Abstract
Tobacco smoking is a risk factor for numerous disorders, including cancers affecting organs outside the respiratory tract. Epidemiological data suggest that smoking is a greater risk factor for these cancers in males compared to females. This observation, together with the fact that males have a higher incidence of, and mortality from, most non-sex-specific cancers, remain unexplained. Loss of chromosome Y (LOY) in blood cells is associated with increased risk of non-hematological tumors. We demonstrate here that smoking is associated with LOY in blood cells in three independent cohorts (TwinGene: odds ratio [OR]=4.3, 95% CI =2.8-6.7; ULSAM: OR=2.4, 95% CI=1.6-3.6; and PIVUS: OR=3.5, 95% CI=1.4-8.4) encompassing a total of 6014 men. The data also suggest that smoking has a transient and dose-dependent mutagenic effect on LOY-status. The finding that smoking induces LOY thus links a preventable risk factor with the most common acquired human mutation.
Tobacco smoking killed ~100 million people during the 20th century and is projected to kill ~1 billion people during the current century, assuming that the current frequency of smoking is retained (1, 2). Lung cancer is the prime cause of cancer-associated death in relation to smoking. However, smoking is also a risk factor for tumors outside the respiratory tract and these are more common in males than females (hazard ratio in males 2.2; 95% CI=1.7-2.8 and in females 1.7; 95% CI=1.4-2.1) (2). Moreover, males have a higher incidence and mortality from most non-sex-specific cancers, disregarding smoking status, and this fact is largely unexplained by known risk factors (3, 4). A recent analysis of non-cancerous blood cells revealed that a male-specific chromosomal aberration, acquired mosaic loss of chromosome Y (LOY), is associated with an increased risk of non-hematological tumors among aging males (5).
Here we analyzed possible causes of LOY by studying 6014 men from three independent prospective cohorts (TwinGene, n=4373 (6, 7); ULSAM, n=1153 (8); PIVUS, n=488 (9)), from which comprehensive epidemiological records are available (tables S2-S4). We included the following environmental, lifestyle and clinical factors in the analyses: smoking, age, hypertension, exercise habits, diabetes, BMI, LDL cholesterol, HDL cholesterol, education level, and alcohol intake. We also included genotyping quality as a confounder in the regression analyses, to adjust for possible influence of experimental noise. Similar definitions of factors were used in all cohorts as outlined in tables S2-S5 and described in detail in Materials and Methods (Supplement). Estimation of LOY was based on SNP-array data using the 2.5MHumanOmni and HumanOmniExpress beadchips, in the ULSAM and PIVUS/TwinGene, respectively (fig. S1). The estimation of the degree of mosaicism and scoring of LOY was undertaken using the continuous mLRR-Y estimate, calculated from SNP-array data as the median of the LogRRatio of all SNP-probes within the male specific part of chromosome Y (MSY), as described previously (5). An mLRR-Y estimate close to zero indicates a normal chromosome Y state, while more negative mLRR-Y values denote an increasing level of blood cells with LOY. To facilitate comparisons between the three cohorts, we corrected the mLRR-Y values for all participants, using cohort-specific correction constants as explained in Materials and Methods (figs. 1 and S2).
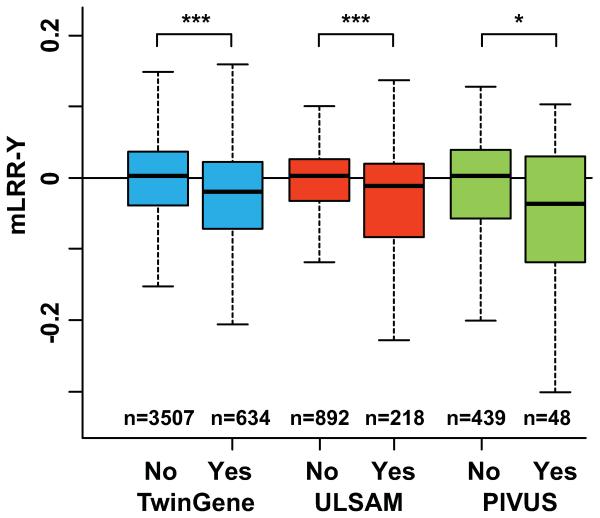
Fig. 1.
