Abstract
Phosphoinositides regulate numerous cellular processes, by recruiting cytosolic effector proteins and acting as membrane signaling entities. The cellular metabolism and localization of phosphoinositides are tightly regulated by distinct lipid kinases and phosphatases. Here, we identify and characterize a unique phosphatidylinositol 3-Kinase (PI3K) in Toxoplasma gondii, a protozoan parasite belonging to the phylum Apicomplexa. Conditional depletion of this enzyme and subsequently of its product, PI(3)P, drastically alters the morphology and inheritance of the apicoplast, an endosymbiontic organelle of algal origin that is a unique feature of many Apicomplexa. We searched the T. gondii genome for PI(3)P binding proteins and identified in total six PX and FYVE-domain containing proteins including a PIKfyve lipid kinase, which phosphorylates PI(3)P into PI(3,5)P2. While depletion of putative PI(3)P binding proteins shows that they are not essential for parasite growth and apicoplast biology, conditional disruption of PIKfyve induces enlarged apicoplasts, as observed upon the loss of PI(3)P. A similar defect of apicoplast homeostasis was also observed by knocking-down the PIKfyve regulatory protein ArPIKfyve, suggesting that in T. gondii, PI(3)P-related function for the apicoplast might mainly be to serve as a precursor for the synthesis of PI(3,5)P2. Accordingly, PI3K is conserved in all apicomplexan parasites whereas PIKfyve and ArPIKfyve are absent in Cryptosporidium species which lack an apicoplast, supporting a direct role of PI(3,5)P2 in apicoplast homeostasis. This study enriches the already diverse functions attributed to PI(3,5)P2 in eukaryotic cells and highlights these parasite lipid kinases as potential drug targets.
Keywords: Apicomplexa, Toxoplasma gondii, Tet-inducible system, Traffic, Apicoplast biogenesis, Delayed death phenotype, Lipid modifications, Phosphoinositides
Introduction
Toxoplasma gondii is a member of the phylum Apicomplexa. T. gondii shares many biological features with species of the genus Plasmodium, the causative agents of malaria, and has emerged as an excellent model in particular for cell biological studies. Most apicomplexan parasites contain an organelle of algal origin named apicoplast, acquired by secondary endosymbiosis and surrounded by four membranes (Kohler et al., 1997, McFadden et al., 1996, Wilson et al., 1996). The apicoplast is essential for parasite survival and harbours different metabolic pathways originating from bacteria (Striepen, 2011). Apicoplast proteins are mainly nuclear encoded and post-translationally imported via the secretory system (Waller et al., 2000); however the mechanism by which proteins are transferred from the secretory system to the apicoplast is poorly understood. The endosymbiotic origin of the apicoplast during which a red algal cell was engulfed by a flagellated protist places the symbiont initially into the endosomal compartment (Janouskovec et al., 2010, van Dooren et al., 2013). Consistent with this idea we recently showed that the outer membranes of the apicoplast are decorated with phosphatidylinositol 3-monophosphate PI(3)P (Tawk et al., 2011), a phosphoinositide (PI) which serves as key regulator of endosomal trafficking in yeast and mammalian cells and is also involved in vacuolar protein sorting (Balla, 2013, Stack et al., 1995).
Indeed, PI(3)P concentrates mainly on endosomes and is mainly known for roles in membrane trafficking from early endosomes to lysosomes (Di Paolo et al., 2006, Volpicelli-Daley et al., 2007). In addition, PI(3)P is also central to autophagy in mammalian cells (Petiot et al., 2000) and yeast (Obara et al., 2011). More recently, its role in cytokinesis has been demonstrated (Sagona et al., 2010). PI(3)P exercises its effects by recruiting a variety of cytosolic proteins that contain mainly FYVE (FAB1-YOTB-Vac1-EEA1) domains (Hayakawa et al., 2004, Katzmann et al., 2003, Kutateladze et al., 1999) or PX domains (Cheever et al., 2001). In yeast and unicellular eukaryotes, PI(3)P is produced by phosphorylation of phosphatidylinositol at position 3 by the class III PI3-Kinase (or Vps34 in yeast) (Figure 1A) (Stack et al., 1994). PI(3)P is also a precursor required for the production of PI(3,5)P2 by a PI(3)P 5-kinase called PIKfyve in mammals (Fab1 in yeast). PIKfyve contains a FYVE domain that binds PI(3)P and phosphorylates it at position 5 of the inositol ring to produce PI(3,5)P2 (Figure 1A) (Gary et al., 1998, McEwen et al., 1999). PI(3,5)P2 is of low abundance yet essential for both yeast and animal cells (Dove et al., 2009). Since its discovery in 1997, the number of pathways found to be regulated by PI(3,5)2 have quickly expanded. It includes major roles in the dynamic between endosomes and lysosomes and between these organelles and other cell compartments, the acidification of the vacuole, autophagy and control of membrane ion transport (de Lartigue et al., 2009, Dong et al., 2010, Ikonomov et al., 2003, Jefferies et al., 2008, Nicot et al., 2006, Rusten et al., 2006, Rutherford et al., 2006).
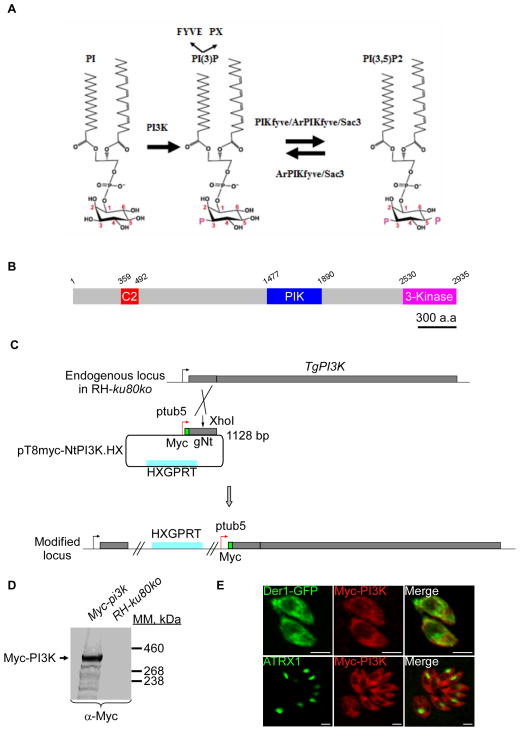
Figure 1. Expression and localization of TgPI3-Kinase in tachyzoites.
(A) Depicted are the two pathways implicated in PI(3,5)P2 biosynthesis and turnover. The PI3Kinase converts PI to PI(3)P and the PAS complex (PIKfyve, ArPIKfyve and Sac3) converts PI(3)P to PI(3,5)P2. ArPIKfyve binds to the Sac3 phosphatase that reverts PI(3,5)P2 back to PI(3)P. The FYVE and PX domains containing proteins are considered as PI(3)P-recognizing effectors. (B) Schematic representation of T. gondii PI3K predicted using SMART EMBL showing the C2 (a calcium dependent membrane-targeting module that, in others proteins, binds a wide variety of molecules), the PIK domain (phosphoinositide 3-kinase family accessory domain) that has been suggested to be involved in substrate presentation, and finally the 3-kinase domain responsible for the kinase activity. Scale bar represents 300 aa. (C) Insertion of a Myc tag at the N-terminus of TgPI3K and promoter exchange by single homologous recombination at the 5′ of the gene (knock-in in RH-ku80ko strain). gNt: genomic N-terminal. (D) Western blot analysis performed on transgenic or RH-ku80ko parasite lysates probed with anti-Myc antibodies. Myc-TgPI3K is found at the expected molecular mass (314 kDa). (E) IFA performed on intracellular transgenic parasites using anti-Myc or anti-ATRX1 (apicoplast) antibodies. Der1-GFP protein was used as ER marker. Scale bars represent 2 μm.
In P. falciparum, PI(3)P has been described as a key player in endocytosis by transporting haemoglobin containing vesicles from the red blood cell to the food vacuole of the malaria parasite (Vaid et al., 2010) and to facilitate the export of host-targeting signal proteins from the intracellular parasite into the host erythrocyte (Bhattacharjee et al., 2012). In addition to the food vacuole, in P. falciparum PI(3)P also localizes to the apicoplast (Tawk et al., 2010). Studies in Toxoplasma confirmed the association of PI(3)P with the apicoplast, and moreover suggested a role for this lipid in apicoplast biology. Interfering with PI(3)P function by over-expressing a PI(3)P-binding module in the parasite or by inhibiting PI3K activity by high concentrations of the inhibitor LY294002, led to the loss of the organelle and ultimately to the death of the parasite (Tawk et al., 2011). While compelling these studies relied solely on indirect observations and the high drug concentrations that were required may have led to off-target effects (Tawk et al., 2011).
In the present study, we addressed the role of PI(3)P in Toxoplasma directly by ablating the parasite enzyme required for its synthesis. We generated conditional knock-out mutants of the Toxoplasma PI3-Kinase and showed the essential contribution of this lipid kinase in the preservation of apicoplast integrity. We searched the T. gondii genome for PI(3)P binding proteins and identify six PX and FYVE-containing proteins including a PIKfyve lipid kinase. Depletion of the PIKfyve lipid kinase disrupted apicoplast morphology and induced a delayed parasites death, suggesting that PI(3)P serves as an intermediate in this pathway. Disruption of Toxoplasma ArPIKfyve, the regulator of PIKfyve, results in a similar apicoplast biogenesis defect, highlighting the essential role of PI(3,5)P2 synthesis in the homeostasis of the apicoplast.
