Abstract
Ureaplasma spp. cause several disorders, such as nongonococcal urethritis, miscarriage, and preterm delivery with lung infections in neonates, characterized by pathological chorioamnionitis in the placenta. Although reports on antibiotic resistance in Ureaplasma are on the rise, reports on quinolone-resistant Ureaplasma infections in Japan are limited. The purpose of this study was to determine susceptibilities to five quinolones of Ureaplasma urealyticum and Ureaplasma parvum isolated from perinatal samples in Japan and to characterize the quinolone resistance-determining regions in the gyrA, gyrB, parC, and parE genes. Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. Among 158 samples, the ParC S83L mutation was found in 37 samples (23.4%), including 1 sample harboring a ParC S83L–GyrB P462S double mutant. Novel mutations of ureaplasmal ParC (S83W and S84P) were independently found in one of the samples. Homology modeling of the ParC S83W mutant suggested steric hindrance of the quinolone-binding pocket (QBP), and de novo prediction of peptide structures revealed that the ParC S84P may break/kink the formation of the α4 helix in the QBP. Further investigations are required to unravel the extent and mechanism of antibiotic resistance of Ureaplasma spp. in Japan.
INTRODUCTION
Ureaplasma spp. are among the smallest self-replicating organisms, in terms of both genome size and cellular dimensions. They lack cell walls and are thus resistant to penicillin and other β-lactams. These organisms exist in association with eukaryotic cells, mainly colonizing mucosal surfaces of the respiratory and urogenital tracts (1, 2). Ureaplasma spp. are common inhabitants of the lower genital tract and can be isolated from 40% to 80% of women of child-bearing age (1, 3). Ureaplasma spp. have been associated with a range of pathologies, including nongonococcal urethritis (NGU), miscarriage, preterm delivery, neonatal pneumonia, and chronic lung disease in preterm neonates (4, 5). Many reports have suggested that Ureaplasma parvum and/or Ureaplasma urealyticum may be associated with urogenital infections, infertility, and adverse pregnancy outcomes (6). Regarding the management of ureaplasmal infections, only a limited number of reports are available on the surveillance of antimicrobial resistance in clinical Ureaplasma strains, which is crucial for providing therapy empirically (6). The treatment of ureaplasmal infections is limited to tetracyclines, macrolides, and quinolones (5, 7). Resistance to all the three antibiotic classes in clinical Ureaplasma isolates has been documented, with unique nucleic acid substitutions identified as potential molecular mechanisms for each (5, 8–10).
Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV (9, 11–14). Beeton et al. determined the role of amino acid substitutions in GyrA, GyrB, ParC, and ParE proteins of Ureaplasma spp. in mediating quinolone resistance (5).
There is only one report of quinolone-resistant Ureaplasma in the field of urology in Japan (6). Quinolone resistance in Ureaplasma spp. may occur to some degree because of the widespread use of these drugs for the treatment of respiratory and urogenital infections. Data on antimicrobial resistance in Ureaplasma in perinatal patients are very limited, and there is no report in the field of perinatal medicine in Japan. We aimed to characterize quinolone susceptibility and identify the quinolone resistance-determining region (QRDR) in isolates obtained in the perinatal field.
MATERIALS AND METHODS
Patients.
Isolates were obtained from patients admitted to four perinatal medical institutions located in two areas in Japan, Kyushu and Kinki, from January 2007 to December 2013. Vaginal and placental swabs and tracheal aspirates were obtained by obstetricians or neonatologists from the patients suspected of having Ureaplasma infections. Informed consent was obtained from the patients or their parents, and the study protocol was approved by the Ethics Committee at the Osaka Medical Center for Maternal and Child Health.
Microbiological laboratory tests for Ureaplasma spp.
Clinical specimens were suspended in UMCHs medium (1) or urea arginine LYO2 medium (bioMérieux). After incubation at 37°C for 48 h, the color of the medium changed from yellow to red because of urea hydrolysis, indicating Ureaplasma positivity. We identified Ureaplasma spp. by colony formation and subsequent PCR-based assays using a modification of the method described by Kong et al. (15). For MIC determination in 28 clinical strains, single-colony cultivation was conducted. In total, 158 genomic DNA samples were extracted by the phenol-chloroform extraction method from 28 clinical strains (including one type strain) and 130 frozen ureaplasma-positive LYO2 culture medium samples. U. parvum and U. urealyticum were identified by the analysis of DNA sequences of the ureB gene and/or 16S rRNA gene (16). The serotypes of U. parvum were identified by PCR targeting the multiple-banded antigen (mba) gene (15).
