Abstract

Comparison of experimental and computational protein folding studies can be difficult because of differences in structural resolution. Isotope-edited infrared spectroscopy offers a direct measure of structural changes involved in protein folding at the single-residue level. Here we demonstrate the increased resolution of site-specific infrared probes to the peptide backbone in the B domain of staphylococcal protein A (BdpA). 13C=18O-labeled methionine was incorporated into each of the helices using recombinant protein expression. Laser-induced temperature jumps coupled with infrared spectroscopy were used to probe changes in the peptide backbone on the submillisecond time scale. The relaxation kinetics of the buried helices, solvated helices, and labeled positions were measured independently by probing the corresponding bands assigned in the amide I region. Using these wavelength-dependent measurements, we observe a fast nanosecond phase and slower microsecond phase at each position. We find at least partial formation of helices 1–3 in the fast intermediate state that precedes the transition state. These measurements provide direct, time-resolved experimental evidence of the early formation of partial helical structure in helices 1 and 3, supporting folding models proposed by computer simulations.
While molecular dynamics simulations allow for the resolution of protein folding dynamics at the atomic level, experiments are often unable to resolve folding at even the single-residue level. Structurally specific experimental characterization of the folding dynamics is necessary to test and further refine computational methods. Nonnatural fluorophores and infrared labels can be incorporated into the protein side chains to increase experimental resolution; however, these methods only indirectly report on backbone conformation. Isotope-edited infrared spectroscopy offers a method of site-specifically resolving protein backbone structure and dynamics.1–4 The amide I mode of the peptide backbone is sensitive to secondary structure; characteristic infrared bands can be assigned to solvated and buried α helix, β sheet, turn, and random coil structures. Isotopic substitution of the carbonyl carbon and carbonyl oxygen shifts the amide I band of the residue of interest to a less congested spectral region. By placing 13C=18O labels at different positions in the protein, it should be possible to determine the folding dynamics at any position of interest. The native fold of a protein is comprised of multiple secondary structure elements stabilized by nonlocal interactions, including the hydrophobic core, salt bridges, and hydrogen bonding. Using isotope labels in each of the secondary structure elements, it should be possible to determine not only the order of formation of secondary structure elements but also the dynamics of the labeled residue.
Here we have compared the folding dynamics of the three helices in an ultrafast folder, the B domain of staphylococcal protein A (BdpA) (Figure 1), using site-specific isotope labeling. Because of its fast folding rate and simple structure, BdpA has been the focus of extensive experimental and computational studies. While experiment and simulation agree that BdpA folds through an intermediate, there are several conflicting models of the transition state structure.5–19 Experimental studies support a model of the transition state that includes a fully formed helix 2, with a partially formed helix 1 and stabilizing interactions between helix 1 and helix 2.5–10 Computational studies have predicted slightly different models that include complete formation of either helix 111–13 or helix 3.14–19 Discrepancies between experimental and computational results may arise from differences in the sequences studied, resolution of experimental probes used, or biases in the computational method. In particular, attempts to resolve dynamics at the single-residue level, time-resolved fluorescence and Förster resonance energy transfer, have relied on incorporation of fluorescent probes, which are not necessarily conservative mutations of the native BdpA sequence and can be difficult to interpret.8,9,20–22 In our approach, 13C=18O-labeled methionine infrared probes were introduced into each of the helices to independently monitor the dynamics of the backbone of each helix.
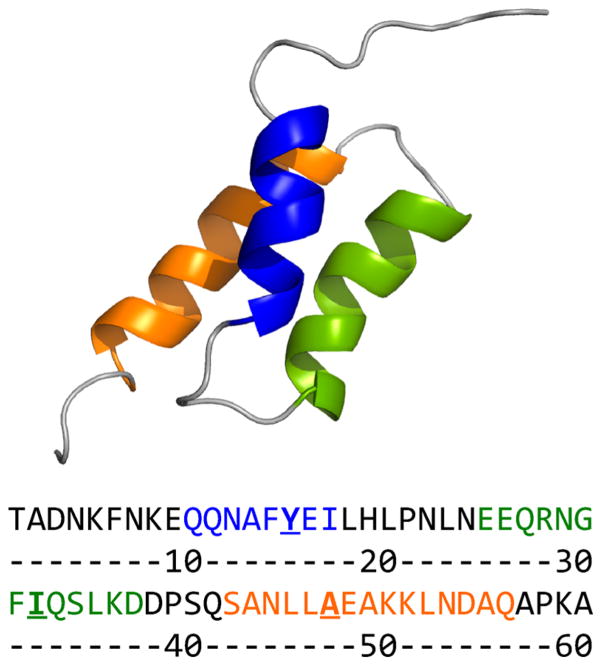
Figure 1.
Cartoon of BdpA (Protein Data Bank entry 1BDD) showing helix 1 (blue), helix 2 (green), and helix 3 (orange). Wild-type BdpA sequence with helices 1–3 colored to match the cartoon. Positions for methionine mutations are underlined. This figure was prepared in PyMOL (www.pymol.org).
An extensive library of point mutations of BdpA has been created for ϕ-value analysis determination of the transition state structure.5,6 In these studies, a Y15W mutation of helix 1 was utilized as a pseudoparent protein to provide an optical probe for ϕ-value analysis. Mutation of I32 in helix 2 to valine or alanine resulted in an ~20 °C decrease in the melting temperature.5 This is likely because I32 is found in the hydrophobic core of BdpA, and mutation disrupts stabilizing interactions between helix 2 and helices 1 and 3. ϕ-value analysis predicted that the N-terminus of helix 3, probed at residue A47, was formed in the transition state but not the C-terminus, K51.5 We selected Y15, I32, and A47 as positions for our labels, because of their respective locations in helices 1–3 and the prediction from ϕ-value analysis that these positions are in nativelike conformations in the transition state. Methionine has a moderately sized side chain, similar to the side chains of tyrosine, isoleucine, and alanine, and we have shown it does not significantly perturb the structure of BdpA. Therefore, we investigated labeled mutants in which Y15, I32, or A47 is replaced with 13C=18O-labeled methionine.
