Abstract
BiP is the endoplasmic reticulum (ER) orthologue of the Hsp70 family of molecular chaperones and is intricately involved in most functions of this organelle through its interactions with a variety of substrates and regulatory proteins. Like all Hsp70 family members, the ability of BiP to bind and release unfolded proteins is tightly regulated by a cycle of ATP binding, hydrolysis, and nucleotide exchange. As a characteristic of the Hsp70 family, multiple DnaJ-like co-factors exist that can target substrates to BiP and stimulate its ATPase activity to stabilize the binding of BiP to substrates. However, only in the past decade have nucleotide exchange factors (NEFs) for BiP been identified, which has shed light not only on the mechanism of BiP assisted folding in the ER but also on Hsp70 family members that reside throughout the cell. We will review the current understanding of the ATPase cycle of BiP in the unique environment of the ER and how it is regulated by the NEFs, Grp170 and Sil1, both of which perform unanticipated roles in various biological functions and disease states.
Keywords: BiP/Grp170/Sil1, protein folding, Nucleotide exchange factors, Hsp70 ATPase, Endoplasmic reticulum
1. Protein Folding and Quality Control in the ER
Approximately one-third of the human genome encodes proteins that reside at the cell surface, are secreted, or populate organelles of the secretory pathway. These proteins are synthesized at the endoplasmic reticulum (ER) membrane and are translocated into the lumen where they acquire their functional tertiary or quaternary structure. The folding of these proteins and their assembly into larger heteromeric complexes is guided by the same principles and processes used throughout the cell, but is further complicated by the addition of large branched oligosaccharide moieties to nascent chains entering the ER, high ER concentrations of calcium, and an oxidizing environment combined with systems that catalyze the formation of intra- and inter-chain disulfide bonds. As a result, tight quality control measures have evolved to monitor the success of secretory pathway protein maturation, a process known as ER quality control (ERQC), which is conserved in most eukaryotic organisms (Fig. 1a and b). If a newly synthesized protein folds properly and passes the scrutiny of the ERQC machinery, it can undergo vesicular-mediated transport through the organelles of the secretory pathway (Fig. 1c), but if the protein fails this inspection it must be targeted for proteasomal degradation in the cytosol via a process known as ER associated degradation (ERAD) (Fig. 1d). A tremendous number of studies have led to the identification of many of the proteins involved in these two outcomes and have provided a detailed understanding of their underlying mechanisms. At the heart of this triage decision are two major chaperone systems; the lectins calnexin/calreticulin (CNX/CRT) (Fig. 1a), which are unique to the ER, and the Hsp70 system (Fig. 1b), which has many aspects common to all Hsp70s. The immunoglobulin heavy-chain binding protein (BiP) is the only conventional Hsp70 chaperone in the ER, but a related large Hsp70 protein with chaperone activity, Grp170, is also present. Unlike the lectins, which monitor both N-linked glycans and unfolded regions on nascent polypeptide chains1, BiP detects only the latter and is the major system used for nonglycosylated proteins or for some glycoproteins in which the most N-terminal glycan occurs relatively late in the linear sequence2. The ability of BiP to bind and release unfolded protein substrates is tightly regulated by a cycle of ATP binding, hydrolysis, and nucleotide exchange, which is controlled by a number of co-factors. They include seven ER-localized DnaJ-like proteins (ERdjs), several of which bind directly to unfolded proteins and transfer them to BiP before stimulating its ATPase activity3; 4, and two nucleotide exchange factors (NEFs) that trigger the release of bound substrates5; 6. Recent data suggest the ERdj co-chaperones play distinct roles in regulating BiP’s involvement in functions that lead to disparate fates for the bound client. These insights have extended our understanding of how BiP’s ATPase cycle is regulated, which together with the NEFs Grp170 and Sil1 is the focus of this review.
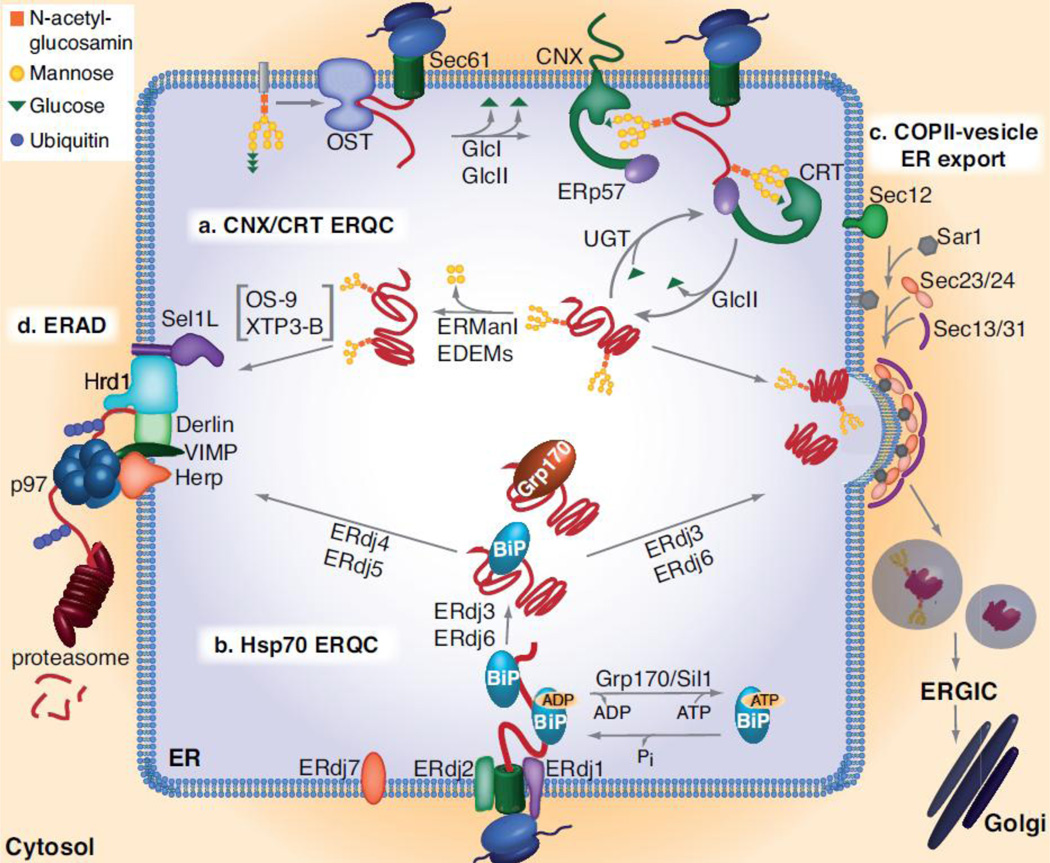
Fig. 1. The ER Quality Control (ERQC) Machinery.
Two main chaperone systems, the lectins CNX/CRT (a) and the Hsp70 chaperone BiP (b), aid the folding proteins for secretion (c) or if folding fails, target them for ERAD (d). (a) The oligosaccharyl transfer (OST) complex attaches a core oligosaccharide from a dolichol donor to the Asn of the Asn-X-Ser/Thr motif on nascent proteins during their translocation into the ER. GlcI and II remove the outer two glucose residues of the oligosaccharide, allowing the remaining glucose to be recognized by CNX/CRT. CNX/CRT assists protein folding in concert with further co-chaperones such as the protein-disulfide isomerase ERp57. Proteins exit the CNX/CRT cycle once the last glucose residue is removed by GlcII. If folded properly, the protein is released from the lectin chaperone cycle and is transported further along the secretory pathway. Incompletely folded intermediates can re-enter the CNX/CRT cycle if a single glucose is re-attached by the folding sensor UGT. If folding ultimately fails, proteins are further trimmed by ERManI and/or an EDEM resulting in removal of 4 mannose residues and recognition by OS-9 and XTP3-B, which then transfer the trimmed glycoprotein to the ERAD machinery for disposal. (b) The Hsp70 chaperone BiP binds hydrophobic patches exposed on nascent or incompletely folded proteins that are often non-glycosylated. BiP possesses low substrate binding affinity in the ATP-bound state and high affinity upon hydrolysis of ATP to ADP. Grp170 and Sil1 facilitate substrate release from BiP by stimulating the release of ADP and allowing ATP to rebind and open the lid on the substrate binding domain. Seven ERdj co-factors have been identified that interact with BiP via their J-domain and assist BiP in its functions during protein translocation (ERdj2), protein folding (ERdj3 and 6) and ERAD (ERdj4 and 5). The functions of ERdj1 and ERdj7 are not well understood, nor is the role of the large Hsp70, Grp170, that also binds to some incompletely folded BiP client proteins. (c) Once the threshold of folding set by the ERQC is met, proteins exit the ER in COPII-coated vesicles, a process that is initiated by Sec12 and driven by a GTPase, Sar1, and four major coat proteins, Sec23, Sec24, Sec13 and Sec31. (d) Once proteins that are clients of either chaperone system are delivered to the ERAD machinery, their retrotranslocation into the cytosol is facilitated by a complex of several transmembrane proteins including Sel1, Derlins, VIMP, Herp and Hrd1, which connect the machinery in the ER lumen to the protein ubiquitination machinery in the cytosol, allowing the ERAD client to be recognized by the p97 hexameric ATPase in the cytosol that provides the energy for extracting a protein from the ER for degradation by the 26S proteasome.
2. The ATPase cycle of BiP in the ER environment
a. A resting state of BiP
Changes in the external environment or different developmental stages of a cell can result in large variations in the load of unfolded or misfolded proteins in the ER. BiP as one of the major ER chaperones must therefore be readily and rapidly available in times of need. Differentially modified and assembled forms of BiP seem to be present in the ER to cope with altering cellular conditions. In the absence of stress, BiP has been shown to be a major ADP-ribosylated protein in mammalian cells7 and is thought to be phosphorylated8; 9. However, whenever the load of unfolded proteins increases in the ER, the amount of modified BiP decreases7; 9–11. Accordingly, it has been suggested that the ADP-ribosylated or phosphorylated forms of BiP represent a pool of inactive oligomeric molecules that can quickly be reactivated when needed7; 12; 13 (Fig. 2a). In agreement with this idea, only monomeric, unmodified BiP is bound to substrates9; 13. Despite this and other14–16 circumstantial evidence suggesting that BiP’s activity in vivo might be affected by post-translational modifications, it remains unclear how these modifications are controlled in response to physiological cues. Although neither the kinase nor ADP-ribosyltransferase has been identified, recently the ADP-ribosylation site of BiP was mapped to two Arg residues within its substrate binding domain (SBD)11. Modification of these residues would interfere with substrate binding and thus explain earlier observations that only unmodified BiP is bound to substrates. Substrates and ATP can induce dissociation of BiP oligomers15; 17, thereby reshuffling BiP into its functional ATPase cycle in the ER. As mutation of these two Arg residues abolished all post-translational modifications of BiP, the role of phosphorylation has been questioned11. Very recently two groups reported that the NBD of drosophila18 and human19 BiP can be modified by AMPylation. Both studies found that the enzymes responsible for the modification, dFIC and HYPE respectively, were induced by ER stress, although the effect of this modification on BiP activity is presently inconclusive. Importantly, possible effects of the various modifications on the interaction of BiP with ERdj co-factors and NEFs have not been determined.
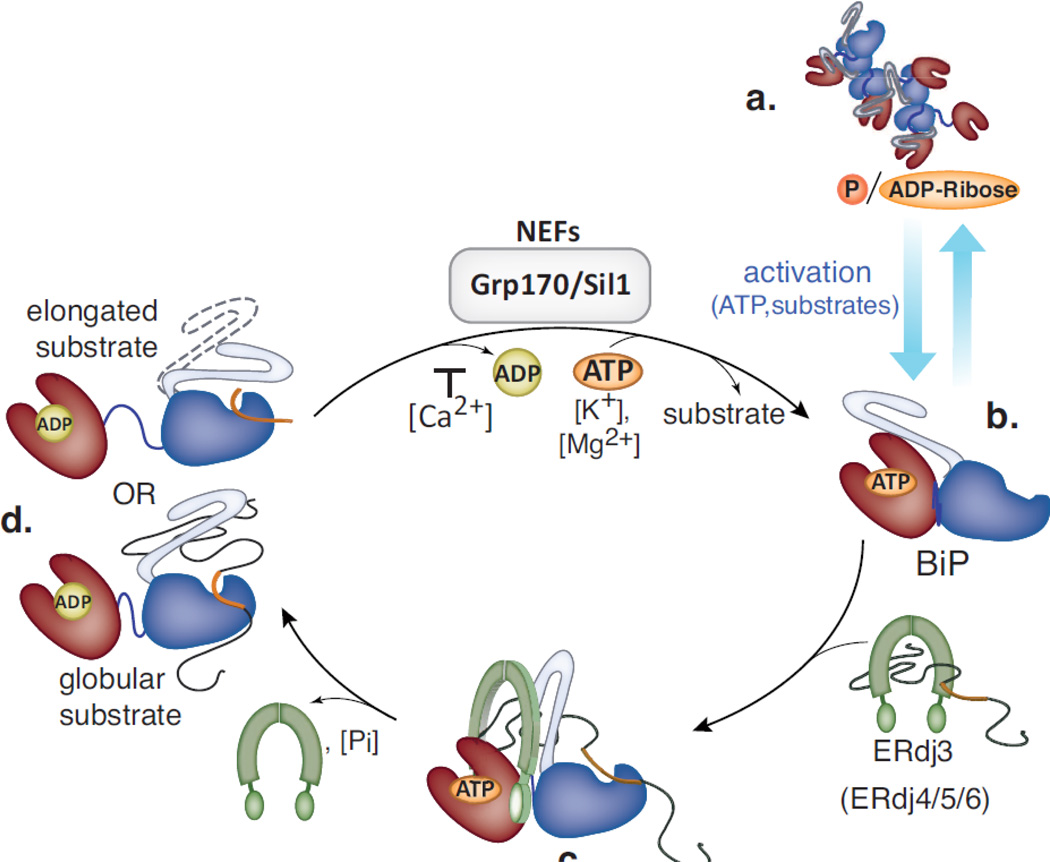
Fig. 2. The ATPase cycle of BiP in the ER.
