increasing concentrations of colchicine for 3 d at given concentrations of test compounds. Cell densities were determined using the sulforhodamine B colorimetric assay (Vichai & Kirtikara, 2006 ▶). 100% is defined as the growth in the absence of colchicine. Parental AUXB1 cells were included as controls.
2.5. Expression, purification, reductive methylation and crystallization of P-gp
Gene-optimized mouse P-gp (ABCB1a; GenBank JF834158) was expressed in Pichia pastoris in 10 l cultures in a BioFlo 415 bioreactor (New Brunswick Scientific) and was induced by the slow addition of methanol (3.6 ml per hour per litre of culture volume) overnight as described previously (Aller et al., 2009 ▶; Bai et al., 2011 ▶). Cells were lysed at 287 MPa by a single pass through a Constant cell disrupter (TS Series, Constant Systems Inc.). Cellular debris was removed by centrifugation (12 500g, 20 min, 277 K) and crude membranes were prepared by centrifugation at 38 000g for 2–3 h at 277 K. P-gp was purified as described previously (Aller et al., 2009 ▶) with modifications. Briefly, membranes containing P-gp were resuspended in cold buffer (100 mM NaCl, 15% glycerol, 20 mM Tris pH 8.0, 23.4 µM leupeptin, 7 µM E-64, 4 µM chymostatin, 14.5 µM pepstatin A, 1 mM PMSF, 25 mM benzamidine) and solubilized with 4.5% Triton X-100 for 1–2 h at 277 K. Centrifugation at 38 400g and 277 K for 30–60 min removed insoluble material, and the supernatant was applied onto Ni–NTA Superflow resin (Qiagen) using FPLC (ÄKTA, GE Life Sciences). The column was then washed with buffer consisting of 0.5 mM TCEP (Thermo Scientific), 0.04% sodium cholate (Sigma), 20 mM imidazole pH 8.0, 4.5% Triton X-100, 100 mM NaCl, 20 mM Tris–HCl pH 8.0, 14% glycerol. Resin-immobilized P-gp was buffer-exchanged into 20 mM HEPES pH 8.0, 0.2 mM TCEP, 100 mM NaCl, 20 mM imidazole, 0.04% sodium cholate, 0.065% β-DDM. P-gp was eluted with 20 mM HEPES pH 7.5, 100 mM NaCl, 0.2 mM TCEP, 0.04% sodium cholate, 200 mM imidazole pH 7.5, 0.065% β-DDM. The eluted protein was then diluted 1:10 with 20 mM HEPES pH 8.0, 100 mM NaCl, 0.2 mM TCEP, 0.04% sodium cholate, 0.065% β-DDM and rebound to Ni–NTA Superflow resin. This resin was washed with the buffer described above containing 20 mM imidazole and eluted with the buffer described above containing 200 mM imidazole. The protein was then concentrated (Centricon YM-50 or YM-100; Millipore), spun at 95 000 rev min−1 (TLA120.1 rotor) for 30–60 min at 277 K and subjected to size-exclusion chromatography (SEC; Superdex 200 16/60, GE Healthcare) at 277 K.
Following SEC, P-gp (at ∼1–2 mg ml−1) was subjected to reductive methylation (Rayment, 1997 ▶). Freshly made borane dimethylamine and formaldehyde were added to the protein solution to final concentrations of 50 and 100 mM, respectively, and then incubated for 2 h at 4°C. The reaction was quenched by the addition of ice-cold 25 mM glycine and was incubated for 30 min at 4°C. Methylated P-gp was then concentrated to 1 ml (Centricon YM-50 or YM-100; Millipore), diluted with 9 ml quench buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 0.2 mM TCEP, 0.01% sodium cholate, 0.035% β-DDM) and this concentration/dilution step was repeated two times. In cases of cyclopeptide co-crystallizations, the appropriate compound was added from a 50 mM stock in 100% DMSO to a final concentration of 2 mM cyclopeptide in a 10 ml aliquot of P-gp at ∼1–2 mg ml−1 and incubated overnight. The following morning, excess cyclopeptide was removed by concentrating to ∼1 ml and dilution to ∼15 ml using quench buffer; this was repeated twice prior to concentrating for crystallization trials.
P-gp crystals were grown in 24-well Cryschem plates (Hampton Research) at a protein concentration of ∼10–12 mg ml−1 using 4 µl sitting drops at a 1:1 protein:mother liquor ratio using a well mother liquor consisting of 0.1 M HEPES pH 7–8, 50 mM lithium sulfate, 10 mM EDTA, 24–29.5%(w/v) PEG 600. Crystals were grown at 277 K; they typically appeared after 1–3 d and continued to grow to full size in approximately two weeks.
Collected crystals were first cryoprotected by soaking in 0.1 M HEPES at a pH identical to the crystal-growth condition, 50 mM lithium sulfate, 10 mM EDTA, 32% PEG 600. Crystals were typically ∼650 × 400 × 300 µm in size.
