Abstract
Pyrazinamide (PZA), an essential component of short-course anti-tuberculosis chemotherapy, was shown by Saturation Transfer Difference (STD) NMR methods to act as a competitive inhibitor of NADPH binding to purified Mycobacterium tuberculosis fatty acid synthase I (FAS I). Both PZA and pyrazinoic acid (POA) reversibly bind to FAS I but at different binding sites. The competitive binding of PZA and NADPH suggests potential FAS I binding sites. POA was not previously known to have any specific binding interactions. The STD NMR of NADPH bound to the mycobacterial FAS I was consistent with the orientation reported in published single crystal X-ray diffraction studies of fungal FAS I. Overall the differences in binding between PZA and POA are consistent with previous recognition of the importance of intracellular accumulation of POA for anti-mycobacterial activity.
Keywords: Pyrazinamide, NADPH, STD NMR, Enzyme inhibition, Fatty acid synthase I (FAS I)
Pyrazinamide (PZA), a nicotinamide analog, is a unique, essential constituent of short-course tuberculosis (TB) chemotherapy that decreases the length of therapy from 9 to 12 months to as little as 6 months. First synthesized in 1936, the anti-tuberculous properties of PZA were not described until 1952.1,2 Despite the crucial role of PZA in the treatment of TB and the remarkable in vivo antimicrobial properties of this drug,3 the mechanism of action of PZA continues to be poorly understood.
PZA is widely assumed to be a prodrug for pyrazinoic acid (POA) which functions as the active anti-tuberculous agent.4,5 PZA enters Mycobacterium tuberculosis (Mtb) by passive diffusion where it is converted to pyrazinoic acid (POA) by a pyrazinamidase (PncA).5 The unique susceptibility of Mtb to PZA has been attributed both to the amidase action and to a deficiency in POA efflux.6 However, the fate of POA as well as the exact biochemical function inhibited by PZA/POA is controversial. A weak acid, POA has been suggested to accumulate at acidic pH and thereby disrupts cellular membrane energetics3,7,8 Alternatively, the demonstration that PZA and POA inhibit M. tuberculosis type-1 fatty acid synthase (FAS I) in whole-cell and cell-free assays9–11 suggests that the disruption might be a consequence of the inhibition of membrane synthesis. Contrary to these results, Boshoff et al.12 reported PZA to be inactive against purified Mtb FAS I and ineffective inhibiting FAS I in whole cell studies.
With conflicting reports on the inhibition of FAS I by PZA and POA, absent identification of a mechanism of inhibition, controversy remains.12 Given the recent communication of the activity of PZA against Leishmania spp.,13 a clearer understanding of the effects of PZA could enable the development of new anti-tuberculous and anti-leishmanial chemotherapeutic agents.
The reversible binding of both PZA and POA to Mtb FAS I has been definitively established by Saturation Transfer Difference NMR spectroscopy (STD-NMR), a NMR technique to characterize ligand–protein interaction. During STD-NMR, the ligand is titrated in the protein solution where ligand binding manifests itself by an increase in the ligand NMR signal intensity.