(a) In the absence of high substrate loads, BiP exists in a multimeric form that is post-translationally modified (ADP-ribosylated and perhaps phosphorylated), which renders the protein inactive. When the demand for BiP increases, the modifications are removed allowing a readily accessable pool of BiP to be reactivated. (b). Once BiP binds potassium and ATP, its NBD (red) and its SBD (blue) come into close proximity to each other and the lid of the SBD (grey) opens, which in combination results in a form that binds substrates with low affinity. (c) Substrates can be introduced into the BiP cycle via their initial binding to DnaJ-like co-chaperones such as ERdj3 (green), which transfers substrate to BiP and increases BiP’s ATPase activity thereby locking the substrate onto BiP. Note that the binding sites within the substrate for ERdj3 and BiP (shown in orange) are probably not identical allowing a transient 3-way complex, which has been detected both in vivo196 and in vitro48. (d) After the magnesium-dependent hydrolysis of ATP, BiP enters a state with low on and off rates for substrates. For elongated/peptide substrates the lid closes over the bound substrate, whereas for globular substrates direct interactions between the lid and the substrate exist but the lid may not close completely. The SBD and NBD become more distant upon substrate binding and ATP hydrolysis, which is less pronounced for globular substrates. To release the substrate and make BiP available for another round of client binding, ADP has to be exchanged against ATP. Calcium increases the affinity for ADP, whereas the NEFs Grp170 and Sil1 facilitate the nucleotide exchange reaction.
b. The ATP-bound state of BiP
Like all Hsp70s BiP is an ATP-dependent molecular chaperone20–23. A large variety of BiP mutants, recent single molecule studies, and a plethora of data on other Hsp70 family members now allow us to describe the ATPase cycle of BiP in quite some detail (Fig. 2). Once ATP binds, BiP enters a state of low substrate affinity with high on and off rates for substrate binding23; 24. A complex allosteric mechanism transmits information on the nucleotide state from the nucleotide binding domain (NBD) to the SBD and on the substrate occupancy state from the SBD back to the NBD25. This reciprocal information transfer is best understood for the E. coli Hsp70 DnaK26–29. ATP binding leads to a closure of the otherwise quite dynamic NBD around the nucleotide30–35. The ATP binding status is then transmitted to the SBD and leads to an opening of the substrate binding cavity by increasing SBD flexibility and lid opening35; 36. It is still not entirely clear, exactly how the nucleotide binding state is transmitted from the NBD to the SBD. Different signal transmission pathways seem to exist between these two domains in the Hsp70 molecule, and details of these pathways may vary for different Hsp70 family members26–29; 37; 38. However, three different themes have emerged for how interdomain communication occurs. In DnaK bound nucleotide is sensed by residues in the NBD that lie in close proximity to the bound ATP, and this information is thought to be transmitted via a Pro residue, which can likely undergo cis/trans isomerization, to an interface at the surface of the NBD that interacts with the SBD39. Interestingly, all residues of the proposed sensor-relay system are conserved in BiP30; 31. However, the impact of mutations of the corresponding residues has not been investigated in BiP. In addition to this putative Pro-focused sensor-relay system, Thr37 in the NBD plays a particularly important role as a nucleotide sensor in BiP40, likely due to a direct interaction of its hydroxyl group with the γ-phosphate oxygen of the bound ATP41. How Thr37 signals to the surface of the NBD is not currently known, but it has been shown that once ATP binding is transmitted to the NBD surface, it influences the interaction of positively charged residues on the NBD with negatively charged residues on the opposing surface of the SBD39; 42. The third known relay that signals nucleotide binding from the NBD to the SBD occurs through the conserved hydrophobic linker, which connects the two domains. Upon ATP binding, the linker binds to a cleft in the NBD, which is important in transmitting the nucleotide state of the NBD to the SBD and increases ATP hydrolysis of the NBD once bound to the cleft35; 36; 43; 44. Although some details of the NBD-SBD interface and regulation by further co-factors vary for different Hsp70s36; 45; 46, the combination of these conserved allosteric signaling pathways, which are most likely interconnected, results in a compaction of the NBD and SBD domains onto each other and an opening of the SBD lid upon ATP binding27; 37; 47; 48 (Fig. 2b). Of note, ATP hydrolysis itself is not necessary for the conformational changes leading to the opening of the SBD, but binding of ATP in combination with potassium ions is sufficient for these changes40; 49; 50. Very important contributions towards understanding the nucleotide-induced conformational changes in Hsp70 molecules came from two recent crystal structures of the ATP-bound form of DnaK51; 52. Both studies show that upon ATP binding, the lid of the SBD docks onto lobe I of the NBD. In addition, the SBD itself docks onto an interface made up of lobe I and II of the NBD, opening up a small crevice at the base of the two lobes where the NBD-SBD linker binds. These binding events are transmitted to the SBD where the substrate binding channel opens up and together with increased flexibility51 or even complete opening of the outer loops of the SBD52 allows substrate release to occur upon ATP binding53.
c. ATP hydrolysis and the ADP state
Like most Hsp70 proteins, BiP has a very low intrinsic Mg2+-dependent ATPase activity23; 54. Once ATP is hydrolyzed, the SBD-NBD interface is broken and both domains behave more independently of each other. The degree of domain separation in the ADP state seems to vary for different Hsp70s. In DnaK the SBD and NBD act independently when in the ADP state with only transient contacts between these two domains36; 47; 55; 56. Conversely, in the case of a bovine Hsc70, extensive contacts between the SBD and the NBD are detected even in the ADP state46, whereas single molecule measurements on mitochondrial Hsp7047 and BiP48 argue for a heterogeneous ensemble of conformations. Taken together, in the ADP state, the NBD and SBD seem to be in a dynamic distance distribution with a general trend towards domain separation. Finally, not only the distance between the SBD and NBD appears to vary in the ADP bound state of Hsp70s, but in the case of DnaK, mitochondrial Hsp70, and BiP the lid also likely populates open and closed conformations with a general trend towards more closed conformations47; 48; 57. Thus, in contrast to the ATP state characterized by close NBD-SBD contact and an open lid, the ADP state seems to be more heterogeneous within a single type of Hsp70 as well as more diverse between individual Hsp70s.
d. Substrates in the ATPase cycle of BiP
The binding of substrates has a very interesting impact on the ATPase cycle of Hsp70 proteins. It has long been appreciated that binding of peptide substrates accelerates the ATP hydrolysis rate of BiP40; 58 and other Hsp70s26–29; 37; 38. Most studies have been performed with small hydrophobic peptides that were thought to be good substrate mimics. In the case of BiP, doubt has been cast on the significance of these results, since although short peptides do stimulate BiP’s ATPase activity, a completely unfolded BiP-binding protein did not54. Indeed, the only known sources of small peptides in the ER are those that enter through TAP transporters to be delivered to MHC I molecules59 and signal peptides that may never fully enter the ER lumen, neither of which have been shown to interact with BiP. Thus, it is more likely that the substrates BiP encounters are elongated polypeptide chains that are in the process of translocation via the Sec61 channel, proteins that are partially folded, and misfolded or even aggregated proteins. In addition, BiP binds to native forms of Sec61 to gate the translocon60; 61 and the UPR transducers to maintain them in an inactive form62; 63. Presumably this involves unfolded or unstructured loops or regions in these proteins that allow them to interact with BiP in a regulated manner. Recent work has shown that in the case of a more globular substrate, in contrast to what has been observed for peptides, complete lid closing over the bound substrate does not necessarily occur48; 57. In this case, direct interactions between the lid and the substrate were observed, leading to even further extension of the conformational space available within the Hsp70s’ SBD. This direct lid-substrate interaction, as well as the recently described interaction between the unstructured C-terminus of Hsp70 chaperones and their substrates64, might aid in conformational remodeling of Hsp70 clients. The ATP-induced opening of the lid51; 52 and the inherent dynamics observed for the lid48; 57 might also have an impact on substrate conformations if lid-substrate interactions occur. Of note, when a peptide binds to the ATP state of DnaK it shifts the structure of this chaperone more towards the ADP state36, whereas binding of a larger polypeptide substrate to the ADP state of BiP induces a conformation that more closely resembles the ATP state in terms of NBD-SBD domain separation and lid opening48 (Fig. 2d). Thus, the substrate itself seems to shift the conformational state of the Hsp70s, playing an active role in the allosteric signaling mechanism. As substrates apparently shift Hsp70s structurally away from the pure nucleotide-regulated states, they might facilitate transitions between them. Important insights into the structural transitions within the Hsp70 DnaK upon nucleotide and substrate binding have recently been obtained by a comprehensive NMR study65. An intermediate between the fully undocked NBD-SBD conformation (ADP-bound state) and the completely docked conformation (ATP-bound state) of DnaK was identified, which showed disruption of the interactions between the NBD and SBD while the binding of the linker to the NBD was retained. Of note, substrate was shown to induce this intermediate, revealing how substrates can directly influence the conformational transitions within an Hsp70 molecule. This highlights the energetic competition between the different possible states of Hsp70s, which vary from Hsp70 member to Hsp7036; 46–48; 55; 56; 65 member and allow extrinsic and intrinsic functional fine-tuning of the ATPase cycle.
In the cell, substrate transfer to Hsp70s is regulated by DnaJ-cochaperones that can further increase the ATPase rate of the Hsp70 molecule and thereby lock a substrate on the Hsp70 molecule in its ADP state28; 66; 67. Thus, in the ER ERdj proteins can help define where substrates are bound by BiP, how fast substrates are bound, and even in the selection of which substrates are bound68. Indeed, some ERdjs directly bind to substrates and deliver them to the Hsp70 molecule4; 68 (Fig. 2c). Mechanistically, they might shift the Hsp70 molecule towards a pre-hydrolysis conformation, perhaps by destabilizing the aforementioned charged SBD-NBD-interface, thus facilitating ATP-hydrolysis. Alternatively, they might either increase interaction of the hydrophobic linker with the NBD or act as a linker mimetic themselves thereby accelerating ATP hydrolysis42; 46; 69. Conflicting models exist for the binding of DnaJ-cochaperones and the detailed molecular mechanism of their action is still unclear36; 45; 46; 69 and may vary between different Hsp70s and their DnaJ co-factors. For BiP, it has been shown that ERdj3, a major co-chaperone in de novo protein folding in the ER4, opens up the lid of the SBD directly thereby facilitating binding of a substrate protein48. Conversely, ERdj3-induced lid opening induced release of small bound peptides, underscoring the difference between peptides and proteins as Hsp70 substrates48. This shows that an ERdj protein can directly alter the lid conformation of BiP to poise it for binding to more globular folding intermediates states, which are likely to be encountered in the lumen of the ER.
e. Re-entering the ATPase cycle
To release a bound substrate and enter a new round in the ATPase cycle, BiP has to be refueled with ATP. The proper functioning of the BiP ATPase cycle is crucial for substrate maturation in the ER70; 71 and the integrity of the entire organelle72; 73. It might thus come as no small surprise that the conditions of the ER are far from optimal for an efficient ATPase cycle of BiP. The optimal pH for BiP’s ATPase activity is quite acidic, and at a more physiological ER pH of around seven, the intrinsic ATPase of BiP is particularly slow23. Even more striking, the presence of calcium, which is present at high concentrations in the ER, almost completely inhibits the ATPase activity of BiP in vitro23; 74. Calcium has been shown to increase the affinity of BiP for ADP almost 1000-fold, thereby inhibiting nucleotide exchange and accordingly BiP´s ATPase activity75 (Fig. 2). In contrast to an in vitro setting, where hydrolysis is the rate-limiting step in the ATPase cycle of BiP54, nucleotide exchange is thus most likely rate-limiting in the ER. Calcium might even poise BiP to enter an inactive phosphorylated state10. Indeed, substrates are not readily released from BiP in vivo, suggesting that BiP does not seem to continuously traverse through its ATPase cycle76. Thus, the chemical environment of the ER, the need for regulated substrate release, and maybe even the reactivation of BiP from its resting state renders another class of accessory proteins for BiP completely indispensable: the NEFs, Grp170 and Sil1. NEFs release ADP allowing ATP to rebind (Fig. 2), thereby decisively influencing how fast substrates are released from BiP, where release in the ER occurs, and maybe even in which folding state substrates are released from BiP.