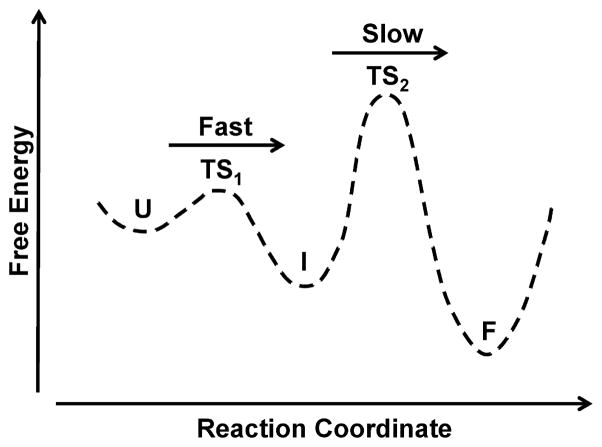
We compared the stability and folding kinetics of wild-type and mutated BdpA. The stability was determined by equilibrium circular dichroism and Fourier transform infrared (FTIR) measurements. The kinetics were measured using temperature jump coupled with time-resolved infrared spectroscopy. Pulsed laser excitation was used to rapidly initiate a shift in the folding equilibrium. The relaxation kinetics of the solvent-exposed helix, buried helix, and labeled helix positions were independently measured by probing the components of the IR amide I band assigned to each structure. As expected, the I32M mutation resulted in destabilization of the protein because of disruption of the hydrophobic core. Mutation of Y15 or A47 had little effect on the protein’s stability. The kinetics of the solvated and buried helix positions of the mutants agree well with those of the parent BdpA. The IR-detected kinetics and melting temperatures of the labeled positions more closely resemble those of the solvated helix than those of the buried helix. All of the mutants exhibit two well-separated kinetic phases when they are probed by infrared spectroscopy, which can be described by a three-state model in which folding proceeds through an intermediate state (Figure 2). The second phase is slow and represents crossing the “global” transition state (TS2) between intermediate and folded states. Interactions present in the intermediate structure will also be present in TS2, because the intermediate occurs prior to TS2 on the reaction coordinate. Contacts at the Y15, I32, and A47 positions are all formed in the fast initial phase, an intermediate structure consistent with the “global” transition state predicted by ϕ-value analysis.5,6
Figure 2.
Folding free energy profile of BdpA and mutants. TS1 and TS2 represent transition states between unfolded (U) and intermediate (I) states and between intermediate (I) and folded (F) states, respectively.
Wavelength-dependent infrared measurements at the labeled position reveal a fast 100 ns phase in addition to a slower microsecond phase. These measurements provide evidence for at least partial formation of helices 1–3 in the transition state. Time-resolved measurements have previously been unable to resolve a fast phase in helix 1 or helix 3. The increased sensitivity of the infrared probes to local structure has aided in the resolution of early formation of helical structure in helices 1 and 3, supporting folding models proposed by computer simulations. These results demonstrate the importance of developing experimental probes of protein folding at the single-residue level in comparing experiments to all-atom molecular dynamics simulations.
EXPERIMENTAL SECTION
Protein Synthesis and Purification
The BdpA gene was incorporated into a pET-30a(+) vector with a histidine affinity tag and TEVprotease cleavage site at the N-terminus. A QuikChange kit was used to generate the mutant BdpA plasmid (Agilent Technologies, Santa Clara, CA). Primer oligonucleotides used to introduce the mutations were obtained from Invitrogen (Life Technologies, Grand Island, NY): Y15M, 5′-ATTTAACAAAGAACAACAAAACGCCTTTATGGAAATCCTCCACTTACCTAATTTAAACG-3′; I32M, 5′-GAAGAACAACGCAATGGTTTTATGCAATCACTCAAAGACGAC-3′; A47M, 5′-ACGACCCATCTCAATCAGCCAACTTACTTATGGAAGCCAAAAAATTAAACG-3′. Mutated plasmid DNA was recovered using a QIAprep Spin Miniprep kit (Qiagen, Valencia, CA), and the sequence was verified by sending the plasmid for sequencing at the Beckman Genomic Center (Danvers, MA). The plasmids containing wild-type and mutated BdpA were introduced into the B834pLysS auxotrophic expression cell line using heat shock (Novagen, Foster City, CA). Cultures were grown in kanamycin- and chloramphenicol-containing media overnight. Glycerol was added to a final concentration of 15% (v/v), and glycerol stocks were stored at −80 °C.
For mutant peptides, an overnight culture of B834pLysS was grown in LB rich medium with kanamycin and chloramphenicol. The starter culture was added to 1 L of M9 minimal medium supplemented with 40 mg of each amino acid, including unlabeled methionine. Mutant cultures were grown to an OD of 0.8 and then harvested again using centrifugation. Cells were resuspended in minimal medium containing the same components, but with 40 mg of 13C=18O-labeled methionine instead of unlabeled methionine. The procedure for 13C=18O labeling methionine has been described elsewhere.23 Protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). (Wild-type cultures in LB medium were grown to an OD of 0.8 and then induced with 1 mM IPTG.) After growing for 24 h, the cells were harvested by centrifugation. Cells were lysed by being stirred and sonicated, and cell debris was removed by centrifugation. Purification was conducted by fast protein liquid chromatography on a Ni His prep FF 16/10 column (GE Healthcare Life Sciences, Pittsburgh, PA). The yield for mutant peptides was 10 mg/L of expression. Expression was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and mass spectrometry.
Isolated proteins containing the histidine purification tag were subjected to a cleavage protocol using AcTEV protease (Invitrogen). Cleaved protein was separated from the free tag using a Ni Sepharose High Performance resin (GE Helathcare Life Sciences, Uppsala, Sweden) in a batch protocol. A mixture of resin and protein were mixed overnight on a rotisserie, and the supernatant was harvested the next day. Clean protein was lyophilized overnight and dissolved in D2O to allow deuterium–hydrogen exchange of the amide protons. The protein was lyophilized a second time and stored at −20 °C prior to use.
Circular Dichroism (CD) Spectroscopy
CD spectra and CD melting curves were recorded on a Jasco J-810 spectropolarimeter equipped with a PFD-425S Jasco temperature controller module (Jasco, Inc., Easton, MD). Peptides were dissolved at a concentration of ~30 μM in 25 mM potassium phosphate and 50 mM NaCl (pH 6.8). All measurements were taken using a 1 mm path length cell. Wavelength scans were recorded over the range from 260 to 190 nm with an average of three repeats. A bandwidth of 2 nm, a response time of 2 s, and a scan rate of 100 nm/min were used for spectral acquisition. Thermal unfolding experiments were performed by monitoring the signal at 222 nm from 20 to 100 °C using a 1 °C interval and a ramp rate of 40 °C/h. After the melt, the temperature was returned to 20 °C and a wavelength scan was obtained to determine the reversibility of folding.