2.6. X-ray data collection, structure determination and refinement of P-gp cyclopeptide co-crystal structures
X-ray diffraction data were collected at 100 K at either the Stanford Synchrotron Radiation Laboratory (SSRL; BL11-1) or the Canadian Light Source (CLS; 08ID-1). Fluorescence scans were taken on P-gp–cyclopeptide co-crystals to maximize the anomalous signal contribution from the incorporated selenium (Table 1 ▶). All diffraction data were processed with MOSFLM (Battye et al., 2011 ▶) and reduced with SCALA (Evans, 2006 ▶) within the CCP4 suite of programs (Winn et al., 2011 ▶). In the case of QZ-Ala, the data from three isomorphous crystals were scaled together to maximize the completeness (Table 1 ▶). The 3.4 Å resolution structure of P-gp was initially solved by molecular replacement (MR) with Phaser (McCoy et al., 2007 ▶) using a previously determined P-gp structure (PDB entry 4ksc; Ward et al., 2013 ▶) as a search model with no modifications. Commensurate with the improved resolution, the new electron-density features guided adjustments of our model when compared with the same more ‘open’ crystal form that we reported in 2013 (Ward et al., 2013 ▶) and are summarized in Supplementary Fig. S2. Residues 30–32 were located in the electron density, and resulted in a subsequent shift in the registration of residues in the first helix (residues 30–43) preceding TM1. Amendments were made to the topology of intracellular helix 1 (IH1; residues 154–168), extracellular loop 3 (ECL3; residues 318–338) and a portion of TM6 leading into the first NBD (residues 358–387). Within NBD1, residues 398–404, 424–427, 520–526 and 597–602 were rebuilt. Elbow helix 2 (EH2) was rebuilt from residues 689 to 708. A registry issue was amended from ECL4 (residue 738) to TM8 (residue 760) and another that constitutes segments of TM9, ECL5 and the beginning of TM10 (residues 826–855). The topology of IH3 was adjusted (residues 795–806), as was ECL6 (residues 961–967) and a portion of TM12 (residues 972–984). Further modifications were made in the region leading into and contributing to NBD2 (residues 1010–1028, 1042–1047, 1129–1137 and 1165–1172). Residues 1272 and 1273 were also located in the electron-density maps at the C-terminus. As for all structures of P-gp determined to date, the ‘linker’ region (residues 627–688) was not located in the electron density. Many of the structural adjustments are in general agreement with the recent corrections (Li et al., 2014 ▶) made to the model of the more ‘closed’ conformation of P-gp first reported in 2009 (Aller et al., 2009 ▶). During the refinement process, the model underwent rigid-body and restrained positional refinement, with H atoms applied in their riding positions, using phenix.refine (Afonine et al., 2012 ▶) against a maximum-likelihood target function with grouped B factors, secondary-structure restraints, reference-model restraints and TLS. Rounds of refinement were interspersed with manual inspection and correction against σA-weighted electron-density maps in Coot (Emsley et al., 2010 ▶) and improvements to model geometry and stereochemistry were monitored using MolProbity (Chen et al., 2010 ▶). Subsequent cyclopeptide co-crystal structures were solved by either MR or rigid-body refinement using the refined 3.4 Å resolution model with residues from TM4 (218–243) and EH2 (689–694) removed to avoid biasing their placement within the electron-density maps. These structures were then refined in a similar fashion to the 3.4 Å resolution structure with an additional round of positional refinement with ligand B factors set to the Wilson B value. Ligand description dictionaries were determined using phenix.elbow (Adams et al., 2010 ▶) and the crystallographic positions of the incorporated seleniums were validated using anomalous scattering methods. The refined structures were judged to have excellent geometry as determined by MolProbity (Chen et al., 2010 ▶). The resulting refinement statistics are listed in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Native | QZ-Ala | QZ-Val | QZ-Leu | QZ-Phe | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength () | 0.9795 | 0.9796 | 0.979 | 0.979 | 0.980 |
| Beamline | 11-1, SSRL | 08ID, CLS | 08ID, CLS | 11-1, SSRL | 11-1, SSRL |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit-cell parameters | |||||
| a () | 88.0 | 85.6 | 88.7 | 91.3 | 86.3 |
| b () | 139.2 | 138.4 | 138.7 | 138.5 | 138.4 |
| c () | 186.0 | 183.7 | 190.0 | 195.6 | 185.0 |
| = = () | 90 | 90 | 90 | 90 | 90 |