3. Regulating the ATPase cycle of BiP in the ER environment by Sil1
a. The identification of a nucleotide exchange factor for the ER lumenal Hsp70
Although BiP’s ATPase cycle would appear to be particularly dependent on co-factors to regulate its binding and release from substrates, no resident ER proteins that possessed nucleotide exchange activity had been identified in any organism until the late 1990s (see Fig. 3 for an overview of cytosolic and ER lumenal Hsp70 NEFs; cytosolic Hsp70 NEFs are reviewed in77). Then several labs independently identified the first BiP nucleotide exchange factor in three different organisms. Sls1p was identified in a synthetic lethal screen to identify genes that interacted with the SCR2-encoded 7S RNA component of SRP in the yeast Yarrowia lipolytica78; 79. It was induced by both heat shock and ER stress and was shown to be a lumenal ER protein that interacted with both Kar2p (yeast BiP) and Sec61, a component of the translocon. Deletion of SLS1 dramatically decreased the synthesis of secretory pathway proteins leading to the hypothesis that Sls1p played a role in the translocation of proteins into the ER lumen. Subsequently, an Sls1p homologue was identified in Saccharomyces cerevisiae and shown to bind preferentially to the ADP bound form of yeast BiP. It stimulated BiP’s ATPase activity in the presence of a DnaJ-like co-factor, Sec63p, by accelerating ADP release, thus making Sls1p the first NEF to be identified for BiP80–82. Independently, studies were being conducted to determine the function of Lumenal Hsp Seventy (Lhs1p), an Hsp70-related chaperone found in the yeast ER that is the homologue of mammalian Grp17083. S. cerevisiae strains that were null for Lhs1p were viable but had a constitutively induced UPR. Blocking UPR activation by constructing a Δlhs1Δire1 mutant resulted in a severe growth defect. A multi-copy suppressor screen was conducted to identify genes whose over-expression would silence the severe growth arrest observed in the IRE1, LHS1 double deleted strains (SIL1)83. This led to the identification of SIL1, which was identical to SLS1. The Δlhs1Δsil1 double mutation was lethal, leading the authors to suggest that both Sil1p and Lhs1p might play a similar role in regulating Kar2p’s activity83. Meanwhile the ATPase domain of human BiP was used as bait in a 2-hybrid screen of a human liver cDNA library to identify proteins that might regulate its activity5. This approach probably would not have succeeded except for a bit of serendipity. Although the bait vector was constructed with both a wild-type and a mutant BiP ATPase domain, the screen was conducted first with the mutant ATPase domain. The BiP associated protein, BAP, was identified and demonstrated to bind preferentially to a variety of BiP ATPase mutants in the mammalian ER. When the wild-type BiP ATPase domain was used as bait, it failed to produce yeast colonies when co-expressed with BAP, suggesting that BAP preferred a particular conformation or nucleotide bound state of BiP. Similar to studies with Sls1p80, BAP was found to interact preferentially with the ADP bound form of BiP and stimulate nucleotide release, thus driving the ATPase cycle forward5. From this point forward we will refer to the proteins of all three species as Sil1/Sil1p.
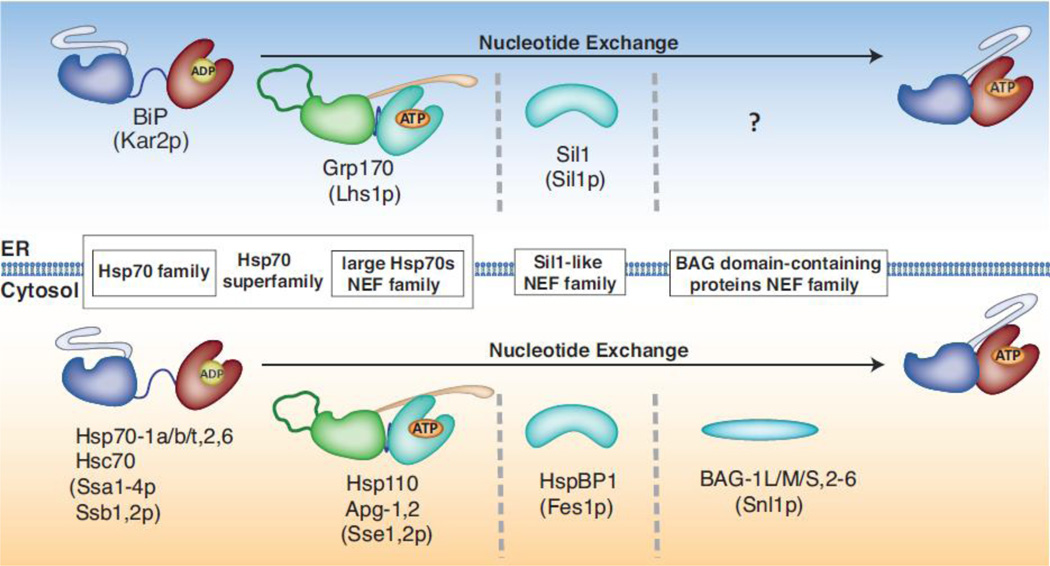
Fig. 3. ER resident and cytosolic Hsp70 family members and their Nucleotide Exchange Factors.
Several members of the Hsp70 protein family exist in the cytosol (bottom) of eukaryotic cells (yeast homologues are indicated in parentheses), whereas a single member, BiP resides in the ER (top). Three different classes of Hsp70 NEFs have been identified in the cytosol: large Hsp70 family members, Sil1-like and BAG-domain proteins that remove ADP from the NBD of Hsp70s, thus allowing ATP to bind and substrates to be released. A single member of the first and second class of NEFs, Grp170 and Sil1 respectively, have been found to act as nucleotide exchange factors for BiP in the ER. While Sil1-like and BAG-domain containing proteins are structurally unrelated to Hsp70 proteins, the large Hsp70 family of NEFs share many structural features with conventional Hsp70. The Hsp70 superfamily comprises conventional as well as large Hsp70s.
b. Mechanism of exchange activity
Amino acid sequence comparisons revealed that yeast and mammalian Sil1 shared significant but low identity with cytosolic Fes1p and HspBP1, respectively. Data were beginning to accumulate at this time demonstrating that these cytosolic proteins regulated the ATPase activity of their respective Hsp70s82; 84, even though they bore no apparent structural relationship to the only previously identified Hsp70 nucleotide exchange factors; E. coli GrpE85 or eukaryotic cytosolic Bag domain proteins86. Both GrpE and Bag-1, although structurally distinct themselves, bind to the cleft at the top surface of the Ib and IIb subdomains of DnaK/Hsp70 NBD and in a similar fashion “push” the IIb domain away from the Ib domain, thereby allowing nucleotide to be released (reviewed in26; 77). The structure of human HspBP1 was solved with only domain II of the Hsp70 NBD. It revealed that HspBP1 is composed of four armadillo-like repeats that wrap around subdomain IIb from the side and bind to it with a higher apparent affinity than Bag-187. When this structure was superimposed on the structure of the Hsc70 nucleotide binding domain crystallized with ADP32, it became clear that this orientation of HspBP1 would have severe steric conflicts with domain Ib in the intact NBD. Thus, the data suggested that HspBP1 served not only to push the IIb subdomain away from Ib, like GrpE and Bag-1 do, but it was likely to additionally displace the Ib lobe, thus further destabilizing this domain and decreasing its affinity for nucleotide87. This argued that unlike Bag-1, HspBP1 induced a major distortion of the Hsp70 ATPase domain to trigger nucleotide release. More recently a structure of yeast Sil1p was solved with the complete nucleotide binding domain of Kar2p88. Similar to HspBP1, the Sil1 protein is composed of four armadillo motifs that wrap around the IIb lobe of the BiP ATPase domain (Fig. 4a). Importantly, this structure further revealed that Sil1p also makes contact with subdomain Ib causing lobe IIb, and to a lesser extent Ib, to rotate away from the bound nucleotide concomitant with its release (Fig. 4b), which confirmed the hypothesis put forward for the HspBP1 structure.
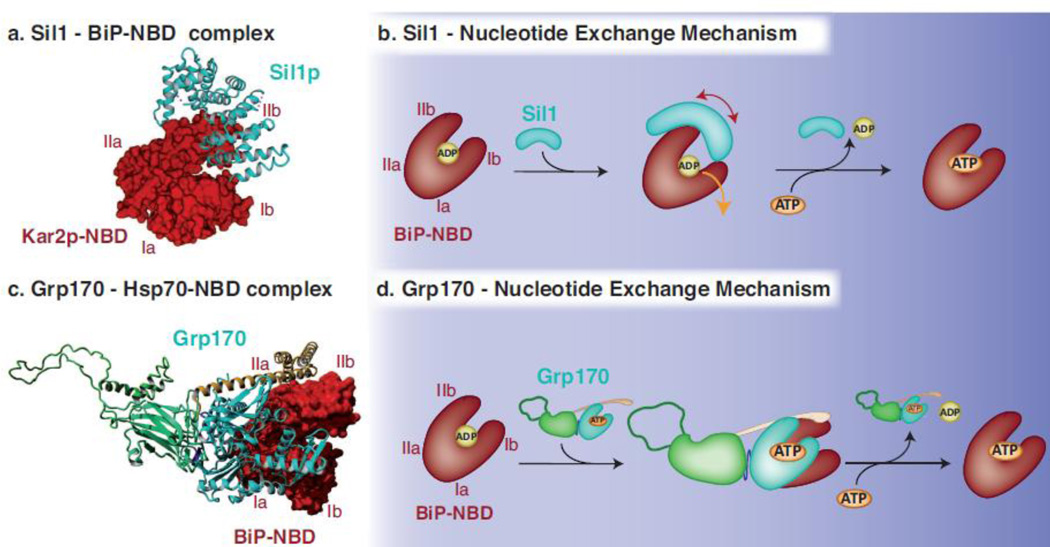
Fig. 4. Mechanisms used by Sil1 and Grp170 to regulate nucleotide exchange for BiP.
(a) The crystal structure obtained for yeast Sil1 bound to the NBD of BiP (PDB: 3QML)88 depicts how the single domain NEF Sil1, consisting of four Armadillo-like repeats (cyan, shown in ribbon), wraps around lobe IIb of BiP’s NBD (red, shown in surface representation) and displaces lobe Ib during the nucleotide exchange reaction. (b) Upon the binding of Sil1 to the ADP-bound form of BiP, the NBD cleft of BiP is opened by tilting lobe IIb and to a lesser extent lobe Ib outwards and thus destabilizing the domain and releasing ADP. ATP can subsequently bind to the NBD and BiP can re-enter its ATPase cycle. (c) The crystal structure of yeast Sse1p bound to the NBD of human Hsp70 (PDB: 3D2F)112 was used to model Grp170 (NBD in cyan, β-sheet and unstructured loop in green and α-helical domain in yellow, shown in ribbon) using Yasara Structure (www.yasara.org). Grp170 is shown bound to the human BiP NBD (3LDL)30 (red, shown in surface representation). Complex formation occurs via multiple contacts between the respective NBDs, and in addition, the C-terminal α-helical domain of Grp170 reaches out to embrace the Hsp70 NBD. (d) For the nucleotide exchange reaction to occur, Grp170 apparently binds to the ADP-bound form of BiP and destabilizes the structure of BiP´s NBD resulting in the release of ADP. Once ATP is bound to BiP, the Grp170-BiP complex would dissociate and BiP can re-enter its ATPase cycle, although key steps in the NEF activity for large Hsp70 proteins remain to be elucidated.
c. Regulation, expression and localization of Sil1
Although Sil1 appears to be present in the ER of all eukaryotic organisms examined, including Arabidopsis and rice plants, studies thus far have only been conducted in a few model organisms (Table 1). Even with this limited characterization, it is clear that species-specific differences exist. One of the first distinctions to be noted is its regulation by conditions that affect protein folding in the ER and activate the unfolded protein response (UPR), which generally regulates the expression of the majority of ER chaperones and their co-factors. For instance in rice, when cells are treated with either DTT or tunicamycin Sil1 is the most dramatically induced UPR target89. In S. cerevisiae, Sil1p (aka PER100) was identified as a UPR-inducible gene90, whereas, in Y. lipolytica, where Sil1 was first discovered, its induction by ER stress is only a very modest ~1.5 fold91. Lastly, in human cells, the Sil1 gene does not appear to be a UPR target, and in fact Sil1 protein levels may even be reduced in response to ER stress in cultured cells5 or primary tissues92. Thus, Sil1 regulation by ER stress appears to run the gamut from super-induced to non-induced or possibly even repressed. The effect this might have on BiP-dependent folding or degradation during ER stress in these different organisms has not been directly addressed. There are only a few studies to examine the levels of Sil1 relative to BiP in various organisms or tissues, although a study using a human multi-tissue blot suggested that the mRNA levels of these two genes are coordinately expressed in a variety of tissues5. Secretory tissues, which have the highest levels of BiP, also express the greatest amounts of Sil1 transcripts, and those tissues with lower levels of BiP have relatively reduced amounts of Sil1. A recent study reported that Sil1 protein was readily detected in mouse pancreas and to a lesser extent in liver, but was undetectable in muscle93, confirming that significant differences in Sil1 protein expression exist between various tissues. The relative quantities of BiP protein and a number of its co-factors were determined in canine pancreatic microsomes, where BiP was found to be expressed at a thousand times higher levels than Sil1 (5µM versus 5nM)94. The effects of ER stress on Sil1 expression in a human cell line suggest the ratio of BiP to Sil1 would be even greater when ER homeostasis was disrupted5.
Table 1.
Species variations in Sil1 characteristics
| UPR↑ | ER Retention | Role in Translocation |
Role in Folding* | |
|---|---|---|---|---|
| Y.lipolytica (Sls1p) | ≤ 2 fold | + RDEL | + + | + + |
| S. cerevisiae (Sil1p) | + + + | + RDEL | + | +/− |
| Human/Mouse (BAP) | − | BiP Assoc. | ND | +# |
| Zebra Fish (Sil1) | + + | ?‡ | ND | ND |
As indicated by evidence of UPR activation in its absence.
Restricted to only certain tissues.
