Abstract
Pseudomonas coronafaciens causes halo blight on oats and is a plant quarantine bacterium in many countries, including the Republic of Korea. Using of the certificated seed is important for control of the disease. Since effective detection method of P. coronafaciens is not available yet, PCR and TaqMan PCR assays for specific detection of P. coronafaciens were developed in this study. PCR primers were designed from the draft genome sequence of P. coronafaciens LMG 5060 which was obtained by the next-generation sequencing in this study. The PCR primer set Pc-12-F/Pc-12-R specifically amplified 498 bp from the 13 strains of P. coronafaciens isolated in the seven different countries (Canada, Japan, United Kingdom, Zimbabwe, Kenya, Germany, and New Zealand) and the nested primer set Pc-12-ne-F/Pc-12-ne-R specifically amplified 298 bp from those strains. The target-size PCR product was not amplified from the non-target bacteria with the PCR and nested primer sets. TaqMan PCR with Pc-12-ne-F/Pc-12-ne-R and a TaqMan probe, Pc-taqman, which were designed inside of the nested PCR amplicon, generated Ct values which in a dose-dependent manner to the amount of the target DNA and the Ct values of all the P. coronafaciens strains were above the threshold Ct value for positive detection. The TaqMan PCR generated positive Ct values from the seed extracts of the artificially inoculated oat seeds above 10 cfu/ml inoculation level. PCR and TaqMan PCR assays developed in this study will be useful tools to detect and identify the plant quarantine pathogen, P. coronafaciens.
Keywords: detection, halo blight, oat, PCR, TaqMan PCR
Halo blight, an important disease of the oats is caused by Pseudomonas coronafaciens (Elliott, 1920; Young et al., 1978) and occurs in relatively cool and moist climates leading to substantial economic losses (Marten et al., 1984). It produces light green, oval spots on the leaves of the plant with dark water-soaked centers (Marten et al., 1984; Harder and Haber, 1992; Wallwork, 1992). P. coronafaciens survives on plant debris, soil and seeds (Martens et al., 1984). One of the effective control measures of the disease is prevention of pathogen transfer to the other plants using of the certificated seed (Collins, 2010). Also, P. coronafaciens is a plant quarantine bacterium in many countries, including the republic of Korea. For the certificated seed program and plant quarantine, the specific and effective detection method must be available.
Various tests can be used to detect the seed-borne pathogens, such as plating on selective media, ELISA, seedling grow-out tests, PCR, real-time PCR and DNA microarrays (Walcott, 2003). Among these techniques, plating on selective media and seedling grow-out tests are laborious and time-consuming and the PCR assay is more sensitive and time-saving than ELISA (Cho et al., 2010). In particular, the TaqMan PCR assay, which detects microorganisms quantitatively with a TaqMan probe, has been shown to be successful in detection and identification of seed-borne pathogens (Bella et al., 2008; Schena et al., 2004; Finetti-Sialer and Ciancio, 2005). For P. coronafaciens, however, effective detection method from oat seeds is not available. Any PCR assay for the specific detection of P. coronafaciens has not been published yet.
In this study highly specific PCR and TaqMan PCR assays for detection of P. coronafaciens have been developed. For development of the P. coronafaciens - specific PCR assays, information for the unique nucleotide sequence of P. coronafaciens must be available. Since there are not many gene sequences of P. coronafaciens available in the GenBank, in the present study, next-generation sequencing analysis was used to obtain a whole draft genome sequence of P. coronafaciens. From the genome sequence, PCR primers were designed for the PCR and TaqMan PCR assays which were specific for the P. coronafaciens.
Materials and Methods
Bacterial strains and cultures
Strains of P. coronafaciens isolated in Canada, Japan, United Kingdom, Zimbabwe, Kenya, Germany and New Zealand were obtained from LMG (Belgian Coordinated Collection of Microorganisms, Laboratory of Microbiology, University of Gent, Belgium) and from KACC (Korean Agricultural Culture Collection, Rural Develpoment Administration, the Republic of Korea). For the non-target bacteria, strains related to P. coronafaciens in genus Pseudomonas or the representative strains of common plant pathogenic bacteria in genus Acidovorax, Pectobacterium, Ralstonia, Rhodococcus, Xanthomonas, and Clavibacter were collected from the various culture collections (Table 1). The bacteria were routinely grown in nutrient agar media containing 8 g of nutrient broth and 15 g of agar per liter.
Table 1.
