Summary
Telomeres protect the ends of cellular chromosomes. We show here that infection with herpes simplex virus 1 (HSV-1) results in chromosomal structural aberrations at telomeres and the accumulation of telomere dysfunction-induced DNA damage foci (TIFs). At the molecular level, HSV-1 induces transcription of telomere repeat-containing RNA (TERRA), followed by the proteolytic degradation of the telomere protein TPP1, and loss of the telomere repeat DNA signal. The HSV-1 encoded E3 ubiquitin ligase ICP0 is required for TERRA transcription and facilitates TPP1 degradation. shRNA depletion of TPP1 increases viral replication, arguing that TPP1inhibits viral replication. Viral replication protein ICP8 forms foci that coincide with telomeric proteins and ICP8 null virus failed to degrade telomere DNA signal. These findings suggest that HSV-1 reorganizes telomeres to form ICP8-associated pre-replication foci and promotes viral genomic replication.
Keywords: telomere, herpes simplex virus, HSV, TERRA, shelterin, TRF, ICP0, ICP8, TPP1
Introduction
Telomeres are the functional genetic elements that protect and monitor the ends of linear chromosomes. The terminal TTAGGG repeats of mammalian telomeres assemble into a nucleoprotein complex that is collectively referred to as shelterin (de Lange, 2005). The core shelterin components include the telomere repeat DNA-binding factors TRF1 and TRF2, the single stranded DNA binding protein Pot1, and their interacting proteins hRap1, TIN2, and TPP1. Shelterin components have essential and distinct roles in telomere length homeostasis and control of DNA damage response (DDR) signaling at the chromosome termini (de Lange, 2010). Loss or damage of the telomere repeat DNA can initiate a DDR and trigger cellular replicative senescence (Deng et al., 2008). Similarly, mutation, deletion, or post-translational modification of shelterin proteins can activate DDR signaling and cause cell cycle arrest (Sfeir and de Lange, 2012). Telomeres also form higher-order chromosomal structures that undergo conformational changes that are important for telomere homeostasis and chromosome integrity (Taddei et al., 2004). The extent to which viruses modify or utilize telomeric factors and structure is not well understood.
Herpesviruses are large double-stranded DNA viruses that yield either a productive lytic infection or establish a long-term latent infection (Roizman and Whitley, 2013). HSV-1 can productively infect many different types of epithelial cells, and establish latent infections in neuronal ganglia in vivo (Knipe and Cliffe, 2008). Productive infection requires the activation of viral transcription and DNA replication, as well as the inactivation of host intrinsic defenses. The HSV-1 ICP0 protein is an immediate early protein that inactivates host intrinsic defense proteins and stimulates viral transcription (Boutell and Everett, 2013). ICP0 has intrinsic E3 ubiquitin-ligase activity mediated by an N-terminal Ring-finger that targets several cellular proteins, directly or indirectly, for proteasome-mediated degradation. ICP0 causes the degradation of the Promyelocytic Leukemia (PML) protein and its associated nuclear body (PML-NB) that are implicated in the host intrinsic defense to viral infection (Everett and Chelbi-Alix, 2007). ICP0 can also target other cellular factors, including centromeric proteins (Lomonte et al., 2001) and DNA damage repair proteins RNF8 and RNF168 (Chaurushiya et al., 2012; Lilley et al., 2010). ICP0 can interact with the chromatin regulatory factors, like BMAL (Kawaguchi et al., 2001) and reverse chromatin mediated silencing (Ferenczy et al., 2011) to promote viral gene expression. How ICP0 transcription activation functions are coordinated with destruction of intrinsic resistance factors is not completely understood.
HSV-1 DNA replication requires the assembly of a viral replisome involving viral-encoded DNA polymerase and accessory factors (Weller and Coen, 2012). Viral replication occurs in large subnuclear structures referred to as replication compartments (Quinlan et al., 1984). Replication compartments form at a subset of pre-replication sites marked by the viral replication protein ICP8 (Lukonis and Weller, 1997; Uprichard and Knipe, 1997). ICP8 is a single-strand DNA binding protein essential for HSV-1 DNA replication. ICP8 promotes strand invasion and homologous recombination that has been implicated in HSV-1 genome replication (Muylaert et al., 2011). Replication compartments form through the coalescing of multiple ICP8 associated pre-replication foci (Taylor et al., 2003) and require large-scale changes in nuclear architecture, including marginalization of the chromatin and eventual breakdown of the nuclear envelope lamina (Simpson-Holley et al., 2005). The molecular mechanisms through which ICP8 contributes to replisome and replication compartment assembly are not completely understood.
Telomere structural maintenance involves complex interactions between shelterin, chromatin, transcription, replication, and recombination factors (Ye et al., 2010). In proliferating cells, shelterin regulates the accessibility to telomerase, a reverse transcriptase that extends the G-rich telomere repeats at 3’ termini. Telomeres can also be transcribed to generate telomere repeat-containing RNA (TERRA) that modulates telomerase activity and telomere DNA accessibility through multiple mechanisms including template competition and heterochromatin formation (Azzalin and Lingner, 2014). TERRA transcription is also elevated in telomerase-negative cells that use homologous recombination for alternative-lengthening of telomeres (ALT) (Ng et al., 2009). In ALT-positive cells, telomeres colocalize with PML-NBs to form ALT-associated PML bodies (APBs) (Wu et al., 2000). PML- NBs are also the site where DNA virus genomes localize prior to replication (Maul et al., 1996), and the destruction of PML-NBs by viral proteins, like ICP0, is necessary for efficient viral replication (Everett and Chelbi-Alix, 2007). It is not yet known whether the alternative lengthening of telomeres at APBs is related to the intrinsic resistance to viral infection mounted by PML-NBs.
The impact of viral replication on telomere structure and regulation has not been explored in molecular detail. Here, we show that HSV-1 is a potent modulator of telomere transcription regulation and DNA structure. We show that ICP0 contributes to the efficient transcriptional activation of TERRA, and promotes ICP8 localization and binding to telomeric DNA. We also found that HSV-1 infection alters TPP1 protein stability and that depletion of TPP1 enhances viral DNA synthesis. We suggest that viral-mediated telomere remodeling overcomes inherent barriers to viral infection in the nucleus and repurposes telomeric molecules or structures to facilitate viral replication.
Results
HSV-1 induces telomere-associated DNA damage and structural aberrations
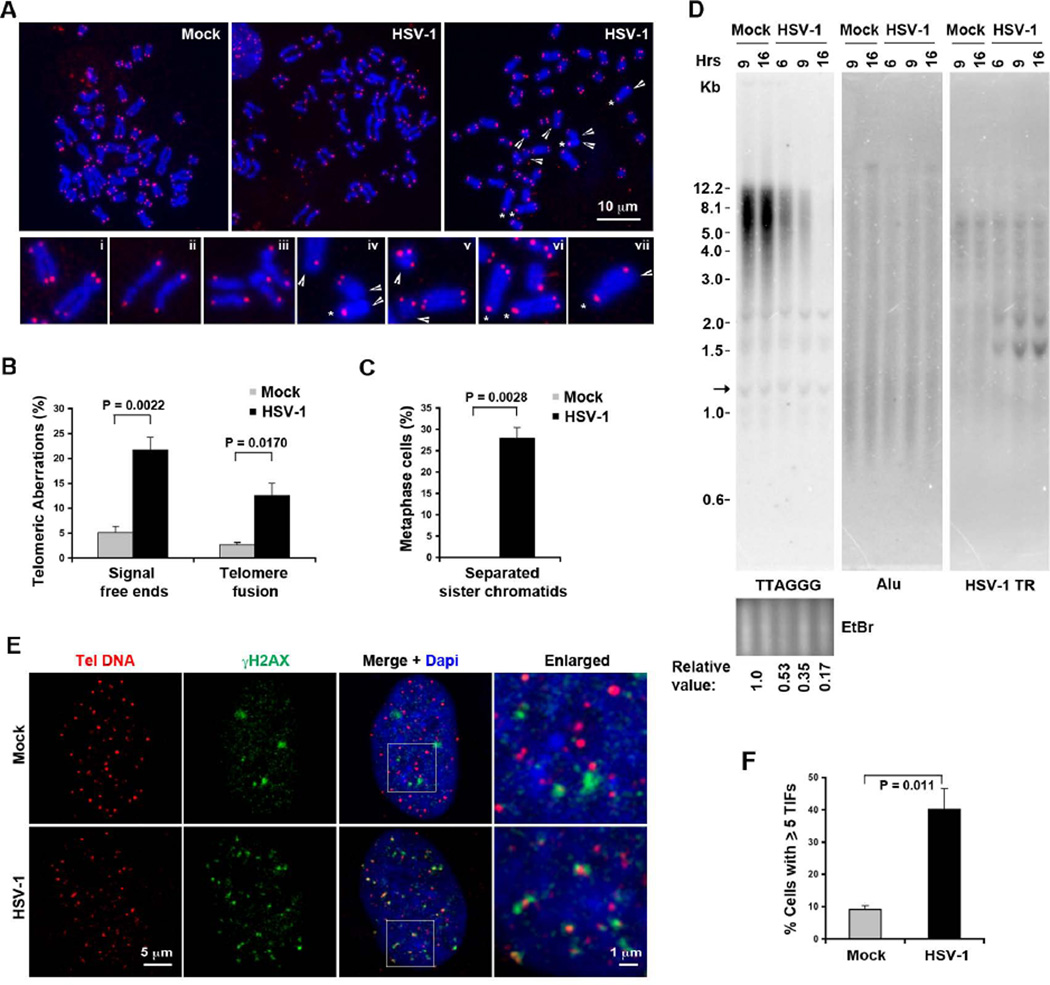
HSV-1 infection is known to induce chromosome aberrations (O'Neill and Rapp, 1971), but specific effects on telomeres have not been reported. To examine the effects of HSV-1 infection on human telomere structure, we performed telomere DNA-FISH on metaphase chromosomes after HSV-1 infection (Figure 1A and S1A). Human diploid fibroblasts (BJ) were infected for 6 hrs and then treated with colcemid to arrest cells in metaphase. We found that HSV-1 infection had a profound effect on metaphase chromosome structure, causing a significant increase in chromosomes with telomere signal free ends (~4 fold) and telomere fusions (~5 fold) (Figure 1B). We also noted an increased appearance of cells with separated sister chromatids (Figure 1C), which is consistent with previous findings that HSV-1 degrades kinetochore protein CENP-C and CENP-A. To determine whether HSV-1 infection affected telomere repeat DNA, we performed Southern blots to measure average telomere length and signal (Figure 1D). We found that HSV-1 infected BJ cells had a significant (~5 fold) loss of telomere repeat DNA signal relative to Alu DNA or bulk DNA (EtBr). As expected, HSV-1 infected cells showed increasing levels of viral DNA signal when the same blot was hybridized with a probe specific for HSV-1 terminal repeats (TR) (Figure 1D, right panel). To determine if the loss of telomeric DNA signal correlated with the appearance of telomere-associated DNA damage, we assayed the colocalization of γH2AX with telomere DNA using immuno-FISH combined with confocal microscopy (Figure 1E). Telomere dysfunction-induced foci (TIFs) can be observed in a few of mock infected BJ cells, whereas the frequency of γH2AX foci colocalization with telomere DNA was increased ~4 fold in HSV-1 infected BJ cells (Figure 1F, S1B).
