Abstract
The Arabidopsis ovate family proteins (AtOFPs) have been shown to function as transcriptional repressors and regulate multiple aspects of plant growth and development. There are 31 genes that encode the full-length OVATE-domain containing proteins in the rice genome. In this study, the gene structure analysis revealed that OsOFPs are intron poor. Phylogenetic analysis suggested that OVATE proteins from rice, Arabidopsis and tomato can be divided into 4 groups (I–IV). Real-time quantitative polymerase chain reaction (RT-qPCR) analysis identified OsOFPs with different tissue-specific expression patterns at all stages of development in the rice plant. Interestingly, nearly half of the total number of OsOFP family was more highly expressed during the seed developmental stage. In addition, seed developmental cis-elements were found in the promoter region of the OsOFPs. Subcellular localization analysis revealed that YFP-OsOFP fusion proteins predominantly localized in the nucleus. Our results suggest that OsOFPs may act as regulatory proteins and play pivotal roles in the growth and development of rice.
Introduction
Development of an organism is based on the temporal and spatial regulation of gene expression, in which transcription factors act as switches of the regulatory cascades [1]. Transcription factor genes comprise a substantial fraction of all eukaryotic genomes [2]. In the Arabidopsis thaliana genome, there are 1922 known and predicted transcription factors, representing >5% of the total number of genes for this genus, distributed across 64 families [2,3]. It has been reported that members of many large transcription factor families play essential roles in plant development [4–7]. The highest representative in plants is the MADS-box transcription factor family. Members of this family have multiple functions in the transition to flowering, floral organ identity, gametophyte and seed development, fruit development, and lateral root formation [8]. Recently the Arabidopsis ovate family proteins (AtOFPs), forming a plant-specific transcription factor family, were found to control multiple aspects of plant growth and development [9–12]. There are 18 genes in the Arabidopsis genome that encode proteins with a predicted OVATE domain. The OVATE gene was originally cloned from the tomato plant in which a single mutation led to a premature stop codon in this gene, thus causing the transition of the tomato fruit from round- to pear-shaped [13]. In addition, overexpression of the ovate gene from the wild-type round-fruited line in pear-shaped fruit producing line conferred unevenly reduced size of floral organs and leaflets [13]. The tomato OVATE protein contains a C-terminal domain of approximately 70 amino acids that are conserved in tomato, Arabidopsis, and rice plants [13]. This domain was designated as the OVATE domain, and the proteins containing this conserved domain were designated as ovate family proteins (OFPs), which can be found exclusively in plant proteins [12].
Hackbusch et al. (2005) reported, for the first time, that 9 members of AtOFPs interact with 3-aa loop extension (TALE) homeodomain proteins, fundamental regulators of plant meristem function and leaf development, and can be cofactors in regulating the subcellular localization of TALE proteins [9]. A systematic function analysis of AtOFPs has been done since 2007 [10–12,14]. It was reported that AtOFP1 plays a key role in suppressing cell elongation by acting as an active transcriptional repressor. The Arabidopsis overexpressing AtOFP1 exhibited a phenotype of reduced length in all aerial organs, including the hypocotyl, rosette leaf, cauline leaf, inflorescence stem, floral organs, and silique. The expression of the gibberellin biosynthesis key enzyme gene AtGA20OX1 was suppressed in AtOFP1 overexpressed Arabidopsis plants [9,11]. Other AtOFPs overexpressed also confer abnormal morphological phenotypes [12]. The Arabidopsis OFP4 plays a role in regulating secondary cell wall formation through its interaction with KNOTTED1-LIKE HOMEODOMAIN PROTEIN7 (KNAT7) [12]. AtOFP5 has been shown to negatively regulate the activity of a BLH1-KNAT3 complex during early embryo sac development [14]. There are 31 OFP genes (SlOFPs) predicted in tomato genome [15]. The expression patterns of 17 SlOFPs were examined tissue-specifically in wild tomato. SlOFP07, SlOFP08, SlOFP14, SlOFP18, SlOFP20, SlOFP29 and SlOFP31 were higher expressed in the reproductive organs than those in the vegetative organs. SlOFP10, SlOFP13 and SlOFP22 were specifically expressed in the seedling stage such as in root, hypocotyl and young leaf [15]. These imply that SlOFPs have a specialized function in plant development [15].
As OFPs are proposed to encode a novel transcription factor class and regulate plant development broadly, further genetic and biochemical studies of the function of these genes in the important food plants are needed to provide a better comprehension for their roles in plant evolution and domestication. In general, transcription factors (TFs) are expected to be excellent candidates for modifying complex traits in crops because they naturally act as master regulators of cellular processes [16]. The functional study of plant transcription factors has become increasingly popular in recent years. Rice (Oryza sativa) is one of the most important food crops worldwide and a model for genomic research in cereals. Some transcription factors have been identified to play prominent roles in many developmental processes in rice [17–19]. However, little is known about the molecular characteristics of OVATE-containing proteins in rice. The results of this study display the bioinformatic characteristics of ovate family proteins in rice (OsOFPs), tissue-specific expression profile, subcellular localization and hormone-responsive expression pattern of each member in this family. Our data provide a very useful reference for the functional analysis of the OFP family in rice.
Results
Chromosomal Distribution of OsOFPs and Evolution of OFPs in Plants
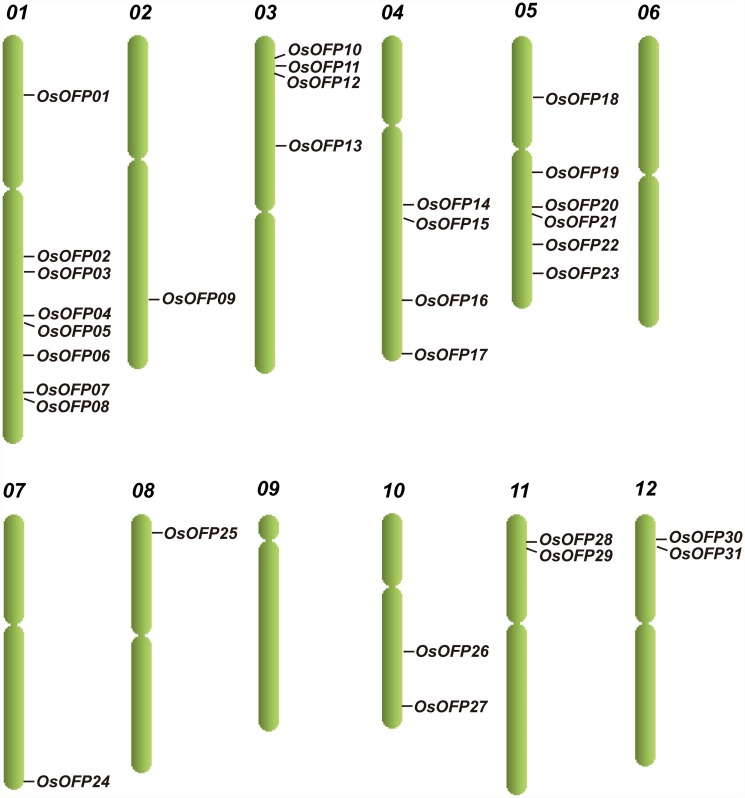
Thirty-one putative OsOFPs were identified in rice genome by searching the plant transcription factors database [20,21]. All 31 putative OsOFPs were designated as OsOFP01–OsOFP31 in accordance with their locations on the chromosomes of rice (Table 1a). Fig. 1 is a diagrammatic representation of the distribution of OsOFPs on the rice chromosomes. Although the OsOFPs are scattered throughout the 12 chromosomes of rice (except on chromosomes 6 and 9), their distribution is not uniform (Table 1b). The chromosomal location of the OsOFPs reveals that certain chromosomes and chromosomal regions have a relatively high density of OsOFPs. For instance, there are 8 and 6 OsOFPs located on chromosomes 01 and 05, respectively; however, only a single OsOFP was present on chromosomes 07 and 08, separately (Fig. 1).