List of Pseudomonas coronafaciens strains, the non-target bacterial strains of Pseudomonas spp. and other plant-pathogenic bacterial strains used in this study
| Strainab | Hostc | Originc | Yearc | |
|---|---|---|---|---|
| Acidovorax | ||||
| avenae subsp. avenae | NCPPB 1011 | Zea mays | USA | 1958 |
| Clavibacter | ||||
| michiganensis subsp. incidiosus | NCPPB 1020 | Medicago sativa | Canada | nk |
| michiganensis subsp. michiganensis | NCPPB 1064 | Lycopersicon esculentum | Italy | 1961 |
| michiganensis subsp. sepedonicus | NCPPB 2137 | Solanum tuberosum | Canada | nk |
| Pectobacterium | ||||
| carotovorum subsp. carotovorum | NCPPB 312 | Solanum tuberosum | Denmark | nk |
| Pseudomonas | ||||
| coronafacines | KACC 12133 | nk | nk | nk |
| KACC 13262 | nk | nk | nk | |
| LMG 2170 | Bromus sp. | Canada | 1962 | |
| LMG 2330 | nk | nk | 1966 | |
| LMG 5030 | Lolium multiflorum | Japan | 1967 | |
| LMG 5060b | Avena sativa | United Kingdom | 1958 | |
| LMG 5061 | Secale cereale | Canada | 1962 | |
| LMG 5081 | Avena sativa | Zimbabwe | 1971 | |
| LMG 5380 | Avena sativa | Kenya | 1970 | |
| LMG 5449 | Avena sativa | Germany | 1959 | |
| LMG 5452 | Avena sativa | New Zealand | 1969 | |
| LMG 5536 | Avena sativa | United Kingdom | 1965 | |
| LMG 13190 | Avena sativa | nk | nk | |
| savastanoi pv. glycinea | NCPPB 1134 | Glycine javanica | Zimbabwe | 1961 |
| savastanoi pv. phaseolicola | KACC 10575 | Phaseolus vulgaris | Poland | nk |
| savastanoi pv. savastanoi | NCPPB 639 | Olea europaea | Yugoslavia | nk |
| syringae pv. actinidiae | KACC 10582 | Actinidia chinensis | Rep. of Korea | 1999 |
| syringae pv. antirrhini | ICMP 4303 | Antirrhinum majus | United Kingdom | 1965 |
| syringae pv. aptata | DSM 50252 | Beta vulgaris | nk | nk |
| syringae pv. atrofaciens | LMG 5095 | Triticum aestivum | New Zealand | 1968 |
| syringae pv. berberidis | NCPPB 2724 | Berberis sp. | New Zealand | 1972 |
| syringae pv. ciccaronei | NCPPB 2355 | Ceratonia siliqua | nk | 1969 |
| syringae pv. delphinii | ICMP 529 | Delphinium sp. | New Zealand | 1957 |
| syringae pv. dysoxyli | ICMP 545 | Dysoxylum spectabile | New Zealand | 1949 |
| syringae pv. eriobotryae | NCPPB 2331 | Eriobotrya japonica | United Kingdom | 1970 |
| syringae pv. helianthi | NCPPB 1229 | Helianthus annuus | Zambia | 1962 |
| syringae pv. japonica | ICMP 6305 | Hordeum vulgare | Japan | 1951 |
| syringae pv. lachrymans | ATCC 11965 | Cucumis sativus | nk | nk |
| syringae pv. lapsa | ATCC 10859 | Triticum aestivum | nk | 1978 |
| syringae pv. maculicola | ICMP 3935 | Brassica oleracea | New Zealand | 1965 |
| syringae pv. mellea | ICMP 5711 | Nicotiana tabacum | Japan | 1968 |
| syringae pv. mori | ICMP 4331 | Morus alba | Hungary | 1958 |
| syringae pv. morsprunorum | ICMP 5795 | Prunus domestica | nk | nk |
| syringae pv. myricae | ICMP 7118 | Myrica rubra | Japan | 1978 |
| syringae pv. panici | NCPPB 1498 | Panicum sp. | nk | nk |
| syringae pv. papulans | ICMP 4040 | Malus domestica | USA | nk |
| syringae pv. passiflorae | NCPPB 1386 | Passiflora edulis | New Zealand | 1962 |
| syringae pv. persicae | NCPPB 2761 | Prunus persica | France | 1974 |
| syringae pv. pisi | ICMP 4433 | Pisum sativum | Canada | 1946 |
| syringae pv. ribicola | NCPPB 963 | Ribes aureum | nk | nk |
| syringae pv. sesame | NCPPB 1016 | Sesamum indicum | Yugoslavia | nk |
| syringae pv. syringae | NCPPB 388 | Oryza sativa | Hungary | nk |
| syringae pv. tabaci | ICMP 2835 | Nicotiana tabacum | Hungary | 1959 |