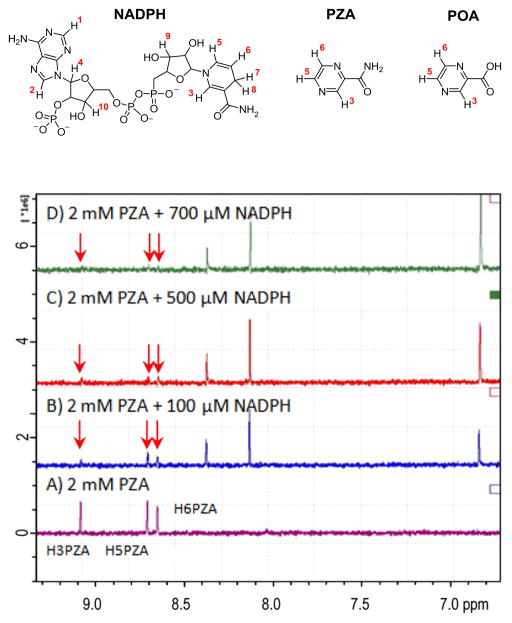
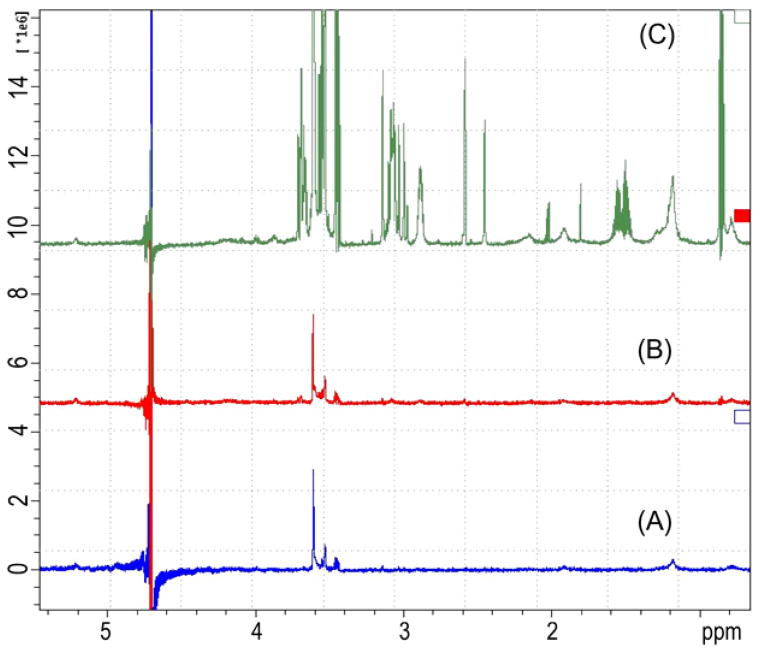
The STD-NMR method of Mayer et al.14 was employed with a sample containing Mtb FAS I and PZA. Resonances derived from the three aromatic protons H3 δ 9.0851 (br s, 1H), H5 δ 8.7086 (d, J = 2.3 Hz, 1H) and H6 δ 8.6524 (br s, 1H) in DMSO-d6 and D2O (1:100) of PZA were detected (see Fig. 1 for assignments). The ligand (PZA) interaction with FAS I can best be evaluated by determination of the absolute magnitude of the STD effect, that is the STD amplification factor.14 Positive STD amplification factors require reversible binding of the ligand to the enzyme and also an excess of ligand relative to the enzyme.14 For instance, a value of 0.14 is equivalent to a saturation of 14% of the ligand’s signal intensity. On binding of PZA to FAS I, the protons (H6, H5 and H3) have a similar degree of saturation, namely 100%, 88% and 90% saturation, that is strongly suggestive of a conformation where PZA either lies flat on Mtb FAS I or inserts deeply in a Mtb FAS I binding pocket.
Figure 1.
PZA competes with NADPH for the same binding sites of Mtb FAS I. Mtb FAS I was isolated as previously described.9,18 Buffer used in STD-NMR experiments was 100 mM NaCl, 1 mM dDTT-d10, 10 mM KPB in D2O at pH 7.2. The intensities of the PZA resonances decrease (see arrows) as the NADPH concentration increases.