Table 1. Information related to the OsOFPs, and performance of the ovate family protein.
| Locus name | Gene name a | Chromosome location b | WoLF PSORT | CELLO | Subcellular localization |
|---|---|---|---|---|---|
| Prediction c | Prediction d | Experiments e | |||
| LOC_Os01g12690.1 | OsOFP01 | Chr1: 7009033–7010505 | nucl(13) | Nuclear(4.3) | Nuclear |
| LOC_Os01g40970.1 | OsOFP02 | Chr1: 23173640–23174380 | nucl(11.5) cyto_nucl(6.5) chlo(1) | Nuclear(3.3) | Nuclear with granular spots |
| LOC_Os01g43610.1 | OsOFP03 | Chr1: 24982666–24981173 | nucl(14) | Nuclear(4.6) | Nuclear with granular spots |
| LOC_Os01g53160.1 | OsOFP04 | Chr1: 30542800–30544190 | chlo(11) nucl(3) | Nuclear(3.6) | Nuclear with granular spots |
| LOC_Os01g54570.1 | OsOFP05 | Chr1: 31391261–31392235 | nucl(13) | Nuclear(3.7) | Nuclear |
| LOC_Os01g60810.1 | OsOFP06 | Chr1: 35169295–35167600 | chlo(7) nucl(2) mito(2) cyto(1) plas(1) | Nuclear(3.4) | Nuclear with granular spots |
| LOC_Os01g64410.1 | OsOFP07 | Chr1: 37381457–37382214 | chlo(6) mito(6) plas(1) | Chloroplast(3.1) | Nuclear |
| LOC_Os01g64430.1 | OsOFP08 | Chr1: 37397573–37396145 | nucl(7) chlo(6) | Nuclear(3.6) | Nuclear |
| LOC_Os02g45620.1 | OsOFP09 | Chr2: 27751407–27752436 | chlo(8) nucl(3) mito(2) | Nuclear(3.1) | Nuclear |
| LOC_Os03g03480.1 | OsOFP10 | Chr3: 1506050–1505340 | chlo(7) mito(6) | Nuclear(4.3) | Nuclear with granular spots |
| LOC_Os03g06350.1 | OsOFP11 | Chr3: 3179590–3178092 | chlo(8) mito(4) nucl(2) | Nuclear(2.5) | Nuclear |
| LOC_Os03g10150.1 | OsOFP12 | Chr3: 5152038–5150966 | chlo(12) mito(2) | Nuclear(2.9) | Nuclear |
| LOC_Os03g21870.1 | OsOFP13 | Chr3: 12510664–12509516 | chlo(6) mito(6) nucl(1) | Nuclear(4.4) | Nuclear |
| LOC_Os04g33870.1 | OsOFP14 | Chr4: 20519152–20520509 | chlo(8) nucl(2) mito(2) plas(1) | Chloroplast(3.4) | Nuclear |
| LOC_Os04g37510.1 | OsOFP15 | Chr4: 22333107–22332676 | chlo(11) mito(2) | Nuclear(1.9) Extracellular(1.3) | Nuclear with granular spots |
| LOC_Os04g48830.1 | OsOFP16 | Chr4: 29121699–29123167 | chlo(6) nucl(6) mito(1) | Nuclear(3.5) | Nuclear |
| LOC_Os04g58820.1 | OsOFP17 | Chr4: 34988834–34990126 | nucl(14) | Nuclear(4.3) | Nuclear with granular spots |
| LOC_Os05g12808.1 | OsOFP18 | Chr5: 7357669–7356782 | nucl(7) mito(5) chlo(2) | Chloroplast(1.9) Nuclear(1.4) | Nuclear |
| LOC_Os05g25910.1 | OsOFP19 | Chr5: 15070481–15069428 | nucl(7) chlo(4) mito(3) | Nuclear(3.7) | Nuclear |
| LOC_Os05g36970.1 | OsOFP20 | Chr5: 21595856–21597025 | chlo(7) nucl(6) | Nuclear(2.4) | Nuclear |
| LOC_Os05g36990.1 | OsOFP21 | Chr5: 21611604–21610390 | nucl(5) mito(4) chlo(3) extr(1) | Nuclear(1.9)Chloroplast(1.3) | Nuclear |
| LOC_Os05g39950.1 | OsOFP22 | Chr5: 23477065–23478850 | nucl(12) chlo(1) | Nuclear(4.3) | Nuclear |
| LOC_Os05g44090.1 | OsOFP23 | Chr5: 25636912–25635962 | nucl(7) chlo(6) | Nuclear(4.0) | Nuclear |
| LOC_Os07g48150.1 | OsOFP24 | Chr7: 28759461–28760724 | chlo(9) mito(3.5) cyto_mito(2.5) | Nuclear(3.4) | Nuclear |
| LOC_Os08g01190.1 | OsOFP25 | Chr8: 138247–139410 | chlo(13) | Nuclear(3.4) | Nuclear |
| LOC_Os10g29610.1 | OsOFP26 | Chr10:15392044–15392805 | chlo(11) mito(2) | Chloroplast(2.7) | Nuclear |
| LOC_Os10g38880.1 | OsOFP27 | Chr10:20714505–20715633 | nucl(13) | Nuclear(2.8) | Nuclear |
| LOC_Os11g05770.1 | OsOFP28 | Chr11: 2659043–2660499 | chlo(9) mito(2) nucl(1) cyto(1) | Chloroplast(2.0) | Nuclear |
| LOC_Os11g05780.1 | OsOFP29 | Chr11: 2666529–2665214 | nucl(7) chlo(6) | Nuclear(4.0) | Nuclear |
| LOC_Os12g06150.1 | OsOFP30 | Chr12: 2898691–2900071 | nucl(5) mito(5) chlo(3) | Nuclear(2.4) | Nuclear |
| LOC_Os12g06160.1 | OsOFP31 | Chr12: 2912589–2911273 | nucl(9) chlo(3) mito(2) | Nuclear(3.3) | Nuclear |
(a) Rice OFPs were designated as OsOFP01–OsOFP31.
(b) Position of gene in the genomic fragment.
(c) The prediction from the subcellular localization program WoLF PSORT. The numbers in parenthesis indicate prior possible protein localization sites of the OsOFPs. Abbreviations of protein localization sites in the dataset are as follows: nucl: nucleus; chlo: chloroplast; cyto: cytosol; cysk: cytoskeleton; chlo_mito: chloroplast and mitochondria; plas: plastids; cyto_nucl: cytosol and nucleus; mito: mitochondria; pero: peroxisomes; and nucl_ plas: nucleus and plastids.
(d) The subcellular localization prediction program CELLO was used to determine the possible localization sites of the OsOFPs.
(e) Subcellular localization of the OsOFPs determined via the experiment in this study.
Fig 1. Genome distribution of the OsOFPs.
