Graphical Abstract
Keywords: Anas acuta, Haematozoa, Haemoproteus, Leucocytozoon, Northern pintail, Pacific Basin, Plasmodium
Highlights
-
•
Northern pintails were sampled in Asia and North America and screened for haematozoa.
-
•
Three parasite genera were detected among 878 samples (apparent prevalence 5–63%).
-
•
Thirty-one unique parasite lineages were identified through genetic sequencing.
-
•
Identical parasite lineages were identified on two continents.
-
•
Results provide evidence for intercontinental genetic exchange of blood parasites.
Abstract
Empirical evidence supports wild birds as playing a role in the interhemispheric exchange of bacteria and viruses; however, data supporting the redistribution of parasites among continents are limited. In this study, the hypothesis that migratory birds contribute to the redistribution of parasites between continents was tested by sampling northern pintails (Anas acuta) at locations throughout the North Pacific Basin in North America and East Asia for haemosporidian infections and assessing the genetic evidence for parasite exchange. Of 878 samples collected from birds in Alaska (USA), California (USA), and Hokkaido (Japan) during August 2011–May 2012 and screened for parasitic infections using molecular techniques, Leucocytozoon, Haemoproteus, and Plasmodium parasites were detected in 555 (63%), 44 (5%), and 52 (6%) samples, respectively. Using an occupancy modeling approach, the probability of detecting parasites via replicate genetic tests was estimated to be high (ρ > 0.95). Multi-model inference supported variation of Leucocytozoon parasite prevalence by northern pintail age class and geographic location of sampling in contrast to Haemoproteus and Plasmodium parasites for which there was only support for variation in parasite prevalence by sampling location. Thirty-one unique mitochondrial DNA haplotypes were detected among haematozoa infecting northern pintails including seven lineages shared between samples from North America and Japan. The finding of identical parasite haplotypes at widely distributed geographic locations and general lack of genetic structuring by continent in phylogenies for Leucocytozoon and Plasmodium provides evidence for intercontinental genetic exchange of haemosporidian parasites. Results suggest that migratory birds, including waterfowl, could therefore facilitate the introduction of avian malaria and other haemosporidia to novel hosts and spatially distant regions.
1. Introduction
The emergence of zoonotic pathogens in wild birds inhabiting North America and Asia has raised global awareness regarding the potential of migratory species to redistribute infectious agents within and between continents (Rappole et al, 2000, Reed et al, 2003, Chen et al, 2005). Empirical evidence supports wild birds as playing a role in the exchange of bacteria and viruses between continental landmasses with examples including Borrelia burgdorferi (Olsen et al., 1995), eastern equine encephalitis virus (Calisher et al., 1971), West Nile virus (Malkinson et al., 2002), influenza A virus (Koehler et al., 2008), and avian paramyxovirus (Ramey et al., 2013a). Data supporting the redistribution of parasites among continents, however, are limited. Hyalomma and Ixodes ticks, including species identified as vectors of human and animal disease, were collected from wild birds in Egypt en route from Europe and Asia to Africa during autumn migration providing evidence for transport of ectoparasites, and their associated pathogens, along migratory flyways (Hoogstraal et al., 1963). Similar results were also recently found at two Mediterranean Islands where large numbers of Rickettsia-positive Hyalomma and Ixodes ticks were collected from seasonal migrants presumably headed from African wintering areas to European breeding grounds during spring (Wallménius et al., 2014). Zonorchis microrchis trematodes, common in Brazil, were identified in pectoral sandpipers (Calidris melanotos) and solitary sandpipers (Tringa solitaria) in Louisiana during spring migration suggesting the acquisition of parasites at wintering areas and subsequent transport to the North America (Tallman et al., 1985). More recently, malarial (Plasmodium) infections were detected in endemic avifauna of the Galapagos Islands and identical genetic lineages identified in North American breeding bobolinks (Dolichonyx oryzivorus) which migrate through this archipelago, suggesting a potential route of introduction (Levin et al., 2013). As such, the redistribution of both ecto- and endo-parasites across large spatial scales by migratory birds appears likely.
Numerous migratory bird species may play some role in the global redistribution of parasites; however, the probability of detecting evidence for parasite exchange may be higher in intercontinentally migratory species such as the northern pintail (Anas acuta). The northern pintail is an abundant species of dabbling duck throughout the Holarctic (Madge and Burn, 1988) and some individuals inhabiting the East Asian-Australasian and Pacific Americas flyways make migratory movements between North America and East Asia (Miller et al, 2005, Hupp et al, 2011) and/or breed sympatrically in northeastern Russia (Flint et al., 2009). Characterization of influenza A viruses and avian paramyxoviruses isolated from northern pintails supports genetic exchange of these viral agents between North America and Eurasia (Koehler et al, 2008, Ramey et al, 2010, Ramey et al, 2013a). Blood parasite infections caused by protozoa of the genera Leucocytozoon, Haemoproteus, and/or Plasmodium have repeatedly been identified in northern pintails sampled at locations throughout the United States and Canada with apparent prevalence previously reported to be 3–49% (Herman, 1951, Bennett et al, 1975, Bennett et al, 1982, Bennett et al, 1991, Williams et al, 1977, Ramey et al, 2013b). In Asia, northern pintails sampled in Bangladesh were also found to be infected with Haemoproteus and Plasmodium parasites with apparent prevalence of 9% and 19%, respectively (Elahi et al., 2014). Given the abundance of northern pintails in both North America and East Asia, the migratory tendencies of this species, the propensity of this taxon to be infected with blood parasites, and the prolonged duration of haematozoa infections in avian hosts (Valkiūnas, 2004), it is plausible that northern pintails facilitate the intercontinental redistribution of blood parasites between North America and Eurasia.
In this study, molecular methodology was used to estimate haematozoa prevalence in northern pintails and to assess the possible role of this species in the intercontinental exchange of blood parasites. Molecular methods and an occupancy modeling statistical approach were used to first test the hypothesis that northern pintails are infected with haematozoa at locations throughout North Pacific Basin at relatively high rates. Parasites were subsequently genetically characterized to assess the evidence for interhemispheric exchange of haematozoa in northern pintails to test the hypothesis that migratory birds contribute to the redistribution of parasites between continents. Sampling efforts were focused at locations throughout the North Pacific Basin in North America and East Asia within the overlapping East Asian-Australasian and Pacific Americas flyways within a single year (i.e. <12 month period). Results of this study build upon existing literature documenting the prevalence and distribution of avian haemosporidia in waterfowl and provide new evidence to assess the global redistribution of parasites by migratory birds.