PZA and NADPH
Since PZA was originally prepared as a nicotinamide analog,15 the hypothesis that PZA acts as a surrogate for the natural protein cofactor NADPH in interactions with Mtb FAS I is logical even if previously unproven. The affinity of PZA for Mtb FAS I in the presence of NADPH was determined using a competition titration STD-NMR experiment (Fig. 1) in which a sample containing 2 mM PZA and 11.2 μM of Mtb FAS I was titrated with increasing NADPH concentrations. Consistent with the binding of NADPH to Mtb FAS I the intensity of the NADPH resonances increases. Most significantly, increasing NADPH concentrations result in decreases in the PZA STD signal confirming that NADPH and PZA compete for binding. Previously we had established the interaction of PZA with FAS I was competitive9 in addition double reciprocal plots of the data utilized in the published experiments are not consistent with cooperative or allosteric binding. NADPH resonances were assigned as H1 δ 8.3607 (s, 1H), H2 δ 8.1176 (s, 1H), H3 δ 6.8107 (s, 1H), H4 δ 6.0925 (d, J = 4.5 Hz, 1H), H5 δ 5.8439 (d, J = 8.32, 1H), H6 δ t, J = 5.09 Hz, 1H), H9–10 δ 3.5353 (dt J = 7.15 Hz, J = 7.43 Hz, 2H), H8 δ 2.713 (dd, J = 18.22 Hz, 1H), and H7 δ 2.6188 (dd, J = 19.332 Hz, 1H) by HSQC and TOCSY NMR.16,17
NADPH is utilized by the β-ketoacyl reductase (KR) and the β-enoyl reductase (ER) catalytic domains of Mtb FAS I. Although it has been suggested that ER may show preference for NADH over NADPH,19 we have shown in our spectrometric assay that Mtb FAS I is functional when only NADPH is available. The competition between PZA and NADPH strongly suggests that PZA targets the KR and/ or the ER catalytic domains, two of the seven functional domains present in Mtb FAS I.20–23
The ratio of the STD signals in the presence and in the absence of an inhibitor is related to the dissociation constant KD and the inhibition constant KI as long as the STD signal intensity is proportional to the concentration of the ligand-bound complex and all other conditions are held constant.24,25 (Eq. 1)
| (1) |
STD(I) and STD(0) are the STD resonance intensities of the ligand in the presence and in the absence of the competitive inhibitor, respectively. [E], [L] and [I] are the total enzyme concentration, the ligand concentration and the inhibitor concentration respectively. The STD binding assay afforded a KD value of 70–123 μM for PZA based on a KI value of 41 μM for NADPH. The NADPH KI value was determined based on our previous spectrophotometric inhibition studies.9
Geometry of binding
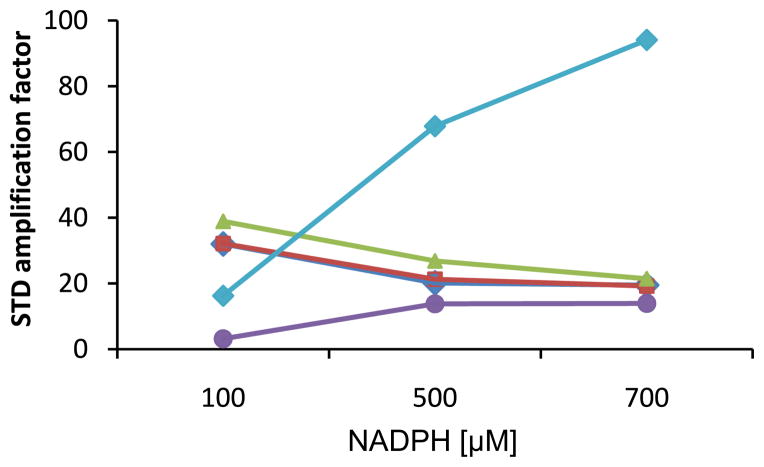
In addition to confirming the relative affinity of PZA and POA for FAS I, the STD experiment can illustrate the geometry of substrate binding. The ligand protons in closest contact with the protein have the most intense NMR resonances, therefore shedding light on the interaction of PZA or POA to FAS I. The STD amplification factors for PZA protons H3, H5 and H6 decrease with a very similar slope on increasing NADPH concentration indicating that these protons are being displaced to the same extent (Fig. 2). However, resonances H3 and H5 on the nicotinamide ring of NADPH increase according to very different slopes suggesting that one of the resonances, H5-NADPH, is consistently in closer contact with FAS 1 than H3-NADPH (Fig. 2).
Figure 2.
Variation of STD amplification factors with NADPH concentration in NADPH and PZA
competition experiments. NADPH resonances H3  and H5
and H5  have very different STD amplification factors,
suggesting that they do not interact equally with the protein whereas H3
have very different STD amplification factors,
suggesting that they do not interact equally with the protein whereas H3
 , H5
, H5  and H6
and H6  of PZA show very comparable declines in
amplification.
of PZA show very comparable declines in
amplification.