Figure 1. HSV-1 infection leads to an increase in chromosomal aberrations and TIFs.
A) Telomeric DNA FISH analysis on metaphase spreads for telomere and chromosomal defects. Infected BJ cells with separated sister chromatids are shown in the middle panel. Representative dysfunctional telomeres are indicated by arrow-head (telomere free ends), or star (sister chromatid fusions). Magnified views of aberrations are shown in panels i–xii: normal telomere ends (i), separated sister chromatids (ii–iii), telomere free ends, and sister chromatid fusions (iv–vii). B) Quantification of telomere defects for telomere free ends and chromatid fusions. Bars represent mean percentage of chromosomes with indicated telomeric aberrations (±SD) obtained from three independent HSV-1 infection and FISH experiments. The total number of counted chromosomes was about 1370 and 1104 for mock and HSV-1 infected cells, respectively. P values were obtained by two-tailed Student’s t –Test. C) Quantification of metaphase cells with separated sister chromatids. The bar graph represents average percentage of metaphase cells (n=37 or 35 for mock or HSV-1 infected cells, respectively) with separated sister chromatids (±SD) from three independent experiments. P values were calculated by two-tailed Student’s t – Test. D) BJ cells were infected with HSV-1 wt (MOI=0.1), or mock for indicated time, and telomere length assay was performed by Southern blot using 32P-labeled (TAACCC)4 probe (left), followed by hybridization with Alu probe (middle) or an oligonucleotide probe specific for HSV-1 TR region (right) under denaturing conditions. Relative telomere signals are shown at the bottom, the arrow indicates the internal band used for normalization. E) Confocal microscopy analysis of cells with TIFs. BJ fibroblasts were infected with wild-type HSV-1 or mock infected, and assayed by immuno-FISH with the CCCTAA PNA probe (red) and antibody to γH2AX (green) at 6 hrs post-infection. Enlarged images derived from the white square in DAPI (blue) and merged images are shown on the right. F) Quantification of telomere associated DNA damage foci as represented in (E). The bar graph shows the mean (±SD) derived from quantification of ~ 100 nuclei for each infection from multiple independent TIF assays (n=4). P value was calculated by a two-tailed Student’s t –Test. See also Figure S1.
Viral infection can induce TERRA accumulation
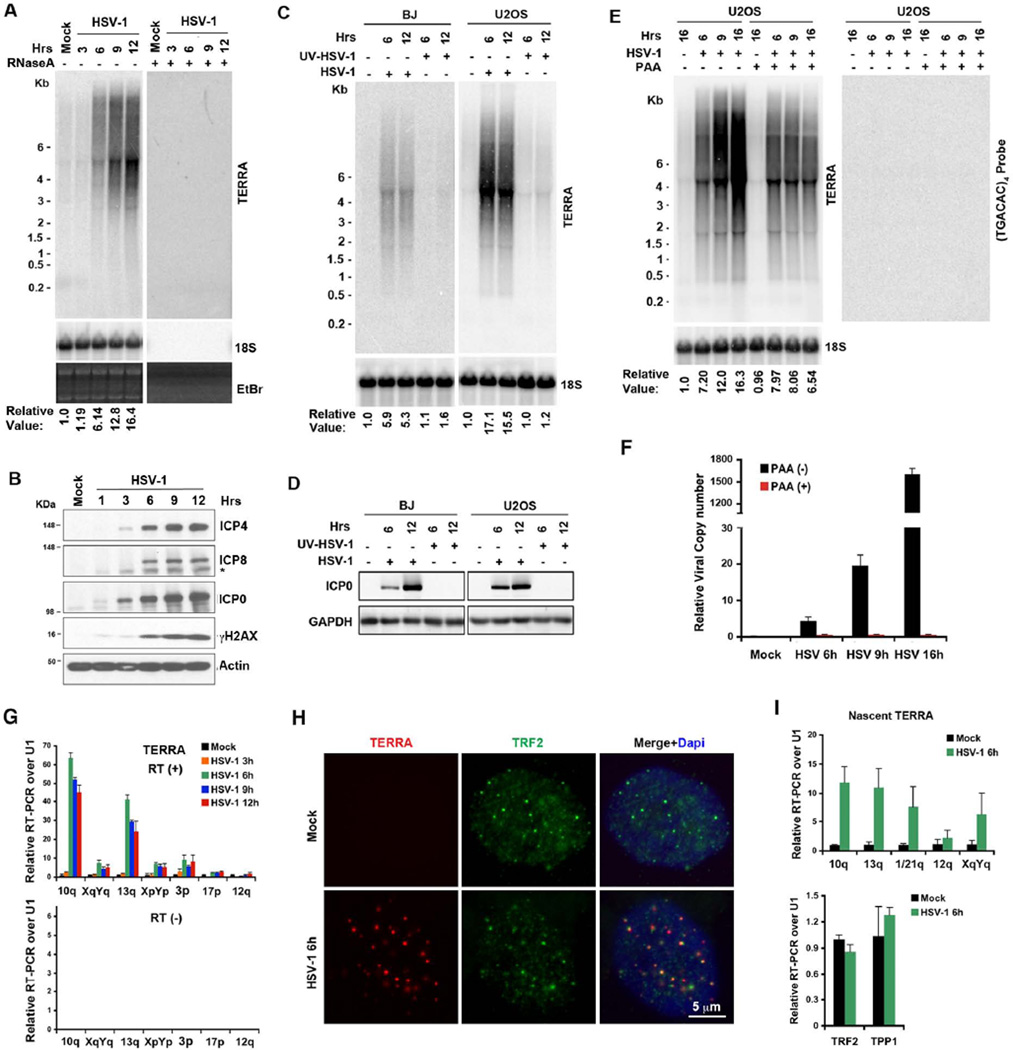
Telomere transcription and non-coding RNA TERRA are known to function in telomere DNA maintenance and stability. Therefore, we assayed the effects of HSV-1 infection on TERRA expression. We found that HSV-1 induced TERRA by 6 fold at 6 hrs, and up to 16 fold at 12 hrs post-infection (Figure 2A). TERRA RNA signal on Northern blots could be eliminated by RNase A treatment (Figure 2A, right panel), indicating that this signal was RNA-specific and not cross-reacting with degraded telomere or viral DNA. HSV-1 induced TERRA in a dose-dependent manner that peaks at ~1 MOI (Figure S2G). HSV-1 induced TERRA expression more rapidly and robustly (17 fold in 6 hrs) in U2OS cells than in BJ cells (Figure 2C). UV-irradiation of viral particles prior to infection eliminated induction of TERRA expression (Figure 2C and D), suggesting that viral gene expression is required for TERRA induction. Phosphonoacetic acid (PAA) treatment, which is a potent inhibitor of HSV-1 DNA replication, had little effect on TERRA levels at 6 hrs post-infection, and only partially reduced TERRA induction at later times (Figure 2E and F). This suggests, that viral DNA replication is not required for the early phase induction of TERRA. No RNA signal was detected with a probe for non-telomere repeat (TGACAC)4 (Figure 2E, right panel) indicating the signal for TERRA was specific for telomere repeat sequence (TAACCC)4.
Figure 2. HSV-1 infection rapidly induces high TERRA levels.