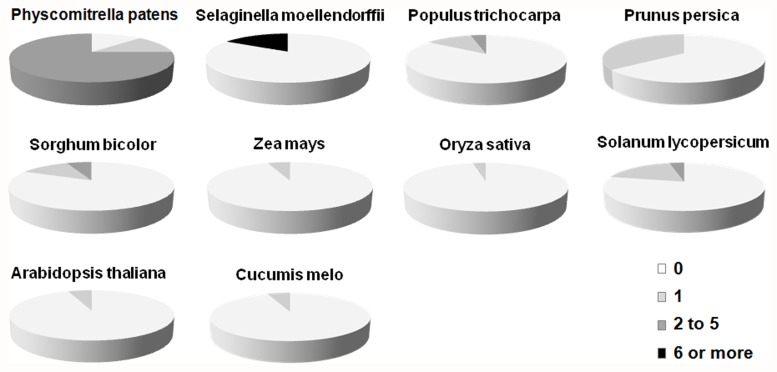
OFPs are found in many species, from the moss to angiosperm, including Physcomitrella patens, Selaginella moellendorffii, Populus trichocarpa, Prunus persica, Sorghum bicolor, Zea mays, Oryza sativa, Solanum lycopersicum, Arabidopsis thaliana and Cucumis melo. The sequences of OFP genes from Physcomitrella patens, Selaginella moellendorffii, Sorghum bicolor, Zea mays, Oryza sativa and Arabidopsis thaliana were downloaded from PlnTFDB (http://plntfdb.bio.uni-potsdam.de/v3.0/). The OFP sequences of Populus trichocarpa and Prunus persica were blasted from Phytozome v10.0.2 (http://phytozome.jgi.doe.gov/pz/portal.html). Solanum lycopersicum OFP sequences were obtained from the tomato WGS Chromosomes (SL2.40) (SGN http://solgenomics.net) and from the reference published by Huang et al. (2013). The sequences of the Cucumis melo OFP were identified and downloaded through the MELONOMICS website (http://melonomics.net). Gene model and locus information (except the same and incomplete gene sequences) for these OFP family genes were listed in S1a-f Table and S2a-d Table. Using the online tool Gene Structure Display Server [22], we found that OFPs in angiosperm plants are inclined to be intron poor. Fig. 2 displays the proportions of OFPs in each species with no introns, 1 intron, 2–5 introns, and ≥6 introns. The final results indicate that 89, 25, 15, 36, 17, 6, 3, 26, 6 and 6% of the total number of OFPs in P. patens, S. moellendorffii, Populus, P. persica, S. bicolor, Z. mays, O. sativa, S. lycopersicum, A. thaliana and C. melo contain introns, respectively. In the OsOFP family, all 31 members (except OsOFP14) do not contain introns. Seven members in SlOFP family are single intron-containing genes. SlOFP06 contains 2 introns [15]. No more than 2 introns appear in the intron-containing OFPs in Populus, P. persica, Z. mays, O. sativa, S. lycopersicum, A. thaliana and C. melo, even though the proportions of the OFPs with introns in P. persica and S. lycopersicum are higher than those in S. moellendorffii. In P. patens and S. moellendorffii, more introns were found in the intron-containing OFPs.
Fig 2. Relative proportions of intron-containing OFPs in Physcomitrella patens, Selaginella moellendorffii, Populus trichocarpa, Prunus persica, Sorghum bicolor, Zea mays, Oryza sativa, Solanum lycopersicum, Arabidopsis thaliana and Cucumis melo.
Proportions are colored as: no introns (white), 1 intron (light gray), 2–5 introns (dark gray), and ≥6 (black).
Multiple Sequence Alignment and Phylogenetic Analysis of the OFP Proteins
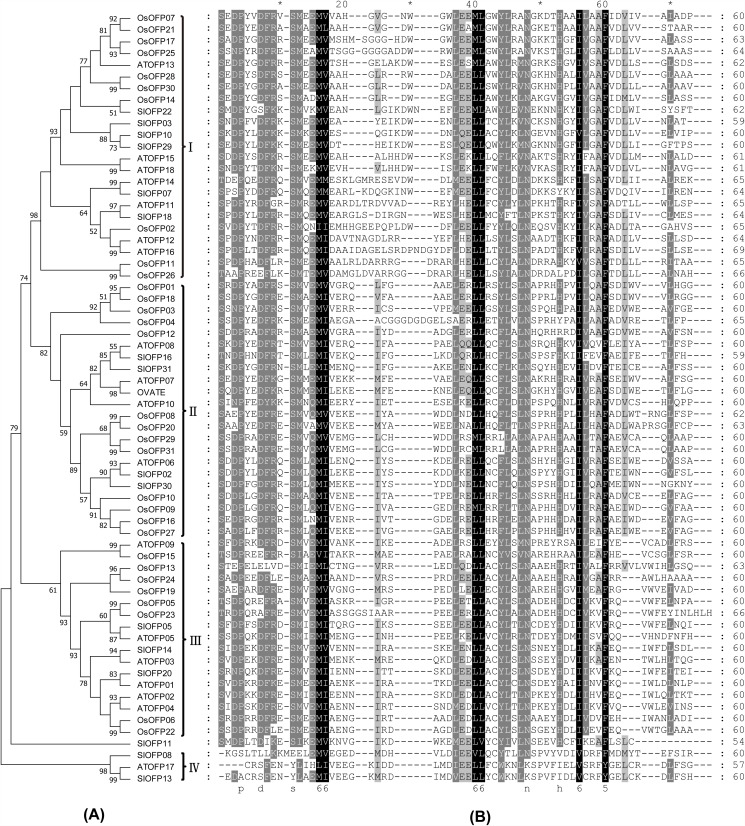
To investigate the phylogenetic relationship among rice, Arabidopsis and tomato OFP genes, the phylogenetic tree was constructed based on their OVATE domain sequences (Fig. 3A). The aligned OVATE domain sequences included 31 OVATE domains from rice, 18 OVATE domains from Arabidopsis and 17 OVATE domains from tomato (Fig. 3B). As shown in Fig. 3, excluding SlOFP11, all the other OFPs was further divided into 4 major subfamilies, designated I to IV. Except the smallest subfamily IV, each subfamily contains OFP members from rice, Arabidopsis and tomato. However, most of the OsOFP members were clustered in species-specific distinct clades. Only two pairs of orthologs, OsOFP15 and AtOFP09 between rice and Arabidopsis, OsOFP14 and SlOFP22 between rice and tomato could be figured out, comparing with 9 pairs of orthologs presented between Arabidopsis and tomato. These results suggest that the main characteristics of plant OFP proteins in rice, Arabidopsis and tomato were formed before divergence between monocots and dicots, and then evolved separately in a species-specific manner.
Fig 3. Multiple sequence alignment and phylogenetic tree of the OVATE domains of OFP proteins from rice, Arabidopsis and tomato.
(A) The phylogenetic trees based on the multiple sequence alignments of the OVATE domains of OFP proteins. Bootstrap values from 1000 replicates are indicated at each node. The proteins on the tree can be divided into four distinct subfamilies (I–IV). Bootstrap values under 50% was not reported. (B) Multiple sequence alignments of the OVATE domains of rice, Arabidopsis and tomato are shown. The black region represents identical amino acids, and the grey region represents similar amino acids.