| Resolution range () | 86.33.4 (3.63.4) | 92.23.8 (4.03.8) | 88.73.6 (3.83.6) | 113.13.8 (4.03.8) | 86.33.8 (4.03.8) |
| No. of crystals | 1 | 3 | 1 | 1 | 1 |
| R merge (%) | 8.4 (73.0) | 9.8 (68.4) | 7.9 (64.9) | 5.2 (73.0) | 5.8 (69.6) |
| R p.i.m. (%) | 4.4 (38.3) | 3.4 (25.8) | 4.1 (35.0) | 3.5 (46.4) | 3.1 (36.6) |
| Observed reflections | 150254 | 198281 | 120622 | 73344 | 97970 |
| Unique reflections | 31984 | 22299 | 26969 | 23166 | 22385 |
| Mean I/(I) | 8.1 (1.8) | 11.9 (2.8) | 8.4 (2.0) | 9.8 (1.7) | 10.9 (2.2) |
| Completeness (%) | 99.3 (100) | 99.6 (99.9) | 97.4 (98.5) | 92.4 (95.8) | 99.4 (100) |
| Multiplicity | 4.7 (4.6) | 8.9 (7.7) | 4.5 (4.3) | 3.2 (3.2) | 4.4 (4.4) |
| Refinement | |||||
| Resolution range () | 77.43.4 (3.53.4) | 91.83.8 (3.93.8) | 78.43.6 (3.73.6) | 49.63.8 (3.93.8) | 76.93.8 (3.93.8) |
| Reflections in working set | 31868 | 22236 | 26927 | 22913 | 22339 |
| Reflections in test set (%) | 5.1 | 5.1 | 5.1 | 5.1 | 5.1 |
| R work/R free (%) | 26.0/29.1 | 26.3/29.5 | 26.1/28.2 | 29.2/32.1 | 25.9/29.3 |
| R.m.s deviations | |||||
| Bond lengths () | 0.007 | 0.004 | 0.004 | 0.004 | 0.004 |
| Bond angles () | 1.026 | 0.765 | 0.801 | 0.758 | 0.755 |
| B (2) | 99.3 | 156.3 | 160.8 | 182.5 | 177.1 |
| Ramachandran statistics | |||||
| Outliers (%) | 0 | 0 | 0 | 0 | 0 |
| Favoured (%) | 95.9 | 95.7 | 95.3 | 95.3 | 95.7 |
| Rotamer outliers (%) | 1.0 | 0.3 | 0.5 | 0.4 | 0.2 |
| C deviations | 0 | 0 | 0 | 0 | 0 |
3. Results
3.1. Rational engineering and functional characterization of cyclopeptide P-gp ligands
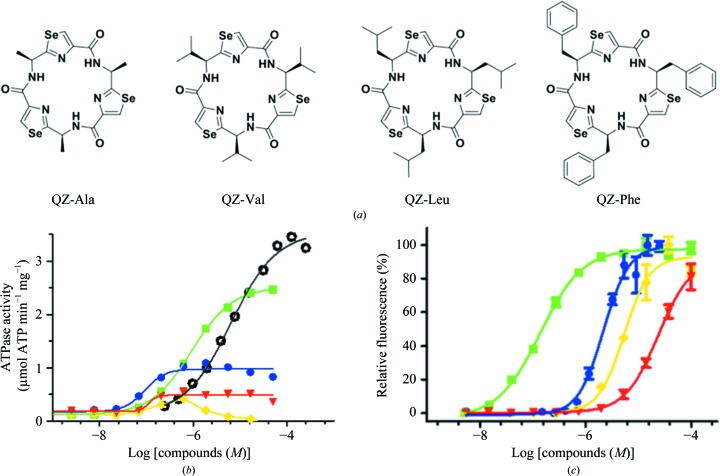
Previous functional studies have identified at least four, and potentially up to seven, sometimes overlapping binding sites for substrates and inhibitors in the greasy, polyspecific binding cavity of P-gp (Shapiro & Ling, 1997 ▶; Martin et al., 2000 ▶). To probe this phenomenon, the cyclic peptide QZ59-SSS (here named QZ-Val), previously reported in a co-crystal structure of mouse P-gp/Mdr1a (Aller et al., 2009 ▶), was taken as a base to engineer a series of selenium-labeled homotrimeric cyclopeptides (Fig. 1 ▶ a). The R groups of this series were systematically varied to generate alanine-, valine-, leucine- and phenylalanine-derived compounds (QZ-Ala, QZ-Val, QZ-Leu and QZ-Phe, respectively) of increasing R-group size and hydrophobicity.
Figure 1.
Structure and function of selenium-labelled homotrimeric cyclopeptides. (a) The chemical structures of the cyclopeptide series bear an identical backbone but with side chains systematically increasing in size and hydrophobicity (from left to right). (b) Stimulation of the basal ATPase activity of purified P-gp. QZ-Ala (green) conferred the highest degree of stimulation (∼16-fold) relative to the basal activity, with an EC50 value of 0.92 µM, followed by QZ-Val (blue; ∼7-fold). Data for verapamil (black), QZ-Leu (yellow) and QZ-Phe (red) are shown. (c) Inhibition of calcein-AM transport in P-gp-overexpressing CR1R12 cells. The same color scheme is applied for each compound as in (b) and the data were fitted using the Hill equation. The mean and SD of triple and quadruplet experiments are shown in (b) and (c), respectively.
We measured the effect of each ligand on the basal level of ATP hydrolysis of purified P-gp. Our results revealed that the smaller compounds were more stimulatory and, most notably, the smallest ligand, QZ-Ala, potently stimulated ATP hydrolysis in a dose-dependent manner at the tested concentrations, similar to verapamil (Fig. 1 ▶ b). Consistent with these substrate-like interactions, P-gp mediated mild resistance to QZ-Ala in cells (Supplementary Fig. S3). These data suggest that the binding of QZ-Ala and, to a lesser degree, QZ-Val at the TMDs generates a signal to the NBDs, accelerating nucleotide hydrolysis. We also characterized the ligands using P-gp-overexpressing CR1R12 cells. A pattern emerged in P-gp-mediated calcein-AM transport out of cells, whereby the potency of the ligand to inhibit export was inversely related to R-group size (Fig. 1 ▶ c). The smaller QZ-Ala inhibited calcein-AM export (IC50 = 140 nM) best compared with QZ-Val (IC50 = 1.7 µM), QZ-Leu (IC50 = 5.4 µM) and QZ-Phe (IC50 = 24 µM) (Fig. 1 ▶ c). Fitting the data to the Hill equation gave Hill coefficients of >1 in each case, suggesting positive cooperativity for their binding to at least two sites. All four compounds also prevented P-gp-mediated export of the anticancer drug colchicine and sensitized CR1R12 cells in a dose-dependent manner (Supplementary Fig. S4).