The similarity in saturation degree percentage of the PZA resonances is indicative of simultaneous dissociation from Mtb FAS I. The differences in saturation degree percentages of the nicotinamide resonances of NADPH (H3-NADPH and H5-NADPH, 35% and 92%, respectively) are consistent with the nicotinamide ring not sitting flat in the binding pocket. The binding motif of NADPH bound to actinorhodin polyketide ketoreductase mutant P94L is clearly consistent with the higher saturation degree of H5 relative to H3 as H5 is directed toward the main chain of the protein while H3 points towards the pyrophosphate.26 Furthermore, the observation of the different saturation degree percentages of H5 and H3 is consistent with the structure of fungal FAS I bound to NADPH.23 In both the KR and ER functional domains, H3 of NADPH is pointed away from the protein and in toward the ribose phosphate. Previously, it has been proposed that the structure of Mtb FAS I is closely related to that of fungal FAS I.23
PZA and POA
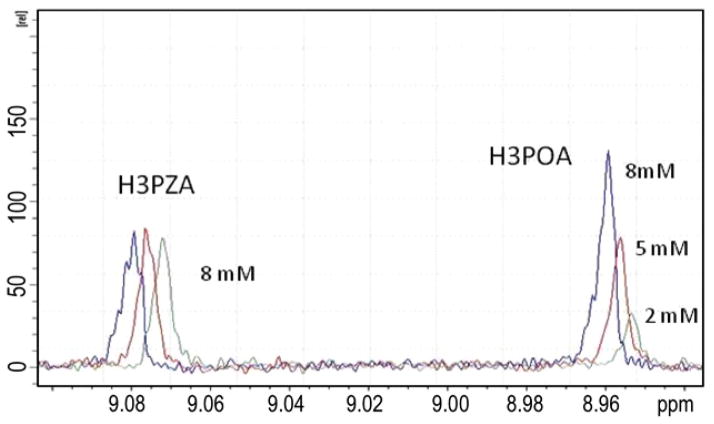
PZA and POA are both modest inhibitors of Mtb FAS I.9–12,27 In a competition titration experiment, a sample containing Mtb FAS I and PZA was titrated with increasing concentrations of POA. Most interestingly, the titration revealed that although POA binds reversibly to Mtb FAS I it does not compete with PZA since PZA is apparently not displaced. This insensitivity is demonstrated by lack of change in the intensity of PZA H-3 resonance with increasing POA concentration (Fig. 3).
Figure 3.
The PZA resonances are unaffected by increasing POA concentrations suggesting that PZA and POA bind at different sites. The greater intensity of POA at 8 mM also indicates that POA binds more tightly to Mtb FAS I than PZA. The slight change in chemical shift is a result of the increasing concentration of the inhibitor solvent d6-DMSO.
Since both PZA and POA interact weakly with FAS I, and that PZA is not active against bacilli that fail to convert PZA to POA, then it is reasonable that the nature of the interaction with FAS I should be different. The failure of PZA to be displaced by increasing concentrations of POA suggests that the biological effect of POA is not inhibition of NADPH binding. Future experiments will enable a detailed examination of this hypothesis.
Control experiments
Two different control experiments were used to verify that the observed STD signals arise as consequence of FAS I binding.