Expression Patterns of OsOFPs
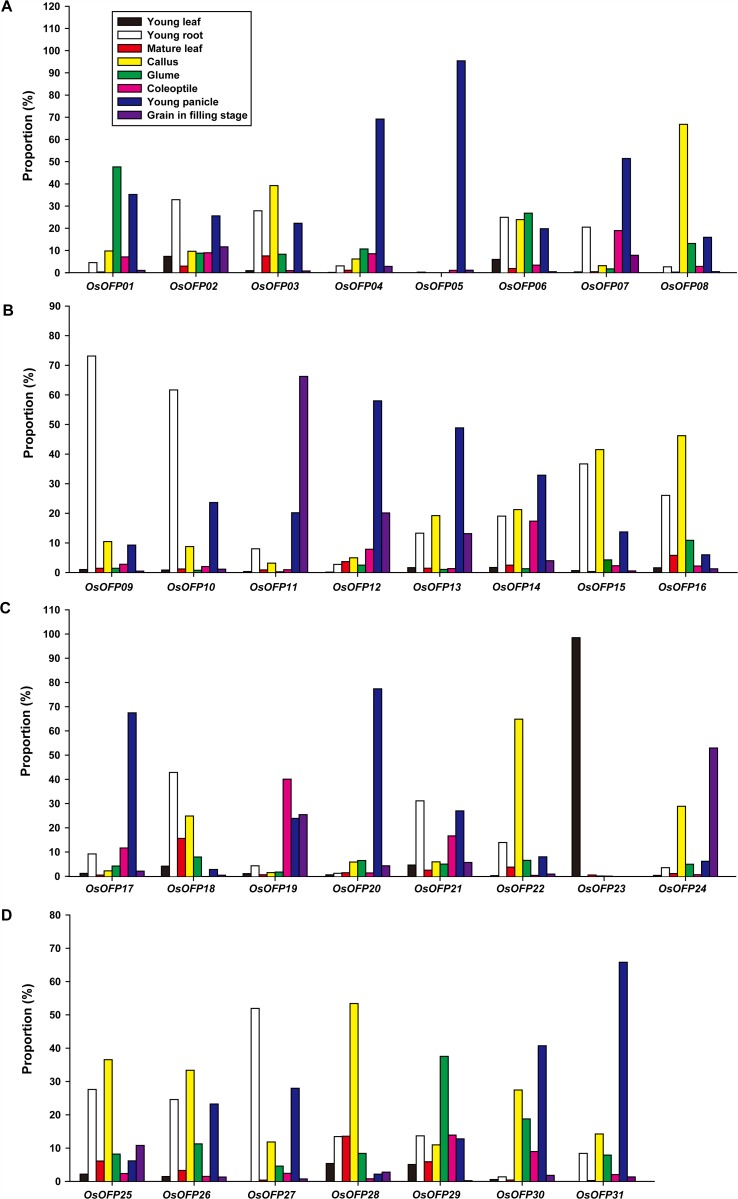
Gene expression pattern provides important clues for investigating gene function. The tissue-specific expression profiles of the 31 OsOFPs were performed by quantitative real-time RT-PCR (qRT-PCR). Total RNA was isolated from calli, coleoptiles, young leaves, young roots, mature leaves, glumes, young panicles and grains in filling stage (Fig. 4). The results showed that OsOFPs exhibited different expression patterns. The transcript levels of OsOFP01, OsOFP06 and OsOFP29 were higher in the glumes than in other organs (Fig. 4). OsOFP02, OsOFP09, OsOFP10, OsOFP18, OsOFP21 and OsOFP27 were prominently higher in the roots than in other organs (Fig. 4). OsOFP03, OsOFP08, OsOFP15, OsOFP16, OsOFP22, OsOFP25, OsOFP26 and OsOFP28 were obviously higher in the calli than in other organs (Fig. 4). The expression patterns of OsOFP04, OsOFP05, OsOFP07, OsOFP12, OsOFP13, OsOFP14, OsOFP17, OsOFP20, OsOFP30 and OsOFP31 were all very similar, and they were preferentially expressed in the young panicles tissue (Fig. 4). OsOFP11 and OsOFP24 were more highly expressed in the grains of filling stage but were only minimally expressed in other organs (Fig. 4B, 4C). The levels of OsOFP19 were notably expressed in the coleoptiles (Fig. 4C). OsOFP23 was abundantly expressed in the young leaves but was minimally expressed in other tissues (Fig. 4C). Interestingly, ~50% of the total number of OsOFPs was more highly expressed during the seed developmental stage than that during other stages of development. This is similar to the expression profile of the tomato OVATE which is expressed in reproductive organs in early stages of flower and fruit development.
Fig 4. Expression profiles of the OsOFPs in various rice tissues at different developmental stages.
The tissue-specific expression profiles were performed by qRT-PCR. The expression profiles of genes from OsOFP01 to OsOFP08 (A), The expression profiles of genes from OsOFP09 to OsOFP16 (B), The expression profiles of genes from OsOFP17 to OsOFP24 (C), The expression profiles of genes from OsOFP25 to OsOFP31 (D).
The seed developmental cis-elements that have been well characterized were investigated in 2-kb 5′-upstream regions of the OsOFPs [23]. The GCN4 motif is found in the promoter region of the glutelin gene, which encodes a major seed storage protein and is involved in endosperm-specific gene expression in rice [24–26]. The Skn-1 motif is a consensus sequence in a number of seed-specific promoters and is associated with endosperm-specific gene expression [24]. The promoter region of each OsOFP contains at least one of the 2 seed development cis-elements (S1 Fig.).
Hormone-Induced Expression Profiles of OsOFPs
AtGA20OX1 is thought to be a direct target gene of AtOFP1 and exogenous gibberellic acid can partially rescue the reduced length in rapidly elongating aerial organs of plants overexpressing AtOFP1, suggesting its important role in suppressing GA biosynthesis and maintaining the balance of this hormone in plants [11]. The expression profiles of OsOFPs responding to exogenous GA3 were investigated in our study. Transcript levels of OsOFPs did not change significantly in response to GA treatment (S2 Fig.), indicating that OsOFPs may not participate in the response to GA in rice.
