Abstract
Acinetobacter baumannii, a Gram-negative multidrug-resistant (MDR) bacterium, is now recognized as one of the more common nosocomial pathogens. Because most clinical isolates are found to be multidrug resistant, alternative therapies need to be developed to control this pathogen. We constructed a bacteriophage genomic library based on prophages induced from 13 A. baumannii strains and screened it for genes encoding bacteriolytic activity. Using this approach, we identified 21 distinct lysins with different activities and sequence diversity that were capable of killing A. baumannii. The lysin (PlyF307) displaying the greatest activity was further characterized and was shown to efficiently kill (>5-log-unit decrease) all tested A. baumannii clinical isolates. Treatment with PlyF307 was able to significantly reduce planktonic and biofilm A. baumannii both in vitro and in vivo. Finally, PlyF307 rescued mice from lethal A. baumannii bacteremia and as such represents the first highly active therapeutic lysin specific for Gram-negative organisms in an array of native lysins found in Acinetobacter phage.
INTRODUCTION
Members of Acinetobacter are soil bacteria that frequently colonize the human skin without harm (1). However, in environments in which individuals are immunocompromised or suffer from a variety of wounds (e.g., in hospital settings or on battlefields), Acinetobacter baumannii can cause severe life-threatening infections (2–4). Symptoms of A. baumannii infections range from mild skin wounds and urinary tract infections to more severe conditions, including pneumonia, meningitis, and sepsis (5). A. baumannii is now one of the most common causes of hospital-acquired pneumonia (2) and sepsis; while not common (only 1.3% of all sepsis cases), it is associated with mortality rates of up to 58% (6).
One of the main threats from A. baumannii is the high rate of resistance to antibiotics commonly used to treat Gram-negative infections. More than 80% of Acinetobacter species are considered to be multidrug resistant (MDR) (i.e., resistant to at least three classes of antibiotics), resulting in infections with poor clinical outcomes, including high rates of morbidity and death, prolonged hospital stays, and substantial health care expenses (3, 7). In addition, several strains of pan-drug-resistant A. baumannii have been isolated, showing resistance to a wide variety of clinically used antibiotics (8). A. baumannii is also capable of surviving treatments with detergents and disinfectants, dehydration, and UV radiation and thus is difficult to eradicate from surfaces in hospital environments (9, 10). The organism not only is intrinsically resistant to many antibiotics (owing to β-lactamases, weak membrane permeability, and efficient efflux systems) but also can readily acquire foreign plasmids and is considered to have a high degree of genetic plasticity (11). Outbreaks caused by MDR Acinetobacter have been reported from hospitals worldwide; more recently, they have become a serious problem in military medical facilities (4). One of the main reasons why A. baumannii is so persistent is its ability to tolerate desiccation and other stresses through its ability to form biofilms on solid surfaces, including medical implants and catheters (12). For these reasons, new and better ways of controlling A. baumannii are needed.
One possible approach to the treatment of Gram-negative infections is based on the use of bacteriophage endolysins (lysins, enzymes that degrade the cell walls of phage-infected bacteria to release their phage progeny) (13, 14). Lysins can be endo-β-N-acetylglucosaminidases or N-acetylmuramidases (lysozymes), which cleave the sugar moiety of peptidoglycan, endopeptidases, which act on the peptide backbone or cross-bridge, or N-acetylmuramoyl-l-alanine amidases, which hydrolyze the amide bond connecting the sugar and peptide moieties of peptidoglycans. Work over the past 14 years has shown that lysins may be recombinantly expressed and added exogenously to sensitive bacteria, resulting in rapid lysis and death. This “lysis from without” is the basis of a novel antibacterial strategy that has proven effective for several different Gram-positive bacterial pathogens (15–18).
Unfortunately, Gram-negative bacteria are largely resistant to the addition of exogenously added lysins, due to their protective outer membranes. A small fraction of lysins, however, display low innate ability to kill Gram-negative bacteria (19), an ability that is improved in the presence of membrane-destabilizing factors (20, 21). This innate ability could be driven by highly positively charged N- or C-terminal domains in the native sequences (19), which enable the lysins to bind to the anionic outer membrane and access their peptidoglycan substrate. Recently, researchers have used this knowledge to create “artilysins,” engineered lysins with added cationic peptides and EDTA for improved antibacterial activity (22). In contrast to this, we have used a broad, expression-based, screening approach to identify the antibacterial lysins present in the bacteriophages of Gram-negative bacteria. Such an approach takes advantage of the vast amount of genetic material accumulated by the phages (i.e., lysins) used to kill Gram-negative bacteria. Here, we identified and isolated several lysins active against A. baumannii, and we used one to demonstrate antibacterial activity for planktonic and biofilm cells both in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. baumannii strains with numbers between 1775 and 1798 were clinical isolates from the clinical laboratory of Weill Cornell Medical Center (New York, NY). Environmental strains 1490 and 1498 were from soil, while strains 2198 and 2204 were from marine sediment; all were verified as Acinetobacter by 16S rRNA sequencing. Bacteria were cultured in trypticase soy broth (TSB) (Thermo Fischer Scientific, Waltham, MA) at 37°C, with shaking at 200 rpm. Strains for determination of the specificity of the lysin were cultured under the same conditions, at the following temperatures: Escherichia coli DH5α, 37°C; Pseudomonas aeruginosa PAO1, 30°C; Staphylococcus aureus RN4220, 30°C.
Generation of phage DNA library.
Thirteen isolates of A. baumannii (isolates 1490, 1498, 2198, 2204, 1775, 1776, 1777, 1788, 1790, 1792, 1794, 1796, and 1798) were treated with 2 μg/ml mitomycin C for 16 h to induce prophages. Growth curves for each strain were monitored, and all 13 were consistent with phage induction and lysis (verified by electron microscopy); uninduced strains yielded no phage. The supernatants were sterile filtered (0.22 μm) and treated with DNase and RNase to remove host nucleic acids. Next, the phages were collected by polyethylene glycol (PEG) precipitation, and DNA was extracted by phenol-chloroform extraction and ethanol precipitation. The purified phage DNA was then used to construct and to screen expressible linker amplified shotgun libraries (E-LASLs) generated on an individual basis for all of the identified phages, to detect lysins (8, 23).
MICs for antibiotics.