3.2. Structures of P-gp in complex with cyclopeptide ligands
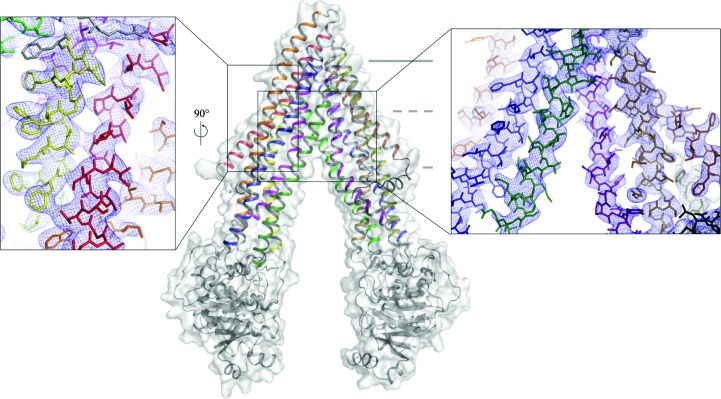
Prior to determining P-gp–ligand co-crystal structures, an improved structure of apo mouse P-gp/Mdr1a was determined to 3.4 Å resolution. To date, this model is the highest resolution structure reported for a mammalian ABCB1/MDR1-type transporter (87% identity to human P-gp; Fig. 2 ▶, Table 1 ▶). This model is similar to those recently described (Ward et al., 2013 ▶) and differs significantly from the original reported in 2009 (Aller et al., 2009 ▶). The increase in resolution has also facilitated electron-density map-driven improvements to the model (see §2 Materials and methods; Fig. 2 ▶, Table 1 ▶), resulting in better refinement statistics.
Figure 2.
Overview of mouse P-gp at 3.4 Å resolution. Magnified insets at different orientations are shown with the resulting 2mF o − DF c electron density (where m is the figure of merit and D is the σA weighting factor) contoured at 1σ; individual transmembrane helices are shown in different colors. TM1 is in red, TM2 is in orange, TM3 is in yellow, TM4 is in light green, TM5 is in sky blue, TM6 is in pink, TM7 is in dark brown, TM8 is in olive, TM9 is in light brown, TM10 is in forest, TM11 is in dark blue and TM12 in deep purple.
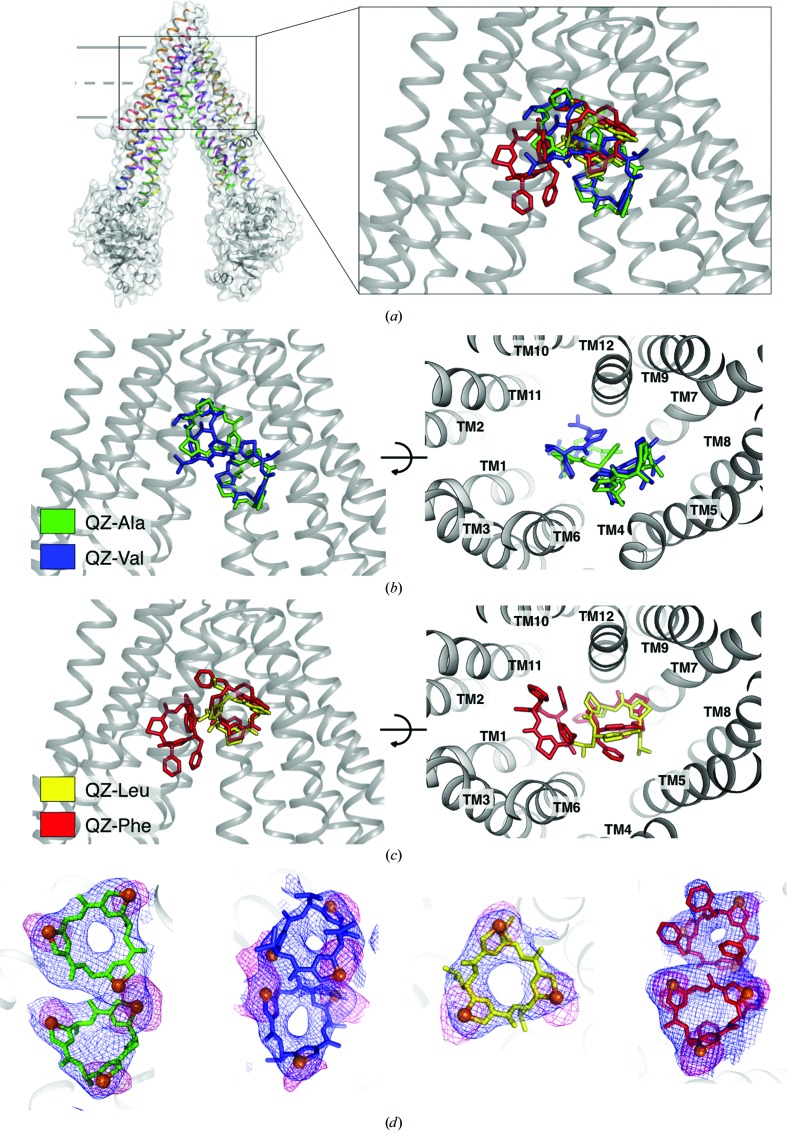
To interrogate R-group-dependent variations in ligand binding, co-crystal structures of P-gp with each ligand were determined (Figs. 3 ▶ a, 3 ▶ b, 3 ▶ c and 4 ▶). Ligands were placed using difference electron densities and strong anomalous scattering from the three triangulated Se atoms (Fig. 3 ▶ d). All ligands bound to P-gp (Fig. 3 ▶ d), and the majority of the coordinating side chains, are well resolved in the electron-density maps. The ligands make a number of interactions with binding-pocket residues (Fig. 4 ▶), burying a high proportion of the solvent-accessible surface area of the ligand (Supplementary Table S1).
Figure 3.
Overview of P-gp–cyclopeptide co-crystal structures. (a) Superposition of homotrimeric cyclopeptide compounds bound to P-gp, showing their relative location and orientation in the substrate-binding pocket. (b) Two orientations of subset A ligands (QZ-Ala and QZ-Val; displayed as sticks) bound in the substrate-binding pocket of P-gp. QZ-Ala is shown in green and QZ-Val in blue. (c) Two orientations of subset B ligands (QZ-Leu and QZ-Phe) bound in the substrate-binding pocket of P-gp. QZ-Leu is shown in yellow and QZ-Phe in red. (d) Close-up view of ligands, colored as in (b) and (c), with the resulting 2mF o − DF c electron density in blue (contour level of 1.0σ) and anomalous difference density peaks in pink (contour levels of 3.0σ for QZ-Ala, QZ-Va and Q-Phe and 4.0σ for QZ-Leu) for the Se atoms (orange spheres).