Results
TgPI3-Kinase is produced at low levels in tachyzoites and is essential for parasite survival and growth
Homology searches in the ToxoDB database (http://www.ToxoDB.org) revealed the presence of one PI3-Kinase protein in T. gondii belonging to the class III and conserved across the Apicomplexa phylum. TgPI3K is a large protein with a predicted molecular weight of 313 kDa, displaying a domain organization typical for class III PI3-Kinases. TgPI3K possesses a C2 domain (a calcium dependent membrane-targeting modules that binds a wide variety of substances including phospholipids, inositol polyphosphates, and intracellular proteins), a PIK domain (phosphoinositide 3-kinase family accessory domain, that has been suggested to be involved in substrate presentation) and a 3-kinase domain located at the extreme carboxy-terminus responsible for the kinase activity (Figure 1B). We tagged the endogenous TgPI3K locus with a triple HA epitope tag but did not detect its expression by IFA or Western blotting (data not shown). This suggested that the endogenous levels of PI3K are low. Detection of TgPI3K was possible when the endogenous promoter of the TgPI3K gene was replaced with the stronger tubulin promoter (Figure 1C). The resulting transgenic parasites expressed the Myc-tagged PI3K protein at the expected mass, as assessed by Western blot (Figure 1D). The tagged protein is detected by IFA as a diffuse punctate cytoplasmic signal in intracellular parasites (Figure 1E) that might possibly correspond in part to the ER as indicated by partial co-localization with the ER-marker Der1-GFP. However, no apparent localization of Myc-PI3K at the apicoplast was observed in these conditions (Figure 1E).
To explore the function of the TgPI3K protein, we exchanged the native promoter with the tetracycline-regulable 7-tet-OpSag4 promoter (Daher et al., 2010), and a 2-HA epitope tag to generate strain pi3ki (Figure 2A). Correct double homologous integration of the cassette at the TgPI3K locus was confirmed by PCR (Figure 2B). We measured the specific modulation of PI3K expression levels by RT-PCR analysis. In samples from mutant parasites grown in the presence of anhydrous tetracycline (ATc) for three days, we observed a significant decrease in TgPI3K mRNA transcript (Figure 2D). A complemented line, pi3kiC, was constructed by introduction of a modified cosmid harbouring the endogenous TgPI3K gene tagged with a C-terminal 3HA epitope (Figure 2C). Transcript levels are unaffected in ATc treated wild-type parasites and are restored in the complemented strain (Figure 2D).
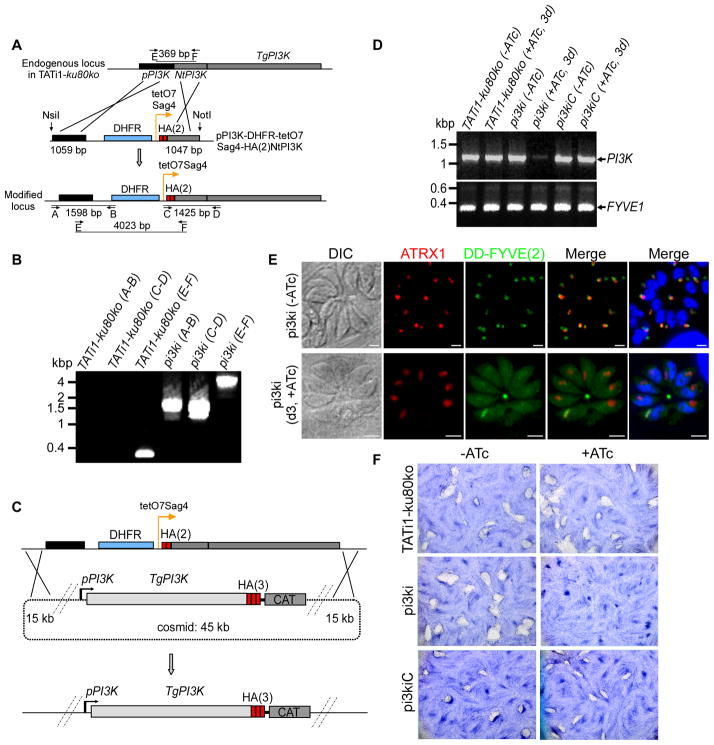
Figure 2. TgPI3-Kinase conditional knock-in by promoter exchange strategy.
(A) Schematic representation of the strategy used to replace the endogenous promoter of TgPI3K with the tetracycline-inducible promoter. The 5′TgPI3KUTR-DHFR-tetO7-SAG4-HA(2)NtTgPI3K plasmid contains the TgPI3K 5′ UTR (in black), the dihydrofolate reductase (DHFR) cassette (in blue) and the N-terminal genomic coding sequence of TgPI3K gene (grey) fused to two HA tags (red) under the control of the inducible tetO7SAG4 promoter (orange arrow). Black arrows represent the primers used for PCR analysis and the length of the PCR fragments is indicated. (B) PCR analysis performed on pi3ki confirming double homologous recombination. Genomic DNA from TATi1-ku80ko parasites was used as negative control. (C) Schematic representation of the crossover event that will result in the replacement of TgPI3K tetracycline inducible locus by the PI3-Kinase gene present in the recombineering modified ToxP331 cosmid in fusion with a 3 HA epitope at the 3′ end of the TgPI3K gene. CAT: chloramphenicol-acetyl-transferase. (D) Semi-quantitative RT-PCR analysis of TgPI3K expression in the wild-type, mutant and complemented parasites, preceded or not by three days of induction by ATc to regulate expression. Primers specific of the gene coding for FYVE1 were used as a loading control. (E) DD-FYVE(2)-GFP expressing pi3ki parasites were processed for IFA ± ATc and in the presence of Shield-1. The DD-FYVE(2)-GFP protein shows a punctuate staining in pi3ki parasites non-treated with ATc. A cytosolic labelling was observed when the pi3ki parasites were cultured in the presence of ATc. (F) Plaque assay of HFF monolayer infected with TATi1-ku80ko or pi3ki or pi3kiC parasites pre-treated first during 48 h with ATc. After 7 days ± ATc, the HFF were stained with Giemsa.
The level and intracellular distribution of PI(3)P (the product of the PI3-Kinase activity) in the pi3ki strain in the presence of ATc, was followed using a DD-FYVE(2)-GFP reporter as described previously (Gillooly et al., 2000, Tawk et al., 2011). For this, DD-FYVE(2)-GFP was introduced into the pi3ki strain by stable transfection and analysed upon short term stabilisation with Shield-1. In absence of ATc, DD-FYVE(2)-GFP labeled the apicoplast as described previously (Tawk et al., 2011), (Figure 2E, upper panel). In contrast, after three days of ATc treatment followed by 20 min of Shield-1 induction, DD-FYVE(2)-GFP staining appeared diffuse throughout the cytoplasm, even when the apicoplast was still detectable using a marker for its outer membrane (Figure 2E, lower panel). We conclude that loss of PI3K leads to a substantial decrease in the level of PI(3)P at the apicoplast and that this reduction precedes the substantial loss of the organelle (see below).
The phenotypic consequences of PI3K knock-down on the parasite lytic cycle were first examined by following the formation of plaques in a monolayer of host cells that result from multiple cycles of the parasite invasion and egress over 7 days. When pi3ki parasites were pre-treated for two days with ATc and further depleted of PI3K by 7 consecutive days of ATc treatment, no plaques were observed, while mock-treated pi3ki parasites formed plaques similar in size to those of the TATi1-ku80ko recipient strain or the pi3kiC complemented strain (Figure 2F). These results show that PI3K is critical for T. gondii parasite growth.
PI(3)P is essential for apicoplast homeostasis
Since PI(3)P has been suggested to be involved in apicoplast biology (Tawk et al., 2011), we assessed the presence of apicoplasts in pi3ki parasites. We noticed that knock-down of PI3K affected the size of the apicoplast (Figure 3A). The organelle became enlarged at day 4 and lost at day 5, leaving behind small structures which might correspond to disintegrated apicoplast fragments or alternatively to trafficking vesicles containing apicoplast proteins which can no longer fuse with their target organelle (Figure 3A, lower panel). The other organelles or sub-cellular structures such as micronemes, dense granules, mitochondria, inner membrane complex (IMC), and rhoptries remained intact (Supplementary figures 1A, 1B, 1C, 1D and 1E respectively), showing that it is a specific effect on the apicoplast. Electron microscopic analysis confirmed that the apicoplast is massively enlarged in the mutant; similar morphological changes were observed after inhibiting PI3K activity pharmacologically (Tawk et al., 2011). The 4 membranes surrounding the organelle remained visible but the interior of the apicoplast appeared electron lucent when compared to the more compact apicoplasts of untreated cells (Figure 3B).
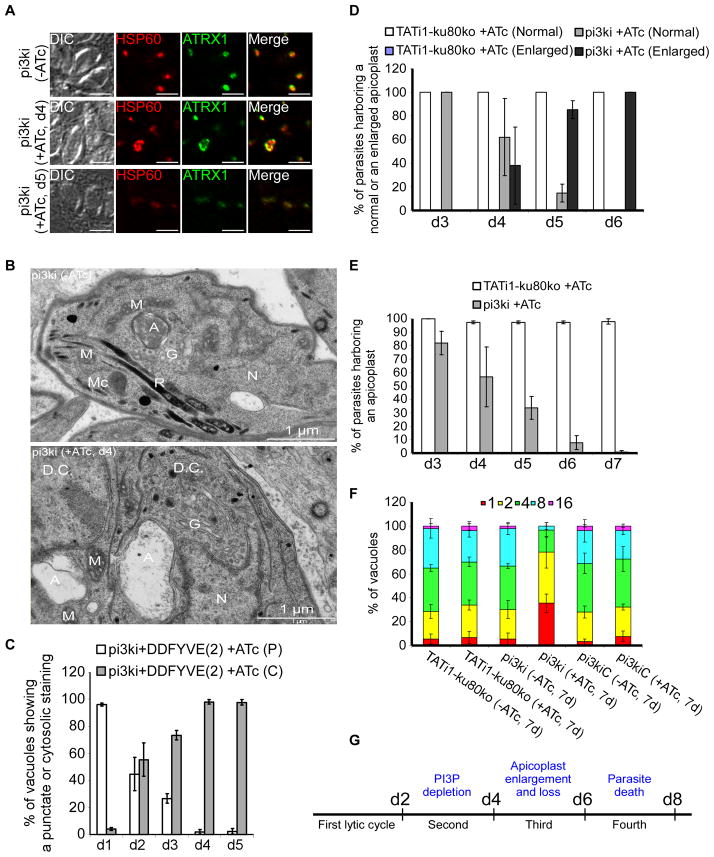
Figure 3. Phenotypic consequences of TgPI3-Kinase knock-down in pi3ki strain.