Non KDEL-like
sequence (KMRQV) at C-terminus. ND = not done
Another significant difference among Sil1 proteins is the mechanism used to ensure that this soluble protein remains a resident of the ER. The majority of soluble ER chaperones and folding enzymes, like BiP and protein disulfide isomerase (PDI), possess a tetrapeptide sequence at their extreme C-terminus that is responsible for their retention in this organelle95. In the case of mammals this is usually KDEL, whereas in birds it is often RDEL, and in yeast it is HDEL. This 4 amino acid tag is recognized by a KDEL receptor that is located in the ER-Golgi intermediate compartment and the cis-Golgi. If a resident ER protein escapes the ER, it is caught by this receptor and returned to the ER lumen96. Indeed, inspection of the Sil1 homologues in S. cerevisiae, Y. lipolytica, several fungi, and mosquitoes reveals the presence of the R/K/DEL tetrapeptide at the C-terminus. However, in most metazoans and plants examined there is not a readily recognizable KDEL-like sequence at the C-terminus of their Sil1 homologue. Instead, a significant number of them, including C. elegans, end with the sequence K/RELR, whereas in others like Xenopus laevis and Arabadopsis thaliana, Sil1 possesses even more divergent C-terminal sequences. More recent studies have suggested that the KDEL receptor can recognize a large number of variations on this sequence97, and thus due to the highly conserved nature of the KELR tetrapeptide, it was assumed that it represented a divergent ER retention sequence5. Instead, it was recently revealed that this conserved sequence plays a critical role in structural integrity of the human Sil1 protein98; 99. These residues are likely to form tertiary interactions with other portions of the protein that serve to stabilize an otherwise weak α-helix at the C-terminus99. The mechanism of ER retention for the human Sil1 protein was shown to occur through its interactions with BiP99, which as mentioned above is present in much higher quantities than Sil194 and possesses a KDEL sequence at its C-terminus.
4. Grp170: a nucleotide exchange factor with a chaperone function
In addition to Sil1, a second NEF for BiP has been identified, the glucose regulated protein of 170 kDa (Grp170). Based on similarities in domain organization, it was assigned to the family of large Hsp70s100–102, which together with the conventional Hsp70s constitute the Hsp70 superfamily (Fig. 3). Although BiP and Grp170 share structural similarities, in contrast to BiP, where much is known about its various functions and how they are regulated103, we are still lacking a clear understanding of Grp170´s functions in the ER. Due to the similar domain organization, it was assumed that Grp170 was also a molecular chaperone that would interact with unfolded proteins in the ER in a similar manner as BiP. Therefore, the discovery that Lhs1p, the Grp170 homologue in yeast, could act a NEF for yeast BiP, Kar2p6, came as a surprise and immediately hinted towards a more complex function for this ER resident protein.
a. Regulation and structural organization of Grp170
Perturbations of ER homeostasis, by deprivation of either glucose in chick fibroblasts104 or oxygen in human and rodent cell lines105, resulted in the induction of a large molecular weight protein thus named Grp170 or oxygen-regulated protein of 150 kDa (ORP150), respectively. These were ultimately found to be the same protein, which will subsequently be referred to as Grp170. Soon after, related proteins were identified in C. elegans100 and S. cerevisiae, Lhs1p,106–108 demonstrating that Grp170 was conserved in eukaryotes. Like BiP, the canonical ER-localized Hsp70 chaperone, Grp170 contains an ER targeting signal sequence, an N-terminal NBD, which binds to ATP even more efficiently than BiP109, followed by a β–sheet domain, an α-helical domain as well as an KNDEL ER retention sequence at its C-terminus100; 110. Even though the NBDs between BiP and Grp170 are well conserved, their C-terminal regions are quite different. Unlike BiP, Grp170 possesses an acidic unstructured loop insertion in its β-sheet domain and a significantly extended and unstructured region at its C-terminus that in part accounts for its much larger size than BiP. Furthermore, inspection of the putative C-terminal SBD of Grp170 reveals significant differences compared to the corresponding region in BiP. This region in BiP forms an α-helical lid that stabilizes substrate binding to the β-sheet portion of the C-terminal SBD. However, structural studies on other cytoplasmic large Hsp70s111–113 reveal that the α-helices comprising this region are too extended to allow the necessary kink to be formed so that this domain could serve as a lid. Instead, the extended α-helical domain in the large Hsp70s reaches out to embrace lobe II of the NBD of Hsp70 to presumably facilitate the nucleotide exchange reaction112; 113 (Fig. 4c). Nevertheless, the recent structures of Hsp70s in their ATP bound state51; 52 resemble the crystal structures of large Hsp70s111–113, as in both cases the α–helical domain is extended and docks onto the NBD. No structural data are available yet on which conformation the α–helical domain of large Hsp70s adapts in the ADP-bound state or when bound to clients. In addition to the structural differences between canonical and large Hsp70s, there are a number of other distinctions. Unlike BiP, which is an unglycosylated chaperone, Grp170 has nine predicted glycosylation sites throughout the protein with the majority of them mapping to the C-terminal region. Furthermore, a highly conserved Arg residue present in the NBD of all known canonical Hsp70s, which is critical for interaction with DnaJ family members42, is missing in all ER-localized large Hsp70 family members that have been identified. Interestingly, this Arg is present in all the cytosolic orthologues, suggesting that the functional regulation of the ER proteins might be distinct from other canonical and large Hsp70-type proteins.
b. A chaperone function of Grp170
In spite of the fact that the more C-terminal regions of Grp170 are quite different from the SBD of BiP, Grp170 was first found in a complex with Ig HCs114, and subsequently shown to bind thyroglobulin upon ER stress115, as well as the non-secreted α1-antitrypsin Z mutant116 and clusterin117. As all of these proteins also interact with BiP and because Grp170 interacts with BiP as its NEF114; 118, it was not clear if the binding of Grp170 to these substrates was direct or occurred indirectly via its association with BiP. In contrast to full-length proteins, radiolabeled peptides that were translocated into microsomes via the TAP transporter were shown to bind to Grp170 directly119; 120. Further support for Grp170’s chaperone function came from a study with purified proteins, where murine Grp170 was significantly more efficient in preventing the aggregation of heat denatured luciferase than Hsc70121. Similar data were obtained with yeast Lhs1p122. However, refolding studies conducted with murine Grp170 revealed that it failed to restore enzymatic activity in denatured luciferase in the presence of ATP and the cytosolic DnaJ family protein, Hdj-1. Only when rabbit reticulocyte lysate was added to the reaction could the activity of luciferase be recovered in an ATP-dependent manner121, suggesting that either some of Grp170´s functions depend on as yet unidentified co-factors or that Grp170 maintains the denatured luciferase in a folding-competent state that can then be folded by another chaperone. Solving the ambiguity whether Grp170 can bind clients in the absence of BiP, a recent study demonstrated that Grp170 continues to bind incompletely folded protein substrates after BiP is released with ATP, arguing for direct client binding123. As opposed to the nucleotide-dependent substrate binding of BiP, the interaction of Grp170 with its substrates is instead modulated by domains unique to large Hsp70s123, suggesting a different regulation of substrate interaction for large Hsp70s compared to conventional Hsp70s. These insights beg the questions of what influence ATP has on binding of Grp170 to its substrates and how substrates are released from the large Hsp70s. The first data on the biological functions of Grp170 came from a study conducted in both yeast and human cells, which revealed that degradation of free α-subunits of the epithelial sodium channel (ENaC) was dependent on Grp170’s chaperone function but did not require its NEF activity124 or even BiP125. Whether Grp170 and BiP together with an ERdj protein also form a protein disaggregation machinery, as recently shown for the cystosolic large Hsp70 orthologs126; 127, remains to be shown.
A number of recent studies have capitalized on Grp170’s ability to bind peptides. Grp170 is being used to present antigens in various vaccine protocols128–134, and the ability of a secreted form of peptide-bound Grp170 to stimulate anti-tumor immunity in cancer treatment has been tested135–138 (reviewed in139). Two peptides were recently identified that bound to the yeast cytosolic large Hsp70 Sse1p, but bound only poorly to the conventional Hsp70s Ssa1p or DnaK, whereas peptides that bound better to the conventional Hsp70s had either dramatically reduced or no binding to Sse1p, which suggested major differences in their binding specificities140; 141. One of these peptides was shown to also bind Sse2p and human Hsp110, and mutational analysis of this peptide revealed that all three chaperones showed a preference for peptides containing aromatic residues140; 141. Notably the other peptide identified to bind Sse1 was also somewhat enriched in aromatic residues140; 141, although their contribution to Sse1 binding was not explored. This in in contrast to BiP, which prefers peptides with alternating hydrophobic residues58; 142, suggesting that conventional Hsp70s and large Hsp70s might each have some unique client proteins or that the two Hsp70 classes bind to different regions within a substrate.
c. Grp170 possesses nucleotide exchange activity
The first indications that Grp170 might possess nucleotide exchange activity for BiP were provided by a yeast genetic screen for suppressors of the severe growth defect observed in the Δire1 Δlhs1 double mutant, which identified Sil1p83. Interestingly, BiP was unable to suppress this phenotype, suggesting that Lhs1p might be functioning in this screen as something other than a molecular chaperone. The fact that yeast deficient either in Lhs1p or Sil1p were viable, while the Δlhs1Δsil1 double mutation was lethal83, led these investigators to suggest a common function for these two proteins. A few years later, Lhs1p was shown to serve as a NEF for Kar2p6, and shortly thereafter NEF activity was established for both Y. lipolitica80 and mammalian5 Sil1, demonstrating that the common function shared by these proteins was exchange activity. In subsequent years, NEF activity was detected for purified canine Grp170143, as well as other cytosolic large Hsp70s from a variety of organisms144–149, thus revealing a completely new and unexpected function for these large Hsp70 proteins.
Insights into the mechanism by which the large Hsp70s stimulate nucleotide exchange were first obtained in studies with yeast where Lhs1p and Sil1p were found to bind to Kar2p in a mutually exclusive manner6; 143. The interaction of Lhs1p with Kar2p is dependent on the presence of a nucleotide in the NBD of Lhs1p, and its exchange activity is enhanced by the addition of ER-localized DnaJ family proteins to stimulate ATP hydrolysis by Kar2p122. Crystallographic studies on Sse1p either alone111 or bound to an Hsp70 NBD112; 113 revealed that the ATP bound form of these large Hsp70s interacts directly with the NBD of Hsp70 in a head to head manner with multiple contacts occurring between domains IIa/b and Ia/b of Sse1p with domains Ia/b and IIb of the Hsp70 protein104–106; 111–113 (Fig. 4c). As noted above, additional interactions with the Hsp70 molecule occur through the α-helical bundle present at the C-terminus of the large Hsp70, which reaches over to embrace the Hsp70 NBD (Fig. 4c). The result of these interactions is to “pin” IIb lobe of Hsp70 between Sse1’s α-helical domain and its own subdomain Ib and rotate Sse1’s it sideways to release the nucleotide (Fig. 4d). Release of ADP from Hsp70 is not sufficient to trigger dissociation of the Hsp70-NBD:Sse1p complex, arguing that rebinding of ATP to the Hsp70 NBD may be required112; 113. This is in keeping with studies showing that Lhs1p can bind to Kar2p in either the apo- or ADP-bound form but not the ATP-bound state122.
Although a crystal structure for the ER large Hsp70 bound to BiP has not been reported, based on the conformational dynamics of the Ssa1p-NBD induced by Sse1p, the mechanism of the nucleotide exchange employed by Lhs1p is likely to be quite similar to that of Sse1p150. Hydrogen-Deuterium exchange assays revealed that Lhs1p formed interactions with the NBD of Ssa1 that were very similar to those occurring with Sse1p, including the interaction of the α-helical bundle with the subdomain IIb. In addition, the residues that are critical for Sse1p’s NEF function112; 113 are conserved in Lhs1p, and mutation of these residues similarly affected exchange activity in Lhs1p150. Together these data make a strong case for the ER-localized large Hsp70s using the same mechanism of nucleotide exchange as has been established for their cytosolic orthologues.
Interestingly, despite the similarity in domain organization and the NBD structure between Grp170 and BiP, Sil1p does not physically interact with Lhs1p6. While both of these proteins promote nucleotide release from BiP, the binding of Lhs1p and Sil1p to Kar2p occur through distinct but mutually exclusive interactions151, suggesting they might compete for Kar2p binding. Studies to quantify levels of the two NEF proteins in the ER of canine pancreas revealed that Grp170 was present at much higher levels (~0.60 µM) than Sil1 (~0.005 µM)94. It is unclear if this is because Grp170 has additional functions or if the two proteins regulate nucleotide exchange to assist BiP in distinct functions or under unique cellular conditions. However, there are little data available to address either of these possibilities and further studies aimed at dissecting how the two NEFs influence the functions of BiP in vivo are needed to understand the significance of the differences in their levels and how they assist BiP in its various functions.
5. Contribution of ER NEFs to biological functions
a. Cellular functions dependent on Sil1
While the ER functions that Sil1 participates in remain rather poorly understood, some insights have been obtained by examining the effects of gene disruption in organisms ranging from yeast to man. In humans, mutations in the SIL1 gene have been found in over half the cases of Marinesco-Sjögren syndrome (MSS)152–154 an autosomal recessive disease characterized by multisystem defects including cerebellar ataxia due to Purkinje cell loss, progressive myopathy, early onset cataracts, skeletal abnormalities, and a variety of developmental abnormalities and intellectual disabilities155–158. MSS-associated mutations occur throughout the SIL1 gene and most lead to the disruption of significant portions of the protein98; 153; 155, including those regions that interact with BiP. However, three MSS-associated mutations have been identified that disrupt only the last 4 or 5 amino acids of Sil1, which affects its solubility and stability resulting in significantly diminished expression of these Sil1 mutants98; 99. Five additional MSS-associated mutations have been identified that result in single amino acid substitutions in Sil198; 154; 155; 157; 159; 160, although the effects of these mutations on Sil1 function or expression have not been examined. It remains unclear why only some tissues appear to be affected by mutations in the ubiquitously expressed Sil1 protein, and in most cases the molecular mechanism(s) underlying the pathology remains unknown. Muscular hypotonia is often the initial symptom observed in children with MSS and presents early in infancy161. A number of electrophysiology162 and ultrastructural studies163–165 have been conducted on muscle biopsies from affected individuals, which reveal evidence of abnormal membrane function and structures, including a dilated sarcoplasmic reticulum, autophagic vacuoles, and the presence of electron dense material suggestive of protein aggregates. A second common feature of MSS is ataxia due to severe atrophy of the cerebellum166–168. However, as most of these studies were done before mutations in SIL1 were linked to MSS, it is not clear if all of the affected individuals examined had SIL1 mutations. To test the potential role of Sil1 in the cerebral cortex, mouse embryos were electroporated in utero with vectors encoding shRNA targeting SIL1169. Defects in neuronal migration were observed at birth in these mice, which were rescued by co-expression of human wild-type Sil1 but not by three MSS-assciated Sil1 mutants.