| syringae pv. tagetis | ICMP 4091 | Tagetes erecta | Zimbabwe | 1972 |
| syringae pv. tomato | NCPPB 2683 | Lycopersicon esculentum | New Zealand | 1972 |
| syringae pv. ulmi | NCPPB 632 | Ulmus sp. | Yugoslavia | 1958 |
| Ralstonia | ||||
| Solanacearum | NCPPB 339 | Solanum tuberosum | Israel | nk |
| Rhizobium | ||||
| radiobacter | DSM 30205 | Malus sp. | nk | 1972 |
| rhizogenes | ATCC 11325 | Malus domestica | nk | nk |
| rubi | NCPPB 1854 | Rubus ursinus var. loganobaccus | USA | 1942 |
| vitis | NCPPB 3554 | Vitis vinifera | Australia | 1977 |
| Rhodococcus | ||||
| fascians | LMG 3601 | Lillium speciosum cv. Rubrum | Belgium | nk |
| Xanthomonas | ||||
| campestris pv. campestris | KACC 10377 | Brassica oleracea var. capitata | Rep. of Korea | nk |
| campestris pv. vesicatoria | KACC 11157 | Capsicum annuum | Rep. of Korea | 1999 |
| oryzae pv. oryzae | KACC 10331 | Oryza sativa | Rep. of Korea | nk |
ATCC, American Type Culture Collection, USA; DSM, DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Germany; ICMP, International Collecti on of Micro-organisms from Plants, Landcare Research, New Zealand; KACC, Korean Agricultural Culture Collection, Rural Development Administration, Rep. of Korea; LMG, Collection of the Laboratorium voor Microbiologie en Microbiele Genetica, Ghent University, Belgium; NCPPB, National Collection of Plant Pathogenic Bacteria, United Kingdom.
This strains was used for genome sequencing by NGS.
nk, not known.
Genome sequencing and PCR primer design
Whole genome shotgun sequencing of P. coronafaciens LMG 5060 was performed using the Roche/454 pyrosequencing method on a genome sequencer with FLX titanium (Margulies et al., 2005). ORF prediction was performed with Glimmer v. 3.02 (Delcher et al., 2007), Prodigal (Hyatt et al., 2010), and GeneMark.hmm-P (Lukashin and Borodovsky, 1998). ORFs from the draft genome sequence of P. coronafaciens LMG 5060 with more than 500 base pairs were BLASTed in the gene bank. To design the primers, candidate ORFs were selected from the ORFs with lower than 80% nucleotide homology to any known genes. The PCR primers were designed with Primer 3 (v. 0.4.0). The specificity of the primers for P. coronafaciens was analyzed by PCR. Nested primers and the nucleotide sequence for the TaqMan probe were designed from amplicon sequence of P. coronafaciens-specific primers. All the primers and the TaqMan probe used in the study were synthesized by Bioneer Co., Ltd (Daejeon, Korea) or NeoProbe Co., Ltd. (Daejeon, Korea). The TaqMan probe was labeled at the 5′ end with FAM and at the 3′ end with TAMRA.
PCR and TaqMan PCR assays
The genomic DNA was extracted from the bacterial cells using the GeneAll ExgeneTM Cell SV kit (GeneAll Biotechnology, Seoul, Korea) according to the manufacturer’s protocol. DNA concentrations were determined with a QubitTM fluorometer (Invitrogen®, Carlsbad, CA, USA).
The PCR assays used 1 μl of template DNA in a 25 μl reaction mixture containing 2 mM of Tris-HCl (pH 8.0), 10 mM of KCl, 10 uM of EDTA, 100 uM of DTT, 0.05% Tween 20, 0.05% Nonidet P-40, 5% glycerol, 2.5 mM of dNTP, 5 units of Ex Taq DNA (TaKaRa Bio Inc., Shiga, Japan), and 10 pM of each primer. The reactions were performed in a T Gradient Thermal Cycler (Biometra, Göttingen, Germany) programmed for one cycle of 10 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C, with a final extension step for 7 min at 72°C. The amplicons were analyzed by electrophoresis in a 2% agarose gel in TAE buffer.