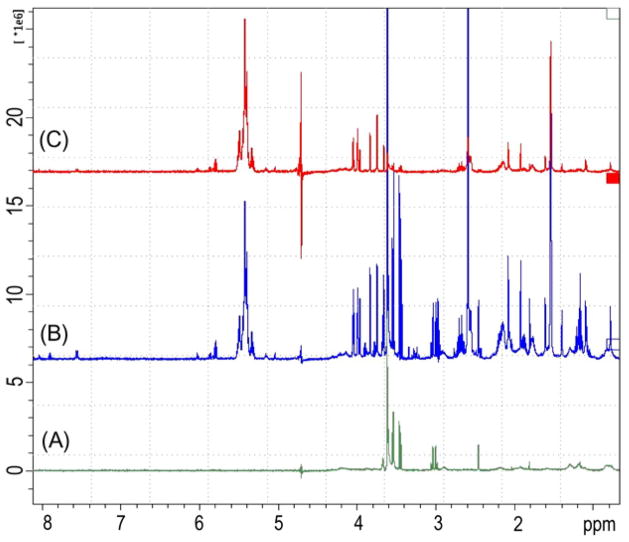
Cerulenin, a well known FAS I inhibitor was used as positive control and the antituberculous agent ethambutol, an inhibitor of arabinosyl transferase, was used as negative control (Figs, 4 and 5). These agents were selected as controls in our earlier FAS I inhibition experiments.9–11 The STD spectrum of FAS I and cerulenin (Fig. 4C) displays signals corresponding to cerulenin resonances that are indicative of cerulenin binding to Mtb FAS I. In the negative control experiments, no binding between FAS I and ethambutol was anticipated and the STD spectrum confirms that expectation. No ethambutol resonances were detected in the STD spectrum of FAS I and ethambutol (500 μM) (Fig. 5B).
Figure 4.
Positive control with cerulenin showing resonances resulting from affinity of cerulenin for FAS I. (A) The STD spectrum of 11.2 μM FAS I. (B) Reference spectrum of FAS I and 350 μM cerulenin. (C) STD spectrum of FAS I and cerulenin.
Figure 5.
Negative control with ethambutol shows no spectral enhancement. (A) STD FAS I. (B) STD FAS I and 500 M ethambutol. (C) Reference spectrum of FAS I and ethambutol.
Previous reports on the inhibition of Mtb FAS I by PZA9–11 have been controversial.12 This study unequivocally establishes that PZA binds reversibly to Mtb FAS I. Furthermore, these experiments in combination with previously published data,9 indicate that PZA competes with NADPH for the same binding site(s) of Mtb FAS I. As only two catalytic domains of Mtb FAS I require NADPH as a cofactor, namely the β-ketoacyl reductase and the β-enoyl reductase, it is plausible that PZA targets either of these domains. Most interestingly, while it was found that both PZA and POA bind to Mtb FAS I with POA binding FAS I with a greater affinity than PZA. However, these two molecules target different Mtb FAS I binding sites. Although it has been postulated that the accumulation of POA has a non-specific effect, it is clear that POA must bind to Mtb FAS I for the measurement of an STD spectrum to be possible. The failure to observe an STD spectrum with ethambutol which has no effect on FAS I, and the clear binding of cerulenin in an STD experiment is consistent with selectivity of the STD technique.
The observation of the interaction of POA with Mtb FAS I is consistent with the studies by Heifets that demonstrated the dose–response of POA anti-tuberculous activity28 and those of Ngo9 where POA inhibition of fatty acid synthesis was dose dependent. Moreover, the greater affinity of POA for Mtb FAS I than PZA is consistent with previously published data in which lower IC50 values were reported for POA.9,11 The fact that PZA and POA bind at different sites to FAS I suggest that each has a different mechanism of FAS I inhibition. This is consistent with the proposal of Zhang and co-workers29 that PZA and POA act differently based on the finding that in contrast with PZA, weak acids are not active under anaerobic conditions. While the findings reported confirm the activity of PZA against FAS I, the potential role of FAS I NADPH binding antagonists in therapy remains to be established.
Acknowledgments
The partial support of the National Science Foundation (CHE0957544) is gratefully acknowledged.