A) Northern blot of TERRA RNA from BJ fibroblasts infected with HSV-1 (MOI=3) over the indicated time course without (−) or with (+) RNase A treatment. Ethidium bromide stained gels are shown to monitor RNA integrity. TERRA signals are shown as normalized relative to 18S rRNA signals and mock infection. B) Western blot of HSV-1 infected cells as shown in (A), using antibodies for viral proteins ICP0, ICP4, or ICP8, and cellular factors phosphorylated H2AX (γH2AX) or β-actin. C) TERRA is not induced by infection with UV-irradiated HSV-1. BJ and U2OS cells infected with wt or UV-irradiated HSV-1 for 6 or 12 hrs were assayed by Northern blot. The numbers below show the relative values for TERRA RNA normalized to 18S RNA and mock infection. D) Western blot of viral infection in infected cells, as shown in (C). Antibody specific for ICP0 was used to monitor HSV-1 infection. E) Inhibition of viral DNA synthesis partially abrogates TERRA induction. U2OS cells were infected with HSV-1 (MOI=1) in the presence or absence of PAA, and assayed by Northern blot using 32P-labeled (TAACCC)4, or (TGACAC)4 or 18S control probes. F) The effects of PAA on viral DNA synthesis were assayed by qPCR to measure viral DNA copy number. The bar graph shows the mean values (±SD) of VP16-p relative to actin region from triplicate samples by ΔCT methods. G) Quantitative RT-PCR analysis of TERRA RNA transcribed from subtelomeres over the course of HSV-1 infections in BJ cells. The bar graph shows the relative values of TERRA signals normalized to U1 snRNA and mock infection by ΔΔCT methods with (upper) or without (lower panel) reverse transcription (RT). The standard errors were derived from triplicate samples. H) BJ cells were infected with HSV-1 or mock infected, and assayed by immuno-RNA FISH for TERRA (red) and TRF2 (green) at 6 hrs post-infection. DAPI staining is shown in blue in merged panels. I) qRT-PCR analysis of nascent TERRA RNA transcribed from indicated subtelomeres in BJ cells infected with HSV-1 for 6 hrs. The bar graph shows the relative value of TERRA values (upper) or TRF2 and TPP1 mRNA (lower panel) normalized to U1 RNA and mock infection (Mean±SD). See also Figure S2.
To further validate TERRA induction by HSV-1, we measured chromosome-specific TERRA molecules by RT-qPCR (Figure 2G). At many chromosomes the TERRA molecules were amplified, with some showing >60 fold induction at 6 hrs post-infection. The heterogeneity of chromosome-specific TERRA transcription is not unexpected because TERRA transcription is subject to single chromosome regulation, including response to changes in telomere length (Arnoult et al., 2012). TERRA RNA could be visualized as discrete foci, the majority of which localize to TRF2 foci by Immuno-RNA FISH analysis at 6 hrs post-infection (Figure 2H). No TERRA foci could be observed in the mock infected BJ cells, possibly because very low levels of TERRA were expressed in the fibroblasts. To understand better the mechanism of HSV-1 induced TERRA, we used ethynyl uridine (EU)-ClickIt chemistry to determine if HSV-1 induced new RNA synthesis of TERRA, rather than stabilized the existing pool of TERRA molecules (Figure 2I). We found that HSV-1 infection stimulated the production of new TERRA RNA by ~2–10 fold, while showing no significant effect on mRNA of cellular genes for TRF2 or TPP1 at 6 hrs post-infection. This indicates that HSV-1 stimulated new RNA synthesis (initiation and/or elongation) of TERRA transcripts. Taken together, these results show that HSV-1 induces new TERRA RNA transcripts that accumulate mainly as telomere-associated foci during viral infection.
We next asked whether TERRA induction was a common feature of viral infection. We compared HSV-1 with human cytomegalovirus (hCMV), adenovirus (AdV), varicella zoster virus (VZV), and influenza virus (PR8) in cell lines permissive for each viral infection (Figure S2A–E). We found that each of these viruses had some effect on TERRA levels, but that HSV-1 was unique for its ability to induce high levels of TERRA in multiple cell types (Figure S2A and B). HSV-1 induced TERRA ~10 fold, while the other viruses had less than 2 fold increase (VZV and PR8), or reduced (hCMV and AdV) TERRA expression levels relative to mock infections. To eliminate the possibility that TERRA induction by HSV-1 was an indirect consequence of the host cell antiviral response we treated cells directly with interferon beta, which produced a strong activation of ISG15, but had no effect on TERRA levels (Figure S2F). These findings suggest that TERRA can be induced by other viruses, but that HSV-1 is unique among those tested for its robust activation of TERRA expression.
HSV-1 requirements for activation of TERRA
To investigate the mechanisms through which HSV-1 induces TERRA transcription and alters telomere regulation, we first tested a viral mutant lacking ICP0. We reasoned that ICP0 was likely to contribute to TERRA activation, because ICP0 is known to stimulate viral transcription. The ICP0 null virus dl1403 (ΔICP0) was compared to mock and wild-type HSV-1 infection in BJ and BJ-hTERT cells (Figure 3A). We found that ΔICP0 was compromised for TERRA induction in all cell types tested, including ALT positive U2OS and neuronal SY5Y cells (Figure S3A–D). In all cases, there was no ICP0 expression in cells infected with ΔICP0, while viral proteins reflective of early stage infection, such as ICP4 and ICP8, were expressed at similar levels to wild-type (Figure 3B, S3B, and S3D). Western blots also indicated that PML was degraded in an ICP0-dependent manner, as expected (Everett et al., 2006), and that γH2AX was induced independently of ICP0 status, as expected (Lilley et al., 2011; Lilley et al., 2010). To test more precisely the requirements of ICP0 in TERRA induction, we compared a mutant virus containing a deletion in the ICP0 ubiquitinligase ring finger domain (FXE) with its revertant (FXER) (Figure 3C and D). We found that the FXE mutant was compromised for TERRA induction, whereas FXER induced TERRA to levels equivalent to wt HSV-1. Western blotting indicated that mutant FXE and wt ICP0 were expressed at comparable levels, and FXE was compromised for PML degradation (Figure 3D). These observations suggested that the E3 ligase activity of ICP0 plays a role in TERRA activation.
Figure 3. TERRA induction by HSV-1 infection is decreased in the absence of viral IE protein ICP0.
A) Northern blot of TERRA RNA in BJ and BJ-hTERT cells infected with either HSV-1 wt or ICP0 null mutant ΔICP0 at 9 or 16 hrs post-infection. Relative TERRA signals were shown at the bottom. B) Western blot of infected cells as shown in (A) using antibodies as indicated. * indicates a non-ICP0 band from a previous blot with ICP8 antibody. C) Northern blot of TERRA RNA in BJ cells infected with HSV-1 wt (MOI=1), FXE (MOI=10), or FXE revertant (MOI=1) at 9 or 16 hrs post-infection. TERRA signals were quantified as values normalized to 18S rRNA signals and mock infection. D) Western blot of infected BJ cells as shown in (C) using indicated antibodies. E) Confocal microscopy analysis of BJ cells infected with HSV-1 wt or mock. The infected cells were assayed by immuno-FISH with CCCTAA PNA probe (red) and antibody to ICP0 (green) at 6 hrs post infection. DAPI (blue) and merged images are shown to the right. F) The same as in (E), except that U2OS cells infected with HSV-1 wt or mock were assayed by immuno-FISH at 6 hrs post-infection. An enlarged image derived from the white square in DAPI (blue) and merged image is shown. G) Quantification of colocalization of telomere DNA foci with ICP0 foci in infected BJ or U2OS cells. The bar graph shows (Mean ± SD) generated from three independent immuno-FISH experiments. See also Figure S3 and S4.
We also made several efforts to test whether ICP0 alone without the context of viral infection is capable of inducing TERRA (Figure S4). We tested whether stable cell lines transduced with lentivirus expressing tetracycline inducible ICP0 or ICP0 (FXE). We also tested stable cell lines with tetracycline-inducible expression of hCMV IE1, which has been shown to degrade PML, similar to ICP0. We found that ICP0 alone induced TERRA expression by only ~2 fold, while ICP0 (FXE) and IE1 had no significant effect on TERRA. Western blots indicated that ICP0 expression was less than HSV-1 infection, but was still sufficient to degrade PML, as efficiently as IE1 (Figure S4A and B). Similar results were observed for U2OS cells expressing YFP-tagged ICP0 or an ICP0 with the ring finger mutation (ICP0fxe) (Figure S4C–E). Confocal microscopy of ICP0 foci with telomere DNA revealed that a fraction of ICP0 foci colocalized with telomere DNA foci in U2OS cells (~30% colocalization rate), and to a lesser extent in BJ cells (~18% colocalization rate) (Figure 3E–G). Both YFP-ICP0 and YFP-ICP0fxe can partially localize to telomere DNA or TRF2 foci, indicating that the failure of TERRA induction by YFP-ICP0fxe was not due to its failure to localize at telomeres (Figure S4F and G). Together, these findings suggest that ICP0 and its E3 ligase activity contribute to the HSV-1 induced increase in TERRA, but that ICP0 alone is not sufficient for TERRA induction.