Modified broth microdilution technique for MIC determination.
The following agents were employed for MIC determination: sitafloxacin (STFX), levofloxacin (LVFX) (Daiichi Sankyo Co., Ltd., Tokyo, Japan), garenoxacin (GRNX), tosufloxacin (TFLX) (Toyama Chemical Co., Ltd., Tokyo, Japan), and ciprofloxacin (CPFX) (Wako Pure Chemical Industries., Ltd., Osaka, Japan). The quinolones STFX, LVFX, GRNX, and TFLX were dissolved appropriately in 0.1 M NaOH, 0.1 M HCl, double-distilled water (DDW), and 0.1 M NaOH, respectively, according to the manufacturer's recommendation. The pH of each UMCHs medium sample containing diluted agents was adjusted to 6.0. CPFX was dissolved in 0.1 M HCl, and the pH of UMCHs culture medium containing diluted agents was immediately adjusted to 6.0 to avoid decarboxylation of the agent. The potency of each quinolone was then assessed via serial dilution tests of susceptibility using McFarland standard number 1 of Escherichia coli K-12 JM109 (TaKaRa Bio) in Mueller-Hinton broth (data not shown).
The purity of each of these agents was above 98.0%. Quinolone susceptibility was determined by a modified breakpoint analysis in a 96-well broth microdilution format that enables a concurrent determination of bacterial load in a sample without prior knowledge of bacterial load. Antibiotic gradients were created for each agent. Ureaplasma organisms (20 μl) from the overnight culture with an unknown number of color-changing units (CCU) were added to each well in the columns (1:10 dilution). Plates were sealed and incubated at 37°C in a humidified cell culture incubator at an ambient CO2 concentration for 48 h. The MIC was defined as the lowest concentration of antibiotic that prevented a color change after 48 h when read at 104 CCU (relative to the growth in the antibiotic-free medium) (9). Samples of UMCHs medium with and without antibiotics were also incubated with no Ureaplasma added to serve as negative color change controls. Although no official breakpoint values are available for Ureaplasma, reference values were based on the normal ranges of MICs reported for Ureaplasma (17).
Quinolone-resistant mutations in the gyrA, gyrB, parC, and parE genes.
Known mutations of ureaplasmal genes associated with quinolone resistance were investigated (5). Using DNA from a single culture colony or extracted DNA, regions of gyrA (nucleotide positions 200 to 535), gyrB (nucleotide positions 1261 to 1570), parC (nucleotide positions 149 to 457), and parE (nucleotide positions 1210 to 1522) of U. parvum were amplified by PCR using previously described primers (9). Primers gyrA-1 and gyrA-2 were used for amplifying the gyrA gene, gyrB-3 and gyrB-4 for the gyrB gene, parC-5 and parC-6 for the parC gene, and parE-7 and parE-8 for the parE gene. The regions of gyrA (nucleotide positions 200 to 536), gyrB (nucleotide positions 1261 to 1570), parC (nucleotide positions 149 to 457), and parE (nucleotide positions 1159 to 1448) of U. urealyticum were amplified by PCR using the following primers: Uu-GyrA200F (5′-TTGCTGCTTTCGAAAATGG-3′) and Uu-GyrA536R (5′-ACCTGATGGCAAAACACTTGG-3′) for gyrA; Uu-GyrB 3F (5′-CCAGGTAAATTAGCTGATTG-3′) and Uu-GyrB 4R (5′-TTCGAATATGGCTACCATC-3′) for gyrB; Uu-ParC149F (5′-ATGCCATGAGCGAATTAGG) and parC-6 for parC; and Uu-ParE2F (5′-CGTGCTCGTGAAGAAACTAA-3′) and Uu-ParE2R 5′-AAATTCAGCACCAATTCCTGT-3′) for parE. PCR products were sequenced using the BigDye Terminator v3.1 cycle sequencing kit and analyzed on an ABI Prism 3130 genetic analyzer (Applied Biosystems), according to the manufacturer's instructions. The sequences of the gyrA, gyrB, parC, and parE genes of U. parvum and U. urealyticum were compared with those of the respective reference strains, U. parvum ATCC 700970 (GenBank AF222894) and U. urealyticum ATCC 33699 (GenBank CP001184.1) (6).
Homology modeling of ureaplasmal ParC S83L, S83W, and de novo prediction of peptide structure at around S84P.