Although a Sil1 null mouse is not available, the gene has been disrupted spontaneously by transposon insertion, Sil1wz, and by gene-trap methodology, Sil1Gt, resulting in the loss of amino acids 261–465 of the Sil1 protein170. Both genetic strains are referred to as woozy mice and have been reported to phenocopy some of the pathologies associated with MSS including cerebellar loss resulting in ataxia170 and a progressive myopathy164, which allow for a more tractable model for understanding both the disease and function of Sil1. Inspection of quadriceps muscle tissue from woozy mice revealed structural changes that are similar to those reported in muscle biopsies from MSS patients. Western blot analyses of muscle lysates detected up-regulation of ER chaperones, including Grp170 and components of the ERAD, as well as the accumulation of ubiquitinated proteins and induction of autophagic responses compared to wild-type muscle lysates164. When the cerebella from these mice were examined, a progressive and profound Purkinje neuron degeneration was observed170. Electron micrographs and immunohistochemistry analyses revealed evidence of autophagosome-like structures, presence of ubiquitinated protein aggregates, and activation of ER stress and apoptotic responses in the Purkinje cells. Although Grp170 is modestly up-regulated in the Purkinje cells in the woozy mice, when they were crossed with transgenic mice overexpressing Grp170 (Tg-Hyou1), the resulting Sil1Gt;Tg-Hyou1 mice showed no obvious Purkinje cell loss or evidence of UPR activation in these cells, and the ataxia was dramatically suppressed171. Correspondingly, when the woozy mice were crossed with mice having decreased levels of Grp170 (Hyou1+/−) Purkinje cell loss, ER stress, and ataxia occurred earlier and were more severe171. Interpretation of the results obtained with increased and diminished Grp170 expression are complicated by the fact that this large Hsp70 protein has both chaperone121; 123 and nucleotide exchange143 activity. Thus it is unclear whether Grp170 can provide the critical missing NEF function in Purkinje cells only when it is super expressed or if under these over-expression conditions it is able to compete for binding BiP clients, thus alleviating the need for a NEF. As an alternative means of examining this point, the woozy mice were crossed with DNAJC3 null mice. DNAJC3/p58IPK/ERdj6 is one of seven identified resident ER DnaJ family members3; 4 and can bind directly to some secretory pathway proteins and deliver them to BiP172. The resulting Sil1−/−; Dnajc3−/− mice showed no signs of ataxia, Purkinje cell loss, or accumulation of protein inclusions. This suggests that the partial restoration of cerebellar function might be due to interfering with the targeting of a particular subset of clients to BiP, although what these clients are remains unknown. Most recently, studies using woozy mouse model demonstrated impaired ER homeostasis in motor neurons. When Sil1 heterozygous mice were crossed with a mouse model of amyotrophic lateral sclerosis, Sil1 levels were progressively depleted in the fast-fatigable motor neurons leading to lowered excitability, a condition that was restored by Adeno-associated virus-mediated delivery of Sil1 to the motor neurons173. In fact, it is unclear whether the associated pathologies in this and other affected tissues arise from reduced maturation of essential secretory pathway proteins, depletion of free BiP stores resulting in activation of the ER stress response, toxicity due to aggregation of secretory pathway proteins, or even perhaps of the mutant Sil1 protein itself.
The woozy mice have also been the focus of recent studies on several additional organ systems. Sil1 is highly expressed in the β islet cells of the pancreas of wild-type mice, and disruption of the SIL1 gene resulted in decreased islet mass, evidence of UPR activation, and reduced plasma insulin levels upon glucose stimulation due to defects in insulin secretion93. This is in keeping with the fact that insulin is a BiP client174; 175 and might suggest that disruption of Sil1 reduces the secretion of insulin by inhibiting the release of BiP, although this was not directly shown. It is noteworthy that individuals with MSS show no evidence of defects in pancreatic function, demonstrating a clear difference between mice and man in the reliance of this organ on Sil1 function. In a separate study using the woozy mice, the contribution of Sil1 to antibody assembly and secretion were investigated. BiP was originally identified in association with free Ig heavy chains in Abelson virus transformed pre-B cells176, which are arguably the best characterized BiP client. Although the co-expression of BiP mutants that cannot be released from clients by ATP inhibits both the assembly and secretion of IgG antibodies76, the Sil1Gt mouse was indistinguishable from wild-type controls in terms of both the kinetics and magnitude of antigen-specific antibody responses and in Ig assembly from LPS stimulated splenic B cells177. This was also confirmed in three human EBV-transformed lymphoblastoid cell lines obtained from individuals with MSS, arguing that the BiP co-factor Sil1 is dispensable for antibody production.
b. Biological functions requiring Grp170
A large variety of biological functions have been associated with Grp170, although molecular insights into the role of Grp170 in these processes are generally lacking (also reviewed in178). Loss or decreased expression of this ER large Hsp70 protein inhibits the translocation of nascent polypeptide chains into the ER in a number of organisms as revealed by both genetic and biochemical experiments106–109. In S. cerevisiae, lhs1 null cells exhibit a selective translocation defect that affects a subset of proteins, including preKar2p, prePDI, prepro-α-factor and preCPY, whereas the translocation of another SRP-dependent protein, dipeptidylaminopeptidase B was not affected106–108. Lhs1p’s NEF activity was shown to be critical to support BiP-mediated protein translocation into the ER, but this activity was not required for its chaperone function122; 124. When mammalian proteoliposomes that were depleted of ATP-binding proteins preprolactin import was inhibited. Translocation was restored when the microsomes were repleted with the ATP-binding proteins but not with purified BiP alone109. Since BiP and Grp170 are the only ATP binding proteins that have been identified, it is reasonable to conclude that Grp170 plays a critical role in the import of this protein into the microsomes. However, since BiP is also required for protein translocation into the yeast ER179; 180, it remains unclear whether Grp170 itself plays a direct role in protein translocation through its chaperone function or whether nucleotide exchange by Grp170 is a rate-limiting factor for BiP’s role in protein translocation. A very recent study demonstrated, however, that the NEF function of Grp170 is critical for BiP release and retrotranslocation of the SV40 virus from the ER into the cytosol181.
Grp170 has also been implicated in a variety of physiologically relevant protein secretion processes linked to disease states. The high expression of Grp170 in pancreatic islets is significantly reduced by fasting, suggesting that it may have a role in insulin biosynthesis182. The effects of both increased and decreased Grp170 expression on insulin production and blood glucose levels has also been examined using a number of normal mice as well as mouse models for type II diabetes. Increasing Grp170 levels with adenovirus delivered constructs increased insulin secretion and decreased blood glucose after feeding, whereas decreasing Grp170 expression with anti-sense constructs inhibited insulin secretion and resulted in significantly higher blood glucose levels183. In another study Grp170 levels were modulated by crossing the diabetes prone Akita mice to either Grp170(+/−) heterozygous mice or Grp170 transgenic mice184. Decreased expression of Grp170 accelerated the onset of diabetes in this model, whereas transgene-enhanced expression of Grp170 resulted in lower body weight and improved glucose tolerance in young mice. Interestingly, there were no changes in the level of insulin in the pancreatic cells of either genetic model, which could suggest that the retained insulin was continuously being degraded in the animal model. It is noteworthy that disruption of Sil1 also results in a similar pancreatic phenotype in mice93, raising the question of why both proteins are required for insulin biosynthesis and secretion.
Similar to its effects on insulin secretion, a number of studies have indicated a role for Grp170 in VEGF secretion, one of the most important proangiogenic factors. Decreased Grp170 levels in C6 glioma cells185 and macrophages186, achieved by expressing an antisense-Grp170 construct, resulted in an inability of both cell lines to secrete VEGF into culture media under hypoxic conditions. In the case of C6 cells, the intracellular levels of VEGF were dramatically increased, arguing for an inhibition of protein transport, possibly due to improper maturation when Grp170 levels were reduced. Conversely, when Grp170 was over-expressed, cells secreted significantly higher levels of VEGF compared to non-transfected cells185; 186. Under hypoxic conditions Grp170 could be co-immunoprecipitated with VEGF and vice versa, indicative of an interaction between these proteins185; 186. The contribution of Grp170 to the expression of this proangiogenic factor may in part explain its contribution to tumor survival and metastasis139.
Grp170 levels have also been associated with cytoprotection during other physiological and disease states. Ischemia and atherosclerotic plaque formation lead to lower oxygen levels in the surrounding tissue, which induces ER stress and hypoxic pathways, resulting in cell death if not resolved. Transgenic mice over-expressing Grp170 lost significantly less brain tissue in response to ischemia187, and upon kainate administration fewer animals suffered from seizures compared to wild type mice188. The survival rate of neurons after hypoxic stress187; 189 or after excitotoxicity upon glutamate administration188; 189 also increased when Grp170 was over-expressed. Regulated cell death is an important event in many developmental phases, including in the brain190. Somewhat counter-intuitively Purkinje cells were shown to significantly up-regulate Grp170 expression during the peak phase of programmed cell death in mouse brain development. However, over-expression of Grp170 in the transgenic mouse model led to increased survival of these cells during development, resulting in a higher number of Purkinje cells compared to wild-type191. Of note, this abnormal brain development resulted in an impaired motor coordination of these mice. In another pathological condition, atherosclerotic plaques, which deprive surrounding tissue of their oxygen supply, lead to up-regulation of hypoxic and ER stress pathways in segments of aortae with severe atherosclerosis192–194. In particular macrophages, one of the key players in the pathogenesis of atherosclerosis within the atherosclerotic plaque195, showed significant induction of stress pathways including up-regulation of Grp170192. When Grp170 levels were reduced in cultured mononuclear phagocytes, the cells became more susceptible to hypoxic stress and even more so when this was combined with oxidized LDL192. Similarly, overexpression of Grp170 in a microvascular endothelial cell line prevented ER stress activation194 and inhibited apoptotic cell death193 triggered by oxLDLs. Hence, Grp170 promotes cell survival under a number of physiological stress conditions.
6. Concluding thoughts
While the past decade has significantly increased our understanding of how the ATPase cycle of BiP is regulated, this information has led to many more questions. First, it is unclear what the relative contributions of Sil1 and Grp170 are to BiP’s requirement for exchange activity, how exchange activity contributes to the various biological functions of BiP, and whether this differs in individual tissues or under various developmental or stress conditions. Mechanistically, it is not completely understood how these NEFs distinguish between the different nucleotide states of BiP. Is the ATP/ADP ratio in the ER together with nucleotide-induced structural changes in the NBD sufficient to control the specificity, or do nucleotide-regulated changes in other regions of the BiP molecule also contribute? Second, although genetic complementation studies reveal that either NEF is able to compensate for loss of the other factor to varying degrees, it is clear in studies ranging from yeast to humans that these factors are not entirely interchangeable and that some defects remain upon complementation or that decidedly non-physiological levels of expression must be used to suppress phenotypes. This is underscored by the fact that the deletion of Grp170 is embryonic lethal in mice188, whereas loss of Sil1 causes a multi-syndrome disease in both humans153 and mouse models170. Lastly, the finding that Grp170 possesses both chaperoning and NEF activity makes it more complicated to determine which activity is critical in a number of the functions for which it has been implicated and how substrate binding and NEF activity of Grp170 contribute to the folding environment of the ER.
Highlights.
Structural and mechanistic insights into the Hsp70 ATPase cycle are reviewed
The influence of the ER environment on the ATPase cycle of BiP is discussed
Mechanisms of nucleotide exchange in BiP by Sil1 and Grp170 are reviewed
Our current understanding of the Grp170 chaperone function is presented
Biological functions and associated pathologies of Sil1 and Grp170 are reviewed
Acknowledgements
JB gratefully acknowledges funding by the Boehringer Ingelheim Fonds, MJF by the German Academy of Sciences Leopoldina, grant number LPDS 2009-32 and LMH by NIH grant number R01 GM054068.
Abbreviations used
- BAP
BiP associated protein
- BiP
immunoglobulin heavy-chain binding protein
- CNX
calnexin
- COPII
coat protein complex II
- CRT
calreticulin
- Derlin
Der1-like protein
- DTT
dithiothreitol
- EDEM
ER-degradation enhancing α-mannosidase like protein
- ERGIC
ER-Golgi intermediate compartment
- ENaC
epithelial sodium channel
- ERManI
ER mannosidase I
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERdj
ER-localized DnaJ-like proteins
- ERQC
ER quality control
- Grp170
glucose-regulated protein of 170 kDa
- Glc I and II
glycosidase I and II
- Hrd1
hydroxymethylglutaryl reductase degradation protein 1
- HspBP1
Hsp70 binding protein 1
- Lhs1p
Lumenal Hsp Seventy
- MHC I
major histocompatibility complex class I
- MSS
Marinesco-Sjögren syndrome
- NBD
nucleotide binding domain
- NEF
nucleotide exchange factor
- ORP150
oxygen-regulated protein of 150 kDa
- OS-9
osteosarcoma amplified 9
- OST
oligosaccharyltransferase
- PDI
protein disulfide isomerase
- SBD
substrate binding domain
- Sel1L
suppressor/enhancer of Lin-12-like
- Sil1
Silence growth defects of IRE1, LHS1 double deleted strain
- Sls1p
SCR2-encoded 7S RNA component of SRP identified in a synthetic lethal screen
- TAP
transporter associated with antigen processing
- UGT
UDP-glucose glycoprotein glucosyltransferase
- UPR
unfolded protein response
- VEGF
vascular endothelial growth factor
- VIMP
valosin-containing protein (VCP)/p97-interacting membrane protein
- XTP3-B
XTP3 transactivated protein B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no competing financial interests.