TPP1 is selectively degraded by HSV-1 infection
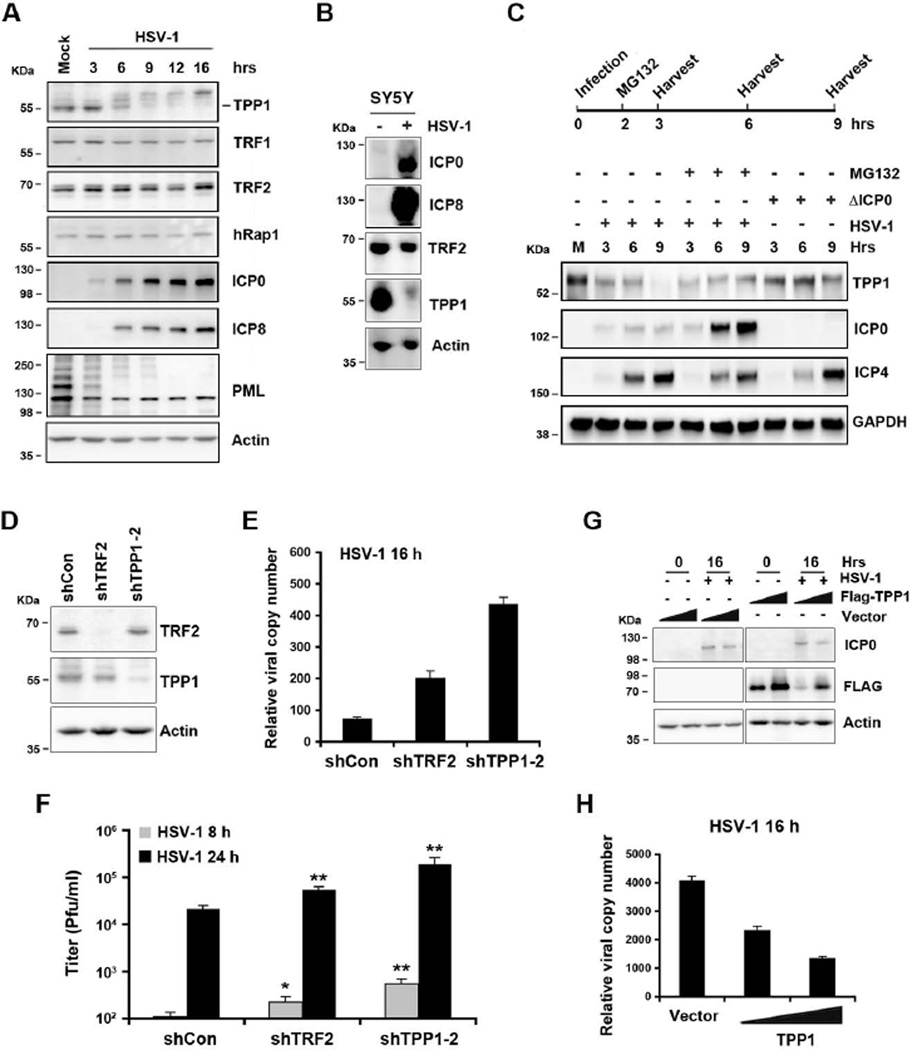
Because ICP0 is known to target numerous cellular proteins for ubiquitin-mediated degradation, we assayed whether HSV-1 infection led to the loss or modification of any shelterin protein (Figure 4). Western blots were performed at different times after HSV-1 infection (Figure 4A). We found that HSV-1 infection led to the loss of TPP1 (Figure 4A and S5A), but no other obvious change in shelterin proteins, including TRF1, TRF2, hRap1, and POT1. TPP1 loss was more dramatic in SY5Y neuronal cells infected with HSV-1 at 24 hrs post infection (Figure 4B). To determine whether the reduction of TPP1 protein level was associated with proteasome-dependent degradation, we assayed whether proteasome inhibitor MG132 prevented the loss of TPP1 during HSV-1 infection (Figure 4C, S5B, and S5C). We found that TPP1 levels were decreased at 3 hrs, and more significantly at 6 and 9 hrs post-infection, and that MG132 largely rescued this reduction (Figure 4C). We also found that TPP1 levels were not altered during the course of infection with ΔICP0 mutant virus (Figure 4C and S5C). These findings suggest that TPP1 is selectively degraded during HSV-1 infection in a proteasome-dependent fashion.
Figure 4. HSV-1 infection leads to TPP1 degradation, and TPP1 depletion enhances HSV-1 lytic replication and viral production.
A) BJ cells were infected with HSV-1 (MOI=1) and assayed by Western blot at indicated times, using antibodies for shelterin proteins TPP1, hRap1, TRF1, TRF2, or control actin. Viral infection was monitored by Western blot for ICP0, ICP8, and PML. B) SY5Y cells were infected with wild-type HSV-1 or mock and assayed by Western blot at 24 hrs post-infection, using antibodies as indicated. C) Top panel, schematic of Western blot for HSV-1 infection and MG132 treatment. Bottom panel, BJ cells were infected with either HSV-1 wt (MOI=3) or ΔICP0 (MOI=3) in the absence or presence of MG132 for the indicated time and assayed by Western blot using antibodies as indicated. D) BJ cells were infected with lentiviruses expressing shRNA against TRF2, TPP1, or control. The infected cells were assayed by Western blot to examine the knockdown efficiency at day 6 post infection. E) Effects of TRF2 or TPP1 depletion on viral DNA synthesis were assayed by qPCR to measure viral copy number. The bar graph shows the mean value (±SD) of ICP0 promoter region normalized to actin region and time 0 from three independent experiments. F) BJ cells depleted for TRF2, TPP1, or control were infected with HSV-1, and viral titers were determined at 8 or 24 hrs post-infection (Mean ± SD) from three independent plaque assays. P values were calculated by a paired two-tailed Student’s t –Test relative to shCon cells (*P< 0.01, **P<0.001). G) U2OS cells transfected with 2 µg or 5 µg of FLAG-TPP1 or control vector were subsequently infected with HSV-1 and assayed by Western blot using indicated antibodies at 0 and 16 hrs post-infection. H) Effects of TPP1 expression on viral DNA synthesis were assayed by qPCR to measure viral copy number. The bar graph represents the mean value (±SD) of qPCR for ICP0 promoter region from three independent experiments. See also Figure S5.
Depletion of TPP1 enhances HSV-1 lytic replication and viral production
To investigate the functional role of TPP1 reduction during HSV-1 infection, we assayed the effects of TPP1 shRNA depletion on HSV-1 DNA replication and virus production (Figure 4D –G). Lentiviral vectors for shControl, shTRF2, or shTPP1 were used to transduce BJ cells followed by puromycin selection and Western blot analysis to verify knock-down efficiency (Figure 4D). We found that shTPP1 depleted ~60% of total TPP1, while shTRF2 depleted ~90% of TRF2 in BJ cells after selection (Figure 4D). These cells were then used for HSV-1 infection and assayed by qPCR for viral DNA copy number (Figure 4E) and plaque-forming units (pfu) at various times post-infection (Figure 4F). We found that TPP1-depleted cells produced ~5 fold more HSV-1 DNA by qPCR than shControl cells. Viral titer was also increased in TPP1-depleted cells relative to control cells at 8 hrs (5 fold) and 24 hrs (9 fold) post-infection. TRF2-depleted cells also enhanced viral DNA replication and pfu, but to a lesser extent than did TPP1-depleted cells. Similar patterns were observed with 4 independent shRNA to TPP1, and the increase levels of viral copy number and pfu in TPP1 depleted cells correlated to knock-down efficiency of shTPP1 (Figure S5D–F). We next tested the effect of ectopic expression of TPP1 on HSV-1 infection (Figure 4G and H). Consistent with HSV-1 induced degradation of TPP1 (Figure 4C), levels of ectopic TPP1 were reduced at 16 hrs post-infection (Figure 4G), but was sufficient to produce a 2–4 fold reduction in HSV-1 intracellular DNA copy number, relative to vector control (Fig. 4H). These results argued that TPP1 functionally inhibits HSV-1 replication and virus production.
HSV-1 infection causes a rapid loss of telomere repeat DNA signal
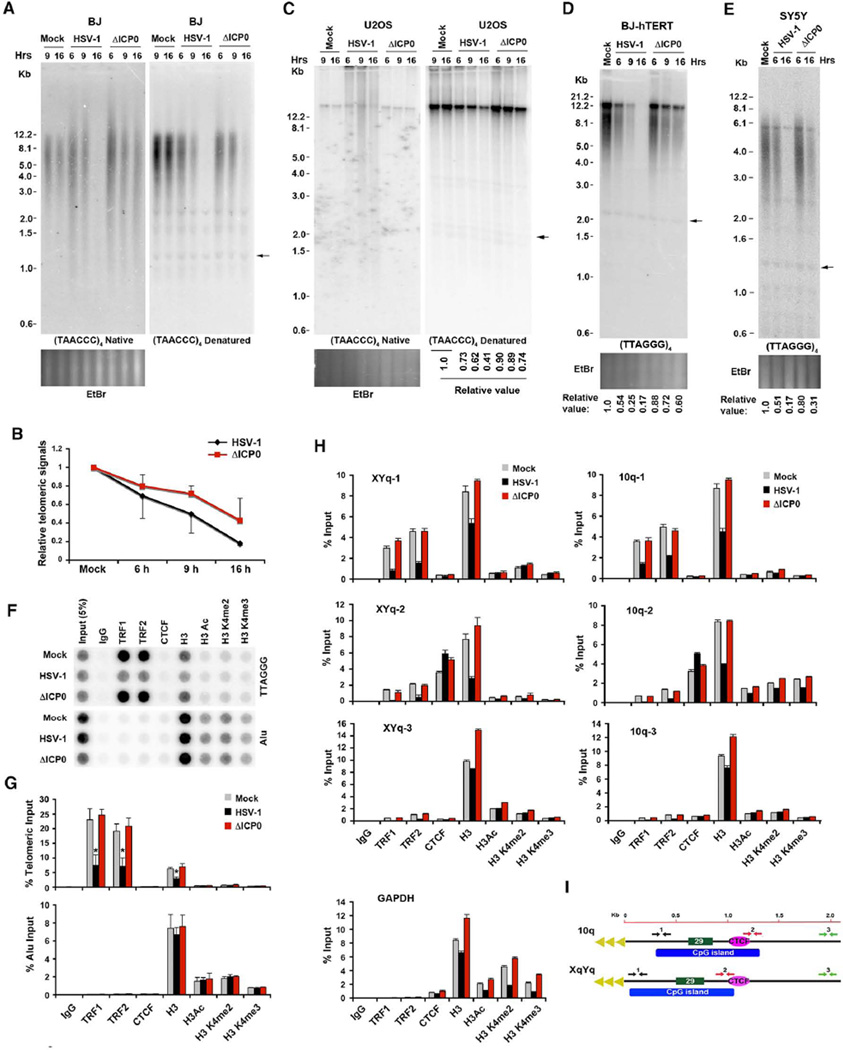
To investigate further the mechanism of HSV-1 induced telomere dysfunction, we analyzed the effect of HSV-1 wt and ΔICP0 mutant virus infection on levels of single-stranded G-rich telomeric DNA and average telomere length (Figure 5A–E). Single stranded G-rich telomere DNA can be measured by native in-gel hybridization without alkaline denaturation of DNA strands, a method that also measures telomeric 3’ G-overhang signal. We found that HSV-1 infection increased the amount and heterogeneity of single stranded G-rich telomere DNA at 6 and 9 hrs post-infection in both BJ and U2OS cells (Figure 5A and C, left panels). As shown in Figure 1, HSV-1 infection also caused a loss of telomere DNA signal at 9 and 16 hrs post-infection when the same gel was denatured by alkaline treatment and re-hybridized with the same C-rich telomeric probe (Figure 5A and C, right panels). This loss of telomere DNA signal was attenuated in ΔICP0 virus infection, in both BJ and U2OS cell infections (Figure 5A–C). HSV-1 did not cause a similar degradation of bulk (as revealed by ethidium bromide staining), indicating that single stranded DNA formation at 6–9 hrs, and DNA loss at 9–16 hrs is selective for telomere DNA (Figure 5A and C, lower panels). The telomere DNA signal loss was not dependent on cell types and status of telomerase activity, as similar telomeric DNA loss was also observed in BJ-hTERT and SY5Y cells infected with HSV-1 viruses (Figure 5D and 5E). These findings indicate that ΔICP0 mutant virus is compromised for telomere single strand formation and the loss of telomeric DNA signal as compared to the wild-type HSV-1 infection.