(A) IFA of a representative pi3ki vacuole non-treated with ATc compared with pi3ki ATc-treated vacuoles after 4 and 5 days. Anti-ATRX1 (in green) and anti-HSP60 (in red) antibodies were used to detect the apicoplast. (B) PI3-Kinase depleted parasites showed an enlarged apicoplast. Thin section electron micrographs were taken from pi3ki that had grown for 4 days in the presence or absence of ATc. A, apicoplast; M, mitochondrion; Mc, micronemes; R, rhoptries; G, Golgi apparatus; N, nucleus; D.C, daughter cell. Scale bar, 1 μm. (C) Quantification of vacuoles showing a punctuate (P) or cytosolic (C) staining upon treatment with ATc for 1 to 5 days by visualizing the DD-FYVE(2)-GFP fluorescence. Values are means ± SD for three independent experiments. (D) Quantification of TATi1-ku80ko or pi3ki parasites harbouring either a normal or an enlarged apicoplast upon treatment with ATc for 3 to 6 days using anti-ATRX1 antibodies. (E) Quantification of TATi1-ku80ko or pi3ki parasites harbouring an apicoplast upon treatment with ATc for 3 to 7 days using anti-ATRX1 and anti-HSP60 antibodies. (F) Intracellular growth of TATi1-ku80ko, pi3ki and pi3kiC parasites cultivated in presence or absence of ATc for 6 days and allowed to invade new HFF cells. Numbers of parasites per vacuole (x-axis) were counted 24 h after inoculation. The percentages of vacuoles containing varying numbers of parasites are represented on the y- axis. (D, E and F) Values are means ± SD for three independent experiments. (G) Schematic diagram showing in the course of time the different steps leading to the delayed death phenotype of pi3ki parasites.
We next studied the kinetics of the events that lead from the knock-down of PI3K to the death of the parasite. Reduction of PI(3)P as measured by aberrant cytosolic staining with DD-FYVE(2)-GFP was first observed at day 2 and appeared complete at day 4 (98% ±2) (Figure 3C). Changes in apicoplast shape became apparent at day 4, subsequent to the loss of PI(3)P, and reached 100% at day 6 (Figure 3D). Loss of the apicoplast was complete at day 7 (Figure 3E). The defects in apicoplast shape observed at day 4 did not significantly affect the intracellular growth of pi3ki parasites at day 5 compared to the wild type or the complemented strains (Supplementary figure 1F). The onset of a severe block of growth only occurred once the apicoplast had been lost, and after the next round of invasion (Figure 3F). The parasites did not progress through cell division as shown by the accumulation of vacuoles with only one, two or four parasites at day 7. These results showed that PI(3)P reduction led to a delay between the loss of the apicoplast and the subsequent death of the parasites (Figure 3G), a classical phenotype associated with the loss of the apicoplast and called “delayed death phenotype” (Fichera et al., 1997) even if disruption of apicoplast functions can also lead to an immediate death (Brooks et al., 2010, Yeh et al., 2011).
Taken together these results demonstrate a role for PI3kinase and its product PI(3)P in apicoplast homeostasis. Importantly, the parasite appears unable to compensate the loss of its PI3K enzyme by salvage of host cell PI(3)P.
Expression and subcellular localization of T. gondii FYVE and PX domain containing proteins
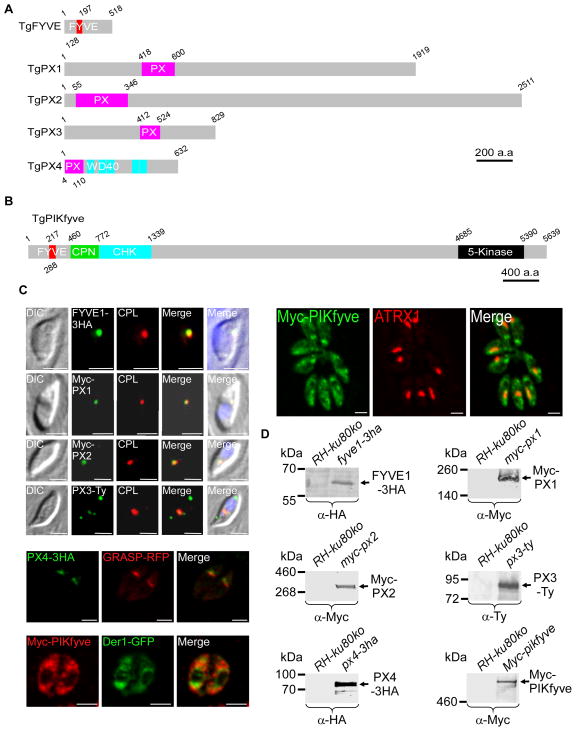
PI(3)P exercises its function by recruiting effector proteins containing FYVE or PX domains (Hayakawa et al., 2004, Katzmann et al., 2003, Kutateladze et al., 1999, Cheever et al., 2001). In order to understand how PI(3)P may control apicoplast integrity, we decided to characterize PI(3)P effector proteins in Toxoplasma. A BLAST homology searches across the available T. gondii genome (ToxoDB version 10) revealed 4 PX and 2 FYVE domain containing proteins (Figures 4A and 4B, Supplementary figure 2), including the lipid kinase PI(3)P 5-kinase (or PIKfyve). We named these proteins TgFYVE1, TgPIKfyve, TgPX1, TgPX2, TgPX3 and TgPX4. RT-PCR and annotation experiments confirmed the transcription of these genes in tachyzoites, but we detected protein only for TgFYVE1 and TgPX4 following endogenous tagging (Figures 4C and 4D, Supplementary figure 3A). We used over-expression under the strong tubulin promoter to detect the remaining proteins (Figures 4C and 4D, Supplementary figures 3B and 3C). Interestingly, TgFYVE1, TgPX1, TgPX2 and TgPX3 were found associated with the recently described vacuolar compartment (VAC), a highly dynamic compartment that houses lysosomal markers such as cathepsin-like cysteine protease (TgCPL) (Figure 4C) (Parussini et al., 2010). TgPX3 protein partially localized to the VAC compartment but was also detected associated with other unknown subcellular structures (Figure 4C). TgPX4 was found next to the cis-Golgi marker GRASP (Figure 4C). Finally, TgPIKfyve displayed a dot-like staining throughout the cytoplasm and the possibility of a partial co-localization with the apicoplast could not be excluded (Figure 4C). We conclude that none of the putative PI(3)P effector proteins is associated specifically to the apicoplast, while the lipid kinase TgPIKfyve could suggest the possibility of partial co-localization with the apicoplast.
Figure 4. Repertoire of proteins containing FYVE and PX domains, their expression and their localization in T. gondii.
(A and B) Schematic representation of the primary structure of the proteins highlighting their different domains; the domains have been searched with SMART (http://smart.embl-heidelberg.de). The FYVE domain is highlighted in red and the PX domain in pink. Only TgPIKfyve and TgPX4 possess additional recognizable domains. TgPIKfyve harbors a chaperonin domain (CPN) (green) known to be engaged in regulatory interactions, a CHK domain (Cys, His and Lys) (blue) and the 5-kinase catalytic domain (black) at the C-terminus responsible for the lipid kinase activity. PX4 contains 5 WD40 (blue) motifs known to coordinate multi-protein complex assemblies and binding to PI(3,5)P2 (Proikas-Cezanne et al., 2007). Scale bar represents 200 a.a or 400 a.a respectively. (C) Subcellular localization of the FYVE and PX proteins. The tagged proteins were detected using anti-HA, anti-Myc or anti-Ty antibodies. Anti-CPL and anti-ATRX1 antibodies were used to stain the vacuole compartment and the apicoplast, respectively; the GRASP-RFP and Der1-GFP proteins were used as a cis-Golgi and ER markers, respectively. Scale bars represent 2 μm. (D) Western blot analysis of transgenic or RH-ku80ko parasite lysates probed with anti-HA, anti-Myc or anti-Ty antibodies. All proteins were detected by western blotting at their expected molecular weight; 58 kDa for TgFYVE1, 205 kDa for TgPX1, 276 kDa for TgPX2, 91 kDa for TgPX3, 70 kDa for TgPX4 and 612 kDa for TgPIKfyve.
PIKfyve plays an essential role in maintaining apicoplast morphology
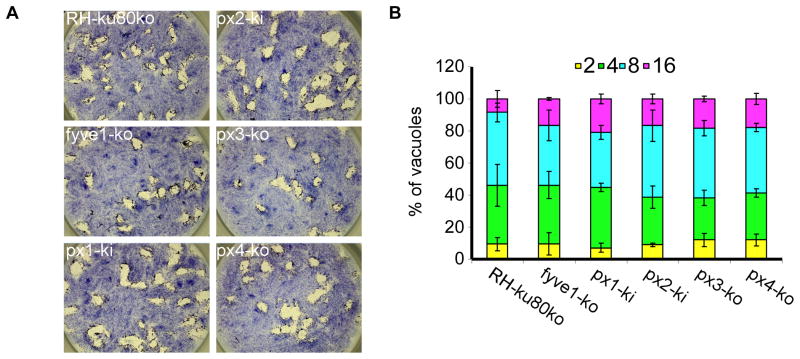
We then ask whether the identified effectors are essential for tachyzoite growth. We disrupted the TgFYVE1, TgPX3 and TgPX4 genes by removing the entire coding sequence (Supplementary figures 4A, 4B, 4I, 4J, 4K and 4L) and TgPX1 and TgPX2 by N-terminal truncation (Supplementary figures 4C–4H). All five manipulations resulted in isolation of viable clones, suggesting that these genes are not crucial for tachyzoite survival in vitro. Plaque and growth assays confirmed that all mutant parasites had a normal growth rate and lytic cycle (Figures 5A and 5B).
Figure 5. TgFYVE1 and all TgPX proteins are not critical for tachyzoite survival.