The association between smoking status and the level of LOY (i.e. mLRR-Y) in three independent cohorts. In all cohorts, these unadjusted analyses indicate that the current smokers (Yes) (table S5) had a significantly higher degree of mosaic loss of chromosome Y (LOY) in blood, compared to non-current smokers (No), composed of never- and previous-smokers. *** and * denotes p-values lower than 0.001 and lower than 0.05, respectively (Kolmogorov–Smirnov tests: TwinGene D=0.15 p=1.131e-11; ULSAM D=0.15 p=0.0006; PIVUS D=0.23 p=0.0203). The definitions used for LOY-scoring and the entire ranges of mLRR-Y data observed in each cohort are shown in figs. S3-S5.
LOY was by far the most common post-zygotic mutation found in the three cohorts. The age range at sampling in ULSAM and PIVUS was 70.7 - 83.6 years and 69.8 - 70.7 years, respectively, and we found LOY in 12.6% of ULSAM participants and 15.6% of PIVUS participants (figs. S3-S4). The age range at sampling in TwinGene was 48 - 93 years and the frequency of LOY in the entire cohort was 7.5% (fig. S5). However, in TwinGene participants aged 70 years or older, 15.4% had LOY, which is similar to the LOY frequency in the other cohorts at the same age range. In TwinGene participants younger than 70 years, only 4.1% was affected by LOY. Thresholds for LOY-scoring, at the lower 99% confidence limit of the distributions of experimental mLRR-Y variation (TwinGene=−0.1324, ULSAM=−0.1024, PIVUS=−0.1182) were used for frequency calculations, as explained in Materials and Methods and figs. S3-S5. At this degree of mosaicism, approximately 10% of the analyzed nucleated blood cells from each sample are affected by LOY. Mosaic LOY in blood detected with SNP-arrays was validated using whole genome next generation sequencing (NGS) in 100 random participants in the ULSAM cohort. There was 100% concordance in LOY-scoring between results from SNP-array and NGS-data (5).
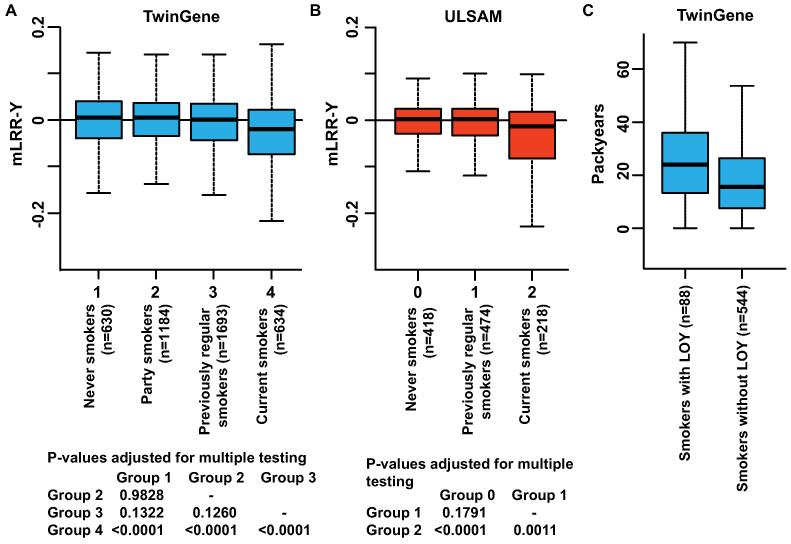
We found a strong association between smoking and LOY-status in the three independent cohorts. Current smokers had a significantly higher degree of LOY-mosaicism, compared to nonsmokers and past smokers, in unadjusted (fig. 1) and in the multivariable models adjusting for the above mentioned potential confounders (table S1, ANCOVA: TwinGene F(1,1666)= 45.4, p=2.225e-11; ULSAM F(1,968)=17.6, p=2.984e-05; PIVUS F(1,385)=9.1, p=0.0028). Apart from smoking, the only other factor significantly associated with LOY was age, which was observed in TwinGene, with higher degree of LOY in older participants. The age range in ULSAM and PIVUS was narrower (see above), which may explain why age had no effect on LOY in these regression models (tables S1 and S5). To assess the risk of LOY in blood cells of smokers, we used logistic regression adjusting for the same confounders as in table S1. Participants were LOY-scored as 1 or 0 prior to these analyses based on the continuous mLRR-Y estimate, using the same threshold as for the estimations of LOY-frequencies, i.e., the lower 99% confidence intervals of the experimentally induced mLRR-Y variation (figs. S3-S5). The adjusted odds ratio estimates from logistic regressions were highest in TwinGene (OR=4.3, 95% CI =2.8-6.7) followed by PIVUS (OR=3.5, 95% CI=1.4-8.4) and ULSAM (OR=2.4, 95% CI=1.6-3.6). The corresponding unadjusted odds ratios are given in table S6. Based on these calculations, we estimate that current smokers in the studied cohorts had a 2.4 – 4.3 times greater risk of displaying LOY compared to non-smokers. Furthermore, among the current smokers in the large TwinGene cohort, we found a strong dose-response effect with more LOY in heavy smokers, i.e. smokers with LOY had been smoking significantly more packyears compared to smokers without LOY (fig. 2C).
Fig. 2.