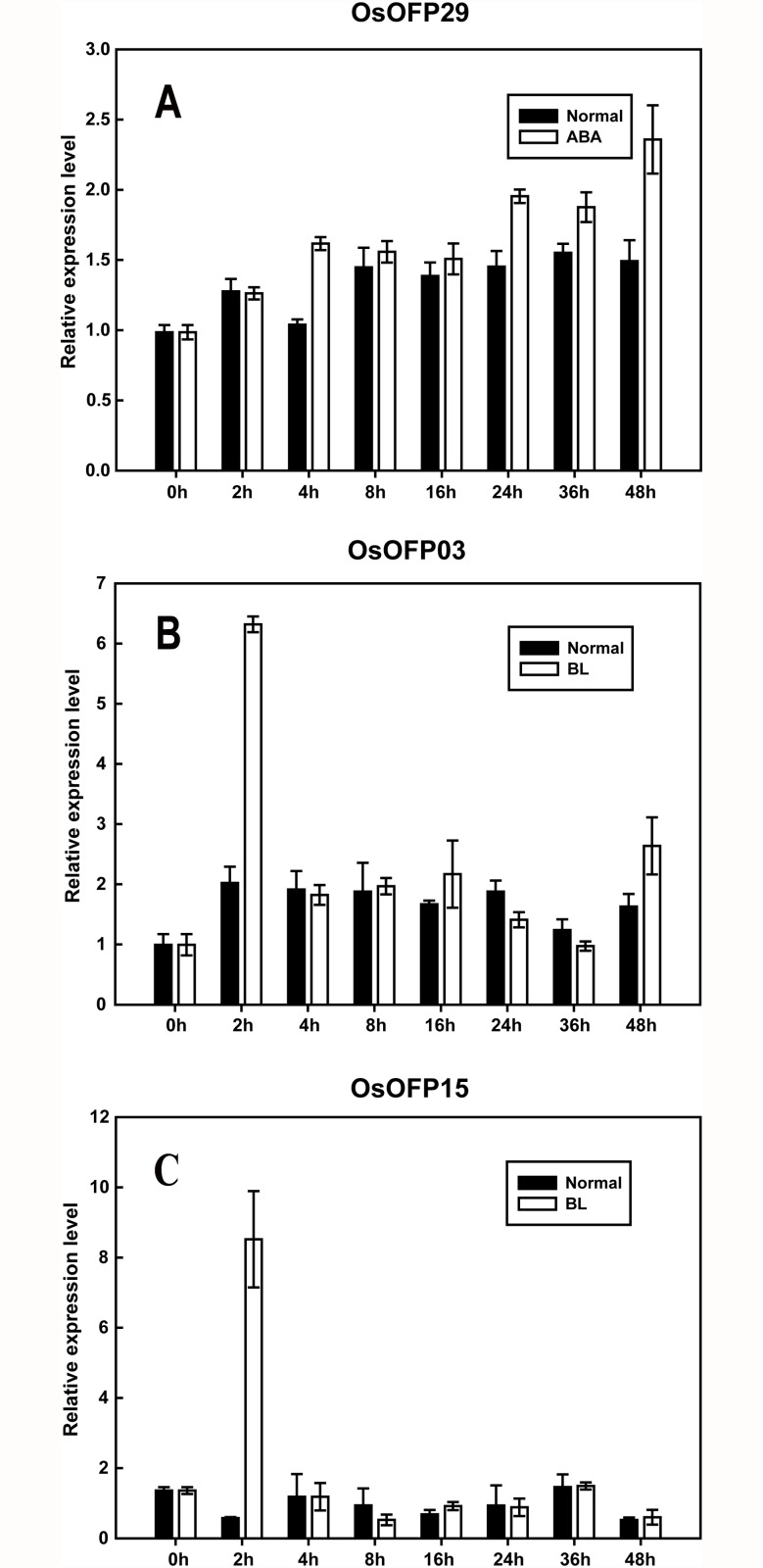
In order to further characterize the relevance between OsOFP family and hormone signaling, the expression patterns of the OsOFP family in response to abscisic acid (ABA) and brassinosteroid (BR), the 2 classes of hormones whose essential function in plant development and stresses resistance were widely appreciated [27–33], were also assessed using the quantitative real-time RT-PCR (qRT-PCR) in the current study (Fig. 5; S3 Fig., S4 Fig.). The results showed that only OsOFP29 was upregulated after 24 h of the ABA treatment (Fig. 5A), whereas the transcript levels of other OsOFPs did not change significantly (S3 Fig.). Interestingly, OsOFP03 and OsOFP15 were upregulated immediately under the brassinolide (BL) treatment. The transcript level of OsOFP03 increased quickly and reached its maximum at 2 h (Fig. 5B). OsOFP15 exhibited a response similar to that of OsOFP03 under the BL treatment (Fig. 5C). The quick responses of the OsOFPs to the BL treatment indicate that they may act as important upstream components in BL signal transduction in rice.
Fig 5. Expression profiles of OsOFPs under phytohormone treatments.
(A) Expression profile of OsOFP29 responding to ABA (50 μM; 0, 2, 4, 8, 16, 24, 36, and 48 h). (B, C) Expression profiles of OsOFP03 and OsOFP15 responding to BR, respectively (1 μM; 0, 2, 4, 8, 16, 24, 36, and 48 h). The expression profiles of OsOFPs responding to ABA and BL were determined by qRT-PCR. Actin was used as an internal control. Error bars indicate the SD based on 3 biological replicates.
Localization of OsOFP Proteins
A previous study has shown that ovate proteins are transcriptional repressors in Arabidopsis [11], but little is known about the molecular function and subcellular localization of OVATE proteins in rice. Usually, transcription regulators are localized in the nucleus to turn a group of target genes on or off. Most of the known AtOFPs are active in the nucleus [9–12]. The predicted target signal peptides for OVATE proteins in rice were checked using the WoLF PSORT [34] and CELLO [35] prediction programs. According to both programs, most proteins in this family were assigned to either the nucleus or chloroplast (Table 1c, d).
Using the WoLF PSORT program, the OsOFPs were predicted to locate in the nucleus or chloroplast (Table 1c). CELLO indicated that most OsOFPs scored relatively higher in the nucleus than in the chloroplast (Table 1d). In total, the differences in the location of OsOFP10, OsOFP12, OsOFP15, OsOFP24, and OsOFP25 were notable between the two prediction programs (Table 1c, d). Although not conclusive, the presented bioinformatics data could serve as a useful reference for further studies on OVATE protein functions in plants.
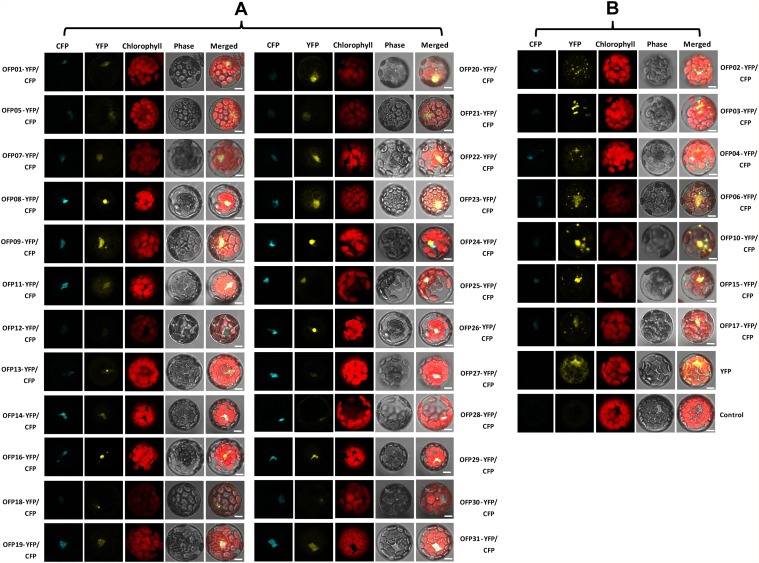
To confirm the subcellular localization of the OsOFP family, the co-localization of OsOFP yellow fluorescent proteins (YFP) and the nuclear marker AHL22-cyan fluorescent protein [36] were analyzed. The corresponding full-length coding sequences were systematically cloned into a pDONR/Zeo gateway entry vector. This recombined cloning system was subsequently used to further subclone OsOFP coding sequences into the YFP fused destination vectors. As shown in Fig. 6A, 24 OsOFPs were shown to target in the nucleus, and the others were shown to target in the nucleus with granular spots (Fig. 6B). The results were, in general, consistent with the bioinformatics prediction of the subcellular localization of OsOFPs (Table 1c, d, e).
Fig 6. Subcellular localization of the OsOFPs.