(A) Plaque assay stained with Giemsa 7 days after invasion of the host cells with RH-ku80ko, fyve1-ko, px1-ki, px2-ki, px3-ko and px4-ko strains. (B) Intracellular growth assay performed by counting the numbers of parasites per vacuole (x-axis) 24 h after invasion of the host cells. The percentages of vacuoles containing varying numbers of parasites are represented on the y-axis. Values are means ± SD for three independent experiments.
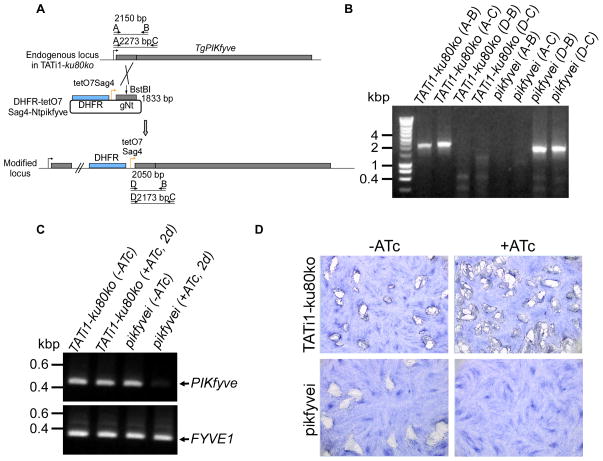
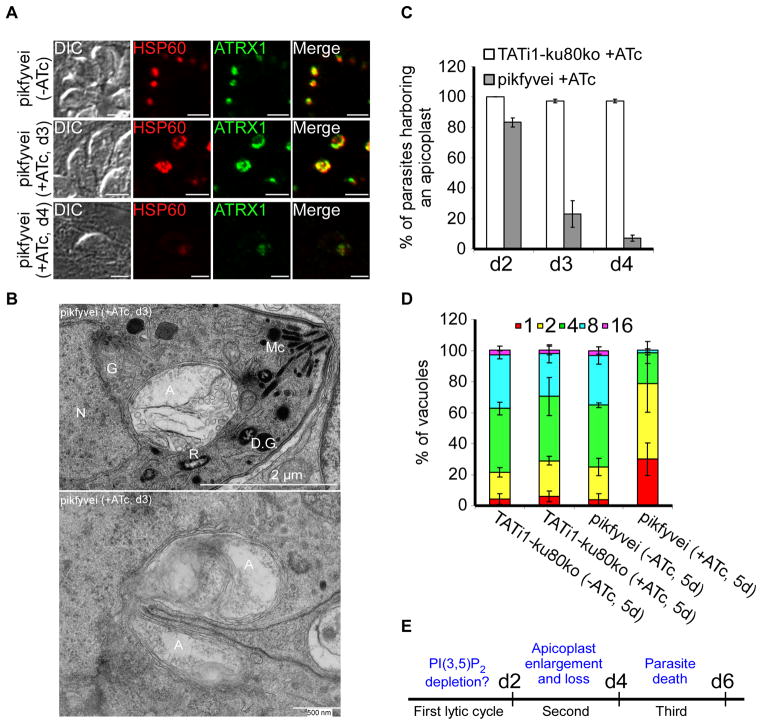
In contrast, PIKfyve, could not be disrupted directly by gene truncation upstream of the 5-kinase domain (data not shown). We therefore generated a conditional knock-down cell line for PIKfyve (pikfyvei), by replacing the endogenous promoter with an ATc-repressible promoter as described previously (Morlon-Guyot et al., 2014) (Figures 6A and 6B). PIKfyve mRNA is down-regulated after 2 days of ATc treatment (Figure 6C) and loss of PIKfyve results in a dramatic effect on parasite growth observed by plaque assays without any ATc pretreatment (Figure 6D). PIKfyve knock-down parasites showed pronounced changes in apicoplast shape (Figure 7A), while other cellular organelles and the inner membrane complex (IMC) remained intact (Supplementary figure 5). As with the pi3ki mutant, the apicoplast appeared enlarged or had disappeared when studied by light and electron microscopy (Figures 7A and 7B). Importantly, electron microscopy also revealed the presence of abnormal enlarged apicoplasts that were dividing in both pi3ki and pikfyvei strains (Figure 7B and Supplementary figure 6). These observations may suggest that the main effect of PI(3)P and PI(3,5)P2 is related to homeostasis control of the apicoplast. Accordingly, a delayed death phenotype (Fichera et al., 1997) was observed when we studied the intracellular growth of pikfyvei mutant parasites in the presence and absence of ATc (Figures 7C, 7D, and 7E).
Figure 6. TgPIKfyve knock-down by promoter replacement strategy.
(A) The endogenous promoter of TgPIKfyve was replaced with the tetracycline-inducible promoter. The DHFR-tetO7-SAG4-NtTgPIKfyve plasmid contains the DHFR resistance cassette (in blue) and the N-terminal genomic coding sequence of TgPIKfyve gene (in grey, 1833 bp) under the control of the inducible tetO7SAG4 promoter (yellow arrow). The primers used for PCR analysis are indicated by black arrows and the length of the PCR fragments is specified. (B) PCR analysis performed on pikfyvei, showing that single homologous recombination had occurred. Genomic DNA from TATi1-ku80ko parasites was used as negative control. (C) Semi-quantitative RT-PCR analysis of TgPIKfyve expression in the wild-type and mutant parasites, preceded or not by two days of ATc-treatment to regulate expression. The FYVE1 gene was used as a loading control. (D) Plaque assays performed in the absence (−) or presence (+) of ATc. Parasite line name is indicated to the left.
Figure 7. Phenotypic consequences of TgPIKfyve knock-down in pikfyvei strain.
(A) Shown are representative vacuoles of the pikfyvei strain treated for 3 and 4 days with ATc and the untreated control. Anti-ATRX1 (in green) and anti-HSP60 (in red) antibodies were used to detect the apicoplast. (B) pikfyvei parasites showed a very enlarged apicoplast in dividing and non-dividing parasites. Thin section electron micrographs were taken from pikfyvei that had grown for 3 days in the presence of ATc. A, apicoplast; Mc, micronemes; R, rhoptries; G, Golgi apparatus; N, nucleus; D.C, daughter cell; D.G., dense granule. Scale bars represent 2 μm and 500 nm respectively. (C) Quantification of TATi1-ku80ko or pikfyvei parasites harbouring an apicoplast upon treatment with ATc at days 2 to 4. (D) Intracellular growth of TATi1-ku80ko and pikfyvei cultivated in presence or absence of ATc for 4 days and allowed to invade new HFF cells. The percentage of vacuoles containing varying numbers of parasites was determined 24 h after inoculation. (C and D) Values are means ± SD for three independent experiments. (E) Schematic diagram summarizing the observed phenotype and parasite survival of the pikfyvei parasites.
In summary, the pikfyvei mutant cell line phenocopies pi3ki mutant parasites suggesting that both lipid kinases act in the same phosphoinositide biogenesis pathway with PI(3)P serving as a precursor for the synthesis of PI(3,5)P2.
Apicoplast homeostasis required the expression of ArPIKfyve, the associated regulator of PIKfyve
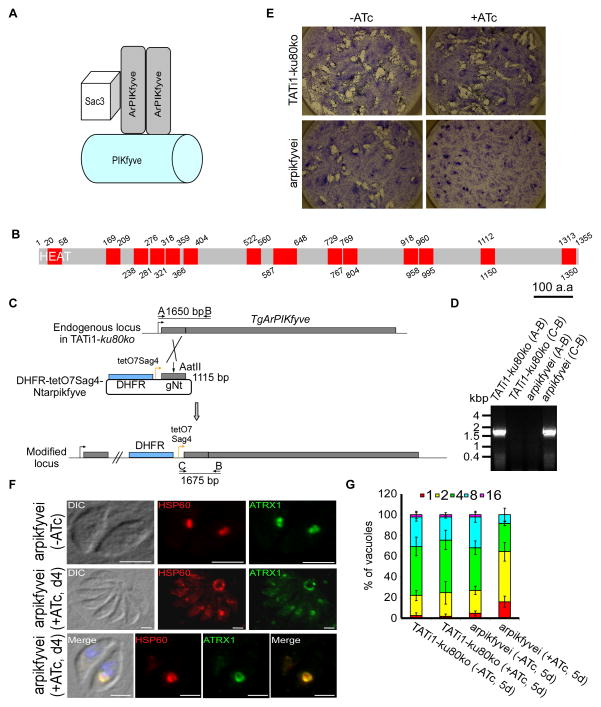
In mammals and yeast, the PI(3,5)P2 steady-state level is controlled by a complex of proteins including the lipid kinase PIKfyve, its regulator ArPIKfyve (or Vac14 in yeast) and the antagonist phosphatidylinositol 3,5-bisphosphate 5-phosphatase Sac3 (or Fig 4 in yeast), that form a complex named PAS (for PIKfyve, ArPIKfyve, Sac3) (Figure 8A) (Ikonomov et al., 2009). A putative protein homolog to ArPIKfyve (TGME49_244040) was found in the T. gondii genome composed of 14 HEAT repeats distributed all along the coding sequence (Figure 8B), while no clear Sac3 homolog was identified. We generated a conditional mutant for this TgArPIKfyve (Figures 8C and 8D) and demonstrated its essentiality (Figure 8E). Importantly, we observed a converging set of phenotypes, similar to mutants of both lipid kinases, with regards to the apicoplast morphology, the loss of the organelle (Figure 8F) and the growth of the mutant parasites (Figure 8G). These data reinforce the critical role played by the PAS complex and the lipid PI(3,5)P2 in maintaining the homeostasis of the apicoplast in T. gondii.
Figure 8. The associated regulator of PIKfyve (ArPIKfyve) interferes with apicoplast morphology.