To generate a homogeneous layer of bacteria, agar plates were overlaid with soft agar (7 g/liter agar) containing A. baumannii strains (100 μl overnight culture mixed with 3 ml soft brain heart infusion [BHI] agar). Etest strips (bioMérieux, Durham, NC) with ampicillin, ceftazidime, levofloxacin, minocycline, or polymyxin B were applied on top of the soft agar layer, and the plates were incubated for 24 h at room temperature. The MIC values were determined from the gradient markings, according to the manufacturer's instructions.
Activity screen of lysins on plates.
All lytic clones (n = 21) were recombinantly expressed in E. coli DH5α. Cells were cultured at 30°C in LB supplemented with 50 μg/ml ampicillin, with shaking at 200 rpm, and induction was performed by adding 0.2% arabinose once the mid-log phase had been reached. Incubation was continued overnight, after which cells were collected by centrifugation, washed with 50 mM sodium phosphate buffer (pH 7.0), and homogenized in an Emulsiflex-C5 homogenizer (Avestin, Ottawa, Ontario, Canada). Cellular debris was removed by centrifugation (16,000 × g for 45 min) and the lysate was passed through a 0.22-μm sterile filter to generate the crude lysate.
A. baumannii grown overnight in TSB was mixed with 50°C TSB soft agar (100 μl bacteria to 5 ml soft agar) and poured onto a TSB agar plate as a top agar layer. The agar was allowed to solidify at room temperature, after which 10 μl of crude lysate was spotted on the plate. Plates were incubated for 1.5 h at 20°C, after which they were stored overnight at 4°C. This procedure (1.5 h at room temperature and then overnight at 4°C) was repeated until a bacterial lawn could be detected on the plates. Lytic activity was scored based on the size of any clearing zone and the general lysis of the clearing zones (e.g., reduction in turbidity).
Expression and purification of PlyF307.
Overnight cultures of E. coli DH5α with the pBAD24-PlyF307 construct were diluted 1:100 in preheated LB containing 50 μg/ml ampicillin and were incubated at 30°C, with shaking at 200 rpm, until reaching an optical density at 595 nm (OD595) of 0.5. The expression of PlyF307 was induced by the addition of 0.2% arabinose, and expression continued overnight at 30°C. The cells were collected, washed in 50 mM sodium phosphate (pH 8.0), and homogenized using an Emulsiflex-C5 homogenizer (Avestin). The lysate was cleared of cellular debris by centrifugation (16,000 × g for 45 min at 4°C) followed by sterile filtration (0.22 μm) to generate a crude lysate.
The crude lysate of PlyF307 in 50 mM sodium phosphate (pH 8.0) was applied to a HiTrap SP FF column (GE Healthcare Life Sciences, Uppsala, Sweden) using the Äkta fast protein liquid chromatography (FPLC) system (GE Healthcare Life Sciences), and fractions were eluted with a linear gradient of 2 M NaCl. Peaks of interest were pooled, concentrated using an Amicon Ultra Ultracel-3K filter (EMD Millipore, Billerica, MA), and then applied to a HiPrep 26/60 Sephacryl S-200 high resolution size exclusion column (GE Healthcare Life Sciences) using 50 mM sodium phosphate buffer (pH 8.0) as the running buffer. Peaks containing the protein of interest were pooled and concentrated using Amicon Ultra Ultracel-10K filters (EMD Millipore).
PlyF307 enzymatic activity.
For all activity measurements, bacteria grown in TSB overnight (stationary phase) or to early exponential phase (OD595 of 0.2 to 0.4) were washed in double-distilled water before being resuspended to approximately 106 CFU/ml. The resuspended bacteria were incubated with 100 μg/ml PlyF307 for 2 h at 37°C. Factors affecting the activity of the lysin, as well as the spectra of activity, were analyzed with early exponentially growing bacteria using the aforementioned conditions, including pH (20 mM sodium phosphate [pH 6.0 to 8.0]) and NaCl (0 to 500 mM). All experiments were performed in triplicate, and results are shown as mean ± standard deviation (SD).
In vitro treatment of catheter-adherent A. baumannii with PlyF307.
Catheter tubing (CareFusion) was cut into 3-cm sections using a sterile scalpel. An overnight culture of A. baumannii strain 1791 was diluted 1:1,000 in TSB supplemented with 0.2% glucose (∼1 × 105 CFU/ml). Each catheter section was seeded with 300 μl of the diluted overnight culture, clamped shut, and incubated for 3 days at 37°C to allow biofilm formation. At that time, the catheters were washed with phosphate-buffered saline (PBS) before 300 μl PlyF307 (1 mg/ml) was added to the tube sections. After incubation at 37°C for 2 h, the catheters were washed with 50 mM sodium phosphate buffer (pH 7.5). Biofilms were mechanically removed from the catheters by being thoroughly resuspended in 50 mM sodium phosphate buffer (pH 7.5) and scraped with a pipette tip. Subsequent crystal violet staining of the catheter confirmed the absence of any remaining attached biofilm. The samples were then vortex-mixed for 1 min, serially diluted, and plated on TSB agar plates to determine the CFU.
Mouse in vivo catheter model.
The Rockefeller University institutional animal care and use committee approved all in vivo protocols. A. baumannii catheter biofilms were formed as described above. The backs of 20 female BALB/c mice (6 to 8 weeks of age; Charles River Laboratories) were shaved and sterilized with three applications of betadine solution and 70% ethanol wipes. A small incision was made in the skin, sufficient to insert a 3-cm catheter section (containing a 2-day preformed biofilm) under the dermis, 1 cm from the opening. The incision was then closed with surgical staples. After 24 h, two doses (4 h apart) of 1 mg PlyF307 (250 μl) or PBS were injected directly into the catheter under the skin. Three hours later, the mice were euthanized with CO2, and the catheters were removed from the mice, washed twice with PBS, and treated as described above to remove residual biofilm bacteria. Serial dilutions were plated on BHI agar plates. The plates were incubated overnight at 37°C, and CFU were counted.
Spreading of A. baumannii in mouse organs.
Two C57BL/6 mice were infected intraperitoneally (i.p.) with a washed overnight culture of A. baumannii strain 1791 (∼108 CFU). The mice were euthanized 2 h postinfection, and the liver, spleen, kidney, and heart were dissected from the mice. The organs were homogenized in PBS, and dilutions were plated on BHI agar plates to determine the number of CFU per organ.
Mouse Acinetobacter sepsis model.