Protoplast transient expression using YFP-OsOFP fusion constructs were used to determine the subcellular localization. AHL22 fused with CFP was used as a nuclear marker. The 35S: YFP-OsOFP and 35S: CFP-AHL22 constructs were transformed into an Arabidopsis protoplast cell. Fluorescence images of CFP, YFP, and chlorophyll autofluorescence (Chl) were captured with confocal laser scanning microscopy and are shown in cyan, yellow, and red, respectively (scale bars, 10 μm).
Discussion
OVATE family proteins were present in 13 sequenced plant genomes that represent the major evolutionary lineages of land plants [37]. During the course of plant evolution, most gene families expanded mainly through large scale segmental duplication or tandem duplication to maintain the large number of family members [38], manner of gene duplication had been described in many large transcription factor families such as the bHLH family [39] and the bZIP family [40] in rice. In this study, the relatively high density chromosomal distribution pattern of the OsOFPs indicates that large-scale segmental duplication or tandem duplication may have been involved in the expansion of the OsOFP family. A recent study had verified that segmental duplication and tandem duplication were two major mechanisms for gene expansion in the OsOFP family. For instance, 24 (72.7%) were located within segmental duplication blocks and eight (24.2%) were tandemly duplicated in the rice OsOFP family [37].
Previous studies have shown that OFPs function as transcriptional repressors and regulate multiple aspects of plant growth and development [9,11–13,41]. The OVATE gene determines the conversion of the tomato fruit from round- to pear-shaped, and overexpression of this gene results in smaller-sized floral organs and leaflets [13]. The tomato OVATE transcripts can be detected in flowers 10 days before anthesis and until 8 days after anthesis in developing fruit [13]. Recently, Huang et al. (2013) reported that the OVATE expression increased in fruit at 33 days post of anthesis, suggesting its important role at the fruit ripening stage [15]. To some extent, there is coincidence between the spatially restricted expression of OVATE and the pear-shaped fruit shape phenotype [13]. Round seeds or blund-end siliques were also observed in AtOFP family members overexpressed A. thaliana respectively [11,12]. Furthermore, an Ovate-like gene in pepper was reported to be involved in determining fruit shape [41]. In this study, tissue-specific expression pattern analysis showed that OsOFPs were expressed in all developmental phases of the rice plant, and nearly half of this gene family was more highly expressed during the seed developmental stage (Fig. 4). Moreover, each OsOFP contained the GCN4 or Skn-1 motifs, 2 of the seed developmental cis-elements that have been well characterized in rice, in the promoter region (S1 Fig.). Results from the functional analysis of the OFPs in the above plant species and from the current study hint at key roles of OsOFPs in controlling important traits related to seed development in rice.
TFs function as key regulators during different stages of seed development [42–44]. A pair of basic helix-loop-helix (bHLH) proteins, PGL1 and APG, are involved in determining grain weight and length antagonistically, in which APG is a negative regulator whose function is inhibited by PGL1 [42]. The homeobox family of genes was found to play a pivotal role throughout the process of seed development. For instance, homeobox gene OSH1 is critical for the regionalization of cell identity during early embryogenesis [43]. Studies in Arabidopsis showed that AtOFPs interact with TALE homeodomain proteins to regulate plant development [9]. Results of most OsOFPs with stronger expression profiles in the mature seed or embryo suggest that OsOFPs may act as important transcriptional regulators during seed development in rice. Further studies are needed to identify the interactions of OsOFPs in order to discover new seed development regulating pathways that include OsOFP at their core.
Many transcriptional factor families were characterized to play important roles in different developmental processes regulated by phytohormones [45–47]. So far, only AtOFP1 was reported to target AtGA20ox1, a key enzyme gene in gibberellin biosynthesis, and to suppress its expression in plants overexpressing AtOFP1 [9,11]. No information about the relevance between plant OFP families and the other phytohormones was provided. In this study, the expression profiles of OsOFPs responding to exogenous GA3, ABA and BL were investigated (Fig. 5; S2 Fig., S3 Fig., S4 Fig.). We found that the transcript level of OsOFP15 increased rapidly in response to BL treatment, meanwhile it was specifically expressed in the mature seed. Brassinolide is the most bioactive form of the growth-promoting plant steroid hormones, termed brassinosteroids. Brassinosteroids are essential for a wide range of developmental and physiological processes [28–31]. BR-deficient and BR-insensitive mutants display dwarfism, erect leaves, and altered panicle length in combination with a reduced seed setting [48–50]. XIAO, a putative LRR receptor-like kinase, is related to BR biosynthesis; and the loss-of-function mutant, xiao, leads to reduced plant size and seed setting due to a decrease in the rate of cell division [49]. Overexpression of the BR biosynthesis related gene Zm-CYP-1 increased grain yield by controlling the seed filling in rice [31]. The spatially restricted and BR-induced expression pattern of OsOFP15 suggest that it may participate in regulating the BR response in a rice seed setting.
Subcellular proteomics are helpful in exploring the biological functions of proteins. In the current study, the subcellular localization analysis clearly revealed that the YFP-OsOFP fusion proteins predominantly localized in the nucleus. With regard to OsOFP05, 28, and 30, only very weak yellow fluorescent signals were observed in the nucleus (Fig. 6), while strong yellow fluorescent signals were clearly observed in the nucleus for other OsOFPs (Fig. 6). The reason for these observations may be that OsOFP05, 28, and 30 were not stable when compared to the other OsOFPs in the nucleus of Arabidopsis protoplast cells. We suspected that the N-terminus-fused YFP may be the reason for the instability of the OsOFPs with weak fluorescent signals. As exhibited in Fig. 6, OsOFP02, 03, 04, 05, 06, 10, 15, and 17 prominently displayed nuclear concentrations with multiple big and small granular spots floating along the nucleus. Some transcription factor proteins were nuclear localization and also present in the cytoplasm [51]. For instance, the KNOX proteins, encoding transcription factors involved in shoot apical meristem development and maintenance, had similar subcellular distribution patterns to these OsOFP proteins [52]. BZR1 and BZR2/BES1, the two key transcription factors of the BR signaling pathway, were found in both the nucleus and cytoplasm [53,54]. BR signal increased the nuclear accumulation of BZR1 and BZR2/BES1 [53–55]. The subcellular distrbution patterns of these OsOFPs suggested that they may function as potential nucleocytoplasmic shuttling proteins and can be regulated by various signals such as hormone and light [56].
The localization prediction programs for OsOFPs on the Website do not support the actual subcellular localization for some proteins. For example, OsOFP07, OsOFP10, OsOFP12, OsOFP14, OsOFP15, OsOFP24, OsOFP25, OsOFP26 and OsOFP28 did not localize in the nucleus according to the predictions of the WoLF PSORT or CELLO programs (Table 1c, d, e). However, OsOFPs were shown to target to the nucleus in the current study. This discrepancy may be explained by the fact that the nuclear localization signals identified and deposited in the WoLF PSORT and CELLO databases are not complete. It is also possible that novel nuclear localization signals, that have not yet been identified, could exist in the OsOFP sequences.
In conclusion, this study provided the genomic framework, intron number, and phylogenetic analysis of the 31 OsOFPs. In addition, we studied the expression profile of the whole family during the entire life cycle of the rice plant and under GA, ABA, and BL treatments. Furthermore, the subcellular localization of OsOFPs was also investigated. Our data provide insight into the roles of OsOFPs during the seed developmental stages and in response to BL. This study provides a useful reference for conducting more detailed functional analyses of these OFPs in rice and will be helpful in the selection of appropriate candidate genes for further studies.