FTIR Spectroscopy
The FTIR absorption spectra and equilibrium melting behavior were monitored on a Varian Excalibur 3100 FTIR spectrometer (Varian Inc., Palo Alto, CA) using a temperature-controlled IR cell. The IR cell consists of two CaF2 windows separated by a 130 μm Teflon spacer split into two compartments, a sample and a reference. The same cells are used for equilibrium FTIR and T-jump experiments. Sample concentrations of 0.5–10 mg/mL were prepared for IR experiments. No aggregation was observed in the infrared at the reported concentrations. The infrared spectrum is highly sensitive to protein aggregation because the amide I band of an aggregated protein is dominated by an intense, sharp band at ~1620 cm−1 and a coupled peak that is also sharp but not as intense at ~1680 cm−1, due to dipolar coupling of the C=O groups in protein aggregates.24 Furthermore, the T-jump experiments are also very sensitive to aggregation, because protein aggregates serve as nucleation sites for cavitation, leading to large cavitation artifacts in the IR transients, which were not observed in our experiments. All spectra shown at a specific temperature were constructed by subtracting the spectrum of the reference buffer solution without protein from that of the sample solution with protein. The temperature-dependent difference spectra were then generated by subtracting the spectrum at the lowest temperature from the spectra at higher temperatures. The second-derivative spectra were computed in IGOR PRO after smoothing the data with a sixth-order binomial algorithm to remove any residual water vapor peaks (WaveMetrics, Lake Oswego, OR).
Time-Resolved Temperature-Jump (T-Jump) Relaxation Measurements
The IR T-jump apparatus has been described previously.25 Pulsed laser excitation is used to rapidly perturb the folding equilibrium on a time scale faster than that of the molecular dynamics of interest. Time-resolved infrared is then used to probe the reaction. The Q-switched GCR-10 Nd:YAG laser (Spectra Physics, Mountain View, CA) fundamental at 1064 nm is Raman-shifted (one Stokes shift in 200 psi H2 gas) to produce a 10 ns pulse at 2 μm. The magnitude of the T-jump is calculated using the change in reference absorbance with temperature. The T-jump reference is taken from D2O buffer with 25 mM potassium phosphate and 50 mM NaCl at pD* 6.8 at the same temperature and probe laser frequency used for the sample. Absorbance changes at the reference frequency are due only to changes in D2O absorption, which is used as an internal thermometer.25
The change in signal induced by the T-jump is probed in real time by a continuous laser with a frequency in the amide I′ band of the IR. The mid-IR probe beam is generated by a continuous wave quantum cascade laser (Daylight Solutions Inc., San Diego, CA) with a tunable output range of 1570–1730 cm−1. The transient transmission of the probe beam through the sample is measured using a fast, 100 MHz, photovoltaic MCT IR detector/preamplifier (Kolmar Technologies, Newburyport, MA). Transient signals are digitized and signal averaged (1000 shots) using a Tektronics digitizer (7612D, Beaverton, OR). Instrument control and data collection are controlled using a LabVIEW computer program.
Analysis of Kinetic Data
The peptide relaxation kinetics must be deconvolved from the observed kinetics. Accurate deconvolution is possible as the instrument response is determined from the reference measurement under the exact conditions of the sample measurements. To minimize detector artifacts, the reference is scaled prior to subtraction from the sample. The decay function is a double-exponential decay with the formula
| (1) |
where A0 is an offset, A1 and A2 are preexponential factors, τ1 and τ2 are relaxation lifetimes of the sample, and x0 is the time offset. The data are fit over the interval from 95 ns to 200 μs. The data analysis was performed in IGOR PRO (Wavemetrics).
RESULTS AND DISCUSSION
Far-UV CD Spectroscopy
13C=18O-labeled methionine mutations were made at single positions in each of the helices of BdpA to determine the folding dynamics of each of the helices independently. Far-UV CD spectra obtained from the wild-type (WT) BdpA and 13C=18O-labeled Y15M (helix 1), I32M (helix 2), and A47M (helix 3) BdpA mutants were almost identical (Figure 3A), suggesting that the methionine mutations do not significantly perturb the native fold. Thermal denaturation was monitored by recording the change in absorbance at 222 nm with temperature (Figure 3B). The melting curves were fit to an apparent two-state equilibrium model:
| (2) |
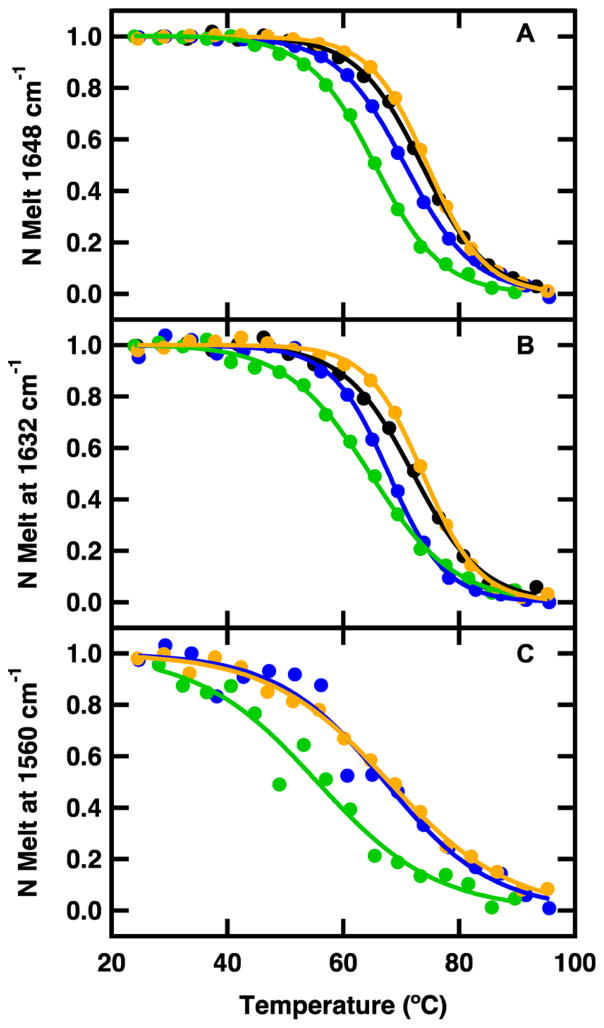
where AO is the observed absorbance, AF and AU are the absorbance contributions from the folded and unfolded populations, respectively, ΔH is the enthalpy change at the midpoint, R is the gas constant, and TM is the transition midpoint.26 The data were then normalized for comparison. The observed melting temperature of WT BdpA is 72.7 ± 0.1 °C (Table 1). This agrees with the previously reported melting temperature of BdpA, 72.5 ± 0.1 °C, also obtained by a thermal melt monitored by far-UV CD of the 222 nm band.9 Each mutant unfolds sigmoidally with unfolding cooperativity similar to that of WT BdpA (Table 1). The Y15M and A47M mutations result in a change in the melting temperature of <1 °C, suggesting that there is little change in protein stability. This is further evidence that the methionine mutations do not affect the fold. The I32M mutation results in a decrease in the melting temperature of ~9 °C (Table 1). I32 is part of the hydrophobic core of BdpA and forms tertiary interactions between helix 2 and helices 1 and 3.5 The methionine mutation likely disrupts these contacts, leading to a decreased melting temperature. Nevertheless, the CD and IR spectra indicate that I32M still adopts the same folded structure as the WT.