Figure 5. HSV-1 infection leads to rapid telomere signal loss, and reduces TRF1 and TRF2 binding at telomeres.
A) BJ cells infected with HSV-1 wt, ΔICP0, or mock were assayed at times indicated by in-gel hybridization assay. Genomic DNA digested with AluI/MboI was hybridized in the gel with 32P-labeled (TAACCC)4 probe under native conditions (left), followed by hybridization with the same probe (right) under denaturing conditions. The arrow indicates the internal band used for normalization of telomeric DNA signals in quantification step. B) Quantification of bulk telomeric DNA signals under denaturing condition relative to internal band (arrow), for two independent experiments (Mean ± errors), shown at the right panel in (A). C) The same as in (A), except that infected U2OS cells were assayed by in-gel hybridization. Quantification is shown below the right panel. D) BJ-hTERT cells were infected with wild-type HSV-1, ΔICP0, or mock for the indicated times, and assayed for telomere length by Southern blot under denaturing conditions. Relative telomere signals are shown at the bottom. E) The same as in (D), except that infected SY5Y cells were used in the assay. F) ChIP assay was performed in BJ cells infected with HSV-1 wt, ΔICP0, or mock at 6 hrs post-infection. ChIP DNA was assayed by dot blotting and hybridization with 32P-labeled probes specific for either TTAGGG or control Alu regions. G) Quantification of three independent ChIP assays represented by (F). ChIP DNA signals were normalized to input DNA signal as a percentage. The bar graph shows mean values (±SD) of the percent input DNA for telomeric (top panel) or Alu (bottom panel) DNA. P values were calculated by a two-tailed Student’s t –Test (*p< 0.005). H) qPCR analysis of ChIP assays shown in (F) at indicated positions of the 10q or XqYq subtelomere relative to the TTAGGG repeat tracts (position 0). The bar graph shows the average value of percentage of input for each ChIP from triplicate samples (Mean ± SD). I) Schematic of the 10q and XqYq subtelomeres showing the relative positions of CpG islands, CTCF binding sites, and telomeric repeats. Arrow pairs 1, 2, or 3 indicate the relative positions of primer sets for 10q or XqYq subtelomere at close (~450 bp) to TTAGGG repeat (black), at CTCF sites (red), or ~2 kb from terminal repeats (green). See also Figure S6.
HSV-1 infection dissociates TRF1, TRF2, and histone H3 from telomere DNA
To measure the effect of HSV-1 infection on shelterin and histone binding to telomere terminal repeat and subtelomeric DNA, we used chromatin immunoprecipitation (ChIP) assays (Figure 5F–I). We found that HSV-1 infection led to a significant loss of core shelterin proteins TRF1 and TRF2 binding to telomere repeat DNA at 6 hrs post-infection. We also found that total histone H3 binding was also reduced at this time. TRF1 or TRF2 protein levels were not altered by HSV-1 infection (Figure 4A and S5A), and comparable levels of TRF2 or H3 protein were pulled down in all ChIP samples (Figure S6C), indicating that loss of telomere ChIP DNA binding was not due to loss of shelterin protein abundance. Importantly, neither TRF1 and TRF2, nor histone H3 binding to telomeres was affected by infection with ΔICP0 mutant virus. Similar losses of TRF1, TRF2, and histone H3 binding were observed at the subtelomeric regions on chromosome 10q and XYq, with proportionally less effect at sites distal from the telomere repeat tracts (Figure 5H). HSV-1 had no measurable loss on CTCF binding at the 10q or XqYq subtelomeres, and no significant effect on histone H3 binding at the Alu repeats. We also examined the effect of HSV-1 infection on nucleosome formation by Southern blotting of micrococcal nuclease I (MNase I) digested nuclei (Figure S6A–B). We found that HSV-1 wt infection led to ~20% and ~40% decrease in di- and tri-nucleosomes assembled on telomere repeat DNA respectively, whereas nucleosome depletion was not observed in ΔICP0 mutant virus when compared to nucleosomes in mock-infected cells. These findings suggest that nucleosomes, along with shelterin, are depleted from telomeres during HSV-1 infection in an ICP0-facilitated manner.
ICP8 pre-replication foci colocalize with telomere DNA foci
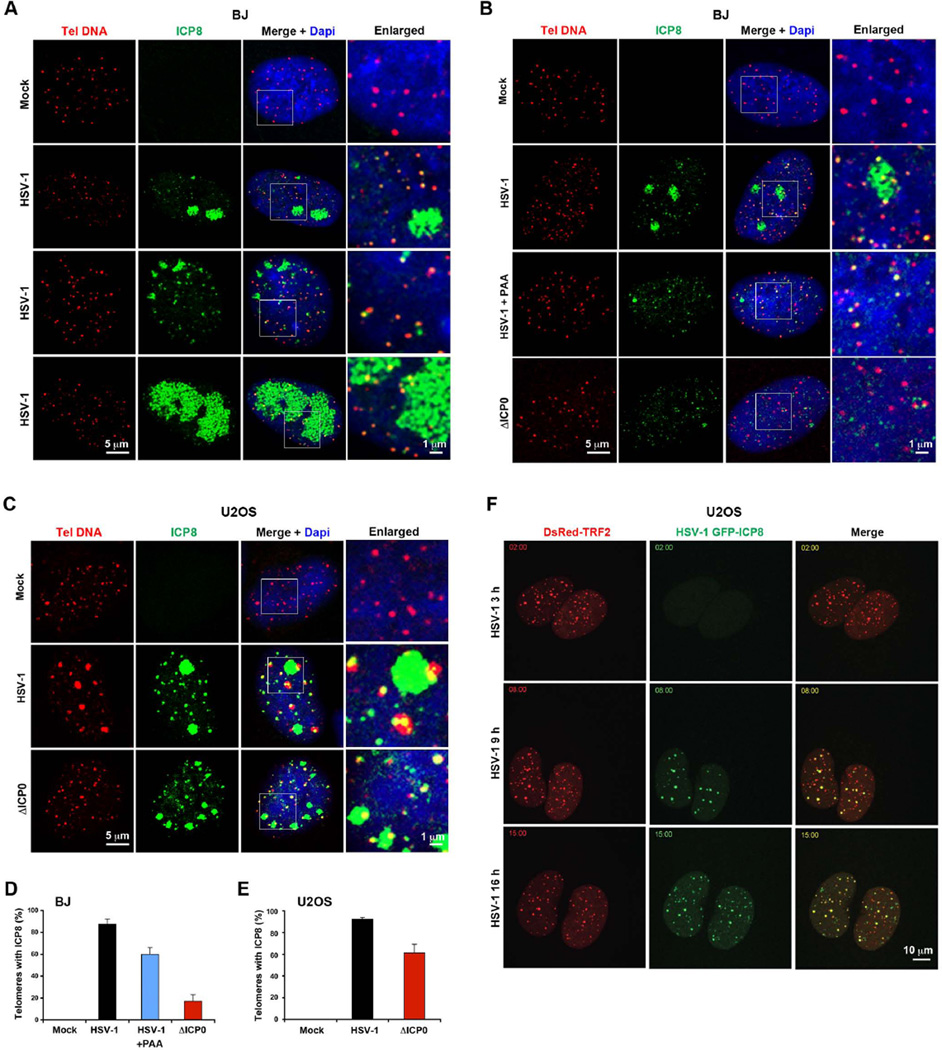
HSV-1 ICP8 is the viral encoded single stranded DNA binding protein that is essential for DNA replication and formation of viral DNA replication compartments. We therefore tested whether ICP8 colocalized with telomere DNA foci during the early stages of HSV-1 replication (6 hrs) (Figure 6 and S7). Confocal imaging of ICP8 foci with telomere DNA revealed a highly significant colocalization of telomere DNA foci (>80%) with ICP8 microfoci (pre-replicative site) in HSV-1 wt infected cells (Figure 6A–E, and S7). We also observed very large ICP8 structures representing viral replication compartments in a subset of infected cells (Figure 6A), and a subset of these contained one or more telomere DNA foci. Treatment of infected cells with PAA prevented the formation of large ICP8 replication compartments, but did not prevent the high frequency of colocalization of telomere DNA with ICP8-associated foci (Figure 6B and D). Colocalization of telomere DNA with ICP8 foci was largely reduced in BJ cells infected with ΔICP0 mutant virus (Figure 6D), whereas there was reduced but still significant colocalization of telomere DNA with ICP8 foci in U2OS cells infected with ΔICP0 mutant virus (Figure 6C and E, and S7A).