Differences in degree of LOY between different smoking categories within TwinGene (A) and ULSAM (B) as defined in table S5. In TwinGene (A) there was a significant difference between four smoking categories (ANOVA; F(3,4137)=22.2, p-value=3.028e-14) and results from post-hoc analysis adjusting p-values for multiple testing using a Tukey HSD test are displayed. In ULSAM (B) there was also a significant difference between three smoking categories (ANOVA; F(2,1107)=12.2, p=5.812e-06) and post-hoc analysis is shown. In both cohorts, the current smokers had a significantly higher degree of LOY compared to all other categories. The average degree of LOY in the previously regular smokers was not significantly different from the average degree of LOY in the never smokers in both cohorts. Panel C visualize a dose response effect within current smokers in TwinGene with men smoking the most packyears also being associated with higher degree of LOY, as defined in fig. S5 (Kolmogorov–Smirnov test: D=0.2244, p=0.0010).
Our results suggest that the association between smoking and LOY is valid for current smokers only (fig. 2A and B). Previous epidemiological studies showed that smoking cessation at any age is associated with dramatically reduced death rates. For smokers who quit at 25 to 34 years of age, survival was nearly identical with those who had never smoked (2, 10). We analyzed the level of LOY after smoking cessation in the ULSAM and the TwinGene cohorts by using LOY data in the past regular smokers (table S5). No difference in LOY frequency between never smokers and previously regular smokers was found (fig. 2A and B). One possible explanation for these results is that the previous smokers with LOY died off faster than the rest of the cohort. Another and perhaps more likely explanation is that LOY is induced and sustained by smoking and that LOY is a dynamic and reversible process.
Whether the LOY induced by smoking plays a direct role in cancer is unclear. One hypothesis is that smoking is clastogenic - that is, it induces many chromosomal abnormalities, including an incidental loss of the Y chromosome. In this scenario, LOY would be a neutral passenger mutation and a reporter of a general tendency of chromosome missegregation in mitosis, which is enhanced by smoking and associated with risk for cancer and mortality. A second hypothesis is that LOY in blood cells is a causative factor in cancer development, possibly through effects on tumor immunosurveillance (11). We conducted a preliminary test of this hypothesis by investigating possible functional consequences of LOY in sorted blood cells from three ULSAM survivors scored with LOY and still alive at the age of 91 years. We sorted cells from three compartments (granulocytes, CD4+ T-lymphocytes and CD19+ B-lymphocytes) in subjects that displayed LOY in earlier serial analyses of whole blood, performed at four time points during two decades (fig. S6). The three subjects were free from cancer diagnoses at the time of blood collection at 91 years. The data from these experiments are tantalizing because: i) the percentage of cells with LOY differed between different compartments of the hematopoietic system; and ii) ULSAM-1412 suggests that LOY might be an oligo-clonal process, as cells derived from myeloid and some (but not all) lymphoid progenitors display LOY. These preliminary results support the second hypothesis. If LOY was a phenotypically neutral passenger mutation, one would expect that LOY cells would be randomly distributed within all components of the haematological system. It was recently shown that LOY-status of blood cells is associated with a higher risk for all-cause mortality as well as a higher risk for non-hematological cancers and that it can be considered as a biomarker of male carcinogenesis (5). We hypothesized that a disrupted tumor-immunosurveillance in LOY-affected cells could help explaining the connection between LOY-status of non-cancerous blood cells and risk for tumors in other tissues (5, 11). These results also support the second hypothesis and the increasingly recognized view that chromosome Y carries many vital functions in biological processes beyond sex determination and sperm production (12-16).