(A) Model for the well established molecular organization of the PAS (PIKfyve-ArPIKfyve-Sac3) complex and its role in PI(3)P-to-PI(3,5)P2 synthesis as described in the literature. The phosphatase Sac3 and the PIKfyve kinase bind to the N-terminal and C- terminal parts of the homodimeric ArPIKfyve respectively to produce PI(3,5)P2. (B) TgArPIKfyve contains 14 HEAT repeats which are highlighted in red. The 14 HEAT repeats were predicted with the repeat finding program REP (http://www.embl-heidelberg.de/~andrade/papers/rep/search.html). (C) TgArPIKfyve knock-down by promoter replacement strategy. The endogenous promoter of TgArPIKfyve was replaced with the tetracycline-inducible promoter. The DHFR-tetO7-SAG4-NtTgArPIKfyve plasmid contains the DHFR resistance cassette (in blue) and the N-terminal genomic coding sequence of TgArPIKfyve gene (in grey, 1115 bp) under the control of the inducible tetO7SAG4 promoter (yellow arrow). The primers used for PCR analysis are indicated by black arrows and the length of the PCR fragments is specified. (D) PCR analysis performed on arpikfyvei, showing that single homologous recombination had occurred. Genomic DNA of TATi1-ku80ko parasites was used as negative control. (E) Growth analysis of either TATi1-ku80ko or arpikfyvei parasites inoculated on HFF cells and cultured for 7 days ± ATc. (F) Immunofluorescence analysis of intracellular arpikfyvei parasites non-treated or cultured for 4 days with ATc and probed with anti-HSP60 (in red) or anti-ATRX1 (in green) antibodies. The arpikfyvei parasites treated with ATc show an effect on the apicoplast shape. The apicoplast was found enlarged in the residual body of the vacuole or within the parasites. Scale bars represent 2 μm. (G) Replication assay of indicated parasites grown for 5 days in presence, or absence of ATc prior to fixation. The number of parasites per PV was determined. The knockdown of ArPIKfyve protein affected parasite replication. Values are means ± SD for three independent experiments.
In T. gondii, the PI3K and PIKfyve kinases seem not to be involved in the import of nuclear-encoded apicoplast proteins
Many apicoplast proteins are nuclear-encoded and traffic to the ER before reaching the apicoplast. We previously suggested a role for PI(3)P in this vesicular trafficking (Tawk et al., 2011). Having now demonstrated that a PI(3)P/PI(3,5)P2 pathway is essential for apicoplast homeostasis, we next examined this hypothesis by quantifying protein import into the apicoplast in PI3-Kinase and PIKfyve mutants. To this end, we studied in real time the maturation of two apicoplast proteins (ACP and PPP1) in the mutants grown in presence or absence of ATc. As most apicoplast proteins, ACP (a stromal protein) and PPP1 (a periplastid compartment protein, (Sheiner et al., 2011)) are N-terminally processed upon reaching the organelle and this modification is used as a reporter for faithful protein import (Agrawal et al., 2013, Glaser et al., 2012, Sheiner et al., 2011, Sheiner et al., 2013). In pi3ki cells grown in the absence of ATc, processing is detected (Figure 9A, upper panel, lane 1), while after five days in the presence of ATc pi3ki parasites showed a pronounced reduction of the mature form with concomitant accumulation of premature ACP-YFP protein (Figure 9A, upper panel, lane 6). This late accumulation of the premature form is downstream of the onset of apicoplast loss (Figure 9A, lower panel), rendering a role for PI3K in apicoplast protein import unlikely. The same conclusions were drawn from import assays in the PIKfyvei mutant strain when assessing the processing of PPP1-HA protein (Figure 9B). In conclusion, these experiments did not point to a probable role for PI(3)P or PI(3,5)P2 in the trafficking of apicoplast proteins from the ER to the apicoplast.
Figure 9. Depletion of PI3-Kinase or PIKfyve did not result in obvious apicoplast protein import defects.
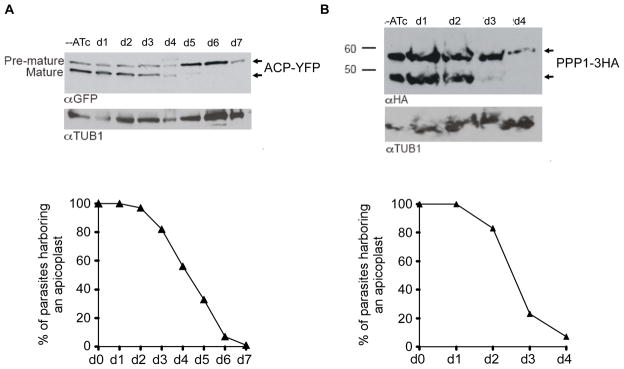
(A and B, upper panels) Western blot analysis following the maturation of either ACP-YFP or PPP1-3HA proteins, in the pi3ki or pikfyvei mutant parasites, respectively. Parasites were grown in ATc for the times indicated. Anti-GFP or anti-HA antibodies were used to detect ACP-YFP or PPP1-3HA proteins. Note the pronounced loss of the mature forms of the proteins at days 5 and 3, respectively. The tubulin protein was used as a loading control. (A and B, lower panels) pi3ki or pikfyvei parasites were grown with (d1 to d7) or without ATc (d0) and apicoplasts were counted in 300 vacuoles for each condition. Only the mean values were plotted on the two graphs. Y-axis shows the percent of parasites that show a clearly identifiable apicoplast.
Discussion
The phosphoinositides PI(3)P and PI(3,5)P2 play evolutionary conserved functions acting as key players of membrane trafficking from early endosomes to lysosomes and of homeostasis of lytic compartments. This study illustrates an original biological role of these two lipids in an apicomplexan parasite. We show here that in T. gondii, PI(3)P and PI(3,5)P2 play an essential role in maintaining the morphology of the apicoplast, an organelle that derived from endosymbiosis of a red algae and that is essential for parasite survival and dissemination. Conditional depletion of TgPI3K or TgPIKfyve, the two lipid kinases responsible for synthesis of PI(3)P and PI(3,5)P2 respectively, drastically altered the shape and the size of the apicoplast. In general, PI(3,5)P2 levels are tightly regulated by coordinated activity of the lipid kinase PIKfyve (Fab1 in yeast) with the scaffolding protein ArPIKfyve (Vac14 in yeast) and the phosphatase SAC3 (Fig 4 in yeast) (Figure 1A) with whom it forms a complex. Accordingly conditional depletion of the T. gondii ArPIKfyve regulator or of PIKfyve resulted in grossly enlarged apicoplasts. Interestingly this phenotype is quite similar to the vacuolation and swelling of the endocytic compartments when PI(3,5)P2 synthesis or its turnover are disrupted in yeast, mammals and plant cells (McCartney et al., 2014). Overall, this study suggests that apicomplexan parasites use the canonical PI(3)P/PI(3,5)P2 endo/lysosomal-dependent functions to perform a specific biological process linked to the apicoplast.
Lipidomic analysis of purified P. falciparum apicoplasts revealed that phosphatidylinositol (PI) which is the precursor for all phosphoinositides is abundant at the apicoplast representing almost 15% of its phospholipids (Botté et al., 2013). We previously showed that PI(3)P is present at the apicoplast in both T. gondii and P. falciparum, and showed that interference with regular PI(3)P function by over-expression of a PI(3)P specific binding module in T. gondii led to the loss of the apicoplast, suggesting that PI(3)P plays a role in the biology of this particular organelle (Tawk et al., 2010, Tawk et al., 2011). Here, using conditional gene ablation of the Toxoplasma PI3K, the lipid kinase responsible for the generation of PI(3)P, we provide further support for the link between PI(3)P and apicoplast homeostasis. Surprisingly, functional analysis of a set of potential PI(3)P binding effector proteins identified in silico detected no association with apicoplast biogenesis. In contrast, we identified the lipid kinase TgPIKfyve and its regulator TgArPIKfyve as key players for maintaining the apicoplast shape, supporting a direct mechanistic role of PI(3,5)P2 in apicoplast biology. Overall, these results support the notion that for apicoplast homeostasis PI(3)P might be mainly acting as substrate for the TgPIKfyve kinase. In this study, we found that the phenotypic consequences of TgPIKfyve and TgArPIKfyve depletions are faster than those of TgPI3K, with an earlier onset of apicoplast loss. These differences might be explained if the pool of PI(3)P is considerably higher than that of PI(3,5)P2, a situation observed in all other organisms in which these lipids have been quantified (Dove et al., 1997, Whiteford et al., 1997). Indeed, the phosphoinositide profile in T. gondii revealed high levels of PI(3)P while we were not able to detect PI(3,5)P2 (Tawk et al., 2011), suggesting that it is produced in minute amounts, below the limit of detection. In this case, it is likely faster to impact on the level of PI(3,5)P2 than of PI(3)P.
We previously defined the apicoplast and surrounding vesicles as the major PI(3)P-containing subcellular compartments in T. gondii (Tawk et al., 2011). But whether the lipid is synthesized at the apicoplast, or reaches the organelle through trafficking of PI(3)P-containing vesicles remains an important unresolved question. Epitope tagging of endogenous TgPI3K failed to detect the kinase in the parasite demonstrating that the protein expression is low convergent with the low number of TgPI3K peptides detected in high throughput proteomic analyses available at ToxoDB. Upon PI3K overexpression no apparent co-localization with the apicoplast was observed, the protein being likely associated with part of the endoplasmic reticulum (Figure 1E). For localizing PIKfyve in T. gondii, we encountered the same technical obstacles that so far prevented formal conclusions about the presence of the kinase at the apicoplast. We only detected the association of PIKfyve after overexpression. In these conditions, the protein is present as dots throughout the cytoplasm, and seems to co-localize partially with the apicoplast. We can therefore speculate that PI(3)P is probably synthesized distant from the apicoplast and reaches the organelle through vesicular trafficking, while PI(3,5)P2 is likely produced directly at the apicoplast by the PIKfyve lipid kinase.