Twenty female C57BL/6 mice (6 to 8 weeks of age; Charles River Laboratories) were injected i.p. with A. baumannii (∼108 CFU). Two hours after injection, animals were treated i.p. with either PlyF307 (1 mg; n = 10) or PBS (n = 10), and survival was tracked for 2 weeks.
Nucleotide sequence accession numbers.
Identified sequences have been deposited in GenBank with accession numbers KJ740393 to KJ740413.
RESULTS
Determination of MICs for clinical A. baumannii isolates.
Since the resistance patterns of our A. baumannii clinical isolates were unknown, we tested them against antibiotics commonly used to treat Gram-negative infections. Thirteen documented clinical strains were tested for their antibiotic resistance profiles to five antibiotics (ampicillin, ceftazidime, levofloxacin, minocycline, and polymyxin B) affecting different molecular targets (the cell wall, topoisomerases, protein synthesis, and the outer membrane). Results revealed that the strains had different degrees of resistance, with some being resistant to more than one drug (Table 1).
TABLE 1.
MICs of different antibiotics against clinical isolates of A. baumannii
| Isolate | MIC (μg/ml)a |
||||
|---|---|---|---|---|---|
| Ampicillin | Ceftazidime | Levofloxacin | Minocycline | Polymyxin B | |
| 1775 | 2 | 12 | 0.125 | 0.125 | 0.064 |
| 1776 | 2 | 12 | 0.125 | 0.125 | 0.5 |
| 1777 | 2 | 8 | 0.125 | 0.125 | 4 |
| 1788 | 16 | 64 | >32 | 8 | 0.75 |
| 1789 | 12 | 48 | >32 | 12 | 1 |
| 1790 | 16 | 96 | 8 | 4 | 0.5 |
| 1791 | 24 | 128 | 8 | 1.5 | 0.75 |
| 1792 | 64 | 64 | 8 | 2 | 0.75 |
| 1793 | 32 | 64 | 12 | 2 | 0.5 |
| 1794 | 32 | 96 | 6 | 1.5 | 0.64 |
| 1795 | 14 | 96 | 4 | 1.5 | 0.125 |
| 1796 | 128 | 96 | 4 | 2 | 0.125 |
| 1797 | 256 | 128 | 4 | 2 | 0.38 |
The antibiotic class, target, dose range tested, and susceptibility breakpoints for the tested drugs were as follows: ampicillin, β-lactam, cell wall, 0.016 to 256 μg/ml, susceptibility at ≤8 μg/ml and resistance at ≥32 μg/ml; ceftazidime, β-lactam, cell wall, 0.016 to 256 μg/ml, susceptibility at ≤16 μg/ml and resistance at ≥64 μg/ml; levofloxacin, fluoroquinolone, topoisomerases, 0.002 to 32 μg/ml, susceptibility at ≤2 μg/ml and resistance at ≥8 μg/ml; minocycline, tetracycline, protein synthesis, 0.016 to 256 μg/ml, susceptibility at ≤4 μg/ml and resistance at ≥16 μg/ml; polymyxin B, polypeptide, outer membrane, 0.064 to 1,024 μg/ml, susceptibility at ≤2 μg/ml and resistance at ≥8 μg/ml. Bold numbers indicate resistance.
Generation and characterization of A. baumannii phage lysin library.
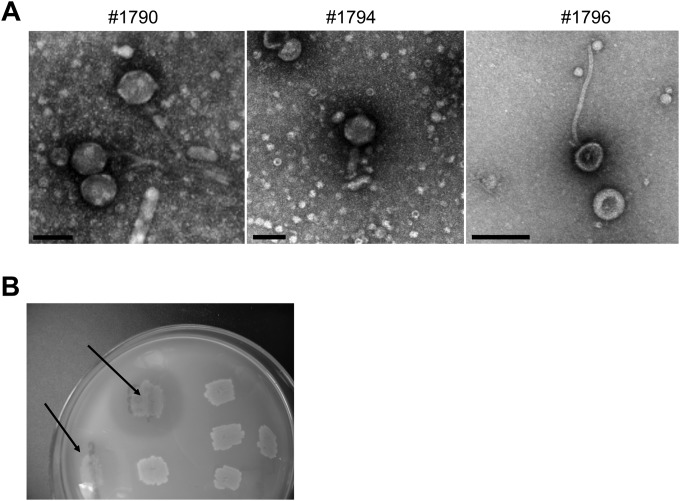
We exploited the knowledge that A. baumannii is polylysogenized (24, 25) by identifying novel phage lysins from prophages induced from 13 clinical and environmental isolates with mitomycin C. Based on electron microscopy, all 13 strains produced phage, and all were tailed (Caudovirales) and could be classified as either siphoviruses or myoviruses based on their distinct morphologies. Examples of three isolated phages are presented in Fig. 1A. Phage DNA was isolated and genomic expression libraries were constructed in E. coli. The libraries were plated, induced with 0.2% arabinose, and overlaid with a top agar layer containing A. baumannii (strain 1794). Clones that generated a clearing zone after overnight incubation at 37°C were subcultured and rescreened, to confirm lytic activity (Fig. 1B). In preliminary experiments, a few libraries were screened with strains 1794, 1490, and 1796; since the three strains yielded similar clearing zones, strain 1794 was used to screen all clones.
FIG 1.
Examples of inducible phages from A. baumannii and generation of a phage lysin library. (A) Inducible bacteriophages from A. baumannii clinical isolates were negatively stained with 2% uranyl acetate and visualized by electron microscopy. They are typical of siphoviruses (strains 1790 and 1796) or myoviruses (strain 1794). Bars, 100 nm. (B) DNA was extracted from the phages, and a phage genomic library was established. Clones were screened for their ability to generate clearing zones on soft agar plates containing A. baumannii (arrows).
Homology and activity of identified lysins.