2. Materials and methods
2.1. Sample collection
Tissue samples were collected from live-captured and hunter-harvested northern pintails in Alaska, USA and Hokkaido, Japan in August 2011–May 2012. Blood samples were also collected from hunter harvested and rocket netted northern pintails in California, USA during September 2011–January 2012 as part of a previous investigation (Ramey et al., 2013b) and previously published data for these samples (n = 157) were incorporated into analyses for this study. In Alaska, blood samples were collected from live birds on Koyukuk-Nowitna National Wildlife Refuge (NWR; n = 200) and on the Yukon-Kuskokwim Delta NWR (n = 201) in August of 2011 (Fig. 1). Wing muscle tissue was also obtained from hunter-harvested northern pintails in Alaska during September–October 2011 at Izembek NWR (n = 206; Fig. 1) as this tissue type has been previously shown to be useful for the detection of avian haemosporidian parasites (Ramey et al., 2013b). In Japan, blood samples (n = 114) were collected from live-captured northern pintails at Kutcharo Lake Waterfowl Observatory, Hokkaido during April–May 2012 (Fig. 1). Live captures were approved by the U.S. Geological Survey Alaska Science Center Institutional Animal Care and Use Committee (ACUC#: 2011–5, 2012–3) and conducted in accordance with authorizations granted by the United States Department of Interior (Federal Bird Banding Permits #09811, #20022, and #22176, Federal Fish and Wildlife Permits #MB122497 and #MB789758) and the Japanese Ministry of the Environment (#21-24-0001–#21-24-0005). Gender and age (adult or juvenile) was determined for most northern pintails in the field by plumage (Carney, 1992); however, in Japan northern pintails were captured in spring after molting into nuptial plumage and therefore juveniles could no longer be identified. Therefore, all birds from Japan were considered to be adults for our analyses. Blood samples were collected from live birds via jugular or brachial venipuncture and preserved in Longmire buffer solution.
Fig. 1.
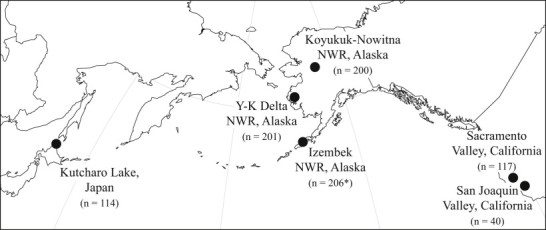
Approximate locations in North America and East Asia at which northern pintail tissue samples were collected during 2011–2012 to test for haemosporidian infection. Regions (i.e. Alaska, California, and Japan) and sub-regions (Koyukuk-Nowitna NWR, Yukon-Kuskokwim Delta NWR, Izembek NWR, Sacramento Valley, San Joaquin Valley) for sampling locations are indicated (NWR = National Wildlife Refuge). The number of tissue samples per location is indicated in parentheses. Sample tissue was whole blood unless indicated by an asterisk (signifying wing muscle tissue).
2.2. Detecting haematozoa infection
Blood parasites were detected using molecular methods as reported by Ramey et al. (2013b). Briefly, DNA was extracted from all samples using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) and screened for haematozoa using a nested PCR as described by Hellgren et al. (2004). A minimum of one negative control was incorporated into PCR reactions for every 24 wells. Reactions were conducted in eight well strip tubes with individual caps which remained closed, except while loading template and reagents, to prevent cross contamination. Amplicons were visualized on 0.8% agarose gels stained with Gel Red Nucleic Acid Gel Stain 10,000× in DMSO (Biotium, Hayward, CA). All samples were screened twice for haematozoa by nested PCR.
The 479 base pair (bp) cytochrome b (cyt b) mitochondrial DNA (mtDNA) target fragment was sequenced for all haematozoa positive samples. PCR products were treated with ExoSap-IT (USB Inc., Cleveland, OH) prior to sequencing. Sequencing was performed with identical primers used for PCR and with BigDye Terminator version 3.1 mix (Applied Biosystems, Foster City, CA) on an Applied Biosystems 3730xl automated DNA sequencer (Applied Biosystems, Foster City, CA). Sequence data were cleaned and edited using Sequencher version 5.1 (Gene Codes Corp., Ann Arbor, MI). Infections were assigned to genera (Leucocytozoon, Haemoproteus, or Plasmodium) using the nucleotide BLAST function available through the National Center for Biotechnology Information (NCBI). Assignment was based on the top NCBI BLAST result with a minimum max identity score of 90%.
A sample was considered as positive for blood parasite infection if one or both nested PCR runs resulted in a double-stranded, target product that was verified through genetic sequencing. Single stranded or otherwise ambiguous products were counted as negative. To confirm the competency of extracted DNA, a 695 bp fragment of the mtDNA cytochrome oxidase I (COI) gene of northern pintail hosts was amplified using PCR protocols described by Kerr et al. (2007). Infections were further classified as co-infected with multiple lineages of one or more haematozoa genera if the cleaned and sequenced, double-stranded target product contained three or more ambiguous nucleotides (Szymanski and Lovette, 2005). Co-infections were classified as infection by parasites of more than one lineage of (1) Leucocytozoon, (2) Haemoproteus/Plasmodium, or (3) a combination of Leucocytozoon and Haemoproteus/Plasmodium based on top NCBI BLAST results. Haemoproteus and Plasmodium were not differentiated for co-infections as both genera were amplified from a single molecular test and samples with large numbers of ambiguities could not be reliably assigned taxonomically.
2.3. Estimating prevalence using occupancy modeling
For some samples, only one of the two nested PCR reactions resulted in detection of parasite mtDNA. Such data indicate that the ability to detect parasites in a sample was imperfect. To statistically account for this imperfect detection, an occupancy modeling approach was used (MacKenzie et al, 2006, McClintock et al, 2010) wherein by evaluating a sample multiple times, the probability of parasite detection can be estimated. The estimated detection probability may then be used to more accurately estimate parasite prevalence in the sample population as has been done in prior studies of blood parasites (Ramey et al, 2012, Ramey et al, 2013b, Ramey et al, 2014).
Using an occupancy modeling framework, variance in detection probability and parasite prevalence was evaluated using covariates for age, sex, region (i.e. Alaska, California, and Japan), and sub-region (Koyukuk-Nowitna NWR, Yukon-Kuskokwim Delta NWR, Izembek NWR, Sacramento Valley, San Joaquin Valley). In this multi-model inference statistical approach (Burnham and Anderson, 2002), models were constructed to evaluate support for variance in parasite detection and prevalence relative to the given covariates. Rather than evaluating all possible models for covariates and interactions, the paradigm offered by Lebreton et al. (1992) was used to reduce the number of models evaluated. As such, a series of models that differed in how detection probability varied, but with full specificity of prevalence parameters, was first evaluated. Once the most parsimonious model of detection was identified, that structure was applied to models constructed to evaluate how prevalence may or may not vary among age classes, sex, and geographic areas with the constraint that there were no juvenile birds identified in Japan which precluded age evaluation for that location. Goodness-of-fit of models was assessed using the bootstrap routine in program MARK (Cooch and White, 2014) and a variation inflation factor was applied to models to account for any observed lack of fit.
2.4. Assessing genetic diversity and evolutionary relationships among parasites
For genetic analyses, double-stranded mtDNA sequences for blood parasites free of ambiguous nucleotides (n = 505) were aligned using Sequencher version 5.1 and cropped to a common length (356 bp) after removal of the shortest 10% of sequences from the dataset (n = 51). Haplotype diversity of the remaining 454 haematozoa mtDNA cyt b sequences was assessed by summarizing the frequency of unique haplotypes by region (through the creation of a median-joining minimum spanning network, MSN; Bandelt et al., 1999) using Network version 4.6 (available at http://www.fluxus-engineering.com). Parasite haplotypes detected in northern pintails and used in the MSN were also compared to lineages reported on the MalAvi (Bensch et al., 2009) and GenBank public databases (accessed 26 April 2014) to identify sequences identical to those previously reported for haematozoa infecting wild birds.