The surprising localization of most of the putative T. gondii PI(3)P effectors proteins containing a FYVE or a PX domain to the vacuolar compartment (Figure 4C) may suggest that a pool of PI(3)P is associated with this compartment. The vacuolar compartment might correspond to the late endosomes of mammalian cells or the vacuole in yeast known to be enriched in PI(3)P, although it should be noted that conventional endo-lysosomal compartments have not yet been precisely delineated in Apicomplexa. PI(3)P might have some functions at the vacuolar compartment, that remain to be elucidated. In P. falciparum, the FYVE(2)-GFP protein was found associated with the apicoplast, the food vacuole and with vesicles accumulating around the food vacuole (Tawk et al., 2010). In contrast, the use of anti-PI(3)P antibodies did not reveal association of PI(3)P with the food vacuole but only with the apicoplast (unpublished data), indicating that in P. falciparum, like in Toxoplasma, the main compartment enriched in PI(3)P is the apicoplast, and supporting a conserved role for a PI(3)P/PI(3,5)P2 pathway related to the apicoplast in both organisms.
The apicoplast is derived from a secondary endosymbiosis of a unicellular red alga by a heterotrophic protist. Consequently, the apicoplast is surrounded by four membranes. Most apicoplast proteins are nuclear-encoded, so they need to be imported into the organelle after translation. The molecular mechanisms underlying the import of proteins from the ER to the outermost apicoplast membrane are poorly understood and we had previously proposed that PI(3)P was involved in this vesicular trafficking (Tawk et al., 2011). This hypothesis was supported by the detection of vesicles in the parasite that contain both PI(3)P and apicoplast peripheral membrane proteins. Our data here fail to support this hypothesis (Figure 9) and suggest a different role for PI(3)P, without withdrawing completely a putative vesicular trafficking pathway dependent on PI(3)P. Indeed, the vesicle-like structures observed inside the enlarged apicoplast by electron-microscopy (Figure 7B) might suggest a defect in membrane recycling or trafficking within the apicoplast.
In other systems, a large number of studies implicate PI(3,5)P2 in several cellular processes, most of them centred around homeostasis of the endosomal/lysosomal system, control of the size and shape of endosomes and/or lysosomes, including control of acidification of the endolysosomal system, regulation of water content and adaptation to osmotic stress (McCartney et al., 2014). The reasons that cause the enlargement of the endolysosomal system in PI(3,5)P2 deficient cells are numerous and emphasize the complexity of the role played by PI(3,5)P2. An emerging body of work reveals that PI(3,5)P2 modulates the activity of many transmembrane channels and transporters which are mostly calcium dependent channels (McCartney et al., 2014). It has been proposed that endolysosome enlargement in PI(3,5)P2-deficient cells may be due to impaired calcium release from endolysosomes (Dong et al., 2010, Onishi et al., 2003). Whether channels associated with the apicoplast are regulated by PI(3,5)P2 in T. gondii remains to be investigated. Overall, understanding of the role of PI(3,5)P2 in apicoplast biology will require detailed information regarding the parasitic protein partners of PI(3,5)P2. PI(3,5)P2 binds directly to certain WD40 domain containing proteins, including Atg18 and Atg21, two proteins involved in autophagy, an essential endomembrane process that requires PI(3)P (Codogno et al., 2011, Mizushima et al., 2011). Interestingly, Toxoplasma and Plasmodium Atg8 localize to the apicoplast (Kitamura et al., 2012, Kong-Hap et al., 2013) and several autophagy-related proteins are required for homeostatic functions at the apicoplast in T. gondii (Besteiro et al., 2011, Kong-Hap et al., 2013). Further investigations are now required to determine a possible crosslink between autophagy and PI(3)P or PI(3,5)P2 in T. gondii.
Both PIKfyve and ArPIKfyve are conserved across the phylum of Apicomplexa. Contrary to what we previously reported (Tawk et al., 2010) we now identified a PIKfyve kinase gene in Plasmodium spp. The PfPIKfyve homolog (PF3D7_1412400) contains a CPN domain and the C-terminal 5-kinase catalytic domain, but the N-terminal FYVE domain is missing. Whether this precludes the binding of PI(3)P by PfPIKfyve remains to be demonstrated as active PIKfyve kinase without FYVE domain have been recently described in Arabidopsis (Serrazina et al., 2014). Fascinatingly, while a PI3-Kinase is present in Cryptosporidium species, PIKfyve and ArPIKfyve are absent in these apicomplexan parasites lacking an apicoplast (Zhu et al., 2000). This underscores a unique role for the PAS complex in apicoplast homeostasis in all apicomplexan parasites harbouring an apicoplast and consequently a critical implication for parasite growth and survival. As highlighted by our results and given the importance of the apicoplast for parasite dissemination and pathogenesis, the PIKfyve lipid kinase is clearly a very interesting target for future drug development.
Experimental procedures
Parasite culture
T. gondii RH strains RH-ku80ko (Huynh et al., 2009) and TATi1-ku80ko (Sheiner et al., 2011) were grown in human foreskin fibroblasts (HFFs) (American Type Culture Collection-CRL 1634) maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; GIBCO, Invitrogen) supplemented with 5% fetal calf serum and 2 mM glutamine. Selection of transgenic parasites were performed with chloramphenicol for CAT selection (Kim et al., 1993); mycophenolic acid (MPA) and xanthine for HXGPRT selection (Donald et al., 1996); pyrimethamine for DHFR-TS selection (Donald et al., 1993). Anhydrotetracycline (ATc) was used at 1.5 μg/ml for the inducible system (Meissner et al., 2001).
Toxoplasma vectors and generation of transgenic T. gondii strains
Primers used in this study are listed in table S1.
Plasmids LIC-DHFR-FYVE1Ctg-3HA and LIC-DHFR-PX4Ctg-3HA were designed to add a sequence coding for a 3HA at the endogenous locus of TGME49_237870 (TgFYVE1) and TGME49_228400 (TgPX4) open reading frames respectively. A 1519 bp and a 1932 bp fragment corresponding to the 3′ of TgFYVE1 and TgPX4 respectively were amplified from genomic DNA using primer sets 20/21 and 22/23 respectively and cloned into LIC-DHFR-3HA vector (Huynh et al., 2009). 40 μg of these plasmids were digested by NarI and AflII respectively, transfected in the RH-ku80ko strain and were subjected to pyrimethamine selection (Fox et al., 2009, Huynh et al., 2009).
Plasmids pT8myc-NtPI3K.HX, pT8myc-NtPX1.HX, pT8myc-NtPX2, and pT8myc-NtPIKfyve.HX were designed to replace the endogenous promoters of TGME49_215700 (TgPI3K), TGME49_243400 (TgPX1), TGME49_262460 (TgPX2) and TGME49_256960 + TGME49_256920 (TgPIKfyve) open reading frames respectively with a tubulin promoter and to add a myc-tag at the N-terminal part of each protein. 1128 bp (TgPI3K), 1146 bp (TgPX1), 1425 bp (TgPX2) and 1261 bp (TgPIKfyve) fragments corresponding to the coding region downstream of the first predicted in-frame ATG codon were amplified by PCR from T. gondii genomic DNA using primers 1/2, 24/25, 26/27, 30/31 respectively and cloned in the pTUB8-mycGFPPftailTy.HX vector. 40 μg of these plasmids were digested by XhoI, StuI, XhoI and SphI respectively, prior to transfection in the RH-ku80ko strain and were subjected to mycophenolic acid and xanthine selection.
Plasmid pT8-PX3-3Ty.HX was designed to insert in the RH strain genome a second or multiple copies of TGME49_309870 (TgPX3) open reading frame under the control of the tubulin promoter. The annotated PX3 open reading frame was amplified by PCR using primers 28/29 and cloned into the EcoRI and NsiI sites in the pTgCtermMLC2g-3Ty.HX vector (Daher et al., 2012). The RH strain was transfected with 100 μg of pT8-PX3-3Ty.HX vector and then subjected to MPA-xanthine selection.
To generate the conditional pi3ki mutant, a genomic fragment of 1047 bp corresponding to the N-terminal coding sequence of TgPI3K gene was amplified by PCR using primers 5/6 and subcloned into BamHI and NotI sites of ptetO7Sag4-HA(2) vector (Sheiner et al., 2011). The 5′ flanking region of the TgPI3K promoter (1059 bp) was amplified by genomic PCR using primers 3/4 and cloned into the NdeI site in ptetO7Sag4-HA(2)NtPI3K vector. The resulting plasmid was named pPI3K-DHFR-tetO7Sag4-HA(2)NtPI3K. TATi1-ku80ko were transfected with 40 μg of this vector (linearized with both NsiI and NotI) and subjected to pyrimethamine selection.
To obtain a complemented pi3kiC cell line, we constructed a ToxP331-HA(3)-CAT cosmid containing the CAT gene to allow selection of parasites that have integrated the cosmid with chloramphenicol. The PI3K open reading frame in cosmid ToxP331 was fused to a triple HA tag using primers 13/14 according to the recombineered cosmid strategy (Brooks et al., 2010). ToxP331-HA(3)-CAT cosmid was then transfected into the conditional pi3ki strain, and clones with the integrated recombineered cosmid were isolated.
Plasmids DHFR-tetO7Sag4-Ntpikfyve and DHFR-tetO7Sag4-Ntarpikfyve were designed to knock-down the level of expression of PIKfyve and ArPIKFYVE proteins respectively using the tet-off system. 1833 bp and 1115 bp fragments corresponding to the 5′ of the coding region of PIKfyve and ArPIKfyve were amplified by PCR from T. gondii genomic DNA using primers 15/16 and 67/68 respectively and cloned BglII/NotI in the DHFR-TetO7SAG4 plasmid (Morlon-Guyot et al., 2014, Sheiner et al., 2011) downstream of the DHFR selection marker, tetO7 operator and pSAG4 promoter. These constructs were linearised by BstBI and AatII, respectively, and transfected TATi1-ku80ko parasites were selected with pyrimethamine and cloned by limiting dilution. Positive clones were verified by PCR to detect the native locus or the single homologous recombination of the inducible vectors in the PIKfyve and ArPIKfyve loci.