Figure 3.
(A) Far-UV CD spectra of 30 μM solutions of WT (black), Y15M (blue), I32M (green), and A47M (orange) BdpA in 25 mM potassium phosphate and 50 mM NaCl (pH 6.8) recorded at 23 °C. The mutants are normalized to the minimum of WT BdpA. (B) Thermal denaturation of WT BdpA and mutants monitored by CD at 222 nm. The solid lines are fit to an apparent two-state model (eq 2) and then normalized.
Table 1.
Summary of Melting Temperatures
| 222 nm (°C) | 1648 cm−1 (°C) | 1632 cm−1 (°C) | 1560 cm−1 (°C) | |
|---|---|---|---|---|
| wild type | 72.7 ± 0.1 | 73.6 ± 0.2 | 72.0 ± 0.4 | not applicable |
| Y15M | 71.9 ± 0.1 | 70.7 ± 0.2 | 67.9 ± 0.3 | 67 ± 2 |
| I32M | 64.2 ± 0.1 | 65.5 ± 0.2 | 64.6 ± 0.5 | 56 ± 3 |
| A47M | 73.3 ± 0.1 | 74.5 ± 0.1 | 73.7 ± 0.3 | 68 ± 1 |
FTIR Spectroscopy
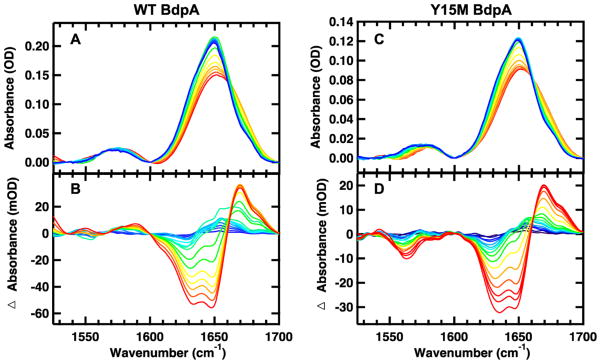
Comparison of the amide I′ (amide I region of peptides in D2O) FTIR absorption spectra of the WT and mutant proteins shows that they are nearly identical (Figure 4A,C; spectra of additional mutants in Figure 1S of the Supporting Information). The amide I′ absorbance arises from C=O stretching vibrations of the polypeptide backbone carbonyls and is an established indicator of secondary structure.27–29 This relatively broad band contains contributions from the entire polypeptide backbone, which in the case of BdpA includes solvated α-helix, buried α-helix, turns, and random coil structure. The FTIR spectra indicate that the secondary structure content is not perturbed by the mutations, consistent with the CD results. The temperature-induced unfolding of WT BdpA and 13C=18O-labeled methionine mutants was studied over the range from 25 to 100 °C in 5 °C intervals. The temperature-dependent absorption spectra of the amide I′ spectral region of WT and Y15M BdpA are shown in panels A and C of Figure 4, respectively. The changes with temperature are highlighted by the difference spectra for each peptide (Figure 4B,D). The difference spectra are generated by subtracting the lowest-temperature spectrum from each absorption spectrum at higher temperatures. There are three main components of the amide I′ band centered at 1632, 1648, and 1670 cm−1. These peaks have been identified previously in α-helical peptides.30 Positive peaks correspond to new interactions with solvent in the unfolded state; the characteristic feature due to the disordered polypeptide is a relatively broad peak, located at 1670 cm−1. Negative peaks correspond to specific structures or interactions present in the folded state. The peak at 1632 cm−1 corresponds to solvated helix (the backbone positions along each helix that are exposed to solvent), and the peak at 1648 cm−1 corresponds to buried helix (the backbone positions protected from solvent in the hydrophobic interior of the structure). Inter-residue coupling of the C=O stretches affects the frequency and intensity of each of these components, particularly for long helices.31 A 13C=18O label placed in the middle of a helix disrupts this coupling (the coupling through the labeled position is inefficient because of the difference in C=O stretching frequency between labeled and unlabeled groups), leading to differences in the intensity of the unlabeled 1632 and 1648 cm−1 bands of the WT and labeled BdpA (Figure 4B,D).
Figure 4.
Temperature-dependent FTIR spectra of 6 mg/mL WT BdpA (A and B) and 3 mg/mL Y15M BdpA (C and D) in 25 mM potassium phosphate and 50 mM NaCl (pH 6.8). (A and C) Absorption spectra in the amide I′ region, where the temperature of the individual traces varies from 25 to 100 °C in 5 °C intervals. (B and D) Difference spectra obtained by subtracting the spectrum at 25 °C from the spectra at higher temperatures.