References
- 1.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci U S A. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 3.Melnyk A, Rieger H, Zimmermann R. Co-chaperones of the Mammalian Endoplasmic Reticulum. Subcell Biochem. 2015;78:179–200. doi: 10.1007/978-3-319-11731-7_9. [DOI] [PubMed] [Google Scholar]
- 4.Otero JH, Lizak B, Hendershot LM. Life and death of a BiP substrate. Semin Cell Dev Biol. 2010;21:472–478. doi: 10.1016/j.semcdb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung KT, Shen Y, Hendershot LM. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem. 2002;277:47557–47563. doi: 10.1074/jbc.M208377200. [DOI] [PubMed] [Google Scholar]
- 6.Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson L, Lazarides E. ADP-ribosylation of the Mr 83,000 stress-inducible and glucose-regulated protein in avian and mammalian cells: modulation by heat shock and glucose starvation. Proc Natl Acad Sci U S A. 1983;80:4664–4668. doi: 10.1073/pnas.80.15.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch WJ, Garrels JI, Thomas GP, Lin JJ, Feramisco JR. Biochemical characterization of the mammalian stress proteins and identification of two stress proteins as glucose- and Ca2+-ionophore-regulated proteins. J Biol Chem. 1983;258:7102–7111. [PubMed] [Google Scholar]
- 9.Hendershot LM, Ting J, Lee AS. Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol Cell Biol. 1988;8:4250–4256. doi: 10.1128/mcb.8.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leustek T, Toledo H, Brot N, Weissbach H. Calcium-dependent autophosphorylation of the glucose-regulated protein, Grp78. Arch Biochem Biophys. 1991;289:256–261. doi: 10.1016/0003-9861(91)90469-y. [DOI] [PubMed] [Google Scholar]
- 11.Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol. 2012;198:371–385. doi: 10.1083/jcb.201202005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leno GH, Ledford BE. ADP-ribosylation of the 78-kDa glucose-regulated protein during nutritional stress. Eur J Biochem. 1989;186:205–211. doi: 10.1111/j.1432-1033.1989.tb15196.x. [DOI] [PubMed] [Google Scholar]
- 13.Freiden PJ, Gaut JR, Hendershot LM. Interconversion of three differentially modified and assembled forms of BiP. EMBO J. 1992;11:63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaut JR. In vivo threonine phosphorylation of immunoglobulin binding protein (BiP) maps to its protein binding domain. Cell Stress Chaperones. 1997;2:252–262. doi: 10.1379/1466-1268(1997)002<0252:ivtpoi>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlino A, Toledo H, Skaleris D, DeLisio R, Weissbach H, Brot N. Interactions of liver Grp78 and Escherichia coli recombinant Grp78 with ATP: multiple species and disaggregation. Proc Natl Acad Sci U S A. 1992;89:2081–2085. doi: 10.1073/pnas.89.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh M, Nakai A, Sokawa Y, Hirayoshi K, Nagata K. Modulation of the phosphorylation of glucose-regulated protein, GRP78, by transformation and inhibition of glycosylation. Exp Cell Res. 1993;205:76–83. doi: 10.1006/excr.1993.1060. [DOI] [PubMed] [Google Scholar]
- 17.Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MJ. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993;268:12730–12735. [PubMed] [Google Scholar]
- 18.Ham H, Woolery AR, Tracy C, Stenesen D, Kramer H, Orth K. Unfolded Protein Response-regulated Drosophila Fic (dFic) Protein Reversibly AMPylates BiP Chaperone during Endoplasmic Reticulum Homeostasis. J Biol Chem. 2014;289:36059–36069. doi: 10.1074/jbc.M114.612515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanyal A, Chen AJ, Nakayasu ES, Lazar CS, Zbornik EA, Worby CA, Koller A, Mattoo S. A Novel Link Between Fic (Filamentation induced by cAMP)-mediated Adenylylation/AMPylation and the Unfolded Protein Response. J Biol Chem. 2015 doi: 10.1074/jbc.M114.618348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clairmont CA, De Maio A, Hirschberg CB. Translocation of ATP into the lumen of rough endoplasmic reticulum-derived vesicles and its binding to luminal proteins including BiP (GRP 78) and GRP 94. J Biol Chem. 1992;267:3983–3990. [PubMed] [Google Scholar]
- 21.Dorner AJ, Wasley LC, Kaufman RJ. Protein dissociation from GRP78 and secretion are blocked by depletion of cellular ATP levels. Proc Natl Acad Sci U S A. 1990;87:7429–7432. doi: 10.1073/pnas.87.19.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro S, Pelham HR. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 23.Kassenbrock CK, Kelly RB. Interaction of heavy chain binding protein (BiP/GRP78) with adenine nucleotides. EMBO J. 1989;8:1461–1467. doi: 10.1002/j.1460-2075.1989.tb03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 25.Zuiderweg ER, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. Allostery in the Hsp70 Chaperone Proteins. Top Curr Chem. 2012 doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Mayer MP. Gymnastics of molecular chaperones. Mol Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Macias AT, Williamson DS, Allen N, Borgognoni J, Clay A, Daniels Z, Dokurno P, Drysdale MJ, Francis GL, Graham CJ, Howes R, Matassova N, Murray JB, Parsons R, Shaw T, Surgenor AE, Terry L, Wang Y, Wood M, Massey AJ. Adenosine-derived inhibitors of 78 kDa glucose regulated protein (Grp78) ATPase: insights into isoform selectivity. J Med Chem. 2011;54:4034–4041. doi: 10.1021/jm101625x. [DOI] [PubMed] [Google Scholar]
- 31.Wisniewska M, Karlberg T, Lehtio L, Johansson I, Kotenyova T, Moche M, Schuler H. Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78. PLoS One. 2010;5:e8625. doi: 10.1371/journal.pone.0008625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–638. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 33.Woo HJ, Jiang J, Lafer EM, Sousa R. ATP-induced conformational changes in Hsp70: molecular dynamics and experimental validation of an in silico predicted conformation. Biochemistry. 2009;48:11470–11477. doi: 10.1021/bi901256y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Zuiderweg ER. The 70-kDa heat shock protein chaperone nucleotide-binding domain in solution unveiled as a molecular machine that can reorient its functional subdomains. Proc Natl Acad Sci U S A. 2004;101:10272–10277. doi: 10.1073/pnas.0401313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J Biol Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- 36.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young JC. Mechanisms of the Hsp70 chaperone system. Biochem Cell Biol. 2010;88:291–300. doi: 10.1139/o09-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sousa R, Lafer EM. Keep the traffic moving: mechanism of the Hsp70 motor. Traffic. 2006;7:1596–1603. doi: 10.1111/j.1600-0854.2006.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel M, Bukau B, Mayer MP. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006;21:359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 40.Wei J, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 41.Sousa MC, McKay DB. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry. 1998;37:15392–15399. doi: 10.1021/bi981510x. [DOI] [PubMed] [Google Scholar]
- 42.Awad W, Estrada I, Shen Y, Hendershot LM. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc Natl Acad Sci U S A. 2008;105:1164–1169. doi: 10.1073/pnas.0702132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci U S A. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 45.Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B, Bertelsen EB, Zuiderweg ER. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc Natl Acad Sci U S A. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mapa K, Sikor M, Kudryavtsev V, Waegemann K, Kalinin S, Seidel CA, Neupert W, Lamb DC, Mokranjac D. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol Cell. 2010;38:89–100. doi: 10.1016/j.molcel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Marcinowski M, Holler M, Feige MJ, Baerend D, Lamb DC, Buchner J. Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat Struct Mol Biol. 2011;18:150–158. doi: 10.1038/nsmb.1970. [DOI] [PubMed] [Google Scholar]
- 49.Palleros DR, Reid KL, Shi L, Welch WJ, Fink AL. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- 50.Wilbanks SM, McKay DB. How potassium affects the activity of the molecular chaperone Hsc70. II. Potassium binds specifically in the ATPase active site. J Biol Chem. 1995;270:2251–2257. doi: 10.1074/jbc.270.5.2251. [DOI] [PubMed] [Google Scholar]
- 51.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 52.Qi R, Sarbeng EB, Liu Q, Le KQ, Xu X, Xu H, Yang J, Wong JL, Vorvis C, Hendrickson WA, Zhou L. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Mayer M, Reinstein J, Buchner J. Modulation of the ATPase cycle of BiP by peptides and proteins. J Mol Biol. 2003;330:137–144. doi: 10.1016/s0022-2836(03)00556-4. [DOI] [PubMed] [Google Scholar]
- 55.Chang YW, Sun YJ, Wang C, Hsiao CD. Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J Biol Chem. 2008;283:15502–15511. doi: 10.1074/jbc.M708992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlecht R, Erbse AH, Bukau B, Mayer MP. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol. 2011;18:345–351. doi: 10.1038/nsmb.2006. [DOI] [PubMed] [Google Scholar]
- 58.Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 59.Chapman DC, Williams DB. ER quality control in the biogenesis of MHC class I molecules. Semin Cell Dev Biol. 2010;21:512–519. doi: 10.1016/j.semcdb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 60.Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- 61.Schauble N, Lang S, Jung M, Cappel S, Schorr S, Ulucan O, Linxweiler J, Dudek J, Blum R, Helms V, Paton AW, Paton JC, Cavalie A, Zimmermann R. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 2012;31:3282–3296. doi: 10.1038/emboj.2012.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 63.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 64.Smock RG, Blackburn ME, Gierasch LM. Conserved, disordered C terminus of DnaK enhances cellular survival upon stress and DnaK in vitro chaperone activity. J Biol Chem. 2011;286:31821–31829. doi: 10.1074/jbc.M111.265835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 68.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hendershot L, Wei J, Gaut J, Melnick J, Aviel S, Argon Y. Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc Natl Acad Sci U S A. 1996;93:5269–5274. doi: 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaut JR, Hendershot LM. Mutations within the nucleotide binding site of immunoglobulin-binding protein inhibit ATPase activity and interfere with release of immunoglobulin heavy chain. J Biol Chem. 1993;268:7248–7255. [PubMed] [Google Scholar]
- 72.Paton AW, Beddoe T, Thorpe CM, Whisstock JC, Wilce MC, Rossjohn J, Talbot UM, Paton JC. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature. 2006;443:548–552. doi: 10.1038/nature05124. [DOI] [PubMed] [Google Scholar]
- 73.Hendershot LM, Wei JY, Gaut JR, Lawson B, Freiden PJ, Murti KG. In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol Biol Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei J, Hendershot LM. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J Biol Chem. 1995;270:26670–26676. doi: 10.1074/jbc.270.44.26670. [DOI] [PubMed] [Google Scholar]
- 75.Lamb HK, Mee C, Xu W, Liu L, Blond S, Cooper A, Charles IG, Hawkins AR. The affinity of a major Ca2+ binding site on GRP78 is differentially enhanced by ADP and ATP. J Biol Chem. 2006;281:8796–8805. doi: 10.1074/jbc.M503964200. [DOI] [PubMed] [Google Scholar]
- 76.Vanhove M, Usherwood YK, Hendershot LM. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity. 2001;15:105–114. doi: 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- 77.Bracher A, Verghese J. GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG Domain Proteins: Nucleotide Exchange Factors for Hsp70 Molecular Chaperones. Subcell Biochem. 2015;78:1–33. doi: 10.1007/978-3-319-11731-7_1. [DOI] [PubMed] [Google Scholar]
- 78.Boisrame A, Beckerich JM, Gaillardin C. Sls1p, an endoplasmic reticulum component, is involved in the protein translocation process in the yeast Yarrowia lipolytica. J Biol Chem. 1996;271:11668–11675. doi: 10.1074/jbc.271.20.11668. [DOI] [PubMed] [Google Scholar]
- 79.Boisrame A, Kabani M, Beckerich JM, Hartmann E, Gaillardin C. Interaction of Kar2p and Sls1p is required for efficient co-translational translocation of secreted proteins in the yeast Yarrowia lipolytica. J Biol Chem. 1998;273:30903–30908. doi: 10.1074/jbc.273.47.30903. [DOI] [PubMed] [Google Scholar]