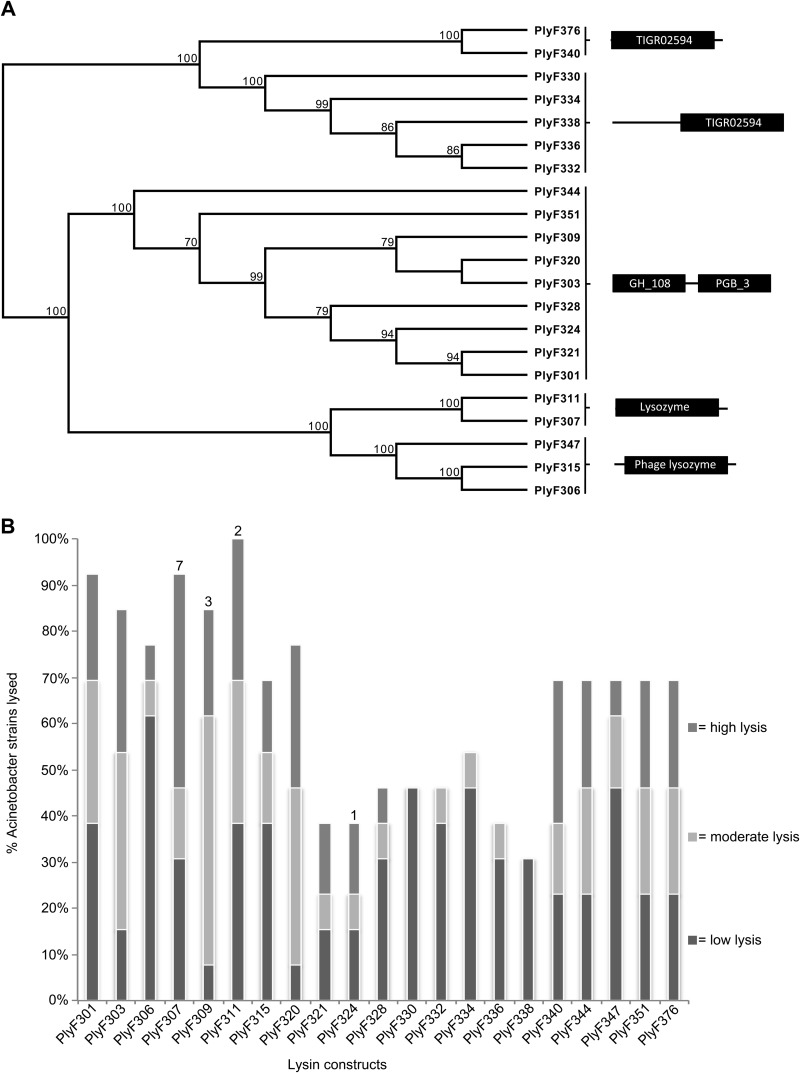
All plasmid inserts from stable clones able to generate clearing zones on A. baumannii (n = 21) were sequenced. The sequences were all annotated as phage lysins and clustered in three distinct groups based on domain organization (Fig. 2A; also see Fig. S1 in the supplemental material). Seven constructs had a single TIGR02594 domain, with or without a preceding region of unknown domain structure. Five constructs had a single lysozyme domain, and nine had a two-domain organization, consisting of an N-terminal catalytic domain (domain GH_108) and a C-terminal binding domain (domain PGB_3) (Fig. 2A) typical of lysins from Gram-positive phages.
FIG 2.
Identified lysins and their activity versus A. baumannii. (A) All constructs with activity against A. baumannii (PlyF301 to PlyF376) were sequenced, and a phylogenetic tree was generated using the software MacVector (unweighted pair group method with arithmetic mean [UPGMA], with Poisson correction). Three main classes of proteins were identified based on domain organization, including proteins with (i) a TIGR02594 domain, (ii) a catalytic domain and a binding domain, and (iii) a lysozyme domain. (B) The 21 different constructs were screened for activity versus 13 different A. baumannii clinical isolates. Crude lysates (10 μl) were added to a soft agar plate with A. baumannii and incubated for 1.5 h at room temperature each day, while being kept at 4°C the rest of the time. Plates were incubated until bacterial growth and clearing zones were visible (4 to 5 days). Clearing zones larger than the original spot of crude lysate were scored. Numbers above the bars, numbers of strains for which the specific lysin was most efficient.
All constructs were expressed, and crude unpurified lysates were generated for an initial screening of activity against all 13 A. baumannii isolates (see Materials and Methods). The lysates were spotted on the surface of soft agar overlays containing A. baumannii, and the plates were allowed to incubate at 20°C for 1.5 h, after which they were kept at 4°C overnight to reduce the growth rate of A. baumannii, allowing better resolution of clearing zones. This procedure was repeated daily until a lawn of bacteria could be seen on the plates, after which clearing zones were analyzed based on size and turbidity. Most lysates generated clearing zones at the site of the initial spot, while others (PlyF307) also generated halo-like clearing zones (see Fig. S2 in the supplemental material). In this setting, PlyF311 was able to clear all 13 isolates of A. baumannii, followed by PlyF307 (its close relative) (Fig. 2A), which cleared 12/13 strains (Fig. 2B). In general, however, PlyF307 displayed greater activity than the other lysins, as judged by the size and turbidity of the clearing zones, and better lytic activity against seven of the tested A. baumannii isolates, while PlyF311 was the most active lysin against only 2 of the 13 strains (Fig. 2B). PlyF307 was derived from phage induced from A. baumannii strain 2198.
Expression and purification of PlyF307.
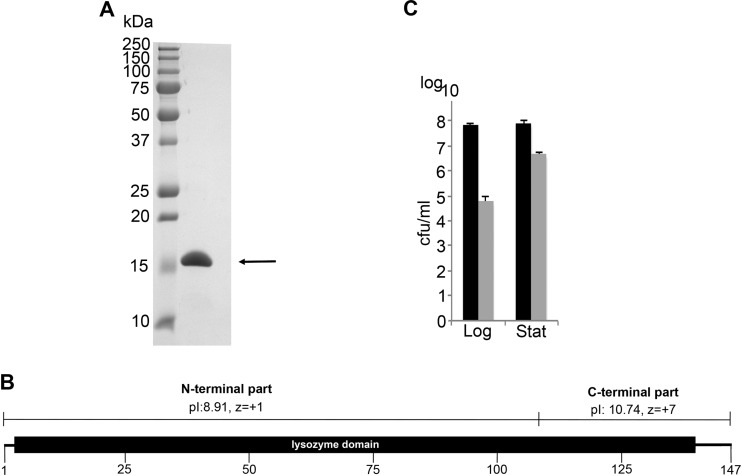
Based on the specificity and lytic activity results, we elected to continue working with PlyF307. Its relatively small size (16.1 kDa) and high pI (pI = 10.12) enabled us to purify it to near homogeneity (>95%, based on visual observation) through a combination of ion-exchange and size exclusion chromatographic steps (Fig. 3A). We consistently produced 4 to 6 mg of purified lysin per liter of induced E. coli clone.