Our results are consistent with a previously described dynamic nature of expanding-contracting non-cancerous cell clones in blood affected with mosaic genetic aberrations; i.e. it appears that the relative frequency of cells from a cell clone can first increase and then decrease later in life (5, 17, 18). In the present analyses, LOY was detected in ≥10% of blood cells from about 15% of elderly males in three cohorts (figs. S3-S5). The cell clones with LOY were likely detectable in our analyses because they are enriched, due to an increased proliferative potential as a consequence of LOY, which is in agreement with chromosome Y containing tumor suppressor genes. Recent analysis of >8200 tumor-normal pairs suggest that two genes (ZFY and UTY, from the male specific part of Y) have properties of tumor suppressors (19). Interestingly, both genes have homologues on chromosome X and escape X inactivation (19, 20). Moreover, other analyses of various tumor collections show that chromosome Y is lost from numerous types of tumors in frequencies ranging from 15 to 80% of cases (21-24). Thus, counting both LOY in non-cancerous blood clones and in transformed tumor cells, nullisomy Y is among the most common, if not the most common, human mutation. The results presented here suggest that this aneuploidy, affecting 1.6% of the genome, is likely induced by smoking. In conclusion, we show that LOY is more common in current smokers compared to non-current smokers in three cohorts (fig. 1 and table S1), that the effect from smoking on LOY is dose dependent, and that this effect might be transient, as it apparently disappears after smoking cessation (fig. 2). Epidemiological observations suggest that smoking could be a greater risk factor for cancer outside the respiratory tract in males compared to females (2, 4, 10). Moreover, males have a higher incidence and mortality from most sex-unspecific cancers (3, 4). The molecular mechanisms behind these observations are not well understood, but LOY, being a male specific, smoking-induced risk factor, could provide a missing link and help explaining these sex differences.
Supplementary Material
Acknowledgments
We thank K. Lindblad-Toh, R.M. Myers, C-H Heldin, D.H. Ledbetter, G.J.B. van Ommen and U. Landegren for critical evaluation of the manuscript. This study was sponsored by the Swedish Cancer Society, the Swedish Research Council, the Swedish Heart-Lung Foundation and Sci-Life-Lab-Uppsala to J.P.D. and the Olle Enqvist Byggmästare Foundation to L.A.F. Genotyping and next-generation sequencing were performed by the SNP&SEQ Technology Platform in Uppsala, Sweden, and supported by Wellcome Trust Grants WT098017, WT064890, WT090532, Uppsala University, Uppsala University Hospital, the Swedish Research Council and the Swedish Heart-Lung Foundation. The SNP&SEQ Technology Platform is part of Science for Life Laboratory at Uppsala University and supported as a national infrastructure by the Swedish Research Council. C.M.L. is a Wellcome Trust Research Career Development Fellow (086596/Z/08/Z). A.P.M. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science. A.P.M. acknowledges funding from the Wellcome Trust under awards WT064890, WT090532 and WT098017. TwinGene was supported by the Swedish Research Council (M-2005-1112), GenomEUtwin (EU/QLRT-2001-01254; QLG2-CT-2002-01254), NIH DK U01-066134, The Swedish Foundation for Strategic Research (SSF), the Heart and Lung foundation no. 20070481. J.P.D and L.A.F are co-founders and share-holders in Cray Innovation AB as well as co-inventors on Patent Application No. PCT/EP2014/071448, protecting the commercial applications of LOY for the assessment of cancer risk. Genetic variants detected in this study are available at dbVar under accession code nstd92 for ULSAM and PIVUS cohorts as well as accession code XXXX (pending submission) for TwinGene cohort, respectively.