The TaqMan PCR assay was carried out using 2 μl of DNA template, 8.5 μl of Premix Ex Taq (TaKaRa Bio Inc., Shiga, Japan), 1 μl of TaqMan probe (10 pmoles/μl), 0.5 μl (10 pmoles/μl) of each nested primer, and 12.5 μl of DEPC-DW (Bioneer, Daejeon, Korea). Real-time PCR amplifications were performed in a Smart CyclerTM System (Cepheid, Sunnyvale, CA, USA) with 40 cycles at 95°C for 30 sec and 60°C for 20 sec after initial incubation for 30 sec at 95°C.
Oat seed preparation and extraction
Prior to inoculation, the oat seeds (cultivar Samhan, provided by the National Honam Agricultural Experiment Station in Korea) were surface sterilized by soaking in 70% ethanol for 30 minute and 1.2% sodium hypochlorite solution for 1 hour. The oats seeds were washed 3 times with sterilized water and dried completely in clean bench. Twelve grams of the surface-sterilized oat seeds were artificially inoculated by shaking (120 rpm) in 45 ml of P. coronafaciens LMG 5060 suspension for 24 h. The seeds were dried by leaving them in a clean bench for 20 h. The bacterial pathogen was extracted from the artificially inoculated oat seeds as described previously (Maes et al., 1996). The seeds (12 g) were shaken in 12 ml of cold aqueous saline (0.85% NaCl) containing a drop of Tween 20 for 5 min on a rotator shaker (180 rpm) at room temperature. The extracts were treated at 95°C for 7 min and used in the PCR assay or stored at −20°C.
Results
Primer and PCR specificity
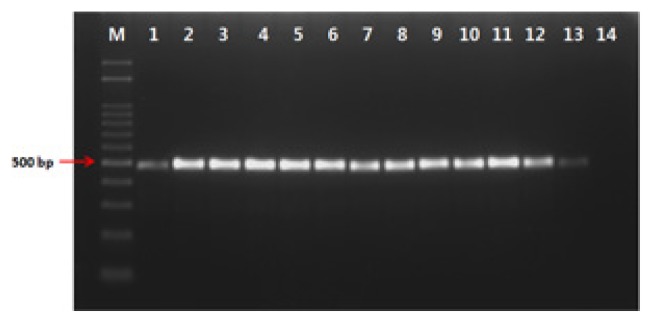
To obtain specific primers of the P. coronafaciens, the draft genome sequence of P. coronafaciens LMG 5060 was constructed by genome shotgun sequencing in this study. The draft genome sequence was deposited at NCBI GenBank (accession number: JSED00000000). The 36 primer sets were designed from the ORFs in total 2028 contigs which showed low homology to that of known genes in GenBank and the primer specificities for P. coronafaciens were checked by PCR assays with target and non-target bacterial strains. Among the primers tested, the Pc-12-F (5′-GATTGCGTCATATGCAACAT-3′) and Pc-12-R (5′-AATAGCAGATCCAGCCAAAG-3′) primer sets amplified a 498 bp in all 13 strains of P. coronafaciens (Fig. 1), whereas target-size DNA was not amplified in non-target bacteria, including 30 P. syringae pathovars, 3 P. savastanoi pathovars, and 16 other pathogenic bacteria (Fig. 2). The nested primers, Pc-12-ne-F (5′-AACGACGGGCTGCAGTTTAT-3′) and Pc-12-ne-R (5′-AACGTGATAGCAGCCCCACT-3′), were designed from the Pc-12-F/Pc-12-R amplicon. The PCR assay was conducted with the Pc-12-ne-F/Pc-12-ne-R nested primer set and genomic DNA amplified the 298 bp in all 13 strains of P. coronafaciens (supplement Fig. 1), whereas target-size DNA was not amplified in any non-target bacteria, including 30 P. syringae pathovars, 3 P. savastanoi pathovars, and 16 other pathogenic bacteria (Supplement Fig. 2).
Fig. 1.
Gel electrophoresis of the polymerase chain reaction products formed with primer Pc-12-F/Pc-12-R and bacterial DNA of Pseudomonas coronafaciens strains. Lanes 1~13, P. coronafaciens LMG 5060, KACC 13262, KACC 12133, LMG 2170, LMG 5030, LMG 5061, LMG 5081, LMG 5380, LMG 5449, LMG 5452, LMG 5536, LMG 13190, LMG 2330; lane 14, water as a negative control.