Figure 6. ICP8-associated viral replication compartments colocalize with telomeres.
A) Confocal microscopy analysis of BJ cells infected with HSV-1 (MOI=0.1) or mock. The infected cells were assayed by immuno-FISH with CCCTAA PNA probe (red) and antibody to ICP8 (green) at 6 hrs post infection. Enlarged images derived from the white square in DAPI (blue) and merge images are shown to the right. Various ICP8 staining patterns are shown to indicate the different stages of the infected cells. B) The same as in (A), except that BJ cells were infected with HSV-1 wt in the absence or presence of PAA, ΔICP0 or mock were assayed at 6 hrs post-infection. C) Confocal microscopy analysis of U2OS cells infected with wild-type HSV-1, ΔICP0 or mock. The infected cells were assayed by immuno-FISH with CCCTAA PNA probe (red) and antibody to ICP8 (green) at 6 hrs post-infection. D) Quantification of colocalization of telomere DNA foci with ICP8 foci in HSV-1 wt, HSV-1 wt in the presence of PAA, or ΔICP0 viruses infected BJ cells, a representative shown in (B). The bar graph shows the percent of telomere DNA signals colocalizing with ICP8 foci (Mean ± SD) generated from three independent experiments. E) Quantification of colocalization of ICP8 foci and telomere DNA foci in HSV-1 wt, or ΔICP0 infected U2OS cells, a representative shown in (C). F) Live cell imaging analysis of colocalization of TRF2 foci (red) and ICP8 foci (green). U2OS cells expressing DsRed-tagged TRF2 were infected with a recombinant HSV-1 virus encoding ICP8-GFP. The images are snapshots of the cells at 3, 9, or 16 hrs post-infection. See also Figure S7, and movie S1 and S2.
Interestingly, HSV-1 induced large telomere DNA foci in U2OS cells, which may correlate to relaxed chromatin and active transcription (Figure 6C and S7A). These enlarged telomere DNA foci were strongly dependent on ICP0, because these were rarely observed in ΔICP0 virus infected cells (Figure S7A and B). In addition, these enlarged telomere DNA foci colocalized with TRF2 foci, and were not affected by PAA treatment (Figure S7D), suggesting that viral DNA synthesis was not essential for relaxed chromatin and active transcription at telomeres.
To further validate the colocalization of ICP8 with telomere foci, we performed live-cell confocal microscopy with virus expressing ICP8-GFP and U2OS cells transfected with Cherry-TRF1 or DsRed-TRF2 (Movie S1 and S2). Still frames from these movies show that ICP8-GFP foci frequently form at DsRed-TRF2 foci (Figure 6F). Additionally, ICP8 foci were shown to colocalize with endogenous TRF2 foci in U2OS cells infected with HSV-1 (Figure S7C). We observed most telomeric foci (>80%) were co-occupied with ICP8 at early times after infection (Figure 6D). At later times, excess ICP8 foci accumulated at additional sites outside of telomeric foci. These findings indicated that telomeres are frequent sites of ICP8 early foci formation, and suggest that telomeres may be sites of HSV-1 pre-replication compartment assembly.
The colocalization of ICP8 at cellular telomeres was confirmed biochemically in BJ cells by ChIP assay (Figure 7A–C). ICP8 showed a substantial enrichment (~10 fold relative to IgG) at G-rich telomere repeat DNA relative to C-rich telomeric DNA or Alu control regions (Figure 7A and B), suggesting that ICP8 preferentially binds to single-stranded, G-rich telomere repeats. Similarly, ICP8 showed a significant enrichment (0.8% input; >10 fold relative to IgG) at cellular subtelomeric DNA (Figure 7C) for chromosome 10q and XYq, which was similar to its binding on the HSV-1 genome (~0.9% input). Only background binding was detected in ΔICP0 virus infected BJ cells, suggesting that ICP0 promotes ICP8 binding. These findings suggested that ICP8 can bind to host telomere repeat and subtelomeric DNA in an ICP0-facilitated manner, and that some ICP8-associated pre-replication compartments form at or near sites of cellular telomeres.
Figure 7. ICP8 binds to telomeres and contributes to rapid loss of telomere signals.
A) A ChIP assay was performed in BJ cells infected with HSV-1 wt, ΔICP0, or mock at 6 hrs post-infection, using antibodies specific to ICP0, ICP8, or control IgG. ChIP DNA was assayed by dot blotting and hybridization with 32P-labeled probes specific for either TTAGGG (top), CCCTAA (middle), or control Alu regions (bottom). B) Quantification of three independent ChIP assays represented by (A). The bar graphs show mean values (±SD) of the percent input DNA for G-rich telomeric (top), C-rich telomeric (middle) or Alu (bottom) DNA. C) ChIP DNA from (A) was assayed by qPCR using primers specific for the 10q and XYq subtelomeres (10q-1 and XYq-1), HSV-1 terminal repeats (TR-4), or control Actin region. The bar graph shows the average value of percentage of input from three independent ChIP assays (Mean ± SD). D) Northern blot of TERRA RNA in BJ cells infected with either KOS1.1 strain of HSV-1 wt (MOI=1) or ICP8 null mutant HD-2 (MOI=1) at 6 or 16 hrs post infection. TERRA signals were normalized to 18S rRNA signals and mock infection, and are shown at the bottom. E) BJ cells were infected with KOS1.1, HD-2, or mock infected for the indicated times, and in-gel hybridization assay was performed with 32P-labeled (TAACCC)4 probe under native conditions (left), followed by hybridization with the same probe under denaturing conditions (right). F) Western blot of infected BJ cells as shown in (E) using antibodies as indicated. G) Quantification of telomeric DNA signals under denaturing condition, shown at the right panel in (E). The graph shows telomeric signals normalized to the internal band (arrow) and the mock. H) Model depicting how HSV-1 infection leads to an increase in telomere transcription, telomere remodeling, and ICP8 pre-replication site formation.
To determine if ICP8 contributes to telomere dysfunction, we assayed the effects of ICP8 mutant virus (HD-2) on TERRA induction (Figure 7D) and telomere signal loss (Figure 7E–G). We found that an ICP8 mutant virus was indistinguishable from its parental wt virus (KOS1.1) in its ability to induce TERRA (Figure 7D). In contrast, ICP8 mutant virus was severely compromised for its ability to induce the heterogeneity of single stranded telomere DNA or reduce telomere repeat DNA signal as measured by in-gel hybridization analysis (Figure 7E–G). These findings indicate that ICP8 functions at a step subsequent to TERRA induction, and is required for the formation of single-stranded DNA (6 hrs) and the subsequent loss of telomeric DNA signal (16 hrs).
Discussion
Viruses typically modulate and repurpose host processes to enable viral functions. Here, we have found that HSV-1 infection modulates and repurposes telomeric processes to promote viral replication. HSV-1 altered multiple aspects of host cell telomeres, including the transcriptional activation of TERRA, the increase in the amount and heterogeneity of single strand telomeric DNA, the loss of total telomeric DNA signal, the selective degradation of TPP1, the reduction of telomere-bound shelterin, and the accumulation of DNA damage foci at telomeres. We show that shRNA depletion of TPP1 increased HSV-1 DNA replication, and that ectopic expression of TPP1 decreased HSV-1 replication, suggesting that TPP1 functions as a host restriction factor for viral DNA replication. We also found that the HSV-1 ICP8 replication protein associates with telomeric DNA and forms pre-replication like structures that colocalize with telomeric factors. Moreover, virus mutants lacking ICP0 are compromised for TERRA expression, and mutants lacking ICP8 are compromised for telomere DNA single strand formation and signal loss. Taken together, our results indicate that HSV-1 extensively remodels host cell telomeres in an ICP0- and ICP8-dependent manner. We propose that telomere remodeling is required for the formation of ICP8-nucleated pre-replication compartments that promote viral genome replication (Figure 7H).