FIG 3.
Purification and activity of A. baumannii phage lysin PlyF307. (A) A. baumannii phage lysin PlyF307 (arrow) was purified by a combination of ion-exchange chromatography and size exclusion chromatography, which resulted in purity of ∼95%, as observed visually. (B) PlyF307 is composed of a single lysozyme domain, involving amino acids 3 to 141. The C-terminal part of the lysin has a high positive net charge (charge of +7), while the N-terminal part is closer to a neutral net charge (charge of +1). (C) The activity of PlyF307 was investigated by determining its effects against both exponentially growing cells (Log) and stationary-phase cells (Stat), by incubating the samples for 2 h at 37°C with 100 μg/ml PlyF307. Black bars, samples without the addition of PlyF307; gray bars, samples with the addition of PlyF307.
PlyF307 is composed of a single lysozyme domain, involving amino acids 3 to 141 of the 147 amino acids constituting the full PlyF307 protein. Its C-terminal region has a considerably higher pI than the N-terminal region (pI values of 10.74 and 8.91, respectively), as well as a higher positive net charge (z values of +7 and +1, respectively) (Fig. 3B).
Activity of PlyF307.
Lysins are generally less active against stationary-phase bacteria than against exponentially growing (log-phase) bacteria, a phenomenon widely seen for Gram-positive lysins (26). With a large inoculum of bacteria (>108 CFU/ml), the addition of PlyF307 (100 μg/ml) was able to reduce the viability of exponentially growing cells (OD600 = 0.4) by >3 log units, while reducing the viability of stationary-phase bacteria by little more than 1 log unit (Fig. 3C). Thus, we continued our further analysis using exponentially growing A. baumannii.
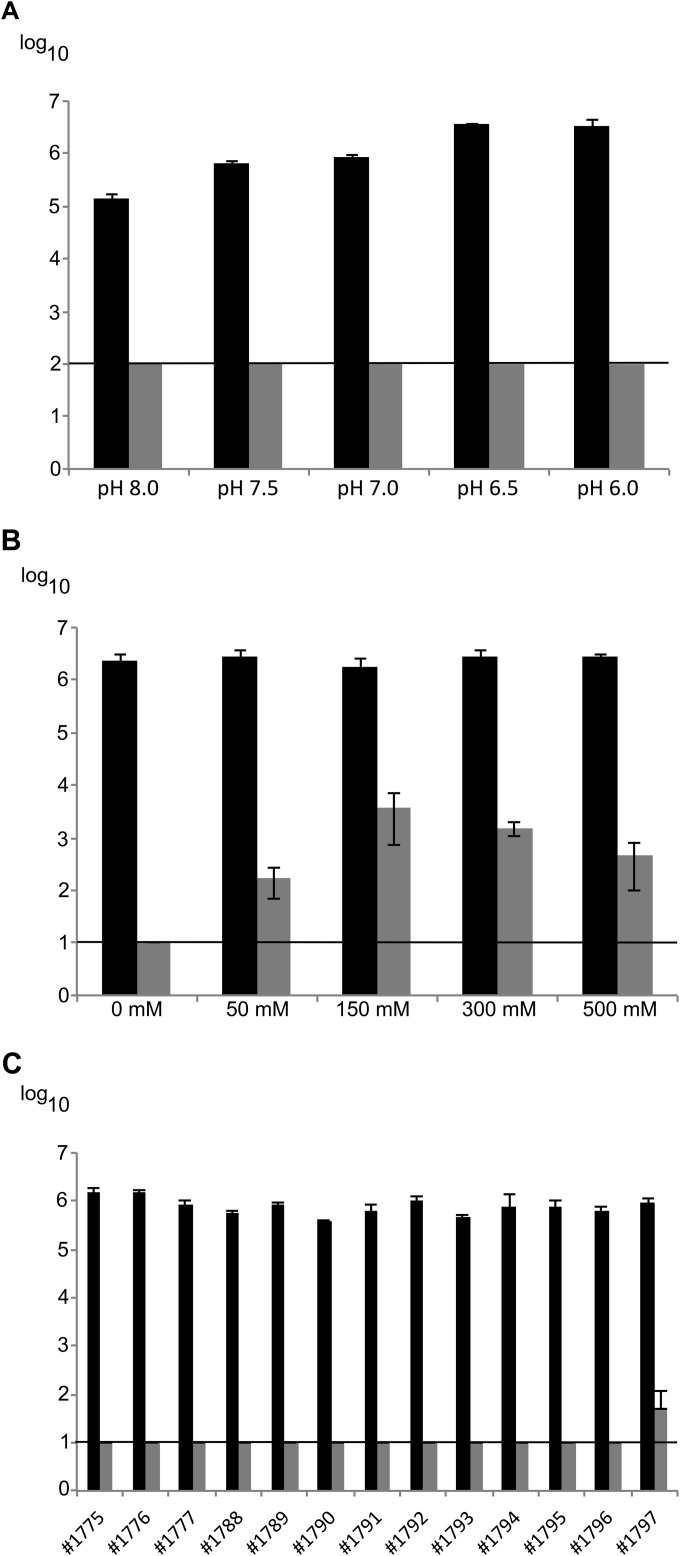
PlyF307 was highly active across a broad pH range (pH 6.0 to 8.0), reducing the amount of A. baumannii isolate 1791 to below the limit of detection (<100 CFU/ml) under all experimental conditions and resulting in a decrease of >4 log units at pH 6.0 (Fig. 4A). Due to the greater stability of the bacteria at lower pH, we continued the experiments at pH 6.0. PlyF307 was most active in the absence of NaCl, resulting in a >5-log-unit decrease to below the limit of detection (<10 CFU/ml) (Fig. 4B). The addition of 50 to 500 mM NaCl reduced the efficiency of PlyF307, although there were still decreases of 3 to 4 log units under all experimental conditions. Finally, PlyF307 was able to clear (>5-log-unit decreases) all clinical strains of A. baumannii (Fig. 4C), with no significant activity against E. coli, Pseudomonas aeruginosa, or Staphylococcus aureus (data not shown). PlyF307 was able to kill clinical strain 1792, the only strain that it did not kill in the initial screen using the plate assay. This may be attributed to both the concentration of lysin (being lower in the plate assay) and the buffer conditions, with the presence of salts in the agar plate.