Plasmids 2854-TgFYVE1-ko, 2854-TgPX3-ko and 2854-TgPX4-ko were generated to remove the TgFYVE1, TgPX3, and TgPX4 genes by double homologous recombination. Genomic fragments of 1048 bp, 1011 bp and 1524 bp corresponding of the 5′ non-coding sequences of the TgFYVE1, TgPX3 and TgPX4 respectively were amplified by PCR using primers 32/33, 40/41 and 44/45 respectively and sub-cloned into the XhoI and HindIII sites of plasmid 2854.HX (Daher et al., 2012). The 3′ non-coding sequences of TgFYVE1, TgPX3 and TgPX4 were amplified from genomic DNA by PCR (969 bp, 995 bp and 1140 bp respectively) using primers 34/35, 42/43 and 46/47 and cloned into the BamHI and XbaI sites of plasmid 2854.HX. The final 2854-TgFYVE1-ko, 2854-TgPX3-ko and 2854-TgPX4-ko constructs were linearized with KpnI and XbaI or XhoI and XbaI at both the 5′ and 3′ flanking sequences prior to transfection.
Disruption of the TgPX1 and TgPX2 genes was performed using the plasmids TgPX1-ki and TgPX2-ki respectively. 1072 bp and 1172 bp respectively upstream or just downstream of the PX1 and PX2 domains that contains a unique StuI or SphI site were PCR amplified using primers 36/37 and 38/39. The PCR products were cloned into pTUB8MycGFPPfMyoAtailTy.HX vector between KpnI and NsiI sites (Daher et al., 2012). The transfections were performed in the RH-ku80ko strain using 40 μg of the linearized vector, and the transfected parasites were subjected to MPA-xanthine selection.
To detect the cis-Golgi apparatus or the ER, the px4-3ha or Myc-pi3k and Myc-pikfyve strains were transfected transiently with 100 μg of GRASP-RFP or Der1-GFP circular vectors respectively (Nishi et al., 2008, Ramakrishnan et al., 2012).
To follow the expression of the DD-FYVE(2)-GFP protein in the pi3ki strain in the presence or absence of ATc, the parasites were stably transfected with 100 μg of pdd-FYVE circular vector and subjected to chloramphenicol selection (Tawk et al., 2011).
Protein detection by western blot
To detect Myc-PI3K or Myc-PIKfyve or Myc-PX2, Myc-PX1, PX3-Ty or PX4-3HA, FYVE1-3HA or ACP-YFP or PPP1-3HA or NtPX1-3Ty or NtPX2-3Ty proteins, parasite lysates were separated on 3–8%, 6%, 8% or 10% acrylamide gels dependent on their size. Upon transfer to nitrocellulose membranes, the blots were probed with appropriate antibodies in 5% non-fat milk powder in TNT buffer (50 mM Tris pH 8.0; 150 mM NaCl; 0.05% Tween20). The primary antibodies used and their respective dilutions were: rat anti-HA (Roche) at 1/300 (Morlon-Guyot et al., 2014), mouse anti-Myc (Santa Cruz Biotechnology) at 1/100, mouse anti-Ty at 1/1000 (Daher et al., 2012), and mouse monoclonal anti-GFP (Roche) at 1/200 (Sheiner et al., 2011). Bound secondary conjugated antibodies were visualized using either the ECL system (Amersham Corp) or using alkaline phosphatase kit according to manufacturer’s instructions (Promega).
Fluorescent staining of cells
Briefly, for IFAs of intracellular parasites, infected confluent HFF monolayers were fixed for 20 min in 4% paraformaldehyde in PBS, permeabilized with 0.2% triton X-100, blocked with 10% FCS in PBS, and then incubated with primary antibodies (anti-HA (Roche) 1:100, anti- IMC1 1:1000 (Mann et al., 2001), anti-HSP60 1:2000 (Agrawal et al., 2009), anti-ATRX1 1:1000 (kindly provided by Dr Peter Bradley), anti-Myc 1:100 (Santa Cruz Biotechnology), anti-MIC3 1:500 (El Hajj et al., 2008), anti-ROP7 1:1000 (kindly provided by Dr Peter Bradley), anti-GRA2 1:500 (Achbarou et al., 1991), anti-GRA3 1:500 (Dubremetz et al., 1993), anti-mitochondrial F1 beta ATPase (P. Bradley, unpublished), anti-Ty 1:1000 (Daher et al., 2012), anti-CPL 1:250 (Larson et al., 2009), followed by goat-anti-rabbit, goat-anti- mouse or goat-anti-rat immunoglobulin G (IgG) conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes, Invitrogen). Coverslips were mounted onto microscope slides using Immu-mount (Calbiochem). Samples were observed with a Zeiss Axioimager epifluorescence microscope equipped with an apotome and a Zeiss Axiocam MRmCCD camera driven by the Axiovision software (Zeiss), at the Montpellier RIO imaging facility.
Electron Microscopy
Parasites were pretreated for 72 h (pi3ki parasites) or 48 h (pikfyvei parasites) with or without 1.5 μg/ml ATc, collected promptly after egress and inoculated onto new HFF monolayers in presence of ATc during 24 hours. Infected HFF monolayers on coverslips were fixed for 4 hours at room temperature with 2.5% glutaraldehyde (EMS) in 0.1M phosphate buffer pH7.2, washed in buffer and post-fixed for 1 hour in 1% OsO4, washed in water and stained overnight in 2% uranylacetate. Coverslips were then dehydrated in ethanol series and embedded in Epon (Embed 812, EMS). Ultrathin sections were prepared with a Leica ultracut E microtome, contrasted with 2% uranylacetate in ethanol and lead citrate and observed with a JEOL 1200E electron microscope.
Plaque assay
Fresh monolayers of HFF on circular coverslips were infected with parasites in the presence or absence of 1.5 μg/ml ATc for 7 days. Fixation, staining and visualization were performed as previously described (Daher et al., 2010).
Intracellular growth assays
Parasites were pretreated or not for 96 h or 144 h with or without 1.5 μg/ml ATc, collected promptly after egress and inoculated onto new HFF monolayers in presence or absence of ATc during 24 h when cultures were fixed with PFA and stained with anti-TgSAG1. The numbers of parasites per vacuole were counted for more than 300 vacuoles for each condition. PI(3)P mis-targeting and apicoplast loss or apicoplast morphological defects were determined by observing the auto-fluorescent DD-FYVE(2)-GFP protein and the staining of the apicoplast using anti-HSP60 or anti-ATRX1 antibodies, respectively. Data are mean values ± s.d. from three independent biological experiments. For each condition, 300 parasites were observed.
Semi-quantitative RT-PCR
Total RNA was extracted from T. gondii tachyzoites using the Nucleospin RNA II kit (Macherey-Nagel, 740955.10). RT-PCR was performed with the Superscript III first-strand synthesis kit (Invitrogen, 18080-051). 3000 ng of total RNA as a template were used per RT-PCR reaction and specific primers of TgFYVE1 (71/72) or TgPI3K (73/74) or TgPIKfyve (75/76) were used. Twenty-eight cycles of PCR were performed.
Apicoplast protein import assay
For western of steady state luminal proteins: clonal pi3ki parasite lines with endogenously tagged ACP with YFP were isolated by FACS sorting (Sheiner et al., 2011). Samples of parasites grown in the presence or absence of ATc were collected, separated by SDS-PAGE and blotted using anti-GFP antibody.
For dynamic import assay: PIKfyvei parasites were grown in ATc for a given period of time, then transiently transfected with pTUB8-PPP1-HA, and let to grow for an additional 24h in the same condition (for example for day 4 +ATc parasites were grown for 3 days in ATc, transfected and grown for an additional 24h in ATc). Samples of transfected and treated parasites were collected, separated by SDS-PAGE and blotted using anti-HA antibody.
Supplementary Material
Acknowledgments
We thank Dr Peter Bradley for providing the anti-ATRX1, anti-ROP7 and anti-α-F1-ATPase β-subunit antibodies, Dr Con Beckers for the anti-IMC1 antibodies, Dr David Roos for the GRASP-RFP vector, Dr Dominique Soldati-Favre for plasmids pTUB8-mycGFPPftailTy.HX, 2854.HX and pTgCtermMLC2g-3Ty.HX, Dr Vern Carruthers for providing the RH-ku80ko strain and anti-CPL antibodies. We are grateful to Eliane Rubio for her technical assistance. We are thankful to Dr Sébastien Besteiro for critical reading of the manuscript. We thank Veronique Richard and Frank Godiard for their technical assistance on the Electron Microscopy Service of the University of Montpellier 2. We are also grateful to the Montpellier RIO imaging facility at the University of Montpellier 2. Dr Wassim DAHER, Dr Juliette Morlon-GUYOT, Dr Maryse LEBRUN and Dr Kai WENGELNIK are INSERM researchers. When this study was initiated, W.D. was the recipient of a long-term fellowship of the FRM (Fondation pour la Recherche Médicale). This work was supported by the Laboratoire d’Excellence (LabEx) (ParaFrapANR-11-LABX-0024) and by the Fondation pour la Recherche Médicale (Equipe FRM DEQ20130326508) to M.L, by ANR APICOPIP (ANR 11 BSV3 015 01) to KW and ML.