HSV-1 induced TERRA transcription could be detected by 3 hrs post-infection and was among the earliest telomeric perturbances observed. A virus containing mutations in the ICP0 ring finger domain was incapable of stimulating TERRA transcription and ICP0 partially colocalized with telomeric foci, suggesting that ICP0 plays a role in transcriptional activation at telomeres. HSV-1 infection also caused the ICP0-enhanced and proteasome-dependent degradation of TPP1, a core subunit of shelterin. However, TPP1 degradation occurs at relatively later stages of HSV-1 infection, and shRNA depletion of TPP1 is not sufficient to induce TERRA. Therefore, TERRA transcription is likely to initiate prior to TPP1 degradation. ICP0 has been implicated in modulating several other cellular factors important for transcription regulation, including PML-NB associated Daxx and ATRX, that have been implicated in TERRA regulation(Goldberg et al., 2010; Lewis et al., 2010). However, individual depletion of PML, ATRX, or Daxx did not significantly induce TERRA (Figure S6D and E). This is consistent with a recent finding that ATRX depletion does not increase TERRA expression in human cells (Episkopou et al., 2014). ICP0 can also disrupt the transcriptional repressor REST/COREST (Gu and Roizman, 2009; Zhou et al., 2011) which has been implicated in the repression of quiescent viral genomes, but has not yet been shown to regulate TERRA transcription. ICP0 can also degrade DNA-PKcs (Parkinson et al., 1999), which has been implicated in telomere end protection and TERRA transcription modulation (Le et al., 2013; Pfeiffer and Lingner, 2012). ICP0 also degrades RNF8 and RNF168 (Chaurushiya et al., 2012), two factors important for stability of TPP1 at telomeres and telomere end protection (Okamoto et al., 2013; Peuscher and Jacobs, 2011; Rai et al., 2011). ICP0 also promotes the degradation of IFI16 (Orzalli et al., 2012), which has been associated with senescence (Song et al., 2010). Thus, ICP0 may target multiple cellular factors that can partially regulate TERRA transcription and telomere protection. However, ICP0 by itself failed to induce TERRA to levels observed during HSV-1 infection, even though the exogenous ICP0 can induce PML degradation and localize with telomeres to the similar extent as ICP0 during infection. Taken together, these data indicate that ICP0 provides a potent, ring finger-dependent transcriptional activation function that robustly increases TERRA expression in the context of HSV-1 infection. However, ICP0 by itself is not sufficient for TERRA transcription induction and no single cellular target of ICP0 appears as the key regulator of telomeric transcription.
ICP8 is an essential component of HSV-1 DNA replication machinery and is implicated in the assembly of viral replication compartments. We found that the majority of cellular telomeric foci were colocalized with ICP8 foci during HSV-1 infection. We also found that ICP8 binds to telomere repeat and subtelomeric DNA by ChIP assay, and that virus lacking ICP8 are compromised for formation of single stranded telomeric DNA, and subsequent loss of telomere repeat DNA at late stages. ICP8 has been shown to bind RNA and stabilize R-loops (Boehmer, 2004), consistent with our observation that it associates with telomeres actively transcribing TERRA. TERRA transcripts are known to form R-loops at telomeres suggesting that TERRA transcription may be sufficient to induce a high-affinity single stranded DNA substrate for ICP8 (Balk et al., 2014; Pfeiffer et al., 2013). ICP8 also associates with components of cellular DNA repair and recombination machinery (de Bruyn Kops and Knipe, 1994; Taylor et al., 2003), that are also known to associate with cellular telomeres (Dejardin and Kingston, 2009). ICP8 assembles into filamentous nucleoprotein structures that promote single strand invasion (Tolun et al., 2013), and it is possible that telomeres transcribing TERRA provide a high-affinity single-strand DNA or R-loop template for early phase ICP8 aggregation. ICP8 forms multiple dynamic foci that congregate to form larger replication compartments that ultimately encompass newly replicating viral DNA (Taylor and Knipe, 2009). However, it is also known that ICP8 associated pre-replication sites are heterogeneous, and only a subset may give rise to replication compartment formation (Lukonis et al., 1997; Uprichard and Knipe, 1997). Thus, while most telomeres are colonized as sites of ICP8 foci formation, it is not yet known whether these telomere-associated ICP8 foci are functional for HSV-1 replication.
Telomere remodeling may provide several functional advantages for HSV-1 replication. One potential function of telomere uncapping and single-strand DNA formation may be to catalyze the formation of ICP8 filaments and foci that serve as the pre-replication scaffold. An additional function may be to create sufficient nuclear space for viral replication compartment and viral capsid assembly. HSV-1 has been shown to cause nuclear marginalization of host chromosomal DNA (Simpson-Holley et al., 2004). In addition, HSV-1 induces changes in nuclear architecture, with cellular and viral transcription complexes interacting with nuclear lamina components, including lamin B (Simpson-Holley et al., 2005). Interestingly, nuclear envelope components, including lamin B, have been implicated in telomere regulation and stability (Burtner and Kennedy, 2010; Gonzalez-Suarez and Gonzalo, 2010).
Telomere remodeling by HSV-1 may also provide the virus with access to telomere-associated factors that dampen the host DDR during viral replication. Our results show that core shelterin components dissociated from telomere repeat DNA during HSV-1 infection, and it is possible that they have alternative functions in viral DNA replication, including recruitment or suppression of host DDR. In support of this model, HSV-1 has been shown to exploit cellular factors involved in homologous recombination, such as ATR and MRE11, that are also implicated in telomere regulation (Schumacher et al., 2012). KU80 and RAD50 are telomere-associated proteins that are also found to associate with ICP8 foci (Taylor and Knipe, 2004), suggesting that ICP8 may recruit these telomere-associated proteins to viral replication compartments. In addition, a telomere-specific poly-ADP ribose polymerase, tankyrase, has been shown to colocalize with HSV-1 replication compartments and facilitate viral DNA replication (Li et al., 2012).
Finally, we consider the scenario that telomere uncapping may arise merely as collateral damage of HSV-1 infection, without any direct functional advantage for HSV-1 infection. Given the parsimony of viral interactions and functions, it seems unlikely that HSV-1 would selectively target key chromosomal structures such as telomeres and centromeres for no functional purpose. Based on the high-frequency colocalization of ICP8 with telomere foci, and the assembly of pre-replication compartments at sites of telomeres, we favor a model that HSV-1 selectively targets telomeres as opportune sites of replication compartment assembly (Figure 7H). Such compartments would be enriched for telomeric factors that promote recombination and limit non-homologous end-joining, conditions which may be necessary for successful HSV-1 replication and virion assembly.
Experimental Procedures
Plasmids and Lentiviral Transduction
Lentiviral plasmids expressing YFP tagged ICP0 or ICP0-fxe has been described previously (Newhart et al., 2012). A FLAG-TPP1 expressing plasmid was generated by PCR amplification and cloning into HindIII-BamHI sites of p3XFLAG-CMV (Sigma). pLKO.1 vector-based shRNA construct for TRF2 was obtained from Open Biosystems (TRCN0000004811). shControl and shTPP1 were generated in pLKO.1 vector with target sequence as followings: shCon, TTATCGCGCATATCACGCG; shTPP1-2, GCAGCTGCTTGAGGTACTACA. Lentiviruses were produced by the use of packaging vectors pMDLg/pRRE, RSV-Rev, and CMV-VSVG as described (Deng et al., 2012a). For shRNA depletion experiments, cells were infected twice on consecutive days, treated with 1 µg/ml Puromycin at 48 hrs after the first infection, and harvested or re-plated at 6 days post-infection for further analysis.
Viral Strains and Infection
HSV-1 wt strain 17+, and 17+ strain-derived ICP0-null mutant dl1403, ICP0 ring finger deletion mutant FXE, and its revertant FXER have been described elsewhere (Lukashchuk and Everett, 2010). HSV-1 wt strain KOS1.1, and KOS1.1-derived ICP8-null mutant HD-2 and ICP8-GFP expressing virus HSV8GFP were described previously (Da Costa et al., 1997; Taylor et al., 2003). For Phosphonoacetic acid (PAA) (Sigma) treatment, infection was performed in the presence of PAA (0.4 mg/ml) during the whole procedures of viral incubation and growth. For MG-132 (Sigma) treatment, a final concentration 5 µM of MG-132 was added into culture medium at 2 hrs post infection to allow efficient viral entry.
RNA Preparation and Analysis
Total RNA was purified with Trizol reagent (Life Technologies) and used for Northern blotting analysis as described previously (Deng et al., 2012a). Quantitative RT-PCR experiments were performed using Super Script III Reverse Transcriptase (Invitrogen) and an ABI Prism 7900 Sequence Detection System (Applied Biosystems), essentially as described (Deng et al., 2012b). Relative RT-PCR was determined using ΔΔCT methods relative to control samples and internal control U1 snRNA. Nascent RNA was prepared with Click-iT Nascent RNA capture Kit (Life Technologies).
Immuno-FISH Assay
Indirect immunofluorescence (IF) combined with fluorescence in situ hybridization (FISH) analysis was performed as described (Rai and Chang, 2011) with minor modifications. Immuno-RNA FISH was performed essentially as described previously (Deng et al., 2009b). Images were captured with a 63 × lens on a Leica SP5 II Confocal microscope (Leica Microsystems) using LAS AF software for image processing and quantification.
Live Cell Imaging
Live cell imaging of HSV-1 infection was performed as described (Taylor and Knipe, 2009), with some modifications. Multiple cells with either DsRed-TRF2 or Cherry-TRF1 foci were positioned for live cell imaging to visualize the colocalization of ICP8-GFP with TRFs starting at around 1 h post-infection. The phase and fluorescent images were captured at 5 min or 10 min intervals for cells transfected with DsRed-TRF2 or Cherry-TRF1, respectively, over a period of ~16 hrs.