FIG 4.
(A and B) Optimal conditions, i.e., pH optimum (A) and NaCl optimum (B), for PlyF307 determined using A. baumannii isolate 1791. (C) Killing activity of PlyF307 against 13 A. baumannii clinical isolates. All experiments were conducted using exponentially growing bacteria that had been washed and resuspended to ∼106 CFU/ml in sodium phosphate buffer (pH 6.0). PlyF307 (100 μg/ml) was added and the samples were incubated for 2 h before serial dilutions were plated for CFU counting. Black bars, samples without the addition of PlyF307; gray bars, samples with the addition of PlyF307. Horizontal lines, limits of detection in the different experiments. All experiments were conducted in triplicate, and results are presented as mean ± SD.
PlyF307 is able to kill A. baumannii in biofilms in vitro and in vivo.
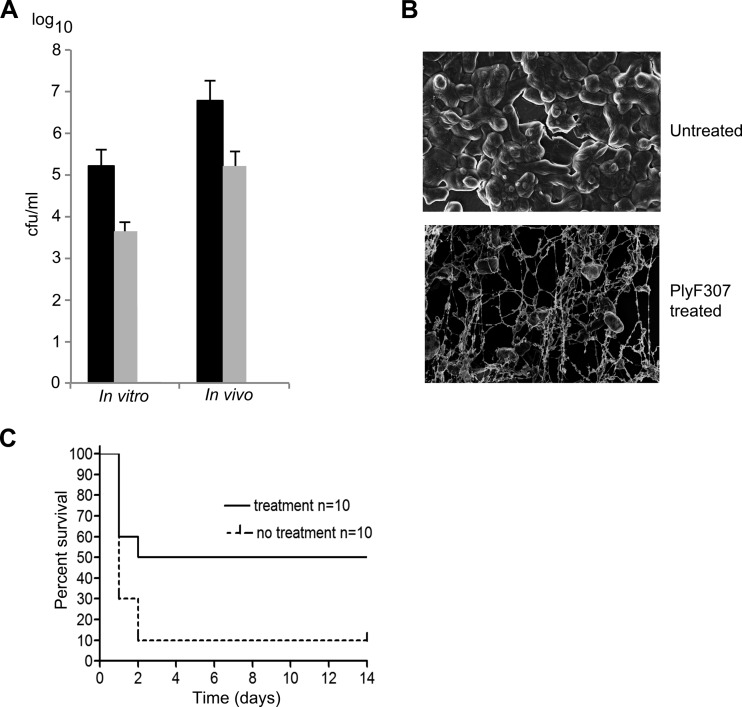
A. baumannii commonly forms biofilms on catheters and replacement joints (27). Therefore, we investigated whether PlyF307 also had antibiofilm activity. Catheter sections were incubated with A. baumannii for 3 days in vitro to establish biofilms, which were then washed and treated with PlyF307. After 2 h, remaining biofilm bacteria were removed and resuspended in buffer for enumeration. We observed an approximately 1.6-log-unit decrease in the number of A. baumannii after treatment with PlyF307 (Fig. 5A). More importantly, a marked reduction in total biofilm biomass on the catheters was confirmed using scanning electron microscopy (Fig. 5B).
FIG 5.
Ability of PlyF307 to degrade A. baumannii biofilms in vitro and in vivo and to rescue mice from lethal bacteremia. (A) A. baumannii biofilms were formed in vitro on catheters for 24 h before being treated in vitro with PlyF307 for 2 h. For the in vivo samples, whole catheter pieces with 2-day-old biofilms were implanted subcutaneously in the backs of mice. After 24 h, two doses of 1 mg of PlyF307 or buffer were administered subcutaneously, 4 h apart, at the implanted site. Two hours after the last dose, the catheter was removed and sonicated, and the dislodged A. baumannii organisms were plated for CFU enumeration. Black bars, controls; light gray bars, samples treated with PlyF307. (B) A 3-day-old A. baumannii biofilm was established on a catheter and treated for 30 min with 250 μg PlyF307 before being analyzed using scanning electron microscopy. Magnification, ×20,000. (C) Mice were infected i.p. with 108 CFU of A. baumannii. They received a single dose of PlyF307 (1 mg) or buffer i.p. 2 hours later, and they were monitored for survival for 14 days.
To better mimic in vivo conditions, we implanted catheter sections, colonized with 2-day-old A. baumannii biofilms, subcutaneously in the backs of mice. The catheters were left undisturbed for 24 h, after which treatment began with 1.0 mg of PlyF307 or PBS control delivered subcutaneously at the site of the implant. A total of two doses were delivered in a 4-h period. The catheters were removed 3 h later, and the residual organisms on the catheter sections were enumerated. In this in vivo setting, we found ∼2-log-unit decreases in bacterial viability (Fig. 5A).
PlyF307 rescues mice from lethal bacteremia.
Since lethal bacteremia is a common outcome of A. baumannii infections, we investigated the ability of the PlyF307 lysin to work systemically and to rescue mice from this type of infection. Mice received 108 CFU of A. baumannii i.p., and they were treated 2 h later with a single dose of 1.0 mg PlyF307 lysin or buffer by the same route. In preliminary experiments, we found that, by 2 h, all organs in the mice were heavily infected with A. baumannii, suggesting that the infection was systemic (see Fig. S3 in the supplemental material). While most (90%) buffer-treated mice died within 1 to 2 days, the F307-treated mice had a significantly higher rate of survival, with 50% being rescued from this highly lethal dose of A. baumannii (Fig. 5C).
DISCUSSION
Antibiotic-resistant A. baumannii is a growing concern, with several strains being resistant to all currently used antibiotics in hospital settings (5). Since bacteriophages have coevolved with bacteria for nearly 1 billion years, we sought to investigate whether phage products (e.g., lysins) could be used to kill A. baumannii. In previous studies, Gram-negative bacteria have been shown to be resistant to exogenous phage lysins because their outer membranes prevent the lysins from reaching the peptidoglycan substrate. However, recent published data support the idea that some lysins (natural or engineered) do have activity against Gram-negative bacteria (14, 19).