Fig. 2.
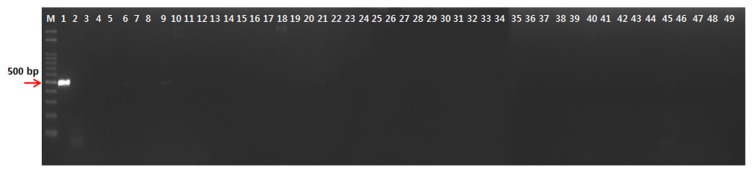
Gel electrophoresis of the polymerase cain reaction products formed with primer Pc-12-F/Pc-12-R and total DNA of lane 1, Pseudomonas coronafaciens LMG 5060, lanes 2–31, P. syringae pvs, actinidiae KACC 10582, antirrhini ICMP 4303, aptata DSM 50252, atrofaciens ICMP 4394, berberidis NCPPB 2724, ciccaronei NCPPB 2355, delphhinii ICMP 529, dysoxyli ICMP 545, eriobotyae NCPPB 2331, helianthi NCPPB 1229, japonica ICMP 6305, lachrymans ATCC 11965, lapsa ATCC 10859, maculicola ICMP 3935, mellea ICMP 5711, mori ICMP 4331, morsprunorum ICMP 5795, myricae ICMP 7118, panici NCPPB 1498, papulans ICMP 4040, passiflorae NCPPB 1386, persicae NCPPB 2761, pisi ICMP 4433, ribicola NCPPB 963, sesami NCPPB 1016, syringae NCPPB 388, tabaci ICMP 2835, tagetis ICMP 4091, tomato NCPPB 2683, ulmi NCPPB 632; lanes 32~34, P. savastanoi pvs. glycinea NCPPB 1134, pahseolicola KACC 10575, and savastanoi NCPPB 639; lane 35, Acidovorax avenae subsp. avenae NCPPB 1011; lanes 36–38, Clavibacter michiganensis subsp. insidiosus NCPPB 1020, michiganensis NCPPB 1064, sepedonicus NCPPB 2137; lane 39, Pectobacterium carotovorum subsp. carotovorum, NCPPB 312; lanes 40–43, Rhizobium radiobacter DSM 30205, R. rhizogenes ATCC 11325, R. rubi NCPPB 1854, R. vitis NCPPB 3554; lane 44, Rhodococcus fascians LMG 3601; lane 45, Ralstonia solanacearum NCPPB 339; lanes 46–47, Xanthomonas campestris pvs. campestris KACC 10377, vesicatoria KACC 11157; lane 48, X. oryzae pv. oryzae KACC 10331; lane 1, P. coronafaciens LMG 5060, as a positive control; lane 49, water as a negative control.
TaqMan PCR specificity
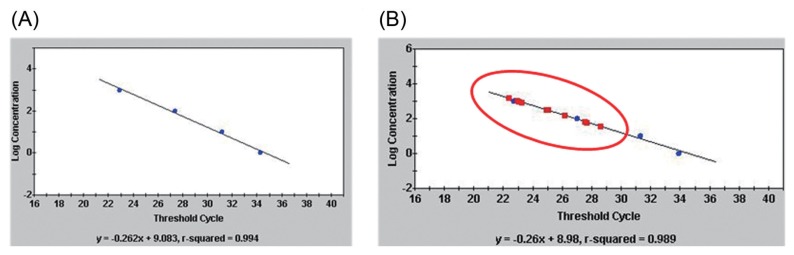
To reduce the rate of false-positives and quantitatively detect P. coronafaciens, a TaqMan probe, Pc-taqman (5′-TGAAACCGCCGAAACGGTCT-3′), was designed from the sequence of the Pc-12-ne-F/Pc-12-ne-R amplicon. The real-time PCR with Pc-12-ne-F/Pc-12-ne-R and Pc-taqman generated Ct values in a dose-dependent for 10 ng-10 pg of P. coronafaciens LMG 5060’ genomic DNA. The r2 of the linear regression was 0.994 (Fig. 3A).
Fig. 3.
Sensitivity and specificity of the TaqMan real-time PCR with primer, Pc-12-ne-F/Pc-12-ne-R and TaqMan probe, Pc-taqman. (A) The linear regression generated by ten-fold dilution of DNA of Pseudomonas coronafaciens LMG 5060 and (B) TaqMan PCR with DNAs (circle dot) of P. coronafaciens LMG 5060, and P. coronafaciens strains (square dot): P. coronafaciens KACC 13262, KACC 12133, LMG 2170, LMG 5030, LMG 5061, LMG 5081, LMG 5380, LMG 5449, LMG 5452, LMG 5536, LMG 13190, and LMG 2330.