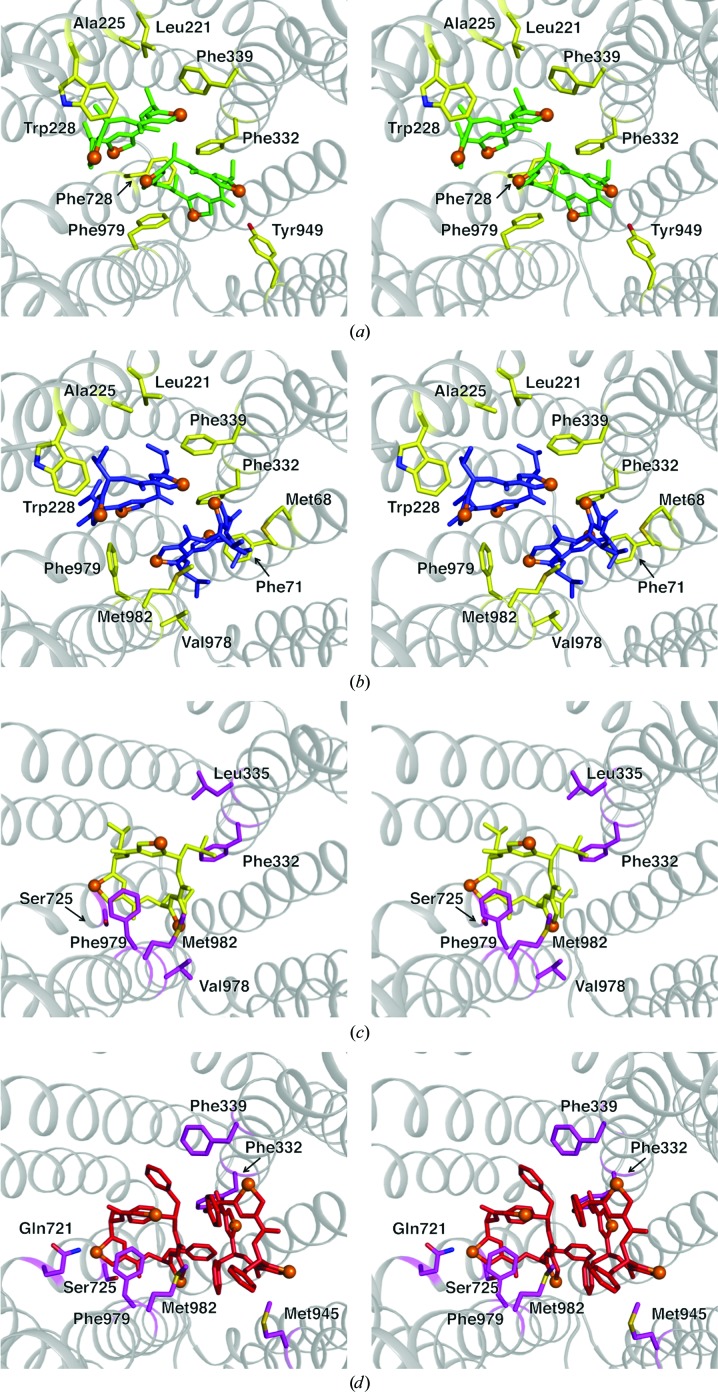
Figure 4.
Key P-gp residues involved in binding homotrimeric cyclopeptides. Stereoviews of the binding pocket are shown perpendicular to the membrane and viewed from the cytosol. P-gp residues are shown as sticks, with those involved in binding subset A ligands colored yellow and those involved in binding subset B ligands colored magenta. The ligand coloring is consistent with that shown in Fig. 3 ▶; QZ-Ala, green; QZ-Val, blue; QZ-Leu, yellow; QZ-Phe, red.
4. Discussion
4.1. Comparison of cyclopeptide ligands in complex with P-gp
Our P-gp–ligand co-crystal structures demonstrate the wide range of possible binding modes for similar substrates (Fig. 3 ▶ a). Within the binding pocket, our structures group the four ligands into two subsets that correlate with size and hydrophobicity (Figs. 3 ▶ b, 3 ▶ c and 4 ▶). The smaller ligands QZ-Ala and QZ-Val (subset A) share an upper and lower binding site (Figs. 3 ▶ b and 4 ▶), while the larger and more hydrophobic ligands QZ-Leu and QZ-Phe (subset B) share a different upper binding site, with QZ-Phe also binding to a second, unique lower site (Figs. 3 ▶ c and 4 ▶). P-gp consists of two pseudo-symmetric halves, each containing six transmembrane (TM) helix bundles. Viewed from the plane of the membrane, the ligands bound in the upper sites of subsets A and B are wedged in the apex of the cavity, engaging TM helices from both pseudo-halves (Figs. 3 ▶ b and 3 ▶ c). In contrast, the ligands in the lower sites interact with distinct pseudo-halves of P-gp that are mutually exclusive to each ligand subset (Figs. 3 ▶ b, 3 ▶ c and 4 ▶).