A phylogeny was constructed to assess genetic structuring of parasites by continent using parasite haplotypes identified in northern pintails throughout the North Pacific Basin, other waterfowl species as identified on public databases (accessed 23–26 April 2014), and tundra-nesting geese in Alaska for which genetic data were unpublished at the time of analysis (Ramey et al., 2014). Haplotypes identified in more than one individual for a given waterfowl species were represented by a single sequence per sampling location (i.e. U.S. state or country). Haplotypes for which host and/or sample location could not be determined (n = 5), that originated from captive hosts (n = 1), and that shared <335 bp of homologous DNA with the final sequence alignment (n = 4) were omitted from phylogenetic analysis. The remaining 110 sequences were aligned using Sequencher version 5.1, cropped to 348 homologous nucleotides, and used to generate a Maximum Likelihood (ML) tree in MEGA version 5.1 (Tamura et al., 2011) using the Nucleotide: Nearest-Neighbor-Interchange method with 10,000 bootstrap replicates.
3. Results
3.1. Molecular detection of parasites and prevalence of infection
A 695 bp fragment from the mtDNA COI gene was successfully amplified from all 878 blood samples collected from northern pintails used in this study confirming the competency of DNA extractions. Leucocytozoon, Haemoproteus, and Plasmodium parasites were detected in 555 (63%), 44 (5%), and 52 (6%) samples, respectively (Table 1). Parasites of all three genera were detected in northern pintails sampled in Alaska and California whereas only Leucocytozoon and Plasmodium infections were identified in birds sampled in Japan (Table 1). Co-infections with multiple genera of haemosporidia were detected at all sample locations as were co-infections with more than one Leucocytozoon lineage (Table 1). Co-infections with more than one Haemoproteus or Plasmodium parasite lineage were only detected in Alaska (Table 1).
Table 1.
Number and percentage (in parentheses) of tissue samples from northern pintails sampled at locations throughout the northern Pacific Basin detected as positive for haemosporidian infection using molecular techniques by sub-region, age (Ad = adult, Juv = juvenile), and sex. The asterisk (*) indicates that wing muscle tissue was collected and used to screen for parasites rather than whole blood. Abbreviations have been used for parasite genera (Leuco = Leucocytozoon, Haemo = Haemoproteus, Plasmo = Plasmodium) and ‘co-infection’ (co-). Co-infected samples (i.e. those from which more than one lineage of blood parasite was detected) are tabulated in all categories describing infections (e.g. a sample in which two Leucocytozoon lineages and one Haemoproteus lineage were detected is tabulated in the following columns: Leuco +, Leuco co-, and Leuco & Haemo/Plasmo co-).
| Location | Year | Age | Sex | n = | Leuco + (%) | Haemo + (%) | Plasmo + (%) | Leuco co- (%) | Haemo/Plasmo co- (%) | Leuco and Haemo/Plasmo co- (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Alaska | ||||||||||
| Izembek NWR* | 2011 | Ad | F | 57 | 41 (72) | 1 (2) | 0 (0) | 6 (11) | 0 (0) | 1 (2) |
| M | 33 | 23 (70) | 0 (0) | 1 (3) | 5 (15) | 0 (0) | 1 (3) | |||
| Juv | F | 66 | 45 (68) | 0 (0) | 0 (0) | 19 (29) | 0 (0) | 0 (0) | ||
| M | 50 | 30 (60) | 0 (0) | 0 (0) | 12 (24) | 0 (0) | 0 (0) | |||
| Koyukuk-Nowitna NWR | Ad | F | 52 | 37 (71) | 9 (17) | 8 (15) | 15 (29) | 1 (2) | 14 (27) | |
| M | 11 | 8 (73) | 1 (9) | 0 (0) | 2 (18) | 0 (0) | 1 (9) | |||
| Juv | F | 88 | 60 (68) | 1 (1) | 18 (20) | 16 (18) | 0 (0) | 15 (17) | ||
| M | 49 | 37 (76) | 1 (2) | 9 (18) | 8 (16) | 0 (0) | 8 (16) | |||
| Yukon-Kuskowim Delta NWR | Ad | F | 36 | 29 (81) | 6 (17) | 1 (3) | 9 (25) | 2 (6) | 6 (17) | |
| M | 36 | 26 (72) | 3 (8) | 2 (6) | 10 (28) | 0 (0) | 3 (8) | |||
| Juv | F | 69 | 47 (68) | 11 (16) | 2 (3) | 14 (20) | 0 (0) | 10 (14) | ||
| M | 60 | 41 (68) | 8 (13) | 4 (7) | 11 (18) | 0 (0) | 9 (15) | |||
| California | ||||||||||
| Sacramento Valley | 2011–2012 | Ad | F | 17 | 7 (41) | 0 (0) | 1 (6) | 1 (6) | 0 (0) | 0 (0) |
| M | 35 | 10 (29) | 2 (6) | 0 (0) | 2 (6) | 0 (0) | 1 (3) | |||
| Juv | F | 29 | 2 (7) | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 1 (3) | ||
| M | 36 | 8 (22) | 1 (3) | 0 (0) | 4 (11) | 0 (0) | 1 (3) | |||
| San Joaquin Valley | Ad | F | 3 | 1 (33) | 0 (0) | 1 (33) | 1 (33) | 0 (0) | 1 (33) | |
| M | 10 | 2 (20) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Juv | F | 7 | 2 (29) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| M | 20 | 2 (10) | 0 (0) | 2 (10) | 1 (5) | 0 (0) | 0 (0) | |||
| Japan | ||||||||||
| Hokkaido | 2012 | Ad | F | 49 | 42 (86) | 0 (0) | 1 (2) | 16 (33) | 0 (0) | 1 (2) |
| M | 65 | 55 (85) | 0 (0) | 1 (2) | 18 (28) | 0 (0) | 1 (2) |
The estimated detection probability for blood parasites in tissues collected from northern pintails was relatively high. The probability of detecting haemosporidian parasites in a single run varied from 0.778 (SE = 0.063) to 0.948 (SE = 0.012; Table 2, Table 3, Table 4 ), depending on parasite genera. The corresponding probability of detecting a parasite given replicate runs for samples was 0.951–0.997. Detection probability was not influenced by age, sex, or sub-region (Table 2, Table 3, Table 4); however, the detection probability of Leucocytozoon parasites was estimated to be higher in samples collected in Alaska (0.948, SE = 0.012) as compared to those collected in California and Japan (0.820, SE = 0.041; Table 2).
Table 2.