The specificity of the TaqMan PCR was checked with the target and non-target bacteria. In the TaqMan PCR assays, 10-fold diluted DNAs (10 ng to 10 pg) of P. coronafaciens LMG 5060 were used as standard, and 10 ng of DNAs of the other bacterial strains were used. The Ct values of the 13 P. coronafaciens strains were 22–28 cycles (Fig. 3B and Table 2), whereas the average Ct values of the DNA of 49 non-target bacterial strains were more than 37 cycles (Table 2). The Ct values of the target and non-target bacteria were well separated into two different groups.
Table 2.
The Ct values of Pseudomonas coronafaciens strains, the non-target bacterial strains and the seed extracts from artificially inoculated oat seeds with Pseudomonas coronafaciens LMG 5060 which were obtained by the TaqMan PCR assays
| Samples | DNA conc. | Mean Cta | |
|---|---|---|---|
| Pseudomonas coronafaciens | LMG 5060 (STD) | 10 ng | 22.8±0.1 |
| 1 ng | 28.7±2.5 | ||
| 100 pg | 31.3±1.5 | ||
| 10 pg | 36.5±3.3 | ||
| KACC 12133 | 10 ng | 22.5±0.1 | |
| KACC 13262 | 10 ng | 22.7±0.2 | |
| LMG 2170 | 10 ng | 23.3±0.1 | |
| LMG 2330 | 10 ng | 27.1±0.7 | |
| LMG 5030 | 10 ng | 22.8±0.3 | |
| LMG 5061 | 10 ng | 28.0±1.0 | |
| LMG 5081 | 10 ng | 25.8±2.1 | |
| LMG 5380 | 10 ng | 24.7±0.4 | |
| LMG 5449 | 10 ng | 25.1±0.4 | |
| LMG 5452 | 10 ng | 25.5±2.0 | |
| LMG 5536 | 10 ng | 23.5±0.6 | |
| LMG 13190 | 10 ng | 27.9±1.5 | |
| Non-target bacterial strains | |||
| Acidovorax avenae subsp. avenae | NCPPB 1011 | 10 ng | 39.1±1.7 |
| Clavibacter michiganensis subsp. incidiosus | NCPPB 1020 | 10 ng | > 40 |
| Clavibacter michiganensis subsp. michiganensis | NCPPB 1064 | 10 ng | > 40 |
| Clavibacter michiganensis subsp. sepedonicus | NCPPB 2137 | 10 ng | > 40 |
| Pectobacterium carotovorum subsp. carotovorum | NCPPB 312 | 10 ng | > 40 |
| Pseudomonas savastanoi pv. glycinea | NCPPB 1134 | 10 ng | > 40 |
| Pseudomonas savastanoi pv. phaseolicola | KACC 10575 | 10 ng | 39.5±0.9 |
| Pseudomonas savastanoi pv. savastanoi | NCPPB 639 | 10 ng | > 40 |
| Pseudomonas syringae pv. actinidiae | KACC 10582 | 10 ng | > 40 |
| Pseudomonas syringae pv. antirrhini | ICMP 4303 | 10 ng | > 40 |
| Pseudomonas syringae pv. aptata | DSM 50252 | 10 ng | > 40 |
| Pseudomonas syringae pv. atrofaciens | LMG 5095 | 10 ng | > 40 |
| Pseudomonas syringae pv. berberidis | NCPPB 2724 | 10 ng | 37.8±1.9 |
| Pseudomonas syringae pv. ciccaronei | NCPPB 2355 | 10 ng | 39.6±0.8 |
| Pseudomonas syringae pv. delphinii | ICMP 529 | 10 ng | 39.5±0.9 |
| Pseudomonas syringae pv. dysoxyli | ICMP 545 | 10 ng | 39.3±1.3 |
| Pseudomonas syringae pv. eriobotryae | NCPPB 2331 | 10 ng | > 40 |
| Pseudomonas syringae pv. helianthi | NCPPB 1229 | 10 ng | 39.9±0.2 |
| Pseudomonas syringae pv. japonica | ICMP 6305 | 10 ng | > 40 |
| Pseudomonas syringae pv. lachrymans | ATCC 11965 | 10 ng | > 40 |
| Pseudomonas syringae pv. lapsa | ATCC 10859 | 10 ng | > 40 |
| Pseudomonas syringae pv. maculicola | ICMP 3935 | 10 ng | > 40 |
| Pseudomonas syringae pv. mellea | ICMP 5711 | 10 ng | 38.7±1.3 |
| Pseudomonas syringae pv. mori | ICMP 4331 | 10 ng | > 40 |