- 80.Kabani M, Beckerich JM, Gaillardin C. Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol Cell Biol. 2000;20:6923–69 34. doi: 10.1128/mcb.20.18.6923-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531:339–342. doi: 10.1016/s0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- 83.Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raynes DA, Guerriero V., Jr Inhibition of Hsp70 ATPase activity and protein renaturation by a novel Hsp70-binding protein. J Biol Chem. 1998;273:32883–32888. doi: 10.1074/jbc.273.49.32883. [DOI] [PubMed] [Google Scholar]
- 85.Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 86.Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 87.Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 88.Yan M, Li J, Sha B. Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor. Biochem J. 2011;438:447–455. doi: 10.1042/BJ20110500. [DOI] [PubMed] [Google Scholar]
- 89.Yi M, Chi MH, Khang CH, Park SY, Kang S, Valent B, Lee YH. The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell. 2009;21:681–695. doi: 10.1105/tpc.107.055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 91.Babour A, Kabani M, Boisrame A, Beckerich JM. Characterization of Ire1 in the yeast Yarrowia lipolytica reveals an important role for the Sls1 nucleotide exchange factor in unfolded protein response regulation. Curr Genet. 2008;53:337–346. doi: 10.1007/s00294-008-0190-1. [DOI] [PubMed] [Google Scholar]
- 92.Liu ZC, Chu J, Lin L, Song J, Ning LN, Luo HB, Yang SS, Shi Y, Wang Q, Qu N, Zhang Q, Wang JZ, Tian Q. SIL1 Rescued Bip Elevation-Related Tau Hyperphosphorylation in ER Stress. Mol Neurobiol. 2015 doi: 10.1007/s12035-014-9039-4. [DOI] [PubMed] [Google Scholar]
- 93.Ittner AA, Bertz J, Chan TY, van Eersel J, Polly P, Ittner LM. The nucleotide exchange factor SIL1 is required for glucose-stimulated insulin secretion from mouse pancreatic beta cells in vivo. Diabetologia. 2014;57:1410–1419. doi: 10.1007/s00125-014-3230-z. [DOI] [PubMed] [Google Scholar]
- 94.Weitzmann A, Baldes C, Dudek J, Zimmermann R. The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum - a quantitative approach. FEBS J. 2007;274:5175–5187. doi: 10.1111/j.1742-4658.2007.06039.x. [DOI] [PubMed] [Google Scholar]
- 95.Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 96.Hardwick KG, Lewis MJ, Semenza J, Dean N, Pelham HR. ERD1, a yeast gene required for the retention of luminal endoplasmic reticulum proteins, affects glycoprotein processing in the Golgi apparatus. EMBO J. 1990;9:623–630. doi: 10.1002/j.1460-2075.1990.tb08154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raykhel I, Alanen H, Salo K, Jurvansuu J, Nguyen VD, Latva-Ranta M, Ruddock L. A molecular specificity code for the three mammalian KDEL receptors. J Cell Biol. 2007;179:1193–1204. doi: 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anttonen AK, Siintola E, Tranebjaerg L, Iwata NK, Bijlsma EK, Meguro H, Ichikawa Y, Goto J, Kopra O, Lehesjoki AE. Novel SIL1 mutations and exclusion of functional candidate genes in Marinesco-Sjogren syndrome. Eur J Hum Genet. 2008;16:961–969. doi: 10.1038/ejhg.2008.22. [DOI] [PubMed] [Google Scholar]
- 99.Howes J, Shimizu Y, Feige MJ, Hendershot LM. C-terminal mutations destabilize SIL1/BAP and can cause Marinesco-Sjogren syndrome. J Biol Chem. 2012;287:8552–8560. doi: 10.1074/jbc.M111.333286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen X, Easton D, Oh HJ, Lee-Yoon DS, Liu X, Subjeck J. The 170 kDa glucose regulated stress protein is a large HSP70-, HSP110-like protein of the endoplasmic reticulum. FEBS Lett. 1996;380:68–72. doi: 10.1016/0014-5793(96)00011-7. [DOI] [PubMed] [Google Scholar]
- 101.Craven AR, Tyson JR, Stirling CJ. A novel subfamily of Hsp70s in the endoplasmic reticulum. Trends Cell Biol. 1997;7:277–282. doi: 10.1016/S0962-8924(97)01079-9. [DOI] [PubMed] [Google Scholar]
- 102.Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shimizu Y, Hendershot LM. Organization of the functions and components of the endoplasmic reticulum. Adv Exp Med Biol. 2007;594:37–46. doi: 10.1007/978-0-387-39975-1_4. [DOI] [PubMed] [Google Scholar]
- 104.Olden K, Pratt RM, Yamada KM. Role of carbohydrates in protein secretion and turnover: effects of tunicamycin on the major cell surface glycoprotein of chick embryo fibroblasts. Cell. 1978;13:461–473. doi: 10.1016/0092-8674(78)90320-3. [DOI] [PubMed] [Google Scholar]
- 105.Heacock CS, Sutherland RM. Induction characteristics of oxygen regulated proteins. Int J Radiat Oncol Biol Phys. 1986;12:1287–1290. doi: 10.1016/0360-3016(86)90155-0. [DOI] [PubMed] [Google Scholar]
- 106.Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- 107.Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hamilton TG, Flynn GC. Cer1p, a novel Hsp70-related protein required for posttranslational endoplasmic reticulum translocation in yeast. J Biol Chem. 1996;271:30610–30613. doi: 10.1074/jbc.271.48.30610. [DOI] [PubMed] [Google Scholar]
- 109.Dierks T, Volkmer J, Schlenstedt G, Jung C, Sandholzer U, Zachmann K, Schlotterhose P, Neifer K, Schmidt B, Zimmermann R. A microsomal ATP-binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]
- 110.Ikeda J, Kaneda S, Kuwabara K, Ogawa S, Kobayashi T, Matsumoto M, Yura T, Yanagi H. Cloning and expression of cDNA encoding the human 150 kDa oxygen-regulated protein, ORP150. Biochem Biophys Res Commun. 1997;230:94–99. doi: 10.1006/bbrc.1996.5890. [DOI] [PubMed] [Google Scholar]
- 111.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 113.Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, Jin S, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–243. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin HY, Masso-Welch P, Di YP, Cai JW, Shen JW, Subjeck JR. The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol Biol Cell. 1993;4:1109–1119. doi: 10.1091/mbc.4.11.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kuznetsov G, Chen LB, Nigam SK. Multiple molecular chaperones complex with misfolded large oligomeric glycoproteins in the endoplasmic reticulum. J Biol Chem. 1997;272:3057–3063. doi: 10.1074/jbc.272.5.3057. [DOI] [PubMed] [Google Scholar]
- 116.Schmidt BZ, Perlmutter DH. Grp78, Grp94, and Grp170 interact with alpha1-antitrypsin mutants that are retained in the endoplasmic reticulum. Am J Physiol Gastrointest Liver Physiol. 2005;289:G444–G455. doi: 10.1152/ajpgi.00237.2004. [DOI] [PubMed] [Google Scholar]
- 117.Bando Y, Ogawa S, Yamauchi A, Kuwabara K, Ozawa K, Hori O, Yanagi H, Tamatani M, Tohyama M. 150-kDa oxygen-regulated protein (ORP150) functions as a novel molecular chaperone in MDCK cells. Am J Physiol Cell Physiol. 2000;278:C1172–C1182. doi: 10.1152/ajpcell.2000.278.6.C1172. [DOI] [PubMed] [Google Scholar]
- 118.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Spee P, Neefjes J. TAP-translocated peptides specifically bind proteins in the endoplasmic reticulum, including gp96, protein disulfide isomerase and calreticulin. Eur J Immunol. 1997;27:2441–2449. doi: 10.1002/eji.1830270944. [DOI] [PubMed] [Google Scholar]
- 120.Spee P, Subjeck J, Neefjes J. Identification of novel peptide binding proteins in the endoplasmic reticulum: ERp72, calnexin, and grp170. Biochemistry. 1999;38:10559–10566. doi: 10.1021/bi990321r. [DOI] [PubMed] [Google Scholar]
- 121.Park J, Easton DP, Chen X, MacDonald IJ, Wang XY, Subjeck JR. The chaperoning properties of mouse grp170, a member of the third family of hsp70 related proteins. Biochemistry. 2003;42:14893–14902. doi: 10.1021/bi030122e. [DOI] [PubMed] [Google Scholar]
- 122.de Keyzer J, Steel GJ, Hale SJ, Humphries D, Stirling CJ. Nucleotide binding by Lhs1p is essential for its nucleotide exchange activity and for function in vivo. J Biol Chem. 2009;284:31564–31571. doi: 10.1074/jbc.M109.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Behnke J, Hendershot LM. The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s. J Biol Chem. 2014;289:2899–2907. doi: 10.1074/jbc.M113.507491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Buck TM, Plavchak L, Roy A, Donnelly BF, Kashlan OB, Kleyman TR, Subramanya AR, Brodsky JL. The Lhs1/GRP170 chaperones facilitate the endoplasmic reticulum-associated degradation of the epithelial sodium channel. J Biol Chem. 2013;288:18366–18380. doi: 10.1074/jbc.M113.469882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell. 2010;21:1047–1058. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mattoo RU, Sharma SK, Priya S, Finka A, Goloubinoff P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J Biol Chem. 2013;288:21399–21411. doi: 10.1074/jbc.M113.479253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang XY, Kazim L, Repasky EA, Subjeck JR. Characterization of heat shock protein 110 and glucose-regulated protein 170 as cancer vaccines and the effect of fever-range hyperthermia on vaccine activity. J Immunol. 2001;166:490–497. doi: 10.4049/jimmunol.166.1.490. [DOI] [PubMed] [Google Scholar]
- 129.Park JE, Facciponte J, Chen X, MacDonald I, Repasky EA, Manjili MH, Wang XY, Subjeck JR. Chaperoning function of stress protein grp170, a member of the hsp70 superfamily, is responsible for its immunoadjuvant activity. Cancer Res. 2006;66:1161–1168. doi: 10.1158/0008-5472.CAN-05-2609. [DOI] [PubMed] [Google Scholar]
- 130.Wang XY, Arnouk H, Chen X, Kazim L, Repasky EA, Subjeck JR. Extracellular targeting of endoplasmic reticulum chaperone glucose-regulated protein 170 enhances tumor immunity to a poorly immunogenic melanoma. J Immunol. 2006;177:1543–1551. doi: 10.4049/jimmunol.177.3.1543. [DOI] [PubMed] [Google Scholar]
- 131.Huo W, Ye J, Liu R, Chen J, Li Q. Vaccination with a chaperone complex based on PSCA and GRP170 adjuvant enhances the CTL response and inhibits the tumor growth in mice. Vaccine. 2010;28:6333–6337. doi: 10.1016/j.vaccine.2010.06.093. [DOI] [PubMed] [Google Scholar]
- 132.Wang XY, Sun X, Chen X, Facciponte J, Repasky EA, Kane J, Subjeck JR. Superior antitumor response induced by large stress protein chaperoned protein antigen compared with peptide antigen. J Immunol. 2010;184:6309–6319. doi: 10.4049/jimmunol.0903891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yuan B, Xian R, Wu X, Jing J, Chen K, Liu G, Zhou Z. Endoplasmic reticulum chaperone glucose regulated protein 170-Pokemon complexes elicit a robust antitumor immune response in vivo. Immunobiology. 2012 doi: 10.1016/j.imbio.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 134.Zuo D, Yu X, Guo C, Yi H, Chen X, Conrad DH, Guo TL, Chen Z, Fisher PB, Subjeck JR, Wang XY. Molecular chaperoning by glucose-regulated protein 170 in the extracellular milieu promotes macrophage-mediated pathogen sensing and innate immunity. FASEB J. 2012;26:1493–1505. doi: 10.1096/fj.11-197707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Manjili MH, Park JE, Facciponte JG, Wang XY, Subjeck JR. Immunoadjuvant chaperone, GRP170, induces 'danger signals' upon interaction with dendritic cells. Immunol Cell Biol. 2006;84:203–208. doi: 10.1111/j.1440-1711.2006.01418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gao P, Sun X, Chen X, Wang Y, Foster BA, Subjeck J, Fisher PB, Wang XY. Secretable chaperone Grp170 enhances therapeutic activity of a novel tumor suppressor, mda-7/IL-24. Cancer Res. 2008;68:3890–3898. doi: 10.1158/0008-5472.CAN-08-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao P, Sun X, Chen X, Subjeck J, Wang XY. Secretion of stress protein grp170 promotes immune-mediated inhibition of murine prostate tumor. Cancer Immunol Immunother. 2009;58:1319–1328. doi: 10.1007/s00262-008-0647-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Arnouk H, Zynda ER, Wang XY, Hylander BL, Manjili MH, Repasky EA, Subjeck JR, Latif Kazim A. Tumour secreted grp170 chaperones full-length protein substrates and induces an adaptive anti-tumour immune response in vivo. Int J Hyperthermia. 2010;26:366–375. doi: 10.3109/02656730903485910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang H, Pezeshki AM, Yu X, Guo C, Subjeck JR, Wang XY. The Endoplasmic Reticulum Chaperone GRP170: From Immunobiology to Cancer Therapeutics. Front Oncol. 2014;4:377. doi: 10.3389/fonc.2014.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goeckeler JL, Petruso AP, Aguirre J, Clement CC, Chiosis G, Brodsky JL. The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 2008;582:2393–2396. doi: 10.1016/j.febslet.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu X, Sarbeng EB, Vorvis C, Kumar DP, Zhou L, Liu Q. Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J Biol Chem. 2012;287:5661–5672. doi: 10.1074/jbc.M111.275057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 143.Weitzmann A, Volkmer J, Zimmermann R. The nucleotide exchange factor activity of Grp170 may explain the non-lethal phenotype of loss of Sil1 function in man and mouse. FEBS Lett. 2006;580:5237–5240. doi: 10.1016/j.febslet.2006.08.055. [DOI] [PubMed] [Google Scholar]