References and notes
- 1.Malone L, Schurr A, Lindh H, McKenzie D, Kiser JS, Williams JH. Am Rev Tuberculosis. 1952;65:511. [PubMed] [Google Scholar]
- 2.Solotorovsky M, Gregory FJ, Ironson EJ, Bugie EJ, O’Neill RC, Pfister K, III, Wombolt TH. Proc Soc Expt Bio Med. 1952;79:563. doi: 10.3181/00379727-79-19447. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Mitchison D. Intl J Tuberculosis Lung Dis. 2003;7:6. [PubMed] [Google Scholar]
- 4.Konno K, Feldmann FM, McDermott W. Am Rev Respir Dis. 1967;95:461. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 5.Scorpio A, Zhang Y. Nat Med. 1996;2:662. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Scorpio A, Nikaido H, Sun Z. J Bacteriol. 1999;181:2044. doi: 10.1128/jb.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. J Antimicrob Chemother. 2003;52:790. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- 8.Wade MM, Zhang Y. J Antimicrob Chemother. 2006;58:936. doi: 10.1093/jac/dkl358. [DOI] [PubMed] [Google Scholar]
- 9.Ngo SC, Zimhony O, Chung WJ, Sayahi H, Jacobs WR, Jr, Welch JT. Antimicrob Agents Chemother. 2007;51:2430. doi: 10.1128/AAC.01458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimhony O, Cox JS, Welch JT, Vilcheze C, Jacobs WR., Jr Nat Med. 2000;6:1043. doi: 10.1038/79558. [DOI] [PubMed] [Google Scholar]
- 11.Zimhony O, Vilcheze C, Arai M, Welch JT, Jacobs WR., Jr Antimicrob Agents Chemother. 2007;51:752. doi: 10.1128/AAC.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boshoff HI, Mizrahi V, Barry CE., III J Bacteriol. 2002;184:2167. doi: 10.1128/JB.184.8.2167-2172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendez S, Traslavina R, Hinchman M, Huang L, Green P, Cynamon MH, Welch JT. Antimicrob Agents Chemother. 2009;53:5114. doi: 10.1128/AAC.01146-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer M, Meyer B. J Am Chem Soc. 2001;123:6108. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 15.Kushner S, Dalalian H, Sanjurjo JL, Bach FL, Safir SR, Smith VK, Williams JH. J Am Chem Soc. 1952:74. [Google Scholar]
- 16.Pravat KM, Ananya M. Concepts in Mag Reson Part A. 2004;20A:1. [Google Scholar]
- 17.Crews P, Rodriquez J, Jaspars M. Organic Structure Analysis; Oxford University Press; Oxford, UK: 1998. p. 544. [Google Scholar]
- 18.Zimhony O, Vilcheze C, Jacobs WR., Jr J Bacteriol. 2004;186:4051. doi: 10.1128/JB.186.13.4051-4055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White HB, 3rd, Mitsuhashi O, Bloch K. J Biol Chem. 1971;246:4751. [PubMed] [Google Scholar]
- 20.Stoops JK, Arslanian MJ, Oh YH, Aune KC, Vanaman TC, Wakil SJ. Proc Natl Acad Sci USA. 1975;72:1940. doi: 10.1073/pnas.72.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood WI, Peterson DO, Bloch K. J Biol Chem. 1978;253:2650. [PubMed] [Google Scholar]
- 22.Maier T, Jenni S, Ban N. Science. 2006;311:1258. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 23.Jenni S, Leibundgut M, Maier T, Ban N. Science. 2006;311:1263. doi: 10.1126/science.1123251. [DOI] [PubMed] [Google Scholar]
- 24.Dalvit C, Fasolini M, Floco M, Knapp S, Peravello P, Veronesi M. J Med Chem. 2002;45:2610. doi: 10.1021/jm011122k. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Liu D, Wyss DF. Magn Reson Chem. 2004;42:485. doi: 10.1002/mrc.1381. [DOI] [PubMed] [Google Scholar]
- 26.Korman TP, Tan YH, Wong J, Luo R, Tsai SC. Biochemistry. 2008;47:1837. doi: 10.1021/bi7016427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cynamon MH, Speirs RJ, Welch JT. Antimicrob Agents Chemother. 1998;42:462. doi: 10.1128/aac.42.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heifets LBF, Marcella A, Lindholm-Levy, Pamela J. Antimicrob Agents Chemother. 1989;33:1252. doi: 10.1128/aac.33.8.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu P, Constantino L, Zhang Y. J Med Microbiol. 2008;57:1129. doi: 10.1099/jmm.0.2008/000786-0. [DOI] [PubMed] [Google Scholar]