Most competitive models describing Leucocytozoon prevalence as assessed using tissue samples collected from northern pintails in 2011–2012 from Alaska, California, and Japan where psi (Ψ) is the estimate of prevalence and rho (ρ) is the estimate of detection probability from occupancy models.
| Predicting Leucocytozoon infection | Parameters for top Leucocytozoon model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | QAICc | ΔQAICc | QAICc weights | Model likelihood | No. of parameters | Deviance | Estimate | s.e. | Lower c.i. | Upper c.i. | |
| {Ψ Region + Age, ρ Alaska vs Others} | 748.293 | 0.000 | 0.419 | 1.000 | 6 | 45.242 | ρ Alaska | 0.948 | 0.012 | 0.920 | 0.967 |
| {Ψ Region, ρ Alaska vs Others} | 748.418 | 0.125 | 0.394 | 0.940 | 5 | 47.395 | ρ California + Japan | 0.820 | 0.041 | 0.727 | 0.886 |
| {Ψ Region + Sex, ρ Alaska vs Others} | 750.393 | 2.100 | 0.147 | 0.350 | 6 | 47.342 | Ψ Alaska adult | 0.747 | 0.040 | 0.662 | 0.816 |
| {Ψ Sub-region + Age, ρ Alaska vs Others} | 753.616 | 5.324 | 0.029 | 0.070 | 9 | 44.455 | Ψ Alaska juvenile | 0.673 | 0.034 | 0.604 | 0.736 |
| {Ψ Sub-region + Sex, ρ Alaska vs Others} | 755.851 | 7.559 | 0.010 | 0.023 | 9 | 46.690 | Ψ California adult | 0.261 | 0.061 | 0.160 | 0.395 |
| {Ψ Sub-region, ρ Alaska vs Others} | 759.989 | 11.696 | 0.001 | 0.003 | 11 | 46.730 | Ψ California juvenile | 0.198 | 0.049 | 0.119 | 0.310 |
| {Ψ Sub-region * Age, ρ Alaska vs Others} | 760.405 | 12.112 | 0.001 | 0.002 | 13 | 43.029 | Ψ Japan adult | 0.879 | 0.052 | 0.737 | 0.950 |
| {Ψ Sub-region * Sex, ρ Alaska vs Others} | 764.483 | 16.190 | 0.000 | 0.000 | 14 | 45.042 | |||||
| {Ψ Sub-region * Age * Sex, ρ Alaska vs Others} | 779.647 | 31.354 | 0.000 | 0.000 | 24 | 39.286 | Variance inflation = 2.102 | ||||
| {Ψ Sub-region * Age * Sex, ρ Region} | 781.732 | 33.439 | 0.000 | 0.000 | 25 | 39.252 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region} | 784.460 | 36.168 | 0.000 | 0.000 | 28 | 35.593 | |||||
| {Ψ Sub-region * Age * Sex, ρ} | 791.674 | 43.381 | 0.000 | 0.000 | 23 | 53.428 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Sex} | 794.998 | 46.705 | 0.000 | 0.000 | 34 | 33.220 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Age * Sex} | 804.649 | 56.356 | 0.000 | 0.000 | 44 | 20.941 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Age * Sex * Run} | 833.859 | 85.566 | 0.000 | 0.000 | 66 | 0.000 | |||||
Table 3.
Most competitive models describing Haemoproteus prevalence as assessed using tissue samples collected from northern pintails in 2011–2012 from Alaska, California, and Japan where psi (Ψ) is the estimate of prevalence and rho (ρ) is the estimate of detection probability from occupancy models. Abbreviations used for sub-regions in Alaska are as follows: NWR = National Wildlife Refuge, Y-K = Yukon-Kuskokwim, K-N = Koyukuk-Nowitna.
| Predicting Haemoproteus infection | Parameters for top Haemoproteus model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | QAICc | ΔQAICc | QAICc weights | Model likelihood | No. of parameters | Deviance | Estimate | s.e. | Lower c.i. | Upper c.i. | |
| {Ψ Alaska sub-region vs Others, ρ} | 287.616 | 0.000 | 0.733 | 1.000 | 5 | 37.605 | ρ | 0.778 | 0.063 | 0.631 | 0.878 |
| {Ψ Alaska sub-region vs Others, ρ Alaska sub-region vs Others} | 289.635 | 2.018 | 0.267 | 0.365 | 6 | 37.595 | Ψ Izembek NWR | 0.005 | 0.006 | 0.001 | 0.049 |
| {Ψ Alaska vs Others, ρ} | 308.816 | 21.200 | 0.000 | 0.000 | 3 | 62.846 | Ψ Y-K Delta NWR | 0.147 | 0.030 | 0.096 | 0.216 |
| {Ψ Alaska vs Others + Age, ρ} | 309.098 | 21.481 | 0.000 | 0.000 | 4 | 61.109 | Ψ K-N NWR | 0.063 | 0.021 | 0.033 | 0.118 |
| {Ψ Alaska vs Others + Sex, ρ} | 310.539 | 22.923 | 0.000 | 0.000 | 4 | 62.551 | Ψ California + Japan | 0.012 | 0.008 | 0.003 | 0.043 |
| {Ψ Alaska vs Others + Age + Sex, ρ} | 310.900 | 23.283 | 0.000 | 0.000 | 5 | 60.888 | |||||
| {Ψ Region * Sub-region * Sex, ρ} | 374.862 | 87.245 | 0.000 | 0.000 | 13 | 108.498 | Variance inflation = 1.364 | ||||
| {Ψ Sub-region * Age * Sex, ρ} | 382.337 | 94.721 | 0.000 | 0.000 | 23 | 95.102 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Age * Sex * Run} | 382.848 | 95.232 | 0.000 | 0.000 | 66 | 0.000 | |||||
Table 4.
Most competitive models describing Plasmodium prevalence as assessed using tissue samples collected from northern pintails in 2011–2012 from Alaska, California, and Japan where psi (Ψ) is the estimate of prevalence and rho (ρ) is the estimate of detection probability from occupancy models. Abbreviations used for sub-regions in Alaska are as follows: NWR = National Wildlife Refuge, Y-K = Yukon-Kuskokwim, K-N = Koyukuk-Nowitna.
| Predicting Plasmodium infection | Parameters for top Plasmodium model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | QAICc | ΔQAICc | QAICc weights | Model likelihood | No. of parameters | Deviance | Estimate | s.e. | Lower c.i. | Upper c.i. | |
| {Ψ Alaska sub-region vs Others, ρ} | 189.771 | 0.000 | 0.775 | 1.000 | 5 | 22.539 | ρ | 0.928 | 0.040 | 0.800 | 0.976 |
| {Ψ Sub-region, ρ} | 192.293 | 2.522 | 0.220 | 0.283 | 7 | 21.002 | Ψ Izembek NWR | 0.005 | 0.007 | 0.000 | 0.081 |
| {Ψ Sub-region * Age, ρ} | 201.016 | 11.245 | 0.003 | 0.004 | 12 | 19.492 | Ψ Y-K Delta NWR | 0.045 | 0.022 | 0.017 | 0.112 |
| {Ψ Sub-region * Sex, ρ} | 201.101 | 11.330 | 0.003 | 0.004 | 13 | 17.517 | Ψ K-N NWR | 0.176 | 0.040 | 0.111 | 0.267 |
| {Ψ Region, ρ} | 210.264 | 20.493 | 0.000 | 0.000 | 4 | 45.055 | Ψ California + Japan | 0.026 | 0.014 | 0.009 | 0.074 |
| {Ψ K-N NWR vs Others, ρ} | 212.223 | 22.452 | 0.000 | 0.000 | 3 | 49.033 | |||||
| {Ψ Sub-region * Age * Sex, ρ} | 216.875 | 27.104 | 0.000 | 0.000 | 23 | 12.419 | Variance inflation = 2.156 | ||||
| {Ψ Sub-region * Age * Sex, ρ Alaska sub-region vs Others} | 218.987 | 29.216 | 0.000 | 0.000 | 24 | 12.417 | |||||
| {Ψ Sub-region * Age * Sex, ρ Region} | 220.764 | 30.993 | 0.000 | 0.000 | 25 | 12.075 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region} | 223.532 | 33.761 | 0.000 | 0.000 | 28 | 8.456 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Sex} | 224.676 | 34.906 | 0.000 | 0.000 | 30 | 5.318 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Age} | 226.972 | 37.201 | 0.000 | 0.000 | 31 | 5.464 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Age * Sex} | 254.518 | 64.747 | 0.000 | 0.000 | 44 | 4.602 | |||||
| {Ψ Sub-region * Age * Sex, ρ Sub-region * Age * Sex * Run} | 300.068 | 110.297 | 0.000 | 0.000 | 66 | 0.000 | |||||
The most parsimonious model of variation in prevalence of Leucocytozoon parasites suggested that the parasite infection prevalence was highest in northern pintails sampled in Japan (87.9%, SE = 5.2%), slightly lower among adults in Alaska (74.7%, SE = 4.0%), and lowest for adults in California (26.1%, SE = 6.1%; Table 2, Fig. 2). Juvenile northern pintails had slightly lower estimated prevalence of Leucocytozoon parasites in Alaska 67.3% (SE = 3.4%) and in California 19.8% (SE = 4.9%) as compared to adult birds (Table 2, Fig. 2).