- 144.Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- 145.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Raviol H, Bukau B, Mayer MP. Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 2006;580:168–174. doi: 10.1016/j.febslet.2005.11.069. [DOI] [PubMed] [Google Scholar]
- 147.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Andreasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 150.Andreasson C, Rampelt H, Fiaux J, Druffel-Augustin S, Bukau B. The endoplasmic reticulum Grp170 acts as a nucleotide exchange factor of Hsp70 via a mechanism similar to that of the cytosolic Hsp110. J Biol Chem. 2010;285:12445–12453. doi: 10.1074/jbc.M109.096735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hale SJ, Lovell SC, de Keyzer J, Stirling CJ. Interactions between Kar2p and its nucleotide exchange factors Sil1p and Lhs1p are mechanistically distinct. J Biol Chem. 2010;285:21600–21606. doi: 10.1074/jbc.M110.111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Anttonen AK, Mahjneh I, Hamalainen RH, Lagier-Tourenne C, Kopra O, Waris L, Anttonen M, Joensuu T, Kalimo H, Paetau A, Tranebjaerg L, Chaigne D, Koenig M, Eeg-Olofsson O, Udd B, Somer M, Somer H, Lehesjoki AE. The gene disrupted in Marinesco-Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet. 2005;37:1309–1311. doi: 10.1038/ng1677. [DOI] [PubMed] [Google Scholar]
- 153.Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, Breitbach-Faller N, Rudnik-Schoneborn S, Blaschek A, Wolf NI, Harting I, North K, Smith J, Muntoni F, Brockington M, Quijano-Roy S, Renault F, Herrmann R, Hendershot LM, Schroder JM, Lochmuller H, Topaloglu H, Voit T, Weis J, Ebinger F, Zerres K. Mutations in SIL1 cause Marinesco-Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet. 2005;37:1312–1314. doi: 10.1038/ng1678. [DOI] [PubMed] [Google Scholar]
- 154.Krieger M, Roos A, Stendel C, Claeys KG, Sonmez FM, Baudis M, Bauer P, Bornemann A, de Goede C, Dufke A, Finkel RS, Goebel HH, Haussler M, Kingston H, Kirschner J, Medne L, Muschke P, Rivier F, Rudnik-Schoneborn S, Spengler S, Inzana F, Stanzial F, Benedicenti F, Synofzik M, Lia Taratuto A, Pirra L, Tay SK, Topaloglu H, Uyanik G, Wand D, Williams D, Zerres K, Weis J, Senderek J. SIL1 mutations and clinical spectrum in patients with Marinesco-Sjogren syndrome. Brain. 2013;136:3634–3644. doi: 10.1093/brain/awt283. [DOI] [PubMed] [Google Scholar]
- 155.Horvers M, Anttonen AK, Lehesjoki AE, Morava E, Wortmann S, Vermeer S, van de Warrenburg BP, Willemsen MA. Marinesco-Sjogren syndrome due to SIL1 mutations with a comment on the clinical phenotype. Eur J Paediatr Neurol. 2013;17:199–203. doi: 10.1016/j.ejpn.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 156.Anttonen AK, Lehesjoki AE. Marinesco-Sjogren Syndrome. 1993 [Google Scholar]
- 157.Ezgu F, Krejci P, Li S, de Sousa C, Graham JM, Jr, Hansmann I, He W, Porpora K, Wand D, Wertelecki W, Schneider A, Wilcox WR. Phenotype-genotype correlations in patients with Marinesco-Sjogren syndrome. Clin Genet. 2014;86:74–84. doi: 10.1111/cge.12230. [DOI] [PubMed] [Google Scholar]
- 158.Sjogren T. Hereditary congenital spinocerebellar ataxia accompanied by congenital cataract and oligophrenia; a genetic and clinical investigation. Confin Neurol. 1950;10:293–308. [PubMed] [Google Scholar]
- 159.Goto M, Okada M, Komaki H, Sugai K, Sasaki M, Noguchi S, Nonaka I, Nishino I, Hayashi YK. A nationwide survey on Marinesco-Sjogren syndrome in Japan. Orphanet J Rare Dis. 2014;9:58. doi: 10.1186/1750-1172-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Riazuddin SA, Amiri-Kordestani L, Kaul H, Butt T, Jiao X, Riazuddin S, Hejtmancik JF. Novel SIL1 mutations in consanguineous Pakistani families mapping to chromosomes 5q31. Mol Vis. 2009;15:1050–1056. [PMC free article] [PubMed] [Google Scholar]
- 161.Superneau DW, Wertelecki W, Zellweger H, Bastian F. Myopathy in Marinesco-Sjogren syndrome. Eur Neurol. 1987;26:8–16. doi: 10.1159/000116305. [DOI] [PubMed] [Google Scholar]
- 162.Torbergsen T, Aasly J, Borud O, Lindal S, Mellgren SI. Mitochondrial myopathy in Marinesco-Sjogren syndrome. J Ment Defic Res. 1991;35(Pt 2):154–159. doi: 10.1111/j.1365-2788.1991.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 163.Sewry CA, Voit T, Dubowitz V. Myopathy with unique ultrastructural feature in Marinesco-Sjogren syndrome. Ann Neurol. 1988;24:576–580. doi: 10.1002/ana.410240416. [DOI] [PubMed] [Google Scholar]
- 164.Roos A, Buchkremer S, Kollipara L, Labisch T, Gatz C, Zitzelsberger M, Brauers E, Nolte K, Schroder JM, Kirschner J, Jesse CM, Goebel HH, Goswami A, Zimmermann R, Zahedi RP, Senderek J, Weis J. Myopathy in Marinesco-Sjogren syndrome links endoplasmic reticulum chaperone dysfunction to nuclear envelope pathology. Acta Neuropathol. 2014;127:761–777. doi: 10.1007/s00401-013-1224-4. [DOI] [PubMed] [Google Scholar]
- 165.Goto Y, Komiyama A, Tanabe Y, Katafuchi Y, Ohtaki E, Nonaka I. Myopathy in Marinesco-Sjogren syndrome: an ultrastructural study. Acta Neuropathol. 1990;80:123–128. doi: 10.1007/BF00308914. [DOI] [PubMed] [Google Scholar]
- 166.Georgy BA, Snow RD, Brogdon BG, Wertelecki W. Neuroradiologic findings in Marinesco-Sjogren syndrome. AJNR Am J Neuroradiol. 1998;19:281–283. [PMC free article] [PubMed] [Google Scholar]
- 167.McLaughlin JF, Pagon RA, Weinberger E, Haas JE. Marinesco-Sjogren syndrome: clinical and magnetic resonance imaging features in three children. Dev Med Child Neurol. 1996;38:636–644. doi: 10.1111/j.1469-8749.1996.tb12128.x. [DOI] [PubMed] [Google Scholar]
- 168.Sakai K, Tada M, Yonemochi Y, Nakajima T, Onodera O, Takahashi H, Kakita A. Marinesco-Sjogren syndrome with atrophy of the brain stem tegmentum and dysplastic cytoarchitecture in the cerebral cortex. Neuropathology. 2008;28:541–546. doi: 10.1111/j.1440-1789.2008.00884.x. [DOI] [PubMed] [Google Scholar]
- 169.Inaguma Y, Hamada N, Tabata H, Iwamoto I, Mizuno M, Nishimura YV, Ito H, Morishita R, Suzuki M, Ohno K, Kumagai T, Nagata K. SIL1, a causative cochaperone gene of Marinesco-Sojgren syndrome, plays an essential role in establishing the architecture of the developing cerebral cortex. EMBO Mol Med. 2014;6:414–429. doi: 10.1002/emmm.201303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet. 2005;37:974–979. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
- 171.Zhao L, Rosales C, Seburn K, Ron D, Ackerman SL. Alteration of the unfolded protein response modifies neurodegeneration in a mouse model of Marinesco-Sjogren syndrome. Hum Mol Genet. 2010;19:25–35. doi: 10.1093/hmg/ddp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Filezac de L'Etang A, Maharjan N, Cordeiro Brana M, Ruegsegger C, Rehmann R, Goswami A, Roos A, Troost D, Schneider BL, Weis J, Saxena S. Marinesco-Sjogren syndrome protein SIL1 regulates motor neuron subtype-selective ER stress in ALS. Nat Neurosci. 2015;18:227–238. doi: 10.1038/nn.3903. [DOI] [PubMed] [Google Scholar]
- 174.Schmitz A, Maintz M, Kehle T, Herzog V. In vivo iodination of a misfolded proinsulin reveals co-localized signals for Bip binding and for degradation in the ER. EMBO J. 1995;14:1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Scheuner D, Vander Mierde D, Song B, Flamez D, Creemers JW, Tsukamoto K, Ribick M, Schuit FC, Kaufman RJ. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 176.Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 177.Ichhaporia VP, Sanford T, Howes J, Marion TN, Hendershot LM. Sil1, a nucleotide exchange factor for BiP, is not required for antibody assembly or secretion. Mol Biol Cell. 2014 doi: 10.1091/mbc.E14-09-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kusaczuk M, Cechowska-Pasko M. Molecular chaperone ORP150 in ER stress-related diseases. Curr Pharm Des. 2013;19:2807–2818. doi: 10.2174/1381612811319150016. [DOI] [PubMed] [Google Scholar]
- 179.Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- 180.Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 181.Inoue T, Tsai B. A Nucleotide Exchange Factor Promotes ER-to-cytosol Membrane Penetration of the Non-enveloped Virus SV40. J Virol. 2015 doi: 10.1128/JVI.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kobayashi T, Ogawa S, Yura T, Yanagi H. Abundant expression of 150-kDa oxygen-regulated protein in mouse pancreatic beta cells is correlated with insulin secretion. Biochem Biophys Res Commun. 2000;267:831–837. doi: 10.1006/bbrc.1999.2052. [DOI] [PubMed] [Google Scholar]
- 183.Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, Ozawa K, Ogawa S, Hori M, Yamasaki Y, Matsuhisa M. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- 184.Ozawa K, Miyazaki M, Matsuhisa M, Takano K, Nakatani Y, Hatazaki M, Tamatani T, Yamagata K, Miyagawa J, Kitao Y, Hori O, Yamasaki Y, Ogawa S. The endoplasmic reticulum chaperone improves insulin resistance in type 2 diabetes. Diabetes. 2005;54:657–663. doi: 10.2337/diabetes.54.3.657. [DOI] [PubMed] [Google Scholar]
- 185.Ozawa K, Tsukamoto Y, Hori O, Kitao Y, Yanagi H, Stern DM, Ogawa S. Regulation of tumor angiogenesis by oxygen-regulated protein 150, an inducible endoplasmic reticulum chaperone. Cancer Res. 2001;61:4206–4213. [PubMed] [Google Scholar]
- 186.Ozawa K, Kondo T, Hori O, Kitao Y, Stern DM, Eisenmenger W, Ogawa S, Ohshima T. Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. J Clin Invest. 2001;108:41–50. doi: 10.1172/JCI11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tamatani M, Matsuyama T, Yamaguchi A, Mitsuda N, Tsukamoto Y, Taniguchi M, Che YH, Ozawa K, Hori O, Nishimura H, Yamashita A, Okabe M, Yanagi H, Stern DM, Ogawa S, Tohyama M. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nat Med. 2001;7:317–323. doi: 10.1038/85463. [DOI] [PubMed] [Google Scholar]
- 188.Kitao Y, Ozawa K, Miyazaki M, Tamatani M, Kobayashi T, Yanagi H, Okabe M, Ikawa M, Yamashima T, Stern DM, Hori O, Ogawa S. Expression of the endoplasmic reticulum molecular chaperone (ORP150) rescues hippocampal neurons from glutamate toxicity. J Clin Invest. 2001;108:1439–1450. doi: 10.1172/JCI12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Miyazaki M, Ozawa K, Hori O, Kitao Y, Matsushita K, Ogawa S, Matsuyama T. Expression of 150-kd oxygen-regulated protein in the hippocampus suppresses delayed neuronal cell death. J Cereb Blood Flow Metab. 2002;22:979–987. doi: 10.1097/00004647-200208000-00009. [DOI] [PubMed] [Google Scholar]
- 190.Hyman BT, Yuan J. Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat Rev Neurosci. 2012;13:395–406. doi: 10.1038/nrn3228. [DOI] [PubMed] [Google Scholar]
- 191.Kitao Y, Hashimoto K, Matsuyama T, Iso H, Tamatani T, Hori O, Stern DM, Kano M, Ozawa K, Ogawa S. ORP150/HSP12A regulates Purkinje cell survival: a role for endoplasmic reticulum stress in cerebellar development. J Neurosci. 2004;24:1486–1496. doi: 10.1523/JNEUROSCI.4029-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tsukamoto Y, Kuwabara K, Hirota S, Ikeda J, Stern D, Yanagi H, Matsumoto M, Ogawa S, Kitamura Y. 150-kD oxygen-regulated protein is expressed in human atherosclerotic plaques and allows mononuclear phagocytes to withstand cellular stress on exposure to hypoxia and modified low density lipoprotein. J Clin Invest. 1996;98:1930–1941. doi: 10.1172/JCI118994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sanson M, Ingueneau C, Vindis C, Thiers JC, Glock Y, Rousseau H, Sawa Y, Bando Y, Mallat Z, Salvayre R, Negre-Salvayre A. Oxygen-regulated protein-150 prevents calcium homeostasis deregulation and apoptosis induced by oxidized LDL in vascular cells. Cell Death Differ. 2008;15:1255–1265. doi: 10.1038/cdd.2008.36. [DOI] [PubMed] [Google Scholar]
- 194.Sanson M, Auge N, Vindis C, Muller C, Bando Y, Thiers JC, Marachet MA, Zarkovic K, Sawa Y, Salvayre R, Negre-Salvayre A. Oxidized low-density lipoproteins trigger endoplasmic reticulum stress in vascular cells: prevention by oxygen-regulated protein 150 expression. Circ Res. 2009;104:328–336. doi: 10.1161/CIRCRESAHA.108.183749. [DOI] [PubMed] [Google Scholar]
- 195.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145:341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]