| Pseudomonas syringae pv. morsprunorum | ICMP 5795 | 10 ng | > 40 |
| Pseudomonas syringae pv. myricae | ICMP 7118 | 10 ng | > 40 |
| Pseudomonas syringae pv. panici | NCPPB 1498 | 10 ng | > 40 |
| Pseudomonas syringae pv. papulans | ICMP 4040 | 10 ng | > 40 |
| Pseudomonas syringae pv. passiflorae | NCPPB 1386 | 10 ng | 39.7±0.6 |
| Pseudomonas syringae pv. persicae | NCPPB 2761 | 10 ng | > 40 |
| Pseudomonas syringae pv. pisi | ICMP 4433 | 10 ng | > 40 |
| Pseudomonas syringae pv. ribicola | NCPPB 963 | 10 ng | > 40 |
| Pseudomonas syringae pv. sesami | NCPPB 1016 | 10 ng | > 40 |
| Pseudomonas syringae pv. syringae | NCPPB 388 | 10 ng | > 40 |
| Pseudomonas syringae pv. tabaci | ICMP 2835 | 10 ng | > 40 |
| Pseudomonas syringae pv. tagetis | ICMP 4091 | 10 ng | > 40 |
| Pseudomonas syringae pv. tomato | NCPPB 2683 | 10 ng | > 40 |
| Pseudomonas syringae pv. ulmi | NCPPB 632 | 10 ng | 39.8±0.3 |
| Ralstonia solanacearum | NCPPB 339 | 10 ng | > 40 |
| Rhizobium radiobacter | DSM 30205 | 10 ng | > 40 |
| Rhizobium rhizogenes | ATCC 11325 | 10 ng | > 40 |
| Rhizobium rubi | NCPPB 1854 | 10 ng | > 40 |
| Rhizobium vitis | NCPPB 3554 | 10 ng | > 40 |
| Rhodococcus fascians | LMG 3601 | 10 ng | > 40 |
| Xanthomonas campestris pv. campestris | KACC 10377 | 10 ng | > 40 |
| Xanthomonas campestris pv. vesicatoria | KACC 11157 | 10 ng | > 40 |
| Xanthomonas oryzae pv. oryzae | KACC 10331 | 10 ng | > 40 |
| Seed extracts from oat seeds inoculated in | |||
| 108 cfu/ml cell suspension | – | 21.2±0.5 | |
| 107 cfu/ml cell suspension | – | 21.4±0.2 | |
| 106 cfu/ml cell suspension | – | 21.3±0.2 | |
| 105 cfu/ml cell suspension | – | 22.5±0.5 | |
| 104 cfu/ml cell suspension | – | 24.2±0.7 | |
| 103 cfu/ml cell suspension | – | 25.1±1.1 | |
| 102 cfu/ml cell suspension | – | 29.2±0.3 | |
| 10 cfu/ml cell suspension | – | 32.2±0.7 | |
| control (water treatment) | – | > 40 | |
TaqMan PCR detection of P. coronafaciens from the artificially inoculated oat seeds
The TaqMan PCR assay developed in this study was applied to detect P. coronafaciens from the oat seeds. Since the seeds naturally infested with the pathogen were not available, the artificially inoculated oat seeds were used. TaqMan PCR with the seed extracts generated a range of mean Ct values from 21.2 for 108 cfu/ml-inoculated seed extracts to 32.2 for 10 cfu/ml-inoculated seed extracts (Table 2). Since Ct values of all the non-target bacteria used in this study were larger than 37 cycles, the TaqMan PCR can detect P. coronafaciens from the seed extract of the artificially inoculated oat seeds above 10 cfu/ml inoculation level.
Estimation of the recovery number of P. coronafaciens from the artificial inoculated seed extract was tried with several different ways, but the accurate enumeration of P. coronafaciens from the seed extract failed because the many non-target-shape colonies were cultured on the culture plate and the selective medium for P. coronafaciens was not available. Surface sterilization of the oat seeds prior to artificial inoculation could not prevent growth of some non-target bacteria on the recovery culture plates.