Fig. 2.
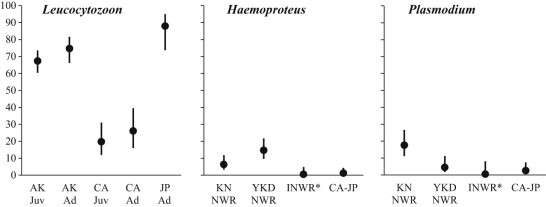
Prevalence estimates and 95% confidence intervals for Leucocytozoon, Haemoproteus, and Plasmodium parasites in northern pintails sampled in Alaska, California, and Japan 2011–2012. Estimates were derived from top supported models. Abbreviations have been used for covariates including region (AK = Alaska, CA = California, JP = Japan), age (Juv = juvenile, Ad = adult), and sub-region (KN NWR = Koyukuk-Nowitna National Wildlife Refuge, YKD NWR = Yukon-Kuskokwim Delta National Wildlife Refuge, INWR = Izembek National Wildlife Refuge). The asterisk indicates that prevalence estimates were obtained by testing samples derived from wing muscle tissue rather than whole blood.
Estimated prevalence of Haemoproteus parasites was highest in northern pintails at locations within Alaska; however, there was substantial variation among sub-regions. Prevalence of Haemoproteus parasites was estimated to be 14.6% (SE = 3.0%) among northern pintails at Yukon-Kuskokwim Delta NWR, 6.3% (SE = 2.1%) at Koyukuk-Nowitna NWR, and 0.5% at Izembek NWR (SE = 0.6%; Table 3, Fig. 2). The collective prevalence of Haemoproteus parasites in northern pintails in California and Japan was 1.2% (SE = 0.8%; Table 3, Fig. 2).
Similar to results for Haemoproteus, prevalence of Plasmodium parasites was also estimated to be highest in birds sampled at locations within Alaska, with substantial variation among sub-regions (Table 4, Fig. 2). An estimated 17.6% (SE = 4.0%) of northern pintails sampled at Koyukuk-Nowitna NWR were infected with Plasmodium parasites in contrast to 4.5% (SE = 2.2%) of ducks sampled at Yukon-Kuskokwim Delta NWR and 0.5% (0.7%) at Izembek NWR (Table 4, Fig. 2). The collective estimated prevalence for Plasmodium parasites in northern pintails sampled in California and Japan was 2.6% (SE = 1.4%; Table 4, Fig. 2).
3.2. Genetic analysis of parasite mtDNA cyt b haplotypes
A total of 31 unique haplotypes were observed among 454 genetic sequences for haematozoa mtDNA cyt b derived from northern pintails sampled throughout the North Pacific Basin (GenBank accession numbers: KJ776796- KJ776826; Fig. 3). Seven haplotypes were shared between samples from North America and Japan including each of the five most common haplotypes (Leucocytozoon 1, 6, 13, and 20; Plasmodium 2; Fig. 3). Twenty-two haplotypes were identified only in samples collected from North America including 17 identified solely in samples originating from Alaska (Fig. 3). Of the 17 haplotypes unique to Alaska, 13 were detected in only a single sample each (Fig. 3). In contrast to the relatively large number of haplotypes identified only in samples originating from Alaska, only two haplotypes were unique to northern pintail samples from Japan and no haplotype was detected only in California.
Fig. 3.
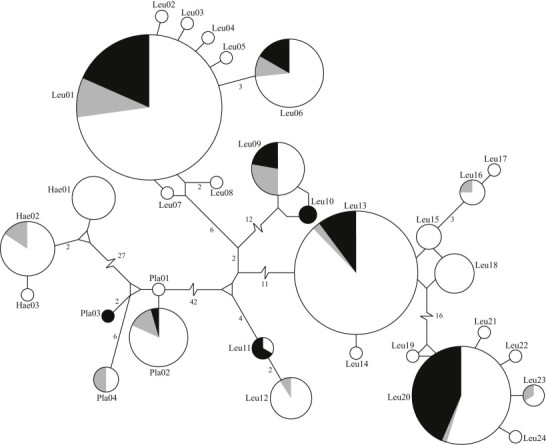
Minimum spanning network for haematozoa mitochondrial DNA cytochrome b haplotypes identified in northern pintails sampled throughout the North Pacific Basin. Circles are drawn proportional to the frequency at which haplotypes were detected. Color represents the location from which haplotypes originated: white (Alaska), grey (California), and black (Japan). A single mutation separates nodes unless indicated by number. Lines separating nodes are drawn to scale unless indicated by a break. Parasite genera have abbreviated haplotype names (Leu = Leucocytozoon, Pla = Plasmodium, and Hae = Haemoproteus).
Fourteen of 31 mtDNA cyt b haemosporidian haplotypes detected in northern pintails had 100% identity as compared to homologous sequences for parasites as reported on the MalAvi and GenBank databases (Table 5). Identical matches for two Haemoproteus and nine Leucocytozoon haplotypes were derived from parasites detected in other species of waterfowl including common/green-winged teal (Anas crecca), northern pintail, and tundra swan (Cygnus columbianus; Table 5). In contrast, three Plasmodium haplotypes originating from northern pintail samples shared 100% identity with parasite lineages identified in a diversity of avian taxa including passerines, waterfowl, shorebirds, raptors, and cranes (Table 5).
Table 5.