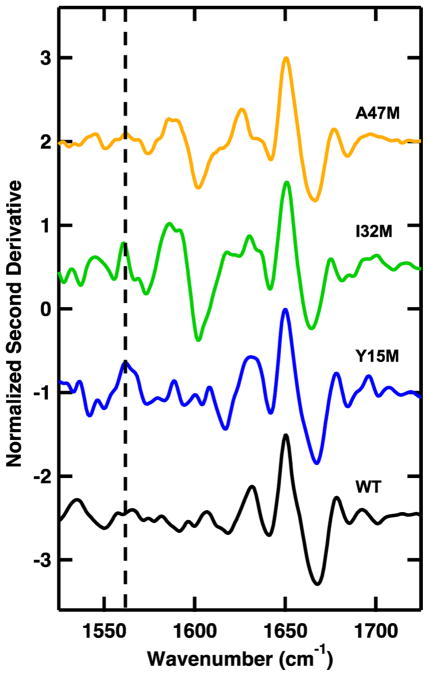
The 13C=18O-labeled methionine is shifted to a lower frequency relative to the unlabeled 12C=16O. The expected shift is 75 cm−1 based on the local harmonic oscillator model of the C=O stretching vibration.23 This peak can be difficult to identify because of its relatively small intensity, overlap with the absorbance of carboxylic acid side chains, and mismatch in HOD content between the sample and reference. The apparent intensity of the labeled peak in Figure 4D is larger than expected, partially because of mismatch in the HOD content of the sample and reference. The second derivative of the FTIR difference spectra between the highest and lowest temperature can be used to eliminate broad contributions to the baseline and better resolve peaks contributing to secondary structure (Figure 5). The positive peaks highlight interactions that are lost at high temperature, the 1632 and 1648 cm−1 peaks corresponding to loss of solvated and buried helices. The I32M and A47M spectra exhibit a temperature-dependent peak at 1586 cm−1, consistent with side chains of carboxylic acid. This peak arises from residual EDTA from the cleavage buffer solution. This contaminant was removed from the Y15M and WT BdpA samples. Comparison of purified samples to samples with residual EDTA demonstrates that the residual EDTA does not affect the folding or stability of BdpA (Figures 2S and 3S of the Supporting Information). The 13C=18O-labeled methionine samples exhibit a peak at 1560 cm−1 that is not present in the WT BdpA sample. Comparison of the His-tagged unlabeled methionine mutant to WT BdpA and the His-tagged 13C=18O-labeled methionine mutant demonstrates that this peak does not arise from differences in side chain packing between the mutant and WT peptide (Figures 4S and 5S of the Supporting Information). The frequency of the labeled position is determined by secondary structure, with contributions from hydrogen bonding and dipole coupling modifying the amide I position.32 The strength of hydrogen bonding between the 13C=18O and neighboring 12C=16O groups will lead to a shift in the labeled amide I position consistent with an α helix. The observed shift is 88 cm−1 relative to the unlabeled buried helix peak at 1648 cm−1 and 72 cm−1 relative to the unlabeled solvated helix position. This is slightly larger or smaller than the expected shift of 75 cm−1, suggesting that the 13C=18O-labeled positions are partially solvated. Partially solvated helices have been demonstrated to have an IR frequency between a fully solvated, 1632 cm−1, or fully buried, 1648 cm−1, helix frequency.33 This assignment is also consistent with changes in the ratio of intensities of the unlabeled 1632 and 1648 cm−1 bands of the labeled proteins. Disruption of the C=O coupling of the solvated position, particularly for the longer helices 2 and 3, results in a decrease in the intensity of the 1632 cm−1 band (solvated helix) relative to that of the 1648 cm−1 band (buried helix) compared to that of WT BdpA (Figure 1S of the Supporting Information). Consistent with previous experiments with α helices with a single 13C=16O-labeled carbonyl, we observe no shift of the 12C=16O amide I modes.34 While incorporation of the 13C=18O labels disrupts the coupling of the amide I mode (Figure 4B,D), such labels can still be described as nonperturbing because the coupling energy is small (a few cm−1) and therefore does not affect the folded structure or thermodynamics of folding.
Figure 5.
Second derivative of the FTIR difference spectrum (100–25 °C) of WT BdpA (black), Y15M BdpA (blue), I32M BdpA (green), and A47M BdpA (orange). The data are normalized at the maximum and offset for the sake of clarity. The dashed vertical line at 1560 cm−1 highlights the 13C=18O-labeled peak.
The normalized melting curves for WT BdpA and 13C=18O-labeled mutants derived from temperature-dependent IR absorption are shown in Figure 6. Infrared melts were obtained at wavelengths corresponding to buried helix (1648 cm−1), solvated helix (1632 cm−1), and the 13C=18O-labeled position (1560 cm−1). The data were normalized after being fit to an apparent two-state equilibrium model using eq 2. There is good agreement between the melting temperature and breadth of the transition probed at 222 nm by circular dichroism and melting temperatures probed at 1632 and 1648 cm−1 by FTIR (Figure 6 and Table 1). The observation of a lower melting temperature of the I32M mutant compared to that of WT BdpA supports the conclusion from the CD measurements that the I32M mutation destabilizes the hydrophobic core. In all four peptides, there is a slight difference in the melting temperature measured at the buried and solvated helix position. The lower melting temperature of the solvated helix band is likely due to fraying of solvent-exposed regions of the individual helices prior to the global unfolding transition, which broadens and shifts the apparent transition midpoint to a lower temperature. This is evidence of a folding intermediate with a reduced helical content, lacking some of the less stable helical regions. The 13C=18O-labeled positions have melting temperatures similar to that of the solvated helix positions, supporting the assignment of this peak as partially solvated. The overall signal change at the 13C=18O position is smaller than in the amide I′ region, leading to the relative increase in noise compared to that of the unlabeled position. The breadth of the transition at the labeled positions is twice as large as the breadth of the transition at the unlabeled positions. The broader transition is a result of the 13C=18O position being a local probe, primarily sensitive to local changes in structure and solvation and only indirectly sensitive to the global, cooperative folding transition.
Figure 6.
FTIR melt curves for WT BdpA (black), Y15M BdpA (blue), I32M BdpA (green), and A47M BdpA (orange) obtained by plotting the change in IR difference spectra at (A) 1648, (B) 1632, and (C) 1560 cm−1 vs temperature. The data are fit to an apparent two-state model (eq 2) and then normalized.
Temperature-Jump Relaxation Kinetics
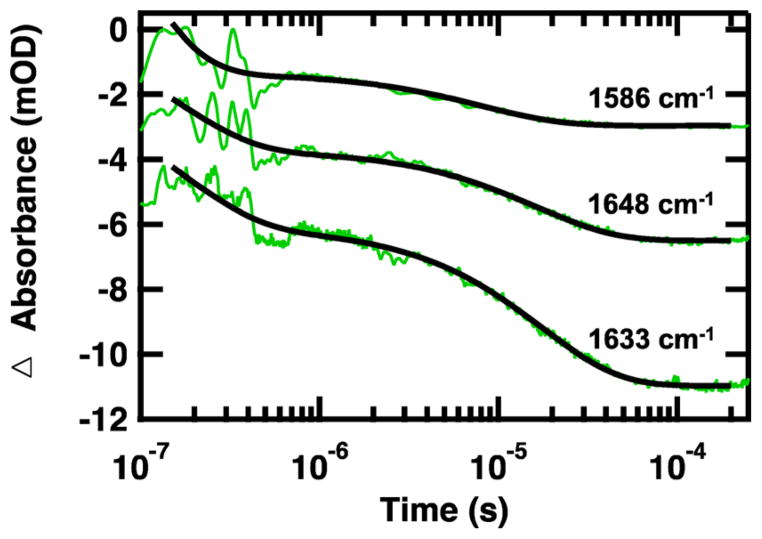
The relaxation kinetics of the folding–unfolding transition following a laser-induced temperature jump were probed using time-resolved infrared spectroscopy of the amide I′ frequency of the buried helix (1648 cm−1), solvated helix (1632 cm−1), and 13C=18O-labeled methionine (1560 cm−1). Jumps were performed slightly off peak center (near the maxima of the difference features) to maximize the transient absorbance signal. Because the amide I subcomponents are relatively broad (Figure 4), the off center probe frequencies employed still correspond to the respective structural assignments. Figure 7 displays the wavelength-dependent relaxation kinetics for the I32M mutant following a jump from 50 to 60 °C. All of the relaxation transients are fit by a double exponential, and the lifetimes are reported in Table 2 (Figures 6S–8S of the Supporting Information). There is good agreement between the kinetics observed for WT BdpA and the mutants at the unlabeled probe frequencies of 1648 and 1632 cm−1 (Table 2). All exhibit a fast ~100 ns phase (τ1) and a slower ~10 μs phase (τ2). These kinetics agree with previous WT BdpA temperature-jump measurements to a final temperature of 70 °C, which reported a fast ~100 ns phase and a slower ~10 μs phase.10 For all of the peptides, kinetics of the slow phase of 1632 cm−1, assigned to the solvated helix, are slightly faster than the kinetics of the slow phase of 1648 cm−1, buried helix.