The molecular structure of ParC from Ureaplasma spp. remains unknown. We attempted to model Ureaplasma parvum ParC by homologous modeling of the quinolone-DNA cleavage complex of Streptococcus pneumoniae (PDB code 3RAE) using the Swiss-Model homology-modeling server (http://swissmodel.expasy.org/) (18). Data for the S83L and S83W mutants were calculated on the basis of our U. parvum ParC model structure using the software MolFeat (FiatLux), and the energy minimization was performed with the software DeepView/Swiss-PdbViewer (v4.1) (http://spdbv.vital-it.ch/) (19). Each de novo prediction of the peptide structure models was constructed via the PEP-FOLD server with the quality assessment by APOLLO (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD/) (20) using the wild-type ParC α3-α4 helices (SARTVGEVIGKYHPHGDSSIYEAMVRMSQDW) and S84P ParC (SARTVGEVIGKYHPHGDSPIYEAMVRMSQDW).
RESULTS
Samples from a total of 820 patients were sent to our laboratory between 2007 and 2013. Of these, 240 were positive for Ureaplasma spp. by culture color change and PCR, and from these, 28 strains could be analyzed for MIC and gene mutations. Patient characteristics are shown in Table 1. Twenty-four isolates were obtained from maternal vaginal swabs, two from placental swabs, and four from tracheal aspirates of premature infants who had respiratory complications with a diagnosis of severe neonatal chronic lung disease (new bronchopulmonary dysplasia). For all 28 strains, 19 were determined as U. parvum (68%) and nine as U. urealyticum (32%). Amniotic fluid samples of the patients with vaginal swabs positive for strains UP12, UU4, and UU6 were also screened for Ureaplasma spp. Among these three amniotic fluid samples, only one, that from the woman who was swab positive for strain UP12, was culture positive for Ureaplasma. Comparative in vitro activities of quinolones (CPFX, LVFX, TFLX, GRNX, and STFX) against Ureaplasma spp. are shown in Table 2. All isolates had high-level resistance to the older quinolones, such as CPFX, with MICs ranging from 4 to 128 μg/ml. Susceptibility to the newer quinolones, such as LVFX and TFLX, depended strongly on the individual isolate, with MICs ranging from 1 to 16 μg/ml and 2 to >16 μg/ml, respectively. The newer quinolones GRNX and STFX exhibited MICs ranging from 0.5 to 4 μg/ml in the wild-type bacterium, whereas their MICs in the S83L strains ranged from 1 to 4 μg/ml, illustrating that the mutation slightly affected MICs. The increase of MICs against GRNX and STFX for strains UP3, UP4, UP10, UP12, UP18, and UU8 were less pronounced than that for strains UP9, UP15, and UU6.
TABLE 1.
Sources of the 28 clinical Ureaplasma isolates from perinatal patients in Japana
| Isolate | Patient sex and ageb | Species (serovar) | Specimen source | Past pregnancies | Pregnancy outcomec | Gestational age (wks) | Presence of Lactobacillus spp. |
|---|---|---|---|---|---|---|---|
| UP1 | F, 31 | U. parvum (SV3) | Placental | G1P1 | 26 | ND | |
| UP2 | F, 30 | U. parvum (SV3) | Vaginal | G1P0 | M | 13 | + |
| UP3 | F, 30 | U. parvum (SV6) | Placental | G1P1 | 39 | ND | |
| UP4 | F, 33 | U. parvum (SV3) | Vaginal | G4P1 | M, P | Not pregnant | − |