Mitochondrial DNA cytochrome b haplotypes for blood parasites identified in northern pintails sampled throughout the northern Pacific Basin with 100% identity to previously reported haemosporidian lineages.
| Haplotype | Previously reported avian host | GenBank accession numbers |
|---|---|---|
| Haemoproteus 1 | Anas crecca (location unreported), Cygnus columbianus (USA), Anas acuta (USA) | GU251990, JQ314225, KC409130 |
| Haemoproteus 2 | Cygnus columbianus (USA), Anas acuta (USA)* | JQ314226, KC409129* |
| Leucocytozoon 1 | Cygnus columbianus (USA), Anas acuta (USA)* | JQ314222, JQ314223, KC409125* |
| Leucocytozoon 6 | Cygnus columbianus (USA) | JQ314223 |
| Leucocytozoon 9 | Anas acuta (USA)* | KC409127* |
| Leucocytozoon 10 | Anas acuta (USA)* | KC409124*, KC409126 |
| Leucocytozoon 12 | Anas acuta (USA)* | KC409119* |
| Leucocytozoon 13 | Anas crecca (Japan), Anas acuta (USA)* | AB743872, KC409120* |
| Leucocytozoon 16 | Anas acuta (USA)* | KC409121* |
| Leucocytozoon 20 | Anas acuta (USA)* | KC409122* |
| Leucocytozoon 23 | Anas acuta (USA)* | KC409123* |
| Plasmodium 2 | Culex sasai (Japan)†, Luscina svecica (Sweden), Pluvialis fulva (USA), Carpodacus erythrinus (South Korea), Cyanistes caeruleus (UK), Geothlypis trichas (USA), Aythya marila (location unreported), Aythya marila (location unreported), Milvus sp. (location unreported), Catharus ustulatus (Costa Rica), Cygnus columbianus (USA), Anas acuta (USA)* | AB458851, AY393793, DQ659583, DQ839065, DQ991069, EU328175, GQ141559, GU252010, HF543643, JN792144, JQ314228, KC409132* |
| Plasmodium 3 | Sylvia borin (location unreported), Emberiza rutila (South Korea), Emberiza spodocephala (South Korea) | DQ368392, DQ659582, DQ839064 |
| Plasmodium 4 | Grus japonensis (Japan), Anas platyrhynchos (Japan), Acrocephalus schoenobaenus (Nigeria), Anas acuta (USA)* | AB601441, AB741486, AB741488, AB741489, AF495574, KC409133* |
Haplotype previously identified from 2011–2012 California northern pintail blood samples reported in Ramey et al. (2013b) and included this study.
Presumed vector.
Phylogenetic analysis of haemosporidian mtDNA cyt b sequences previously identified in waterfowl worldwide provided relatively strong support for structuring of parasite haplotypes by genus (bootstrap support values ≥94; Fig. 4). The sub-tree containing Leucocytozoon haplotypes could be characterized as being comprised of four major clades, all of which contained parasite lineages identified in northern pintails sampled in Alaska, California, and Japan (Fig. 5). The largest major clade was relatively poorly supported and was also comprised of Leucocytozoon haplotypes previously identified in waterfowl from Europe. In contrast, smaller major Leucocytozoon clades were better supported (bootstrap support values ≥95) and comprised entirely of sequences originating from waterfowl sampled in North America and Asia (Fig. 5).
Fig. 4.
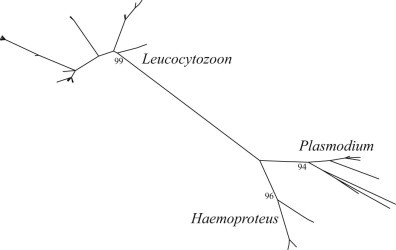
Maximum likelihood phylogenetic tree depicting inferred relationship among haematozoa mitochondrial DNA cytochrome b haplotypes obtained from waterfowl. Bootstrap support values for structuring of parasite haplotypes by genus are shown.
Fig. 5.
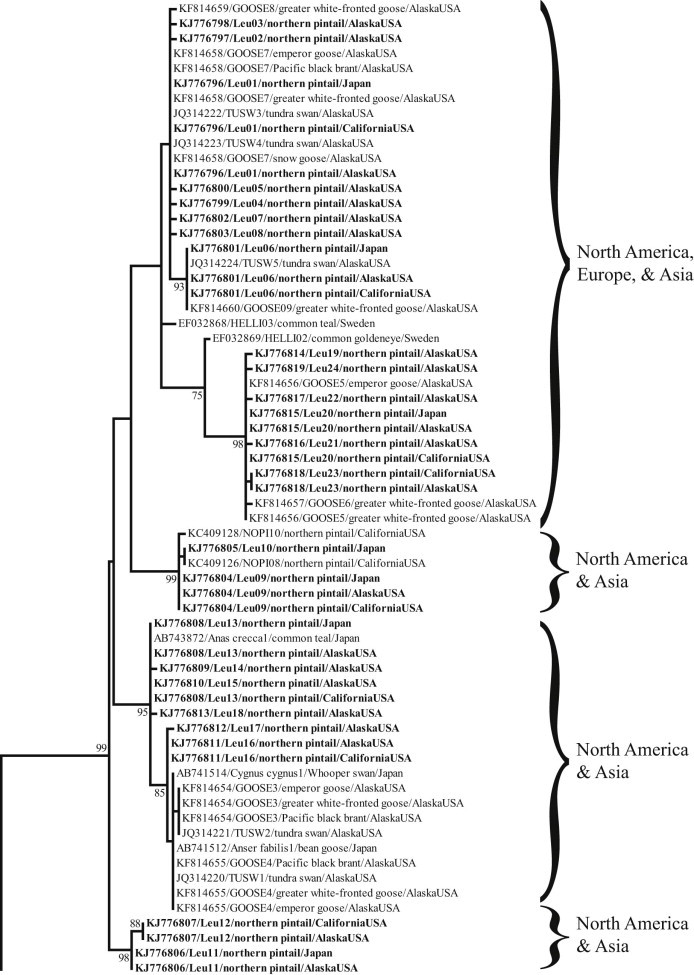
Partial maximum likelihood phylogenetic tree depicting inferred relationship among Leucocytozoon mitochondrial DNA cytochrome b haplotypes obtained from waterfowl. Branch tips are labeled with GenBank accession number, lineage name, host taxon, and sample location (U.S. state and/or country). Haplotypes identified in northern pintails in Alaska, California, and Japan as part of this study are indicated in bold font. A summary of continent of origin for parasite haplotypes comprising major clades is provided to the right of brackets. Bootstrap support values >70 are shown.
The sub-tree containing Haemoproteus haplotypes was comprised of two well supported major clades (bootstrap support values ≥97; Fig. 6), only one of which contained parasite haplotypes identified in northern pintails as part of this study. One clade contained only three parasite haplotypes, all of which originated from mute swans from the United Kingdom. A second clade was comprised of parasite haplotypes detected in a diversity of ducks, swans, and geese from throughout North America including Haemoproteus lineages identified in northern pintails sampled in Alaska and California (Fig. 6).
Fig. 6.

Partial maximum likelihood phylogenetic tree depicting inferred relationship among Haemoproteus mitochondrial DNA cytochrome b haplotypes obtained from waterfowl. Branch tips are labeled with GenBank accession number, lineage name, host taxon, and sample location (U.S. state and/or country). Haplotypes identified in northern pintails in Alaska and California as part of this study are indicated in bold font. A summary of continent of origin for parasite haplotypes comprising major clades is provided to the right of brackets. Bootstrap support values >70 are shown.
Clear structuring of parasite haplotypes into major clades was generally less well supported within the Plasmodium sub-tree as compared to the other two parasite genera analyzed in this study (Fig. 7). Plasmodium mtDNA haplotypes originated from waterfowl samples collected in North America, Europe, and Asia, including identical parasite lineages shared among samples from Alaska, California, and Japan (Fig. 7). Relatively low bootstrap support (<64) was observed for all Plasmodium clades comprised of non-identical sequences (Fig. 7).
Fig. 7.