Discussion
Highly specific PCR and TaqMan PCR assays have been developed in this study to detect P. coronafaciens, the halo blight pathogen of oats. Since P. coronafaciens is a plant quarantine bacterium in many countries and using of the certificated seed is important for the disease control, developments of these assays are significant for the management of halo blight of oats.
To obtain specific primers of the P. coronafaciens, a genome-wide search was conducted from the draft genome sequence of P. coronafaciens which was constructed by genome shotgun sequencing in this study. A specific primer, Pc-12-F/Pc-12-R, was designed from ORF 7 in contig 7 of the draft genome sequence of P. coronafaciens (NCBI GenBank accession number: JSED00000000). ORF 7 was 1341 nucleotides long, and no significant homologous gene was found in the NCBI GenBank database. PCRs with Pc-12-F/Pc-12-R and Pc-12-ne-F/Pc-12-ne-R generated the target size DNA from all 13 strains of P. coronafaciens isolated in seven countries. The target-size DNA was not amplified in 49 strains of non-target bacteria. The Ct values of the target and non-target bacteria were well separated in the TaqMan PCR. The homology of the ORFs in which PCR was designed and the results of the PCR and TaqMan PCR indicate that the primer sequences and PCR assays developed in this study are highly specific to P. coronafaciens.
TaqMan PCR was applied to detect the target pathogen from artificially inoculated oat seeds. TaqMan PCR generated the P. coronafaciens–positive Ct values in the seed extracts obtained from oat seeds inoculated in 10 cfu/ml and above. Although detection sensitivity of this TaqMan PCR cannot compared to the previously published results because detection of P. coronafaciens from oat seeds has not been published, positive detection of the seed extracts obtained from oat seeds inoculated in 10 cfu/ml P. coronafaciens LMG 5060 suspension and above is thought to be a quite high sensitivity and can apply to seed test and plant quarantine service.
Acknowledgements
This work was supported by the research grant of Chungbuk National University in 2012.
References
- Bella P, Licciardello G, Tessitori M, Catara V. A real-time PCR quantitative detection assay for Pseudomonas savastanoi pv. nerii in Nerium oleander. Phytopathol Mediterr. 2008;47:204–213. [Google Scholar]
- Cho JH, Jeong MJ, Song MJ, Yim KO, Lee HI, Kim JH, Baeg JH, Cha JS. Development of PCR Primers to Detect Pseudomonas savastanoi pv. phaseolicola from the Bean Seeds. Res Plant Dis. 2010;16:129–135. doi: 10.5423/RPD.2010.16.2.129. [DOI] [Google Scholar]
- Collins DJ. Oat Diseases in Alabama: Circular ANR-631. Alabama Cooperative Extension Service 2010 [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott C. Halo-blight of oats. J Agric Res. 1920;19:139–172. [Google Scholar]
- Finetti-Sialer MM, Ciancio A. Isolate-specific detection of Grapevine fan-leaf virus from Xiphinema index through DNA-based molecular probes. Phytopathology. 2005;95:262–268. doi: 10.1094/PHYTO-95-0262. [DOI] [PubMed] [Google Scholar]
- Harder DE, Haber S. Oat diseases and pathologic techniques. Oat Science and Technology. 1992;33:307–402. [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119–129. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin A, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M, Garbeva P, Kamoen O. Recognition and detection in seed of the Xanthomonas pathogens that cause cereal leaf streak using rDNA spacer sequences and polymerase chain reaction. Phytopathology. 1996;86:63–69. doi: 10.1094/Phyto-86-63. [DOI] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated highdensity picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JW, Seaman WL, Atkinson TG. Diseases of field crops in Canada. Canadian Phytopathological Society 1984 [Google Scholar]
- Schena L, Nigro F, Ippolito A, Gallitelli D. Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. Eur J Plant Pathol. 2004;110:893–908. doi: 10.1007/s10658-004-4842-9. [DOI] [Google Scholar]
- Walcott RR. Detection of seedborne pathogens. HortTechnology. 2003;13:40–47. [Google Scholar]
- Wallwork H. Cereal leaf and stem diseases. Grains Research and Development Corporation 1992 [Google Scholar]
- Young JM, Dye DW, Wilkie JP. Genus VII. Pseudomonas Migula 1894. New Zealand J Agric Res. 1978;21:153–177. doi: 10.1080/00288233.1978.10427397. [DOI] [Google Scholar]