4.2. Movement of TM4 upon ligand binding
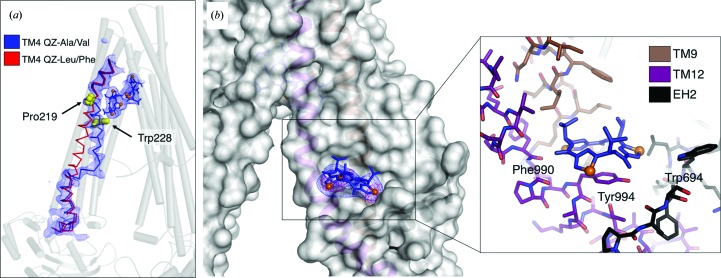
In previous structures of apo mouse P-gp, and also for ligands pertaining to subset B, TM4 adopts a mostly straight-helical conformation characterized by weaker electron density, indicative of regional flexibility (Ward et al., 2013 ▶). Subset A ligands not only revealed an ordered TM4, but a large conformational change (of up to 11 Å when comparing corresponding Cα positions; Fig. 5 ▶ a). This structural kink is not likely to be a consequence of lattice contacts as the crystal form is the same (Table 1 ▶). Thus, we must conclude that the binding of subset A ligands induces these changes in TM4. This ligand binding-induced kinking of TM4 begins at Pro219 before returning to the apo wild-type topology at Tyr243 in the ball-and-socket region (Loo et al., 2013 ▶) close to NBD2.
Figure 5.
Overview of the EH2 ligand-binding site and the ligand-induced movement of TM4. (a) The kinking of TM4 in response to subset A ligands is shown (blue ribbon) in comparison to its ‘straight’ topology in the subset B co-crystal structures (red ribbon). The position of QZ-Val ligands and the resulting 2mF o − DF c electron density (contoured at 1.0σ) are displayed. Residues Pro219 and Trp228 are shown as yellow spheres. (b) QZ-Val bound at the EH2 site with P-gp rendered as a molecular-surface representation. The resulting 2mF o − DF c electron density for the ligand is shown at a contour level of 1.0σ and the resulting anomalous difference peaks for the Se atoms (orange spheres) are shown at 3.5σ. The close-up inset view depicts the surrounding TM9 and TM12 as brown and purple sticks, respectively, and EH2 as black sticks. Residues in close vicinity to the ligand (Trp694, Phe990 and Tyr994) are labeled.
The observed movement of TM4 upon substrate/ligand binding may have significant biological implications. TM4 and TM6 comprise an intramembranous portal for substrate entry to the binding cavity, and a conformational change in these helices may influence the entry and the binding of ligands (Loo & Clarke, 1994 ▶, 2005 ▶; Woebking et al., 2008 ▶). In the recent structure of Cyanidioschyzon merolae P-gp, a portion of TM4 in the wild-type protein was inherently disordered (Kodan et al., 2014 ▶). Mutations in this region not only resulted in a well ordered, straight-helical conformation, but also functionally disrupted substrate transport, suggesting a role for TM4 in facilitating substrate entry and/or binding (Kodan et al., 2014 ▶). For co-crystal structures pertaining to subset A ligands, the movement of TM4 brings residues 221–228 closer to the bound ligands in the lower binding sites, fostering an intermolecular interaction with Trp228, a residue that has been implicated in steroid binding to P-gp (Gruol et al., 2002 ▶). We propose that the movement of TM4 upon binding subset A compounds provides a structural glimpse of the induced-fit model of drug binding proposed for P-gp nearly a decade ago (Loo et al., 2003b ▶).
Several ligands stimulate the basal rate of ATP hydrolysis of P-gp by severalfold (Al-Shawi & Senior, 1993 ▶; Ambudkar et al., 1992 ▶; Scarborough, 1995 ▶; Fig. 1 ▶ b). However, the mechanisms coupling ligand binding in the TMDs to increased ATP turnover at the NBDs are not fully understood. Cross-linking and FRET studies on P-gp and the bacterial homolog MsbA have suggested that ligand binding induces closure of the NBDs in the presence of nucleotides, leading to the increase in catalysis (Szabo et al., 1998 ▶; Eckford & Sharom, 2008 ▶; Liu & Sharom, 1996 ▶; Loo et al., 2003a ▶; Siarheyeva & Sharom, 2009 ▶; Wang et al., 1998 ▶; Scarborough, 1995 ▶; Doshi & van Veen, 2013 ▶). Specifically, conformational changes in TM4 have been linked to this TMD–NBD coupling (Doshi & van Veen, 2013 ▶), consistent with our models. In our structures, ligands that function more as activators of ATPase (Fig. 1 ▶ b; QZ-Ala and QZ-Val; subset A) kink TM4, while those that function more as inhibitors of ATPase activity (QZ-Ile and QZ-Phe; subset B) maintain straight TM4 helices as also observed in the apo structure. Here, we only demonstrate two distinct changes in TM4 (kinked or straight helical) caused by two different subsets of ligands. Other ligand-binding sites in the TMD are possible, resulting in corresponding structural changes for TM4 or perhaps other TM helices extending to the NBDs. Variations in the degree of structural kink of TM helices, for example, are likely for other compounds.
In this study, we cannot resolve long-range structural changes of the intracellular helix (residues 242–256) extending to NBD2 that would provide further insights regarding the exact nature of any potential coupling mechanism between TM4 and NBD2. Several possibilities could explain this observation. For example, the structural changes in TM4 extending to NBD2 might only occur as the transporter goes from a wide-open to a closed inward-facing conformation where the NBDs begin to contact. These changes may also necessitate the presence of ATP, which is absent in these structures. Another possibility is that these structural changes may be too small to resolve using our current data. If so, computational studies using these models incorporating other biochemically derived restraints could be very valuable for understanding how substrate-stimulated ATP hydrolysis in P-gp may be initiated. Taken together with other complementary studies (Loo et al., 2003a ▶; Doshi & van Veen, 2013 ▶), our structures provide a starting molecular-structural framework for a ligand induced-fit mechanism transmitting structural changes from the TMDs to the NBDs.