Partial maximum likelihood phylogenetic tree depicting inferred relationship among Plasmodium mitochondrial DNA cytochrome b haplotypes obtained from waterfowl. Branch tips are labeled with GenBank accession number, lineage name, host taxon, and sample location (U.S. state and/or country). Haplotypes identified in northern pintails in Alaska, California, and Japan as part of this study are indicated in bold font. A summary of continent of origin for parasite haplotypes comprising major clades is provided to the right of brackets. Bootstrap support values >70 are shown.
4. Discussion
Estimates of detection probabilities for haemosporidian parasites in northern pintail tissues were generally high, particularly when using duplicate runs of nested PCR. For Leucocytozoon parasites, detection probability varied by location and was slightly higher in Alaska as compared to California and Japan. The difference in detection probability may be related to timing of sampling. Samples were collected from northern pintails in Alaska during summer/autumn which may correspond to seasonal peaks in parasitemia. Samples collected from northern pintails in California and Japan occurred during autumn/winter and early spring when chronic infections, characterized by lower parasitemia, may be more common.
Using multi-model inference, there was no evidence for effects of host sex on parasite prevalence; however, higher estimates of infection with Leucocytozoon parasites were derived for adult northern pintail ducks at locations in Alaska and California as compared to juvenile birds. Higher estimates of parasite prevalence for adult birds may be a function of additive effects of seasonal transmission of parasites plus relapses of infections obtained in prior seasons/years. In contrast, juvenile birds had only limited time for parasite exposure prior to sample collection (i.e. months rather than years).
Geographic variation in parasite prevalence was also observed among northern pintails sampled in East Asia and North America. Leucocytozoon infections were common at all sampling locations in contrast to Plasmodium parasites which were identified less frequently in Alaska, California, and Japan. Haemoproteus infections were also less frequently identified as compared to Leucocytozoon parasites; however, parasites of this genus were only identified in northern pintails sampled in North America. Although Haemoproteus infections were not identified among samples originating from Japan, this may be an artifact of the timing of sampling events or another unidentified sampling bias rather than absence of this parasite genus from the region. Further sampling at other times of the year and/or other taxa may result in molecular detection of haemoproteids in East Asian waterfowl.
The five most common parasite haplotypes were each found in northern pintails sampled in Alaska, California, and Japan and there was little evidence of genetic structuring by continent in the ML phylogeny, particularly among cyt b sequences in Leucocytozoon and Plasmodium sub-trees. Shared mtDNA haplotypes and support for common ancestry of parasites infecting northern pintails on two continents provide evidence for genetic exchange of haemosporidians by migratory birds among widely distributed geographic locations. Although it cannot be determined if northern pintails have contributed to exchange of parasites between North America and Eurasia; the relative abundance of this species, intercontinental migratory tendencies of some individuals, high prevalence of infections at locations in overlapping flyways during spring and autumn migration periods, and apparent waterfowl-limited host range of Leucocytozoon haplotypes detected in this study collectively suggest that this species has likely played some role. Structuring of the Haemoproteus sub-tree into well supported major clades by continental affiliation does suggest that gene flow could be limited for parasites of this genus between North America and Eurasia; however, the limited genetic information from Europe and lack of data for Asia precludes rigorous inference.
Although a relatively large number of parasite mtDNA cyt b haplotypes were only detected in North America, most of these were only found in samples originating in Alaska which may reflect a sampling bias. Nearly 70% of samples from northern pintails analyzed as part of this study originated from Alaska where parasite prevalence was relatively high. Thus, the detection of greater genetic diversity in samples from this location was expected. Two haplotypes, one Leucocytozoon and one Plasmodium, were only detected in northern pintails sampled in Japan. The reason for the lack of detection of these haplotypes in North America is cryptic but could be related to geographic differences in the distribution and relatively frequency of specific parasite lineages.
Results of this study provide evidence that haemosporidian parasitic infections are relatively common in northern pintails throughout the North Pacific Basin and support the hypothesis that migratory birds redistribute parasites between North America and Asia. These findings therefore suggest that birds could facilitate the introduction of avian malaria and other haemosporidia to novel hosts and distant regions via migratory movements. Additional research to explore the redistribution of haemosporidia between other regions (e.g. North America and South America) and among diverse sympatric taxa would be useful to better understand the global extent of parasite exchange.
Conflict of Interest
None of the authors have any financial interests or conflict of interest with this article.
Acknowledgements
We would like to thank M. Gabrielson (U.S. Fish and Wildlife Service; USFWS), K. Spragens (USFWS), J. Kohl (U.S. Geological Survey, USGS), S. Oldenberger (California Department of Fish and Wildlife), T. Hara, and Y. Murayama for assistance with live-captures of northern pintails. We appreciate the collection of blood samples from hunter-harvested northern pintails by J. Kay and T. Kay. We are grateful for critical reviews provided by J. Pearce (USGS), J. Hupp (USGS), and two anonymous reviewers. This work was funded by the USGS through the Wildlife Program of the Ecosystem Mission Area and by the USFWS through the Avian Health and Disease Program. Any use of trade names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
References
- Bandelt H.-J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bennett G.F., Smith A.D., Whitman W., Cameron M. Hematozoa of the Anatidae of the Atlantic Flyway. II. The maritime provinces of Canada. J. Wildl. Dis. 1975;11:280–289. doi: 10.7589/0090-3558-11.2.280. [DOI] [PubMed] [Google Scholar]
- Bennett G.F., Nieman D.J., Turner B., Kuyt E., Whiteway M., Greiner E.C. Blood parasites of prairie anatids and their implication in waterfowl management in Alberta and Saskatchewan. J. Wildl. Dis. 1982;18:287–296. doi: 10.7589/0090-3558-18.3.287. [DOI] [PubMed] [Google Scholar]
- Bennett G.F., Stotts V.D., Bateman M.C. Blood parasites of black ducks and other anatids from Labrador and insular Newfoundland. Can. J. Zool. 1991;69:1405–1407. [Google Scholar]
- Bensch S., Hellgren O., Pérez-Tris J. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Res. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. [DOI] [PubMed] [Google Scholar]
- Burnham K.P., Anderson D.R. Springer-Verlag; New York: 2002. Model Selection and Multi-Model Inference: A Practical Information-Theoretic Approach. [Google Scholar]