A. baumannii strains are polylysogenic in nature, usually harboring several inducible prophages in their genomes (24, 25). We took advantage of this and extracted DNA from mitomycin C-induced prophages from several clinical and environmental isolates. We developed an expressible recombinant genomic library with this DNA to screen for phage-encoded proteins with bacteriostatic or bactericidal activity against A. baumannii (23). In doing so, we identified 21 unique lytic clones from 13 strains with three distinct domain organizations (Fig. 2A), highlighting the abundance of distinct prophages in this limited group of A. baumannii isolates and the strength of using such an approach to identify lysins. The two-domain organization found in several of the lysins, with a C-terminal catalytic domain and an N-terminal binding domain, is worth noting. While this domain organization is dominant among lysins active against Gram-positive bacteria, it is rarely found among lysins active against Gram-negative bacteria, for which single catalytic domains (lacking binding domains) are usually found (28).
One striking aspect of the identified lysins is their diversity. Although a limited number of strains were examined (most from New York) and only inducible phages were tested, we still generated 21 unique clones in our library screen. BLAST analysis of the publically available A. baumannii genomes confirmed that a wide variety of lysins are present in this bacterial species (data not shown). With the exception of mycobacteriophage endolysins, which also display wide diversity in terms of modular domain organization (29), such diversity of phage lysins is rarely seen within a species.
In Acinetobacter, it seems rather unusual that the lysogens would evolve a large diverse group of lysins for the sole purpose of releasing their phage progeny. We speculate that these lysins might in some way be harnessed by the bacteria to control their environment. Being a soil organism, A. baumannii shares a highly competitive niche with other bacteria, including Bacillus and Pseudomonas. Those bacteria do have a distinct advantage over A. baumannii, however, with their ability to produce several bacteriocins, molecules that are used to efficiently kill bacteria in close proximity and usually target strains of the same species (30, 31). Although no bacteriocins from A. baumannii have been identified, we suspect that they might in some way use phage lysins to control their niche with respect to other A. baumannii strains. How this is accomplished is under investigation.
To evaluate and compare the abilities of the lysins to kill A. baumannii, we employed a soft agar plate assay in which we spotted crude E. coli lysates (containing the induced lysins) on top of a soft agar plate containing A. baumannii. Based on the comparison of activities, lysin PlyF307 was distinguished not only by clearing A. baumannii exceedingly well on agar plates but also by the halo-like semiclear zone around the initial spot on the plate (see Fig. S2 in the supplemental material). These halo-like clearing zones have been attributed to an ability of cell wall hydrolases to degrade extracellular polymeric substances (e.g., biofilms) (32); therefore, this finding was of particular interest for a novel therapeutic agent.
PlyF307 had a significant lethal effect on exponentially growing cells in vitro, resulting in a >3-log-unit decrease over 2 h when a large inoculum (108 CFU/ml, with 100 μg/ml) was used, while it was less effective against stationary-phase cells, resulting in an ∼1.5-log-unit decrease (Fig. 3C). However, optimizing the conditions (20 mM sodium phosphate buffer [pH 6] and 106 CFU/ml) resulted in >5-log-unit decreases for all A. baumannii clinical isolates tested (Fig. 4C). Thus, this enzyme is comparable to or more effective than other characterized A. baumannii lysins (e.g., LysAB2) or artilysins (19, 22). The mechanism behind this lytic effect is for further studies to determine; however, one study suggested that the presence of a potential positively charged outer membrane-destabilizing domain found within the C-terminal region of these lysins could be playing a role (19). Importantly, PlyF307 is highly positively charged in its C-terminal region, which suggests that this part may be involved in interacting with the outer membrane (Fig. 3B).
One of the reasons A. baumannii is a widespread nosocomial pathogen involves its ability to form biofilms (33). The ability of PlyF307 to kill A. baumannii and its ability to form halo-like clearing zones suggested that PlyF307 could be used to clear biofilms. In an in vitro setting using A. baumannii biofilms on catheters, the addition of PlyF307 resulted in decreases in colonizing bacteria of approximately 1.6 to 1.7 log units (Fig. 5A). Similar decreases (∼2 log units) could be seen in an in vivo model in which 2-day-old biofilms on catheter sections were placed beneath the skin of mice (Fig. 5A) and treated in situ. Along with the reduction in bacterial viability, much of the extracellular polymeric matrix was degraded after this treatment (Fig. 5B), suggesting that direct lysin treatment of infected implants in human patients, without surgical removal, may be a possible strategy.
To study the lysin's ability to function under physiological conditions to control infection, we developed a mouse bacteremia model based on intraperitoneal injection of 108 CFU of A. baumannii. The mice developed systemic infection within 2 h (see Fig. S3 in the supplemental material), at which time treatment was initiated by injecting 1 mg PlyF307 (or buffer) intraperitoneally. Due to the rapid onset and progression of the disease, almost all control animals died within 24 h, whereas treatment with PlyF307 was able to rescue 50% of the infected mice (Fig. 5C). The success rate in rescuing mice from systemic infection is somewhat lower than rates for lysins active against Gram-positive pathogens, which usually rescue 80 to 95% of infected mice under similar conditions (15, 18). This effect is commensurate with the in vitro activity of PlyF307, which decreased the bacterial burden by ∼2 log units in the presence of physiological concentrations of NaCl, while the in vitro effects of Gram-positive lysins are 4 to 5 log units under similar conditions (15, 18). Nevertheless, this is the first study successfully using an intact native lysin (without additional factors) for Gram-negative infections in a mammalian infection model (22).
In conclusion, we have shown here that bacteriophage lysins from Acinetobacter prophages can be used to efficiently reduce the bacterial burden of Gram-negative multidrug-resistant A. baumannii, both in vitro and in vivo. Our studies emphasize the potential therapeutic role of phage lysins for the treatment of Gram-positive and now Gram-negative bacterial infections. We are currently exploring the mode of action of PlyF307 in order to engineer more effective Gram-negative lysins. In one such study, we have managed to use the information we have acquired to date to create a significantly more effective agent against Acinetobacter (R. Lood, M. Thandar, B. Y. Winer, D. R. Deutsch, C. W. Euler, and V. A. Fischetti, unpublished data).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants from ContraFect to V.A.F., from the Tegger Foundation to R.L., and from the Rockefeller University Center for Clinical and Translational Science to R.S.
The Electron Microscopy Resource Center at the Rockefeller University is acknowledged for expert technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04641-14.