4.3. Ligand-binding site at the membrane interface
Previous biochemical experiments have led to the proposal that P-gp extrudes drugs from the inner leaflet of the plasma membrane, functioning as a so-called ‘hydrophobic vacuum cleaner’ (Raviv et al., 1990 ▶; de Graaf et al., 1996 ▶; Bolhuis et al., 1996 ▶). In our QZ-Val co-crystal structure, we observed an additional binding site on the exterior of P-gp bounded by residues from TM9, TM12 and EH2 (Fig. 5 ▶ b). The site faces away from the transporter, but lies close to the predicted membrane–water interface and intramembranous substrate-entry portal. Drug binding near the EH has been reported using electron paramagnetic resonance on a bacterial P-gp homolog that transports lipids (Smriti et al., 2009 ▶). These data lend strong credence to the proposal of an initial lower-affinity ‘ON-site’ for a ligand near the inner leaflet of the lipid bilayer preceding the higher-affinity ‘ON-site(s)’ within the central binding cavity (Dey et al., 1997 ▶; Al-Shawi & Omote, 2005 ▶).
5. Conclusions
Understanding drug–transporter interactions is indispensable for engineering drugs to inhibit or evade P-gp. The findings presented here complement and extend the related X-ray structures (Hohl et al., 2012 ▶; Ward et al., 2007 ▶, 2013 ▶; Shintre et al., 2013 ▶; Kodan et al., 2014 ▶; Jin et al., 2012 ▶; Aller et al., 2009 ▶), as well as many biochemical/biophysical studies (Juliano & Ling, 1976 ▶; Al-Shawi & Omote, 2005 ▶). We have conducted the first detailed structure–activity relationship (SAR) study on how altering the size and hydrophobicity of the R groups in a known P-gp ligand can impact its interactions with this clinically important protein. The mechanistic concepts delivered through this structural study include (i) ligand entry via the elbow helix binding site, (ii) distinct and shared binding modes and (iii) ligand binding-induced fit that could cause transmission coupled to ATP catalysis. These structural observations provide an excellent basis to biochemically and computationally test hypotheses in further studies on this transporter.
6. Related literature
The following reference is cited in the Supporting Information for this paper: Krissinel & Henrick (2007 ▶).
Supplementary Material
PDB reference: P-glycoprotein, 4q9h
PDB reference: cocrystallized with QZ-Ala, 4q9i
PDB reference: cocrystallized with QZ-Val, 4q9j
PDB reference: cocrystallized with QZ-Leu, 4q9k
PDB reference: cocrystallized with QZ-Phe, 4q9l
Supporting Information.. DOI: 10.1107/S1399004715000978/cb5079sup1.pdf
Acknowledgments
We thank the support staff at beamlines 11-1 at Stanford Synchrotron Radiation Laboratory and 08ID at the Canadian Light Source. Data contributing to this work were also collected at the Advanced Light Source and the Advanced Photon Source. This work was funded by National Institutes of Health (NIH) grants R01 GM94367, R01 ES021985 (to GC), R01 GM098538 (to QZ) and RGM102928 (to ILU). APM is supported by an NHMRC CJ Martin Postdoctoral Research Fellowship.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Aller, S. G., Yu, J., Ward, A., Weng, Y., Chittaboina, S., Zhuo, R., Harrell, P. M., Trinh, Y. T., Zhang, Q., Urbatsch, I. L. & Chang, G. (2009). Science, 323, 1718–1722. [DOI] [PMC free article] [PubMed]
- Al-Shawi, M. K. & Omote, H. (2005). J. Bioenerg. Biomembr. 37, 489–496. [DOI] [PMC free article] [PubMed]
- Al-Shawi, M. K. & Senior, A. E. (1993). J. Biol. Chem. 268, 4197–4206. [PubMed]
- Ambudkar, S. V., Lelong, I. H., Zhang, J., Cardarelli, C. O., Gottesman, M. M. & Pastan, I. (1992). Proc. Natl Acad. Sci. USA, 89, 8472–8476. [DOI] [PMC free article] [PubMed]
- Bai, J., Swartz, D. J., Protasevich, I. I., Brouillette, C. G., Harrell, P. M., Hildebrandt, E., Gasser, B., Mattanovich, D., Ward, A., Chang, G. & Urbatsch, I. L. (2011). PLoS One, 6, e22577. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Bolhuis, H., van Veen, H. W., Molenaar, D., Poolman, B., Driessen, A. J. & Konings, W. N. (1996). EMBO J. 15, 4239–4245. [PMC free article] [PubMed]
- Cascorbi, I. (2006). Pharmacol. Ther. 112, 457–473. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Dey, S., Ramachandra, M., Pastan, I., Gottesman, M. M. & Ambudkar, S. V. (1997). Proc. Natl Acad. Sci. USA, 94, 10594–10599. [DOI] [PMC free article] [PubMed]
- Doshi, R. & van Veen, H. W. (2013). J. Biol. Chem. 288, 21638–21647. [DOI] [PMC free article] [PubMed]
- Eckford, P. D. & Sharom, F. J. (2008). J. Biol. Chem. 283, 12840–12850. [DOI] [PubMed]
- Eckford, P. D. & Sharom, F. J. (2009). Chem. Rev. 109, 2989–3011. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Falasca, M. & Linton, K. J. (2012). Expert Opin. Investig. Drugs, 21, 657–666. [DOI] [PubMed]
- US Food and Drug Administration (2012). Drug Interaction Studies – Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm292362.pdf.