- Calisher C.H., Maness K.S.C., Lord R.D., Coleman P.H. Identification of two South American strains of eastern equine encephalomyelitis virus from migrant birds captured on the Mississippi Delta. Am. J. Epidemiol. 1971;94:172–178. doi: 10.1093/oxfordjournals.aje.a121309. [DOI] [PubMed] [Google Scholar]
- Carney S.M. US Department of the Interior; Washington, DC: 1992. Species, Age, and Sex Identification of Ducks Using Wing Plumage. [Google Scholar]
- Chen H., Smith G.J.D., Zhang S.Y., Qin K., Wang J., Li K.S. Avian flu: H5N1 outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- Cooch E.G., White G.C. thirteenth ed. 2014. Program MARK: A Gentle Introduction.http://www.phidot.org/software/mark/docs/book/ accessed 27.03.14. [Google Scholar]
- Elahi R., Islam A., Hossain M.S., Mohiuddin K., Mikolon A., Paul S.K. Prevalence and diversity of avian haematozoan parasites in wetlands of Bangladesh. J. Parasitol. Res. 2014;2014:493754. doi: 10.1155/2014/493754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint P.L., Ozaki K., Pearce J.M., Guzzetti B., Higuchi H., Fleskes J.P. Breeding-season sympatry facilitates genetic exchange among allopatric wintering populations of northern pintails in Japan and California. Condor. 2009;111:591–598. [Google Scholar]
- Hellgren O., Waldenström J., Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J. Parasitol. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- Herman C.M. Blood parasites from California ducks and geese. J. Parasitol. 1951;37:280–282. [PubMed] [Google Scholar]
- Hoogstraal H., Kaiser M.N., Traylor M.A., Guindy E., Gaber S. Ticks (Ixodidae) on birds migrating from Europe and Asia to Africa, 1959–61. Bull. Wildl. Health Org. 1963;28:235–262. [PMC free article] [PubMed] [Google Scholar]
- Hupp J.W., Yamaguchi N., Flint P.L., Pearce J.M., Tokita K., Shimada T. Variation in spring migration routes and breeding distribution of northern pintails Anas acuta that winter in Japan. J. Avian Biol. 2011;42:289–300. [Google Scholar]
- Kerr K.C.R., Stoeckle M.Y., Dove C.J., Weigt L.A., Francis C.M., Hebert P.D.N. Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. 2007;7:535–543. doi: 10.1111/j.1471-8286.2007.01670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A.V., Pearce J.M., Flint P.L., Franson J.C., Ip H.S. Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta) Mol. Ecol. 2008;17:4754–4762. doi: 10.1111/j.1365-294X.2008.03953.x. [DOI] [PubMed] [Google Scholar]
- Lebreton J.-D., Burnham K.P., Clobert J., Anderson D.R. Modeling survival and testing biological hypotheses using marked animals: a unified approach with case studies. Ecol. Monogr. 1992;62:67–118. [Google Scholar]
- Levin I.I., Zwiers P., Deem S.L., Geest E.A., Higashiguchi J.M., Iezhova T.A. Multiple lineages of avian malaria parasites (Plasmodium) in the Galapagos Islands and evidence for arrival via migratory birds. Conserv. Biol. 2013;27:1366–1377. doi: 10.1111/cobi.12127. [DOI] [PubMed] [Google Scholar]
- MacKenzie D.I., Nichols J.D., Royle J.A., Pollock K.H., Bailey L.L., Hines J.E. Academic Press; Burlington: 2006. Occupancy Estimation and Modeling: Inferring Patterns and Dynamics of Species Occurrence. [Google Scholar]
- Madge S., Burn H. Houghton Mifflin Company; New York: 1988. Waterfowl: An Identification Guide to the Ducks, Geese, and Swans of the World. [Google Scholar]
- Malkinson M., Banet C., Weisman Y., Pokamunski S., King R., Drouet M. Introduction of West Nile virus in the Middle East by migrating white storks. Emerg. Infect. Dis. 2002;8:392–397. doi: 10.3201/eid0804.010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B.T., Nichols J.D., Bailey L.L., MacKenzie D.I., Kendall W.L., Franklin A.B. Seeking a second opinion: uncertainty in disease ecology. Ecol. Lett. 2010;13:659–674. doi: 10.1111/j.1461-0248.2010.01472.x. [DOI] [PubMed] [Google Scholar]
- Miller M.R., Takekawa J.Y., Fleskes J.P., Orthmeyer D.L., Casazza M.L., Perry W.M. Spring migration of northern pintails from California's Central Valley wintering area tracked with satellite telemetry: routes, timing, and destinations. Can. J. Zool. 2005;83:1314–1332. [Google Scholar]
- Olsen B., Duffy D.C., Jaenson T.G., Gylfe A., Bonnedahl J., Bergstrom S. Transhemispheric exchange of Lyme disease spirochetes by seabirds. J. Clin. Microbiol. 1995;33:3270–3274. doi: 10.1128/jcm.33.12.3270-3274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey A.M., Pearce J.M., Flint P.L., Ip H.S., Derksen D.V., Franson J.C. Intercontinental reassortment and genomic variation of low pathogenic avian influenza viruses isolated from northern pintails (Anas acuta) in Alaska: examining the evidence through space and time. Virology. 2010;401:179–189. doi: 10.1016/j.virol.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Ramey A.M., Ely C.R., Schmutz J.A., Pearce J.M., Heard D.J. Molecular detection of hematozoa infections in tundra swans relative to migration patterns and ecological conditions at breeding grounds. PLoS ONE. 2012;7:e45789. doi: 10.1371/journal.pone.0045789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey A.M., Reeves A.B., Ogawa H., Ip H.S., Imai K., Nghia Bui V. Genetic diversity and mutation of avian paramyxovirus serotype 1 (Newcastle disease virus) in wild birds and evidence for intercontinental spread. Arch. Virol. 2013;158:2495–2503. doi: 10.1007/s00705-013-1761-0. [DOI] [PubMed] [Google Scholar]
- Ramey A.M., Fleskes J.P., Schmutz J.A., Yabsley M.J. Evaluation of blood and muscle tissues for molecular detection and characterization of hematozoa infections in northern pintails (Anas acuta) wintering in California. Int. J. Parasitol. Parasites Wildl. 2013;2:102–109. doi: 10.1016/j.ijppaw.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey A.M., Reed J.A., Schmutz J.A., Fondell T.F., Meixell B.W., Hupp J.W. Prevalence, transmission, and genetic diversity of blood parasites infecting tundra-nesting geese in Alaska. Can. J. Zool. 2014;92:699–706. [Google Scholar]
- Rappole J.H., Derrickson S.R., Hubálek Z. Migratory birds and spread of West Nile virus in the Western Hemisphere. Emerg. Infect. Dis. 2000;6:319–328. doi: 10.3201/eid0604.000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K.D., Meece J.K., Henkel J.S., Shukla S. Birds, migration and emerging zoonoses: west Nile virus, Lyme disease, influenza A and enteropathogens. Clin. Med. Res. 2003;1:5–12. doi: 10.3121/cmr.1.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M.M., Lovette I.J. High lineage diversity and host sharing of malarial parasites in a local avian assemblage. J. Parasitol. 2005;91:768–774. doi: 10.1645/GE-417R1.1. [DOI] [PubMed] [Google Scholar]
- Tallman E.J., Corkum K.C., Tallman D.A. The trematode fauna of two intercontinental migrants: Tringa solitaria and Calidris melanotos (Aves: Charadriiformes) Am. Midl. Nat. 1985;113:374–383. [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkiūnas G. CRC Press; Boca Raton: 2004. Avian Malaria Parasites and other Haemosporidia. [Google Scholar]
- Wallménius K., Barboutis C., Fransson T., Jaenson T.G.T., Lindgren P.-E., Nyström F. Spotted fever Rickettsia species in Hyalomma and Ixodes ticks infesting migratory birds in the European Mediterranean area. Parasit. Vectors. 2014;7:318. doi: 10.1186/1756-3305-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N.A., Calverley B.K., Mahrt J.L. Blood parasites of mallard and pintail ducks from Central Alberta and the Mackenzie Delta, Northwest Territories. J. Wildl. Dis. 1977;13:226–229. doi: 10.7589/0090-3558-13.3.226. [DOI] [PubMed] [Google Scholar]