Figure 7.
Representative IR T-jump relaxation kinetics of 13C=18O-labeled I32M BdpA monitored in the amide I′ spectral region at 1648, 1633, and 1586 cm−1 following a T-jump from 50 to 60 °C. A double-exponential fit is overlaid over each kinetic trace (black solid line). Data are offset for the sake of clarity.
Table 2.
Relaxation Kinetics following a Jump from 50 to 60 °C
| ~1648 cm−1
|
~1632 cm−1
|
~1560 cm−1
|
||||
|---|---|---|---|---|---|---|
| τ1 (ns) | τ2 (μs) | τ1 (ns) | τ2 (μs) | τ1 (ns) | τ2 (μs) | |
| BdpA | 115 ± 5 | 13.2 ± 0.2 | 185 ± 8 | 9.4 ± 0.1 | ||
| Y15Ma | 127 ± 5 | 46.0 ± 0.7 | 139 ± 4 | 39.1 ± 0.8 | 130 | 25 ± 1 |
| I32M | 160 ± 10 | 16.6 ± 0.3 | 170 ± 10 | 16.4 ± 0.3 | 77 ± 6 | 8.2 ± 0.3 |
| A47M | 53 ± 2 | 18.7 ± 0.3 | 110 ± 4 | 7.4 ± 0.6 | 120 ± 20 | 8.0 ± 0.5 |
Relaxation kinetics following a jump from 20 to 30 °C. No confidence interval is reported for the fast phase of 1560 cm−1 because of early time scale artifacts.
Temperature-jump measurements at 1560 cm−1 (Figure 8) reveal the kinetics at each of the 13C=18O-labeled positions. Similar to the solvated and buried helix wavelengths, the kinetics of the labeled position fit to a double exponential with a fast ~100 ns phase and a slower ~10 μs phase. There is relatively more noise in this data compared to that in the data for the unlabeled solvated and buried helix because of the smaller overall amplitude of absorbance at the labeled position (Figure 4). The kinetics at the labeled position correspond most closely with the kinetics at the solvated position, supporting the FTIR assignment of these positions to partially solvated helix (Table 2). Because of early time scale artifacts in the Y15M BdpA data, no confidence interval is reported for the fast phase. Despite the large artifacts present at early times in the Y15M data, the fast phase is clearly present, and its lifetime is on the same order of magnitude as that observed for the other mutants. The kinetics probed at 1648 and 1632 cm−1 are dominated by the slow phase but also exhibit a smaller amplitude fast phase. In contrast, the kinetics probed at the labeled frequency of 1560 cm−1 show nearly equal amplitudes in the fast and slow phases.
Figure 8.
Representative IR T-jump relaxation kinetics of Y15M BdpA (blue), I32M BdpA (green), and A47M BdpA (orange) monitored at the 13C=18O-labeled amide I′ spectral position at ~1560 cm−1 following a T-jump from 50 to 60 °C (20 to 30 °C for Y15M). A double-exponential fit is overlaid on each kinetic trace (black solid line). Data are normalized and offset for the sake of clarity.
The two well-separated relaxation phases make it clear that BdpA folds through an intermediate state as described in the legend of Figure 2. Both the folded and intermediate states are populated at the starting temperature of the T-jump experiments, such that relaxation is observed over both barriers. The fast phase corresponds to relaxation over the first barrier between states U and I, whereas the slower phase is assigned to the relaxation over the global transition state barrier between I and the fully folded state. The equilibrium CD and FTIR melts are also consistent with a folding intermediate despite reasonable fits to a two-state model, because the Tm and transition width depend on the probe, whether CD or the different IR probe frequencies. A previous infrared study of WT BdpA studied this phenomenon extensively using T-jumps to multiple temperatures to deconvolve a second broad melt component with a Tm of 58 °C corresponding to the fast kinetics phase.10 This phase was assigned to fast helical formation preceding formation of long-range tertiary contacts on the microsecond time scale. Differences in the FTIR equilibrium melting temperatures of the solvated and buried helices (Figure 6) suggest that this intermediate has a largely intact hydrophobic core with fraying at the ends of the helices. Whereas the transients of the solvated and buried helix have a small intensity in the fast phase, the transients of the 13C=18O-labeled position have nearly half of their intensity in the fast phase. The 13C=18O labels are local probes, whereas the solvated and buried helix positions are more sensitive to global changes. The larger relative intensity in the fast phase when labels are incorporated at positions in the hydrophobic core is indicative of an intermediate in which these positions are helical. The infrared melt probed at the 13C=18O-labeled position is more broad than the melt at the buried or solvated helix position, supporting our conclusion that the absorbance includes a relatively greater contribution from the broad melt component. Because of the small overall absorbance intensity at this position, it is not possible to deconvolve the two melts; however, the breadth of the infrared melt combined with the observations of two phases from kinetic measurements supports the assignment of the fast helical formation phase at the 13C=18O-labeled position.