Abbreviations
- TATi-1
Trans-Activator Trap identified
- PI3-Kinase
PhosphatidylInositol 3-Kinase
- PI(3)P
PhosphatidylInositol 3-MonoPhosphate
- PIKfyve
PhosphoInositide Kinase for five position containing a Fyve finger
- ArPIKfyve
Associated regulator of PIKfyve
- PI(3,5)P2
PhosphatidylInositol 3,5-biPhosphate
- PX
PhoX homologous domain
- FYVE
For Fab1, YOTB, Vac1 and EEA1 proteins
References
- Achbarou A, Mercereau-Puijalon O, Autheman JM, Fortier B, Camus D, Dubremetz JF. Characterization of microneme proteins of Toxoplasma gondii. Mol Biochem Parasitol. 1991;47:223–233. doi: 10.1016/0166-6851(91)90182-6. [DOI] [PubMed] [Google Scholar]
- Agrawal S, Chung DW, Ponts N, van Dooren GG, Prudhomme J, Brooks CF, et al. An apicoplast localized ubiquitylation system is required for the import of nuclear-encoded plastid proteins. PLoS Pathog. 2013;9:e1003426. doi: 10.1371/journal.ppat.1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, van Dooren GG, Beatty WL, Striepen B. Genetic evidence that an endosymbiont-derived endoplasmic reticulum-associated protein degradation (ERAD) system functions in import of apicoplast proteins. J Biol Chem. 2009;284:33683–33691. doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besteiro S, Brooks CF, Striepen B, Dubremetz JF. Autophagy protein Atg3 is essential for maintaining mitochondrial integrity and for normal intracellular development of Toxoplasma gondii tachyzoites. PLoS Pathog. 2011;7:e1002416. doi: 10.1371/journal.ppat.1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Stahelin RV, Speicher KD, Speicher DW, Haldar K. Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the host cell. Cell. 2012;148:201–212. doi: 10.1016/j.cell.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botte CY, Yamaryo-Botte Y, Rupasinghe TW, Mullin KA, MacRae JI, Spurck TP, et al. Atypical lipid composition in the purified relict plastid (apicoplast) of malaria parasites. Proc Natl Acad Sci U S A. 2013;110:7506–7511. doi: 10.1073/pnas.1301251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CF, Johnsen H, van Dooren GG, Muthalagi M, Lin SS, Bohne W, et al. The toxoplasma apicoplast phosphate translocator links cytosolic and apicoplast metabolism and is essential for parasite survival. Cell Host Microbe. 2010;7:62–73. doi: 10.1016/j.chom.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- Daher W, Klages N, Carlier MF, Soldati-Favre D. Molecular characterization of Toxoplasma gondii formin 3, an actin nucleator dispensable for tachyzoite growth and motility. Eukaryot Cell. 2012;11:343–352. doi: 10.1128/EC.05192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher W, Plattner F, Carlier MF, Soldati-Favre D. Concerted action of two formins in gliding motility and host cell invasion by Toxoplasma gondii. PLoS Pathog. 2010;6:e1001132. doi: 10.1371/journal.ppat.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Donald RG, Carter D, Ullman B, Roos DS. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- Donald RG, Roos DS. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc Natl Acad Sci U S A. 1993;90:11703–11707. doi: 10.1073/pnas.90.24.11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XP, Shen D, Wang X, Dawson T, Li X, Zhang Q, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- Dubremetz JF, Achbarou A, Bermudes D, Joiner KA. Kinetics and pattern of organelle exocytosis during Toxoplasma gondii/host-cell interaction. Parasitol Res. 1993;79:402–408. doi: 10.1007/BF00931830. [DOI] [PubMed] [Google Scholar]
- El Hajj H, Papoin J, Cerede O, Garcia-Reguet N, Soete M, Dubremetz JF, Lebrun M. Molecular signals in the trafficking of Toxoplasma gondii protein MIC3 to the micronemes. Eukaryot Cell. 2008;7:1019–1028. doi: 10.1128/EC.00413-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichera ME, Roos DS. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot Cell. 2009;8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, van Dooren GG, Agrawal S, Brooks CF, McFadden GI, Striepen B, Higgins MK. Tic22 is an essential chaperone required for protein import into the apicoplast. J Biol Chem. 2012;287:39505–39512. doi: 10.1074/jbc.M112.405100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa A, Hayes SJ, Lawe DC, Sudharshan E, Tuft R, Fogarty K, et al. Structural basis for endosomal targeting by FYVE domains. J Biol Chem. 2004;279:5958–5966. doi: 10.1074/jbc.M310503200. [DOI] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem. 2009;284:35794–35806. doi: 10.1074/jbc.M109.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Foti M, Carpentier JL, Shisheva A. PIKfyve controls fluid phase endocytosis but not recycling/degradation of endocytosed receptors or sorting of procathepsin D by regulating multivesicular body morphogenesis. Mol Biol Cell. 2003;14:4581–4591. doi: 10.1091/mbc.E03-04-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Horak A, Obornik M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Soldati D, Boothroyd JC. Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science. 1993;262:911–914. doi: 10.1126/science.8235614. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Kishi-Itakura C, Tsuboi T, Sato S, Kita K, Ohta N, Mizushima N. Autophagy-related Atg8 localizes to the apicoplast of the human malaria parasite Plasmodium falciparum. PLoS One. 2012;7:e42977. doi: 10.1371/journal.pone.0042977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJ, et al. A plastid of probable green algal origin in Apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- Kong-Hap MA, Mouammine A, Daher W, Berry L, Lebrun M, Dubremetz JF, Besteiro S. Regulation of ATG8 membrane association by ATG4 in the parasitic protist Toxoplasma gondii. Autophagy. 2013;9:1334–1348. doi: 10.4161/auto.25189. [DOI] [PubMed] [Google Scholar]
- Kutateladze TG, Ogburn KD, Watson WT, de Beer T, Emr SD, Burd CG, Overduin M. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol Cell. 1999;3:805–811. doi: 10.1016/s1097-2765(01)80013-7. [DOI] [PubMed] [Google Scholar]
- Larson ET, Parussini F, Huynh MH, Giebel JD, Kelley AM, Zhang L, et al. Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl. J Biol Chem. 2009;284:26839–26850. doi: 10.1074/jbc.M109.003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- McCartney AJ, Zhang Y, Weisman LS. Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. Bioessays. 2014;36:52–64. doi: 10.1002/bies.201300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen RK, Dove SK, Cooke FT, Painter GF, Holmes AB, Shisheva A, et al. Complementation analysis in PtdInsP kinase-deficient yeast mutants demonstrates that Schizosaccharomyces pombe and murine Fab1p homologues are phosphatidylinositol 3-phosphate 5-kinases. J Biol Chem. 1999;274:33905–33912. doi: 10.1074/jbc.274.48.33905. [DOI] [PubMed] [Google Scholar]
- McFadden GI, Reith ME, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- Meissner M, Brecht S, Bujard H, Soldati D. Modulation of myosin A expression by a newly established tetracycline repressor-based inducible system in Toxoplasma gondii. Nucleic Acids Res. 2001;29:E115. doi: 10.1093/nar/29.22.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Morlon-Guyot J, Berry L, Chen CT, Gubbels MJ, Lebrun M, Daher W. The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell Microbiol. 2014;16:95–114. doi: 10.1111/cmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot AS, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3062–3074. doi: 10.1091/mbc.E05-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M, Hu K, Murray JM, Roos DS. Organellar dynamics during the cell cycle of Toxoplasma gondii. J Cell Sci. 2008;121:1559–1568. doi: 10.1242/jcs.021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Ohsumi Y. PtdIns 3-Kinase Orchestrates Autophagosome Formation in Yeast. J Lipids. 2011;2011:498768. doi: 10.1155/2011/498768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Nakamura Y, Koga T, Takegawa K, Fukui Y. Isolation of suppressor mutants of phosphatidylinositol 3-phosphate 5-kinase deficient cells in Schizosaccharomyces pombe. Biosci Biotechnol Biochem. 2003;67:1772–1779. doi: 10.1271/bbb.67.1772. [DOI] [PubMed] [Google Scholar]
- Parussini F, Coppens I, Shah PP, Diamond SL, Carruthers VB. Cathepsin L occupies a vacuolar compartment and is a protein maturase within the endo/exocytic system of Toxoplasma gondii. Mol Microbiol. 2010;76:1340–1357. doi: 10.1111/j.1365-2958.2010.07181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007;581:3396–3404. doi: 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S, Docampo MD, Macrae JI, Pujol FM, Brooks CF, van Dooren GG, et al. Apicoplast and endoplasmic reticulum cooperate in fatty acid biosynthesis in apicomplexan parasite Toxoplasma gondii. J Biol Chem. 2012;287:4957–4971. doi: 10.1074/jbc.M111.310144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Rodahl LM, Pattni K, Englund C, Samakovlis C, Dove S, et al. Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol Biol Cell. 2006;17:3989–4001. doi: 10.1091/mbc.E06-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, et al. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci. 2006;119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- Serrazina S, Dias FV, Malho R. Characterization of FAB1 phosphatidylinositol kinases in Arabidopsis pollen tube growth and fertilization. New Phytol. 2014;203:784–793. doi: 10.1111/nph.12836. [DOI] [PubMed] [Google Scholar]
- Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, et al. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011;7:e1002392. doi: 10.1371/journal.ppat.1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiner L, Striepen B. Protein sorting in complex plastids. Biochim Biophys Acta. 2013;1833:352–359. doi: 10.1016/j.bbamcr.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Emr SD. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem. 1994;269:31552–31562. [PubMed] [Google Scholar]
- Striepen B. The apicoplast: a red alga in human parasites. Essays Biochem. 2011;51:111–125. doi: 10.1042/bse0510111. [DOI] [PubMed] [Google Scholar]
- Tawk L, Chicanne G, Dubremetz JF, Richard V, Payrastre B, Vial HJ, et al. Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localizes to the food vacuole membrane and the apicoplast. Eukaryot Cell. 2010;9:1519–1530. doi: 10.1128/EC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk L, Dubremetz JF, Montcourrier P, Chicanne G, Merezegue F, Richard V, et al. Phosphatidylinositol 3-monophosphate is involved in toxoplasma apicoplast biogenesis. PLoS Pathog. 2011;7:e1001286. doi: 10.1371/journal.ppat.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid A, Ranjan R, Smythe WA, Hoppe HC, Sharma P. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood. 2010;115:2500–2507. doi: 10.1182/blood-2009-08-238972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren GG, Striepen B. The algal past and parasite present of the apicoplast. Annu Rev Microbiol. 2013;67:271–289. doi: 10.1146/annurev-micro-092412-155741. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley L, De Camilli P. Phosphoinositides’ link to neurodegeneration. Nat Med. 2007;13:784–786. doi: 10.1038/nm0707-784. [DOI] [PubMed] [Google Scholar]
- Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323 (Pt 3):597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- Yeh E, DeRisi JL. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol. 2011;9:e1001138. doi: 10.1371/journal.pbio.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Marchewka MJ, Keithly JS. Cryptosporidium parvum appears to lack a plastid genome. Microbiology. 2000;146 (Pt 2):315–321. doi: 10.1099/00221287-146-2-315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.