References and notes
- 1.Jha P. Nature reviews. 2009 Sep;9:655. doi: 10.1038/nrc2703. [DOI] [PubMed] [Google Scholar]
- 2.Jha P, et al. N Engl J Med. 2013 Jan 24;368:341. doi: 10.1056/NEJMsa1211128. [DOI] [PubMed] [Google Scholar]
- 3.Cook MB, McGlynn KA, Devesa SS, Freedman ND, Anderson WF. Cancer epidemiology, biomarkers & prevention. 2011 Aug;20:1629. doi: 10.1158/1055-9965.EPI-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgren G, Liang L, Adami HO, Chang ET. Eur J Epidemiol. 2012 Mar;27:187. doi: 10.1007/s10654-011-9647-5. [DOI] [PubMed] [Google Scholar]
- 5.Forsberg LA, et al. Nature Genetics. 2014 Jun;46:624. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtenstein P, et al. Twin Res Hum Genet. 2006 Dec;9:875. doi: 10.1375/183242706779462444. [DOI] [PubMed] [Google Scholar]
- 7.Magnusson PK, et al. Twin Res Hum Genet. 2013 Feb;16:317. doi: 10.1017/thg.2013.57. [DOI] [PubMed] [Google Scholar]
- 8.Hedstrand H. Ups J Med Sci Suppl. 1975;19:1. [PubMed] [Google Scholar]
- 9.Lind L, Fors N, Hall J, Marttala K, Stenborg A. Arterioscler Thromb Vasc Biol. 2005 Nov;25:2368. doi: 10.1161/01.ATV.0000184769.22061.da. [DOI] [PubMed] [Google Scholar]
- 10.Thun MJ, et al. N Engl J Med. 2013 Jan 24;368:351. doi: 10.1056/NEJMsa1211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Nat Immunol. 2002 Nov;3:991. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 12.Lemos B, Araripe LO, Hartl DL. Science. 2008 Jan 4;319:91. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 13.Lemos B, Branco AT, Hartl DL. Proc Natl Acad Sci U S A. 2010 Sep 7;107:15826. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark AG. Nature. 2014 Apr 24;508:463. doi: 10.1038/508463a. [DOI] [PubMed] [Google Scholar]
- 15.Bellott DW, et al. Nature. 2014 Apr 24;508:494. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez D, et al. Nature. 2014 Apr 24;508:488. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg LA, et al. Am J Hum Genet. 2012 Feb 10;90:217. doi: 10.1016/j.ajhg.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg LA, Absher D, Dumanski JP. J Med Genet. 2013 Jan;50:1. doi: 10.1136/jmedgenet-2012-101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davoli T, et al. Cell. 2013 Nov 7;155:948. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider-Gadicke A, Beer-Romero P, Brown LG, Nussbaum R, Page DC. Cell. 1989 Jun 30;57:1247. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhang LJ, Shin ES, Yu ZX, Li SB. Chin Med J (Engl) 2007 Nov 20;120:2002. [PubMed] [Google Scholar]
- 22.Bianchi NO. Mutation research. 2009 Jul-Aug;682:21. doi: 10.1016/j.mrrev.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Veiga LCS, Bergamo NA, Reis PP, Kowalski LP, Rogatto SR. Braz J Med Biol Res. 2012 Feb;45:172. doi: 10.1590/S0100-879X2012007500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duijf PH, Schultz N, Benezra R. Int J Cancer. 2013 May 15;132:2316. doi: 10.1002/ijc.27924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichtenstein P, et al. J Intern Med. 2002 Sep;252:184. doi: 10.1046/j.1365-2796.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- 26.Chee-Seng K, Xueling S, Kee-Seng C. Technical Note: Illumina DNA analysis. Illumina, Inc; San Diego: 2008. [Google Scholar]
- 27.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.