While ϕ-value analysis has been extensively used to identify specific interactions formed in the transition state, there have been few time-resolved studies that resolve a fast-forming intermediate, which can indirectly report on the transition state structure. The studies that have been conducted rely on incorporation of tryptophan or nonnatural fluorescent probes, and most do not resolve the nanosecond time scale.5,6,8,9,20–22 Many utilize tryptophan as a reporter of global BdpA folding, with probes located only in helix 1.6,9,21,22 Previous work from the Dyer group used nanosecond T-jump fluorescence measurements of tryptophan probes at positions similar to those of the 13C=18O methionine mutations in helices 1–3.8 A fast phase was observed only for the helix 2 mutant, suggesting that the initial step in BdpA formation is the formation of helix 2. Radford and co-workers also did not observe a fast phase in their nanosecond T-jump fluorescence measurements of a fast-folding mutant of BdpA with a probe in helix 1.21 However, interpretation of tryptophan fluorescence is difficult, and the lack of a fast phase for the helix 1 and helix 3 mutants does not preclude formation of some helical structure. If, as in the model proposed by infrared measurements, the fast phase arises from helical formation and the slow phase from tertiary contact formation, changes in the environment necessary for changes in tryptophan fluorescence may not occur until tertiary contacts are formed.10 Our results support this model, as we observe a fast phase for infrared probes located within one residue of the tryptophan mutants. We incorporated our infrared probes at locations where ϕ-value analysis performed by Fersht and coworkers determined transition state structure formation in helices 1–3.5,6 Our measurements demonstrate that there is an intermediate with helical structure formed at these positions, providing the first time-resolved evidence of structure formation in helix 1 and helix 3 prior to forming the global transition state.
While kinetic rates of BdpA derived from computer simulations agree with experimental results,35–37 the models do not necessarily capture the details of the folding mechanism correctly. One group of all-atom simulations predicts early formation of helix 3.14–19 Results from ϕ-value analysis and fluorescence T-jump conflict with this model; they predict formation of helix 2 with at most partial formation of contacts in helices 1 and 3. They agree better with a second group of simulations that predict early formation of helices 1 and 2.11–13 The distributed computing approach employed by the Pande group predicts that while helix 2 is most likely to form first and lead to the native state, there is an alterative pathway in which helix 3 forms first.37 Our kinetic measurements provide evidence for this model, as we observe at least partial formation of each of the helices in the intermediate state that precedes formation of the global transition state. Further measurements of BdpA labeled at additional positions in the helices would be necessary to determine fully the degree of helix formation prior to the global transition state. The results demonstrate that 13C=18O-labeled methionine infrared labels provide a powerful tool for studying protein folding at the single-residue level.
CONCLUSION
Using 13C=18O-labeled methionine infrared labels, we are able to resolve folding of individual residues within BdpA. We propose a transition state model in which helices 1–3 are all at least partially structured, similar to that proposed by ϕ-value analysis.5,6 This is the first direct observation of fast helix formation in helices 1 and 3, supporting computational models that predict formation of these helices in the transition state. 13C=18O-labeled methionine infrared probes are sensitive to structural changes of a specific residue in the protein. This provides information that is complementary to unlabeled infrared spectroscopy, which is sensitive to changes in secondary structure of the protein, and fluorescence spectroscopy, which is sensitive to environmental changes around the fluorophore. Recombinant expression in a methionine auxotroph provides an efficient and in this case minimally perturbing method for incorporating isotopic labels. In principle, any position may be isotopically labeled using recombinant expression, provided it will tolerate a methionine substitution. By utilizing isotope-edited infrared probes at multiple positions in the protein, we have established an experimental methodology to approach an all-atom view of the folding landscape.
Supplementary Material
Acknowledgments
Funding
This work was supported by a grant from the National Institutes of Health (R01 GM53640) to R.B.D.
We thank Charu Kumar for her assistance in the development of the BdpA expression protocol.
Footnotes
Notes
The authors declare no competing financial interest.
A van’t Hoff analysis, temperature-dependent FTIR spectra of I32M and A47M BdpA mutants, the protocol for removing EDTA contamination, and figures of relaxation kinetics of WT, Y15M, and A47M BdpA. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zimmermann J, Thielges MC, Seo YJ, Dawson PE, Romesberg FE. Cyano groups as probes of protein microenvironments and dynamics. Angew Chem, Int Ed. 2011;50:8333–8337. doi: 10.1002/anie.201101016. [DOI] [PubMed] [Google Scholar]
- 2.Nagarajan S, Taskent-Sezgin H, Parul D, Carrico I, Raleigh DP, Dyer RB. Differential ordering of the protein backbone and side chains during protein folding revealed by site-specific recombinant infrared probes. J Am Chem Soc. 2011;133:20335–20340. doi: 10.1021/ja2071362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer SH, Song B, Raleigh DP, Dyer RB. Residue specific resolution of protein folding dynamics using isotope-edited infrared temperature jump spectroscopy. Biochemistry. 2007;46:3279–3285. doi: 10.1021/bi602372y. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann J, Thielges MC, Yu W, Dawson PE, Romesberg FE. Carbon-deuterium bonds as site-specific and nonperturbative probes for time-resolved studies of protein dynamics and folding. J Phys Chem Lett. 2011;2:412–416. [Google Scholar]
- 5.Sato S, Fersht AR. Searching for multiple folding pathways of a nearly symmetrical protein: Temperature dependent ϕ-value analysis of the B domain of protein A. J Mol Biol. 2007;372:254–267. doi: 10.1016/j.jmb.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 6.Sato S, Religa TL, Daggett V, Fersht AR. Testing protein-folding simulations by experiment: B domain of protein A. Proc Natl Acad Sci USA. 2004;101:6952–6956. doi: 10.1073/pnas.0401396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxa MC, Freed KF, Sosnick TR. Quantifying the structural requirements of the folding transition state of protein A and other systems. J Mol Biol. 2008;381:1362–1381. doi: 10.1016/j.jmb.2008.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyer RB, Vu DM. 3.3 fast events in protein folding. In: Egelman EH, editor. Comprehensive biophysics. Elsevier; Amsterdam: 2012. pp. 34–42. [Google Scholar]