- Giacomini, K. M. et al. (2010). Nature Rev. Drug Discov. 9, 215–236. [DOI] [PMC free article] [PubMed]
- Gottesman, M. M. & Ling, V. (2006). FEBS Lett. 580, 998–1009. [DOI] [PubMed]
- Graaf, D. de, Sharma, R. C., Mechetner, E. B., Schimke, R. T. & Roninson, I. B. (1996). Proc. Natl Acad. Sci. USA, 93, 1238–1242. [DOI] [PMC free article] [PubMed]
- Gruol, D. J., King, M. N. & Kuehne, M. E. (2002). Mol. Pharmacol. 62, 1238–1248. [DOI] [PubMed]
- Hohl, M., Briand, C., Grütter, M. G. & Seeger, M. A. (2012). Nature Struct. Mol. Biol. 19, 395–402. [DOI] [PubMed]
- Jin, M. S., Oldham, M. L., Zhang, Q. & Chen, J. (2012). Nature (London), 490, 566–569. [DOI] [PMC free article] [PubMed]
- Juliano, R. L. & Ling, V. (1976). Biochim. Biophys. Acta, 455, 152–162. [DOI] [PubMed]
- Kodan, A., Yamaguchi, T., Nakatsu, T., Sakiyama, K., Hipolito, C. J., Fujioka, A., Hirokane, R., Ikeguchi, K., Watanabe, B., Hiratake, J., Kimura, Y., Suga, H., Ueda, K. & Kato, H. (2014). Proc. Natl Acad. Sci. USA, 111, 4049–4054. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Li, J., Jaimes, K. F. & Aller, S. G. (2014). Protein Sci. 23, 34–46. [DOI] [PMC free article] [PubMed]
- Liu, R. & Sharom, F. J. (1996). Biochemistry, 35, 11865–11873. [DOI] [PubMed]
- Loo, T. W., Bartlett, M. C. & Clarke, D. M. (2003a). J. Biol. Chem. 278, 1575–1578. [DOI] [PubMed]
- Loo, T. W., Bartlett, M. C. & Clarke, D. M. (2003b). J. Biol. Chem. 278, 13603–13606. [DOI] [PubMed]
- Loo, T. W., Bartlett, M. C. & Clarke, D. M. (2013). J. Biol. Chem. 288, 20326–20333. [DOI] [PMC free article] [PubMed]
- Loo, T. W. & Clarke, D. M. (1994). Biochemistry, 33, 14049–14057. [DOI] [PubMed]
- Loo, T. W. & Clarke, D. M. (2005). Biochem. Biophys. Res. Commun. 329, 419–422. [DOI] [PubMed]
- Martin, C., Berridge, G., Higgins, C. F., Mistry, P., Charlton, P. & Callaghan, R. (2000). Mol. Pharmacol. 58, 624–632. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Raviv, Y., Pollard, H. B., Bruggemann, E. P., Pastan, I. & Gottesman, M. M. (1990). J. Biol. Chem. 265, 3975–3980. [PubMed]
- Rayment, I. (1997). Methods Enzymol. 276, 171–179. [PubMed]
- Scarborough, G. A. (1995). J. Bioenerg. Biomembr. 27, 37–41. [DOI] [PubMed]
- Shapiro, A. B. & Ling, V. (1997). Eur. J. Biochem. 250, 130–137. [DOI] [PubMed]
- Shintre, C. A. et al. (2013). Proc. Natl Acad. Sci. USA, 110, 9710–9715.
- Siarheyeva, A. & Sharom, F. J. (2009). Biochem. J. 419, 317–328. [DOI] [PMC free article] [PubMed]
- Smriti, Zou, P. & Mchaourab, H. S. (2009). J. Biol. Chem. 284, 13904–13913. [DOI] [PMC free article] [PubMed]
- Szabo, K., Welker, E., Bakos, É., Muller, M., Roninson, I., Varadi, A. & Sarkadi, B. (1998). J. Biol. Chem. 273, 10132–10138. [DOI] [PubMed]
- Tao, H., Weng, Y., Zhuo, R., Chang, G., Urbatsch, I. L. & Zhang, Q. (2011). Chembiochem, 12, 868–873. [DOI] [PMC free article] [PubMed]
- Urbatsch, I. L., Sankaran, B., Weber, J. & Senior, A. E. (1995). J. Biol. Chem. 270, 19383–19390. [DOI] [PubMed]
- Vichai, V. & Kirtikara, K. (2006). Nature Protoc. 1, 1112–1116. [DOI] [PubMed]
- Vogel, G. & Steinhart, R. (1976). Biochemistry, 15, 208–216. [DOI] [PubMed]
- Wang, G., Pincheira, R. & Zhang, J.-T. (1998). Eur. J. Biochem. 255, 383–390. [DOI] [PubMed]
- Ward, A., Reyes, C. L., Yu, J., Roth, C. B. & Chang, G. (2007). Proc. Natl Acad. Sci. USA, 104, 19005–19010. [DOI] [PMC free article] [PubMed]
- Ward, A. B. et al. (2013). Proc. Natl Acad. Sci. USA, 110, 13386–13391.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Woebking, B., Velamakanni, S., Federici, L., Seeger, M. A., Murakami, S. & van Veen, H. W. (2008). Biochemistry, 47, 10904–10914. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: P-glycoprotein, 4q9h
PDB reference: cocrystallized with QZ-Ala, 4q9i
PDB reference: cocrystallized with QZ-Val, 4q9j
PDB reference: cocrystallized with QZ-Leu, 4q9k
PDB reference: cocrystallized with QZ-Phe, 4q9l
Supporting Information.. DOI: 10.1107/S1399004715000978/cb5079sup1.pdf