Materials and Methods
Phylogenetic analysis and sequence alignment
Multiple sequence alignment were performed using the ClustalX version 1.83 [57] and were manually corrected. The obtained sequence alignments were used as input to construct phylogenetic tree with the neighbor—joining algorithm in MEGA 6.06 [58]. Bootstrap analysis was performed using 1000 replicates.
Plant Material and Growth Conditions
Wild-type Nipponbare (O. sativa L. ssp. japonica) was used in the current study. The seeds were imbibed for 48 h at 30°C and cultivated in the test field or growth chamber under diurnal conditions (day phase: 10 h/30°C; night phase: 14 h/24°C). For tissue-specific expression analysis, materials were collected at different developmental stages. Young leaves, young roots, calli and coleoptiles were collected from the growth chamber; and mature leaves, glumes, young panicles and grains in filling stage were collected from the test field of Jilin University. All plant materials were collected in liquid nitrogen and stored at -80°C for RNA extraction.
Arabidopsis seeds of the Columbia ecotype (Col-0) were used in our study. Seeds were surface-sterilized, vernalized, and sown and grown on 1/2 MS media until the leaves reached the four-leaf stage. Then, the seedlings were transplanted to a growth chamber containing peat moss mixed with vermiculite (1/1, v/v), under diurnal conditions (day phase: 8 h/22°C; night phase: 16 h/18°C). The relative humidity was maintained at 60–70%. Leaves were collected from 3- to 5-week-old seedlings for protoplast isolation and transfection assays.
RNA Isolation and cDNA Synthesis
Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The integrity of the RNA was checked electrophoretically, and complementary DNA (cDNA) was synthesized using 4 μg of RNA with the Oligo d(T)18 primers and M-MLV reverse transcriptase (TaKaRa Bio, Tokyo, Japan), according to the manufacturer’s protocol.
Real-time Quantitative PCR Analysis
Relative transcript level expression profiles of OsOFPs were evaluated by real-time quantitative PCR (RT-qPCR) on Mx3005P (Stratagene, La Jolla, CA, USA) using the SYBR Premix ExTaq polymerase (TaKaRa, Bio Inc.). Each reaction contained 12.5 μl of the 2× SYBR Premix ExTaq, 50–100 ng of the cDNA template, 0.5 μl of 10 mM of each primer, and 10.5 μl of double-distilled H2O for a final volume of 25 μl. The PCR reaction parameters were 95°C for 30 s (1 cycle), 95°C for 5 s, and 60°C for 20 s (40 cycles), which was followed by a melting curve analysis at 95°C for 60 s, 55°C for 30 s, and 95°C for 30 s. The relative fold differences were calculated based on the comparative 2-ΔΔ Ct method. Approximately 100–200-bp PCR products, unique to each OsOFP, were amplified; the housekeeping gene ACTIN (X15863.1) was used to normalize the transcript level of each OsOFP in the samples. Expression profile analysis of OsOFP genes in each type tissue was displayed using relative percentage [15] in this study. The specific primer pairs used were listed in S3 Table.
Expression Profiles of OsOFPs in Response to ABA and BL
The rice seeds of the wild-type Nipponbare were sterilized with 3% NaOCl for 30 min, washed extensively with distilled water, and placed in petri dishes with wetted filter papers at 30°C in the dark. After 3 d of incubation, germinated seeds were sown in a 96-well plate, with the bottom removed, and the plate was fixed in a 1/2 MS culture solution at 26°C under continuous light. After 14 d, the seedlings were subjected to ABA and BL treatments. For the ABA or BL treatments, seedlings were transferred to a culture solution with 50 μM of ABA, 1 μM of BL, or without ABA and BL (i.e., the control). Subsequently, young leaves were collected at 0, 2, 4, 8, 16, 24, 36, and 48 h after treatment. Then the expression profiles of OsOFPs responding to ABA and BL were evaluated by real-time quantitative PCR (RT-qPCR).
Construction of the Subcellullar Localization Plasmid
The expression vectors of subcellullar localization were constructed as follows: the coding sequences of the OsOFPs were cloned into an entry vector (pDONR/Zeo; Invitrogen) by using the BP-clonase, according to the manufacturer’s instructions, and subsequently cloned into the destination vector (pENSG-YFP) of the N-terminus fused the YFP reporter gene by an LR reaction (Gateway recombination, Invitrogen). All of the fusion constructs were driven by the 35S promoter. The recombinant plasmids vector pENSG-CFP-AHL22, carrying a nuclear marker gene AHL22 [36], was co-transformed with each target gene. The specific primer pairs used were listed in S4 Table.
Protoplast Isolation, Transformation, and Confocal Microscopy
Leaves of 3- to 5-week-old plants were used in the subcellular localization analysis. The leaves were cut into 0.5–1-mm strips with fresh razor blades (without wounding) and incubated in an enzyme solution that included 1% cellulase R10, 0.25% macerozyme R10, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, and 20 mM MES for 3–4 h, with 40–50 rpm slow shaking. After incubation, the protoplast suspension was filtered through a metal sieve and centrifuged at 100 × g for 5 min. The pelleted protoplasts were suspended in 5 mL of a W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES/KOH; pH 5.7) and centrifuged for 5 min at 100 × g. The protoplasts were transferred to a tube containing 5 mL of the W5 solution. The protoplasts were pelleted again by centrifugation at 100 × g for 5 min and resuspended in 5 mL of the W5 solution. The protoplasts were incubated on ice for 30 min. The protoplasts were again resuspended in 5 mL of MMg buffer (400 mM mannitol, 15 mM MgCl2, and 4 mM MES/KOH; pH 5.7).
For cotransformation, 10 μg of each plasmid DNA was added to 100 μL of the protoplast suspension. An equal volume of 40% (w/v) PEG3350, freshly prepared with 0.1 M CaCl2 and 0.8 M mannitol solution, was added. Then, the mixture was incubated at room temperature for 30 min. After incubation, the mixture was diluted with 500 μL of the W5 solution. The solution was fully mixed, and the protoplasts were pelleted by centrifugation at 100 × g for 5 min. Then, the protoplasts were washed twice using the W5 solution, resuspended gently in 1 mL of the W5 solution, and incubated in 12-well plates at room temperature for 18–20 h in darkness.
All microscopic observations were performed using a confocal laser scanning microscope. The fluorescence of the YFP was visualized with excitation and emission wavelengths of 488 and 505–530 nm, respectively. For CFP, the excitation and emission wavelengths were 458 and 465–530 nm, respectively. Chloroplast autofluorescence was visualized in a detection channel with excitation and emission wavelengths of 488 and 650–710 nm, respectively. Image processing was performed with ImageJ (http://rsb.info.nih.gov/ij/).
Supporting Information
Two figures were used to represent different seed developmental cis-elements; the lines represent promoter sequences. The elements located in the forward and reverse strands are indicated as ‘+’ and ‘-’, respectively.
(TIF)
(TIF)
(TIF)
(TIF)
(a, b, c, d, e, f) Gene model of OFP family genes in physcomitrella patens, selaginella, sorghum, arabidopsis, maize and rice, respectively.
(DOC)
(a, b, c, d) Gene locus of OFP family genes in populus trichocarpa, prunus persica, solanum lycopersicum and cucumis melo, respectively.
(DOC)
(DOC)
(DOC)
Acknowledgments
The authors would like to thank Prof. Chentao Lin for the advice on the experiments.