- 9.Vu DM, Peterson ES, Dyer RB. Experimental resolution of early steps in protein folding: Testing molecular dynamics simulations. J Am Chem Soc. 2004;126:6546–6547. doi: 10.1021/ja048416q. [DOI] [PubMed] [Google Scholar]
- 10.Vu DM, Myers JK, Oas TG, Dyer RB. Probing the folding and unfolding dynamics of secondary and tertiary structures in a three-helix bundle protein. Biochemistry. 2004;43:3582–3589. doi: 10.1021/bi036203s. [DOI] [PubMed] [Google Scholar]
- 11.Garcia AE, Onuchic JN. Folding a protein in a computer: An atomic description of the folding/unfolding of protein A. Proc Natl Acad Sci USA. 2003;100:13898–13903. doi: 10.1073/pnas.2335541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Z, Brooks CL, III, Boczko EM. Exploring the folding free energy surface of a three-helix bundle protein. Proc Natl Acad Sci USA. 1997;94:10161–10166. doi: 10.1073/pnas.94.19.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng S, Yang Y, Wang W, Liu H. Transition state ensemble for the folding of B domain of protein A: A comparison of distributed molecular dynamics simulations with experiments. J Phys Chem B. 2005;109:23645–23654. doi: 10.1021/jp0517798. [DOI] [PubMed] [Google Scholar]
- 14.Alonso DO, Daggett V. Staphylococcal protein A: Unfolding pathways, unfolded states, and differences between the B and E domains. Proc Natl Acad Sci USA. 2000;97:133–138. doi: 10.1073/pnas.97.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolinski A, Skolnick J. Monte Carlo simulations of protein folding. II Application to protein A, rop, and crambin. Proteins. 1994;18:353–366. doi: 10.1002/prot.340180406. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh A, Elber R, Scheraga HA. An atomically detailed study of the folding pathways of protein A with the stochastic difference equation. Proc Natl Acad Sci USA. 2002;99:10394–10398. doi: 10.1073/pnas.142288099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao Q, Gao YQ. The relative helix and hydrogen bond stability in the B domain of protein A as revealed by integrated tempering sampling molecular dynamics simulation. J Chem Phys. 2011;135:135102. doi: 10.1063/1.3630127. [DOI] [PubMed] [Google Scholar]
- 18.Shao Q. Probing sequence dependence of folding pathway of α-helix bundle proteins through free energy landscape analysis. J Phys Chem B. 2014;118:5891–5900. doi: 10.1021/jp5043393. [DOI] [PubMed] [Google Scholar]
- 19.Lei H, Wu C, Wang ZX, Zhou Y, Duan Y. Folding processes of the B domain of protein A to the native state observed in all-atom ab initio folding simulations. J Chem Phys. 2008;128:235105. doi: 10.1063/1.2937135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang F, Lerner E, Sato S, Amir D, Haas E, Fersht AR. Time-resolved fluorescence resonance energy transfer study shows a compact denatured state of the B domain of protein A. Biochemistry. 2009;48:3468–3476. doi: 10.1021/bi801890w. [DOI] [PubMed] [Google Scholar]
- 21.Dimitriadis G, Drysdale A, Myers JK, Arora P, Radford SE, Oas TG, Smith DA. Microsecond folding dynamics of the f13w g29a mutant of the B domain of staphylococcal protein A by laser-induced temperature jump. Proc Natl Acad Sci USA. 2004;101:3809–3814. doi: 10.1073/pnas.0306433101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arora P, Oas TG, Myers JK. Fast and faster: A designed variant of the B-domain of protein A folds in 3 μs. Protein Sci. 2004;13:847–853. doi: 10.1110/ps.03541304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marecek J, Song B, Brewer S, Belyea J, Dyer RB, Raleigh DP. A simple and economical method for the production of 13C,18O-labeled Fmoc-amino acids with high levels of enrichment: Applications to isotope-edited IR studies of proteins. Org Lett. 2007;9:4935–4937. doi: 10.1021/ol701913p. [DOI] [PubMed] [Google Scholar]
- 24.Jackson M, Mantsch HH. Protein secondary structure from ft-ir spectroscopy: Correlation with dihedral angles from 3-dimensional Ramachandran plots. Can J Chem. 1991;69:1639–1642. [Google Scholar]
- 25.Williams S, Causgrove TP, Gilmanshin R, Fang KS, Callender RH, Woodruff WH, Dyer RB. Fast events in protein folding: Helix melting and formation in a small peptide. Biochemistry. 1996;35:691–697. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 26.Manning MC, Woody RW. Theoretical determination of the CD of proteins containing closely packed antiparallel β-sheets. Biopolymers. 1987;26:1731–1752. doi: 10.1002/bip.360261007. [DOI] [PubMed] [Google Scholar]
- 27.Yang WJ, Griffiths PR, Byler DM, Susi H. Protein conformation by infrared-spectroscopy: Resolution enhancement by fourier self-deconvolution. Appl Spectrosc. 1985;39:282–287. [Google Scholar]
- 28.Arrondo JLR, Blanco FJ, Serrano L, Goni FM. Infrared evidence of a β-hairpin peptide structure in solution. FEBS Lett. 1996;384:35–37. doi: 10.1016/0014-5793(96)00261-x. [DOI] [PubMed] [Google Scholar]
- 29.Susi H, Byler DM. Resolution-enhanced fourier-transform infrared-spectroscopy of enzymes. Methods Enzymol. 1986;130:290–311. doi: 10.1016/0076-6879(86)30015-6. [DOI] [PubMed] [Google Scholar]
- 30.Susi H, Timasheff SN, Stevens L. Infrared spectra and protein conformations in aqueous solutions. I The amide I band in H2O and D2O solutions. J Biol Chem. 1967;242:5460–5466. [PubMed] [Google Scholar]
- 31.Huang R, Kubelka J, Barber-Armstrong W, Silva RA, Decatur SM, Keiderling TA. Nature of vibrational coupling in helical peptides: An isotopic labeling study. J Am Chem Soc. 2004;126:2346–2354. doi: 10.1021/ja037998t. [DOI] [PubMed] [Google Scholar]
- 32.Barth A, Zscherp C. What vibrations tell us about proteins. Q Rev Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 33.Turner DR, Kubelka J. Infrared and vibrational CD spectra of partially solvated α-helices: DFT-based simulations with explicit solvent. J Phys Chem B. 2007;111:1834–1845. doi: 10.1021/jp0666840. [DOI] [PubMed] [Google Scholar]
- 34.Barber-Armstrong W, Donaldson T, Wijesooriya H, Silva RAGD, Decatur SM. Empirical relationships between isotope-edited IR spectra and helix geometry in model peptides. J Am Chem Soc. 2004;126:2339–2345. doi: 10.1021/ja037863n. [DOI] [PubMed] [Google Scholar]
- 35.Myers JK, Oas TG. Preorganized secondary structure as an important determinant of fast protein folding. Nat Struct Biol. 2001;8:552–558. doi: 10.1038/88626. [DOI] [PubMed] [Google Scholar]
- 36.Islam SA, Karplus M, Weaver DL. Application of the diffusion-collision model to the folding of three-helix bundle proteins. J Mol Biol. 2002;318:199–215. doi: 10.1016/S0022-2836(02)00029-3. [DOI] [PubMed] [Google Scholar]
- 37.Jayachandran G, Vishal V, Garcia AE, Pande VS. Local structure formation in simulations of two small proteins. J Struct Biol. 2007;157:491–499. doi: 10.1016/j.jsb.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.