REFERENCES
- 1.Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, Vaneechoutte M. 1997. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol 35:2819–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visca P, Seifert H, Towner KJ. 2011. Acinetobacter infection: an emerging threat to human health. IUBMB Life 63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 3.Karageorgopoulos DE, Falagas ME. 2008. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis 8:751–762. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 4.Murray CK, Roop SA, Hospenthal DR, Dooley DP, Wenner K, Hammock J, Taufen N, Gourdine E. 2006. Bacteriology of war wounds at the time of injury. Mil Med 171:826–829. [DOI] [PubMed] [Google Scholar]
- 5.García-Quintanilla M, Pulido MR, López-Rojas R, Pachón J, McConnell MJ. 2013. Emerging therapies for multidrug resistant Acinetobacter baumannii. Trends Microbiol 21:157–163. doi: 10.1016/j.tim.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 7.Livermore DM. 2009. Has the era of untreatable infections arrived? J Antimicrob Chemother 64(Suppl 1):i29–i36. doi: 10.1093/jac/dkp255. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Yong D, Jeong SH, Chong Y. 2011. Multidrug-resistant Acinetobacter spp.: increasingly problematic nosocomial pathogens. Yonsei Med J 52:879–891. doi: 10.3349/ymj.2011.52.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerqueira GM, Peleg AY. 2011. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63:1055–1060. doi: 10.1002/iub.533. [DOI] [PubMed] [Google Scholar]
- 10.Wendt C, Dietze B, Dietz E, Rüden H. 1997. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol 35:1394–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel L, Bonnin RA, Nordmann P. 2011. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 63:1061–1067. doi: 10.1002/iub.532. [DOI] [PubMed] [Google Scholar]
- 12.Gayoso CM, Mateos J, Méndez JA, Fernández-Puente P, Rumbo C, Tomás M, Martínez de Ilarduya O, Bou G. 2014. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Res 13:460–476. doi: 10.1021/pr400603f. [DOI] [PubMed] [Google Scholar]
- 13.Pastagia M, Schuch R, Fischetti VA, Huang DB. 2013. Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol 62:1506–1516. doi: 10.1099/jmm.0.061028-0. [DOI] [PubMed] [Google Scholar]
- 14.Lukacik P, Barnard TJ, Keller PW, Chaturvedi KS, Seddiki N, Fairman JW, Noinaj N, Kirby TL, Henderson JP, Steven AC, Hinnebusch BJ, Buchanan SK. 2012. Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc Natl Acad Sci U S A 109:9857–9862. doi: 10.1073/pnas.1203472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lood R, Raz A, Molina H, Euler CW, Fischetti VA. 2014. A highly active and negatively charged Streptococcus pyogenes lysin with a rare d-alanyl-l-alanine endopeptidase activity protects mice against streptococcal bacteremia. Antimicrob Agents Chemother 58:3073–3084. doi: 10.1128/AAC.00115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuch R, Nelson D, Fischetti VA. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 17.Loeffler JM, Nelson D, Fischetti VA. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 18.Gilmer DB, Schmitz JE, Euler CW, Fischetti VA. 2013. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:2743–2750. doi: 10.1128/AAC.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M-J, Lin N-T, Hu A, Soo P-C, Chen L-K, Chen L-H, Chang K-C. 2011. Antibacterial activity of Acinetobacter baumannii phage φAB2 endolysin (LysAB2) against both Gram-positive and Gram-negative bacteria. Appl Microbiol Biotechnol 90:529–539. doi: 10.1007/s00253-011-3104-y. [DOI] [PubMed] [Google Scholar]
- 20.Díez-Martínez R, de Paz H, Bustamante N, García E, Menéndez M, García P. 2013. Improving the lethal effect of Cpl-7, a pneumococcal phage lysozyme with broad bactericidal activity, by inverting the net charge of its cell wall-binding module. Antimicrob Agents Chemother 57:5355–5365. doi: 10.1128/AAC.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walmagh M, Briers Y, dos Santos SB, Azeredo J, Lavigne R. 2012. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201φ2-1 and PVP-SE1. PLoS One 7:e36991. doi: 10.1371/journal.pone.0036991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay J-P, Miller S, Volckaert G, Lavigne R. 2014. Engineered endolysin-based “artilysins” to combat multidrug-resistant Gram-negative pathogens. mBio 5(4):e01379-14. doi: 10.1128/mBio.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz JE, Daniel A, Collin M, Schuch R, Fischetti VA. 2008. Rapid DNA library construction for functional genomic and metagenomic screening. Appl Environ Microbiol 74:1649–1652. doi: 10.1128/AEM.01864-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hare JM, Ferrell JC, Witkowski TA, Grice AN. 2014. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi. PLoS One 9:e93861. doi: 10.1371/journal.pone.0093861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, NISC Comparative Sequence Program, Murray PR, Segre JA. 2011. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc Natl Acad Sci U S A 108:13758–13763. doi: 10.1073/pnas.1104404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oechslin F, Daraspe J, Giddey M, Moreillon P, Resch G. 2013. In vitro characterization of PlySK1249, a novel phage lysin, and assessment of its antibacterial activity in a mouse model of Streptococcus agalactiae bacteremia. Antimicrob Agents Chemother 57:6276–6283. doi: 10.1128/AAC.01701-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obeidat N, Jawdat F, Al-Bakri AG, Shehabi AA. 2014. Major biologic characteristics of Acinetobacter baumannii isolates from hospital environmental and patients' respiratory tract sources. Am J Infect Control 42:401–404. doi: 10.1016/j.ajic.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Fischetti VA. 2011. Exploiting what phage have evolved to control Gram-positive pathogens. Bacteriophage 1:188–194. doi: 10.4161/bact.1.4.17747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payne KM, Hatfull GF. 2012. Mycobacteriophage endolysins: diverse and modular enzymes with multiple catalytic activities. PLoS One 7:e34052. doi: 10.1371/journal.pone.0034052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleem F, Ahmad S, Yaqoob Z, Rasool SA. 2009. Comparative study of two bacteriocins produced by representative indigenous soil bacteria. Pak J Pharm Sci 22:252–258. [PubMed] [Google Scholar]
- 31.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes KA, Sutherland IW, Clark J, Jones MV. 1998. Bacteriophage and associated polysaccharide depolymerases: novel tools for study of bacterial biofilms. J Appl Microbiol 85:583–590. doi: 10.1046/j.1365-2672.1998.853541.x. [DOI] [PubMed] [Google Scholar]
- 33.Longo F, Vuotto C, Donelli G. 2014. Biofilm formation in Acinetobacter baumannii. New Microbiol 37:119–127. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.