Funding Statement
This work was supported by the National Major Project, Crop Yield Improvement Using Synthetic Transcription Factor (2010ZX08010-002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Scott MP. Development: the natural history of genes. Cell. 2000;100: 27–40. [DOI] [PubMed] [Google Scholar]
- 2. Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290: 2105–2110. [DOI] [PubMed] [Google Scholar]
- 3. Guo A, He K, Liu D, Bai S, Gu X, et al. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21: 2568–2569. [DOI] [PubMed] [Google Scholar]
- 4. Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14: 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hay A, Tsiantis M. KNOX genes: versatile regulators of plant development and diversity. Development. 2010;137: 3153–3165. 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- 6. Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, et al. Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell. 2004;16: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vollbrecht E, Reiser L, Hake S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development. 2000;127: 3161–3172. [DOI] [PubMed] [Google Scholar]
- 8. Smaczniak C, Immink RG, Muino JM, Blanvillain R, Busscher M, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci U S A. 2012;109: 1560–1565. 10.1073/pnas.1112871109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci U S A. 2005;102: 4908–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li E, Wang S, Liu Y, Chen JG, Douglas CJ. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 2011;67: 328–341. 10.1111/j.1365-313X.2011.04595.x [DOI] [PubMed] [Google Scholar]
- 11. Wang S, Chang Y, Guo J, Chen JG. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007;50: 858–872. [DOI] [PubMed] [Google Scholar]
- 12. Wang S, Chang Y, Guo J, Zeng Q, Ellis BE, et al. Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One. 2011;6: e23896 10.1371/journal.pone.0023896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci U S A. 2002;99: 13302–13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pagnussat GC, Yu HJ, Sundaresan V. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell. 2007;19: 3578–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genomics. 2013;288: 111–129. 10.1007/s00438-013-0733-0 [DOI] [PubMed] [Google Scholar]
- 16. Century K, Reuber TL, Ratcliffe OJ. Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008;147: 20–29. 10.1104/pp.108.117887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai M, Hu Y, Zhao Y, Liu H, Zhou DX. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007;144: 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu QH, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM. The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol. 2004;135: 1514–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, et al. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to Rice dwarf virus. Plant J. 2009;57: 615–625. 10.1111/j.1365-313X.2008.03712.x [DOI] [PubMed] [Google Scholar]
- 20. Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, et al. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38: D822–827. 10.1093/nar/gkp805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagamura Y, Antonio BA, Sato Y, Miyao A, Namiki N, et al. Rice TOGO Browser: A platform to retrieve integrated information on rice functional and applied genomics. Plant Cell Physiol. 2011;52: 230–237. 10.1093/pcp/pcq197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29: 1023–1026. [PubMed] [Google Scholar]
- 23. Rombauts S, Dehais P, Van Montagu M, Rouze P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27: 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Washida H, Wu CY, Suzuki A, Yamanouchi U, Akihama T, et al. Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol Biol. 1999;40: 1–12. [DOI] [PubMed] [Google Scholar]
- 25. Muller M, Knudsen S. The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J. 1993;4: 343–355. [DOI] [PubMed] [Google Scholar]
- 26. Yoshihara T, Washida H, Takaiwa F. Assessment of common regulatory regions required for the endosperm-specific expression of rice storage protein glutelin genes by hybrid promoters. Plant Science. 1996;121: 63–73. [Google Scholar]
- 27. Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14 Suppl: S15–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mandava NB. Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol. 1988;39: 23–52. [Google Scholar]
- 29. Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10: 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61: 681–704. 10.1146/annurev.arplant.043008.092057 [DOI] [PubMed] [Google Scholar]
- 31. Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20: 2130–2145. 10.1105/tpc.107.055087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53: 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hetherington AM. Guard cell signaling. Cell. 2001;107: 711–714. [DOI] [PubMed] [Google Scholar]
- 34. Horton P, Park K-J, Obayashi T, Fujita N, Harada H, et al. WoLF PSORT: protein localization predictor. Nucleic acids Res. 2007;35: W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64: 643–651. [DOI] [PubMed] [Google Scholar]
- 36. Xiao C, Chen F, Yu X, Lin C, Fu YF. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol. 2009;71: 39–50. 10.1007/s11103-009-9507-9 [DOI] [PubMed] [Google Scholar]
- 37. Liu D, Sun W, Yuan Y, Zhang N, Hayward A, et al. Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann Bot. 2014;113: 1219–1233. 10.1093/aob/mcu061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X, Duan X, Jiang H, Sun Y, Tang Y, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146: 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsaballa A, Pasentsis K, Darzentas N, Tsaftaris AS. Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC plant Biol. 2011;11: 46 10.1186/1471-2229-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heang D, Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PloS one. 2012;7: e31325 10.1371/journal.pone.0031325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sato Y, Hong SK, Tagiri A, Kitano H, Yamamoto N, et al. A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc Natl Acad Sci U S A. 1996;93: 8117–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xue LJ, Zhang JJ, Xue HW. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PLoS One. 2012;7: e31081 10.1371/journal.pone.0031081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yaish MW, El-Kereamy A, Zhu T, Beatty PH, Good AG, et al. The APETALA-2-like transcription factor OsAP2–39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010;6: e1001098 10.1371/journal.pgen.1001098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang C, Xu Y, Guo S, Zhu J, Huan Q, et al. Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 2012;8: e1002686 10.1371/journal.pgen.1002686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49: 387–400. [PubMed] [Google Scholar]
- 48. Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, et al. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17: 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang Y, Bao L, Jeong SY, Kim SK, Xu C, et al. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 2012;70: 398–408. 10.1111/j.1365-313X.2011.04877.x [DOI] [PubMed] [Google Scholar]
- 50. Nakagawa H, Tanaka A, Tanabata T, Ohtake M, Fujioka S, et al. SHORT GRAIN1 decreases organ elongation and brassinosteroid response in rice. Plant physiol. 2012;158: 1208–1219. 10.1104/pp.111.187567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH. High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 2005;41: 162–174. [DOI] [PubMed] [Google Scholar]
- 52. Kuijt SJ, Lamers GE, Rueb S, Scarpella E, Ouwerkerk PB, et al. Different subcellular localization and trafficking properties of KNOX class 1 homeodomain proteins from rice. Plant Mol Biol. 2004;55: 781–796. [DOI] [PubMed] [Google Scholar]
- 53. Ryu H, Kim K, Cho H, Park J, Choe S, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109: 181–191. [DOI] [PubMed] [Google Scholar]
- 55. Gampala SS, Kim TW, He JX, Tang W, Deng Z, et al. An essential role for 14–3–3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alinsug MV, Chen FF, Luo M, Tai R, Jiang L, et al. Subcellular localization of class II HDAs in Arabidopsis thaliana: nucleocytoplasmic shuttling of HDA15 is driven by light. PLoS One. 2012;7: e30846 10.1371/journal.pone.0030846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two figures were used to represent different seed developmental cis-elements; the lines represent promoter sequences. The elements located in the forward and reverse strands are indicated as ‘+’ and ‘-’, respectively.
(TIF)
(TIF)
(TIF)
(TIF)
(a, b, c, d, e, f) Gene model of OFP family genes in physcomitrella patens, selaginella, sorghum, arabidopsis, maize and rice, respectively.
(DOC)
(a, b, c, d) Gene locus of OFP family genes in populus trichocarpa, prunus persica, solanum lycopersicum and cucumis melo, respectively.
(DOC)
(DOC)
(DOC)