Viral Copy Number and Plaque Assay
BJ cells depleted for TRF2, TPP1, or Control were infected with HSV-1 at an MOI of 0.1. For quantification of viral genome copy number, genomic DNA isolated from infected cells at time 0 and 16 or 24 hrs post-infection was assayed by qPCR using primers specific for either HSV-1 ICP0 promoter region or VP16 promoter region. The relative numbers of viral genomes at 16 hrs post-infection was determined using ΔΔCT relative to time 0 samples and an actin control. For plaque assay, cell culture supernatants from infected BJ cells at time 0, 8, or 24 hrs post-infection were used to infect Vero cells in 24 – well plates for progeny virus titers. After virus adsorption for 1 h, the cells were overlaid with medium containing 0.5% carboxymethylcellulose. Plaques were stained with crystal violet at 3 days post-infection. For viral copy number analysis in TPP1-expressing cells, U2OS cells were first transfected with increasing amount of vectors expressing Flag-TPP1 or control vector. After 20 hrs post-transfection, cells were infected with HSV-1 at an MOI of 0.1 and assayed by qPCR using primers specific for either VP16 promoter region or control actin region at time 0 and 16hrs post-infection.
Supplementary Material
Acknowledgements
We thank Dr. Roger Everett for kindly providing antibodies, cell lines, and viral strains. We also thank Fred Keeney and James Hayden at the Wistar Microscopy Core for imaging analysis, Andreas Wiedmer for technical assistance, and Caroline Lilley for comments on the manuscript. This work was supported by an American Heart Association grant to ZD (11SDG5330017), and NIH grants to PML (RO1CA140652), to MDW (RO1NS082240), and to DMK (AI063106). This work was also supported by the Wistar Cancer Center core grant (P30 CA10815) and the Commonwealth Universal Research Enhancement Program, PA Department of Health. ZD, ETK, MDW, and PL designed the experiments for the project. AN and SJ carried out experiments for lentiviral expression of YFP-ICP0s, DL and JM5 for VZV infection, JM2 and SH for influenza virus infection, ZD, OV, ETK, JD, and ZW performed other experiments and generated data for the figures. ZD, ETK, NWF, DMK, MDW and PL analyzed and interpreted data, assembled the figures and wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nature structural & molecular biology. 2012;19:948–956. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends in cell biology. 2014 doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Balk B, Dees M, Bender K, Luke B. The differential processing of telomeres in response to increased telomeric transcription and RNA-DNA hybrid accumulation. RNA biology. 2014;11:95–100. doi: 10.4161/rna.27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer PE. RNA binding and R-loop formation by the herpes simplex virus type-1 single-stranded DNA-binding protein (ICP8) Nucleic acids research. 2004;32:4576–4584. doi: 10.1093/nar/gkh797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. The Journal of general virology. 2013;94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- Burtner CR, Kennedy BK. Progeria syndromes and ageing: what is the connection? Nature reviews. Molecular cell biology. 2010;11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- Chaurushiya MS, Lilley CE, Aslanian A, Meisenhelder J, Scott DC, Landry S, Ticau S, Boutell C, Yates JR, 3rd, Schulman BA, et al. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Molecular cell. 2012;46:79–90. doi: 10.1016/j.molcel.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa XJ, Bourne N, Stanberry LR, Knipe DM. Construction and characterization of a replication-defective herpes simplex virus 2 ICP8 mutant strain and its use in immunization studies in a guinea pig model of genital disease. Virology. 1997;232:1–12. doi: 10.1006/viro.1997.8564. [DOI] [PubMed] [Google Scholar]
- de Bruyn Kops A, Knipe DM. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. Journal of virology. 1994;68:3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes & development. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- de Lange T. Telomere biology and DNA repair: enemies with benefits. FEBS letters. 2010;584:3673–3674. doi: 10.1016/j.febslet.2010.07.030. [DOI] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nature reviews. Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Wang Z, Stong N, Plasschaert R, Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei MS, et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. The EMBO journal. 2012a;31:4165–4178. doi: 10.1038/emboj.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Wang Z, Stong N, Plasschaert R, Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei MS, et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. The EMBO journal. 2012b doi: 10.1038/emboj.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Episkopou H, Draskovic I, Van Beneden A, Tilman G, Mattiussi M, Gobin M, Arnoult N, Londono-Vallejo A, Decottignies A. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic acids research. 2014 doi: 10.1093/nar/gku114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. Journal of virology. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczy MW, Ranayhossaini DJ, Deluca NA. Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. Journal of virology. 2011;85:4993–5002. doi: 10.1128/JVI.02265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez I, Gonzalo S. Nurturing the genome: A-type lamins preserve genomic stability. Nucleus. 2010;1:129–135. doi: 10.4161/nucl.1.2.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. Journal of virology. 2009;83:181–187. doi: 10.1128/JVI.01940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Tanaka M, Yokoymama A, Matsuda G, Kato K, Kagawa H, Hirai K, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1877–1882. doi: 10.1073/pnas.041592598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nature reviews. Microbiology. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- Le PN, Maranon DG, Altina NH, Battaglia CL, Bailey SM. TERRA, hnRNP A1, and DNA-PKcs Interactions at Human Telomeres. Frontiers in oncology. 2013;3:91. doi: 10.3389/fonc.2013.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yamauchi Y, Kamakura M, Murayama T, Goshima F, Kimura H, Nishiyama Y. Herpes simplex virus requires poly(ADP-ribose) polymerase activity for efficient replication and induces extracellular signal-related kinase-dependent phosphorylation and ICP0-dependent nuclear localization of tankyrase 1. Journal of virology. 2012;86:492–503. doi: 10.1128/JVI.05897-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Everett RD, Weitzman MD. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS pathogens. 2011;7:e1002084. doi: 10.1371/journal.ppat.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. The EMBO journal. 2010;29:943–955. doi: 10.1038/emboj.2009.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte P, Sullivan KF, Everett RD. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. The Journal of biological chemistry. 2001;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
- Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. Journal of virology. 2010;84:4026–4040. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukonis CJ, Burkham J, Weller SK. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. Journal of virology. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukonis CJ, Weller SK. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. Journal of virology. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- Muylaert I, Tang KW, Elias P. Replication and recombination of herpes simplex virus DNA. The Journal of biological chemistry. 2011;286:15619–15624. doi: 10.1074/jbc.R111.233981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhart A, Rafalska-Metcalf IU, Yang T, Negorev DG, Janicki SM. Singlecell analysis of Daxx and ATRX-dependent transcriptional repression. Journal of cell science. 2012;125:5489–5501. doi: 10.1242/jcs.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic acids research. 2009;37:1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill FJ, Rapp F. Early events required for induction of chromosome abnormalities in human cells by herpes simplex virus. Virology. 1971;44:544–553. doi: 10.1016/0042-6822(71)90368-0. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. Journal of virology. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuscher MH, Jacobs JJ. DNA-damage response and repair activities at uncapped telomeres depend on RNF8. Nature cell biology. 2011;13:1139–1145. doi: 10.1038/ncb2326. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. The EMBO journal. 2013;32:2861–2871. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Lingner J. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS genetics. 2012;8:e1002747. doi: 10.1371/journal.pgen.1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- Rai R, Chang S. Probing the telomere damage response. Methods Mol Biol. 2011;735:145–150. doi: 10.1007/978-1-61779-092-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Li JM, Zheng H, Lok GT, Deng Y, Huen MS, Chen J, Jin J, Chang S. The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection. Nature structural & molecular biology. 2011;18:1400–1407. doi: 10.1038/nsmb.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Whitley RJ. An inquiry into the molecular basis of HSV latency and reactivation. Annual review of microbiology. 2013;67:355–374. doi: 10.1146/annurev-micro-092412-155654. [DOI] [PubMed] [Google Scholar]
- Schumacher AJ, Mohni KN, Kan Y, Hendrickson EA, Stark JM, Weller SK. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS pathogens. 2012;8:e1002862. doi: 10.1371/journal.ppat.1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M, Baines J, Roller R, Knipe DM. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. Journal of virology. 2004;78:5591–5600. doi: 10.1128/JVI.78.11.5591-5600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M, Colgrove RC, Nalepa G, Harper JW, Knipe DM. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. Journal of virology. 2005;79:12840–12851. doi: 10.1128/JVI.79.20.12840-12851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song LL, Ponomareva L, Shen H, Duan X, Alimirah F, Choubey D. Interferon-inducible IFI16, a negative regulator of cell growth, down-regulates expression of human telomerase reverse transcriptase (hTERT) gene. PloS one. 2010;5:e8569. doi: 10.1371/journal.pone.0008569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Gasser SM. The function of nuclear architecture: a genetic approach. Annual review of genetics. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. Journal of virology. 2004;78:5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TJ, Knipe DM. The use of green fluorescent fusion proteins to monitor herpes simplex virus replication. Methods Mol Biol. 2009;515:239–248. doi: 10.1007/978-1-59745-559-6_16. [DOI] [PubMed] [Google Scholar]
- Taylor TJ, McNamee EE, Day C, Knipe DM. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology. 2003;309:232–247. doi: 10.1016/s0042-6822(03)00107-7. [DOI] [PubMed] [Google Scholar]
- Tolun G, Makhov AM, Ludtke SJ, Griffith JD. Details of ssDNA annealing revealed by an HSV-1 ICP8-ssDNA binary complex. Nucleic acids research. 2013;41:5927–5937. doi: 10.1093/nar/gkt266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprichard SL, Knipe DM. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- Weller SK, Coen DM. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harbor perspectives in biology. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. The Journal of biological chemistry. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- Ye J, Wu Y, Gilson E. Dynamics of telomeric chromatin at the crossroads of aging and cancer. Essays in biochemistry. 2010;48:147–164. doi: 10.1042/bse0480147. [DOI] [PubMed] [Google Scholar]
- Zhou G, Te D, Roizman B. The CoREST/REST repressor is both necessary and inimical for expression of herpes simplex virus genes. mBio. 2011;2:e00313–e00310. doi: 10.1128/mBio.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.