| UP5 | F, 36 | U. parvum (SV6) | Vaginal | G5P2 | M | Not pregnant | ND |
| UP6 | M, 12 days | U. parvum (SV6) | Tracheal | ND | P | ND | ND |
| UP7 | M, 91 days | U. parvum (SV6) | Tracheal | G2P1 | P | ND | ND |
| UP8 | F, 26 | U. parvum (SV3) | Vaginal | G3P1 | M | 32 | − |
| UP9 | F, 25 | U. parvum (SV6) | Vaginal | G1P1 | 20 | + | |
| UP10 | F, 31 | U. parvum (SV6) | Vaginal | G7P1 | M, P | 33 | − |
| UP11 | F, 35 | U. parvum (SV6) | Vaginal | G3P0 | M | 18 | + |
| UP12 | F, 44 | U. parvum (SV3) | Vaginal | G1P0 | M | 8 | + |
| UP13 | M, 3 days | U. parvum (SV3) | Tracheal | G5P4 | P | ND | ND |
| UP14 | F, 42 | U. parvum (SV6) | Vaginal | G3P0 | M | ND | − |
| UP15 | F, 37 | U. parvum (SV6) | Vaginal | G2P1 | M | Not pregnant | + |
| UP16 | F, 37 | U. parvum (SV6) | Vaginal | G3P1 | M | Not pregnant | − |
| UP17 | F, 33 | U. parvum (SV6) | Vaginal | G2P1 | M, P | Not pregnant | − |
| UP18 | F, 34 | U. parvum (ND) | Vaginal | G4P1 | M | Not pregnant | + |
| UP19 | F, 30 | U. parvum (SV6) | Vaginal | G2P0 | M | 18 | − |
| UU1 | F, 43 | U. urealyticum | Vaginal | G5P0 | M | ND | − |
| UU2 | F, 1day | U. urealyticum | Tracheal | G1P1 | P | ND | ND |
| UU3 | F, 36 | U. urealyticum | Vaginal | G3P0 | M | 33 | + |
| UU4 | F, 26 | U. urealyticum | Vaginal | G2P2 | P | 9 | + |
| UU5 | F, 27 | U. urealyticum | Vaginal | G4P2 | P | 10 | + |
| UU6 | F, 20 | U. urealyticum | Vaginal | G0P0 | 25 | − | |
| UU7 | F, 28 | U. urealyticum | Vaginal | G1P1 | 11 | + | |
| UU8 | F, 22 | U. urealyticum | Vaginal | G5P2 | P | 34 | + |
| UU9 | F, 29 | U. urealyticum | Vaginal | G1P0 | P | 6 | − |
Age is given in years unless otherwise specified.
ND, not determined.
M, miscarriage; P, preterm delivery.
TABLE 2.
Characteristics of 28 Ureaplasma clinical isolates from perinatal patients in Japan
| Strain | MIC (μg/ml) |
Amino acid change at the indicated position ina: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GyrA |
GyrB, 462 | ParC |
ParE, 457 | ||||||||||
| CPFX | LVFX | TFLX | GRNX | STFX | 100 | 104 | 82 | 83 | 87 | 88 | |||
| ATCC 700970 | 4 | 2 | 2 | 0.5 | 1 | ||||||||
| UP1 | 8 | 2 | 4 | 1 | 1 | ||||||||
| UP2 | 8 | 2 | 4 | 1 | 2 | ||||||||
| UP5 | 8 | 2 | 4 | 0.5 | 0.5 | ||||||||
| UP6 | 16 | 4 | 16 | 4 | 1 | ||||||||
| UP7 | 4 | 2 | 4 | 0.5 | 0.5 | ||||||||
| UP8 | 8 | 2 | 4 | 1 | 1 | ||||||||
| UP11 | 16 | 2 | 4 | 0.5 | 0.5 | ||||||||
| UP13 | 4 | 1 | 2 | 0.5 | 0.5 | ||||||||
| UP14 | 32 | 8 | 16 | 2 | 2 | ||||||||
| UP16 | 8 | 1 | 2 | 0.5 | 0.5 | ||||||||
| UP17 | 8 | 2 | 4 | 1 | 1 | ||||||||
| UP19 | 8 | 2 | 4 | 1 | 0.5 | ||||||||
| UU1 | 8 | 1 | 8 | 0.5 | 1 | ||||||||
| UU2 | 16 | 2 | 8 | 0.5 | 1 | ||||||||
| UU3 | 16 | 4 | 8 | 0.5 | 1 | ||||||||
| UU4 | 16 | 4 | 8 | 0.5 | 1 | ||||||||
| UU5 | 16 | 4 | 8 | 0.5 | 1 | ||||||||
| UU7 | 16 | 4 | 16 | 0.5 | 2 | ||||||||
| UU9 | 16 | 4 | 8 | 1 | 1 | ||||||||
| S83L strains | |||||||||||||
| UP3 | 128 | 16 | >128 | 2 | 1 | Leu | |||||||
| UP4 | 64 | 16 | 32 | 2 | 1 | Leu | |||||||
| UP9 | 64 | 16 | 64 | 4 | 2 | Leu | |||||||
| UP10 | 32 | 8 | 16 | 2 | 1 | Leu | |||||||
| UP12 | 64 | 8 | 32 | 2 | 1 | Leu | |||||||
| UP15 | 64 | 16 | 64 | 4 | 2 | Leu | |||||||
| UP18 | 32 | 8 | 32 | 1 | 1 | Leu | |||||||
| UU6 | 64 | 8 | >128 | 2 | 4 | Leu | |||||||
| UU8 | 64 | 8 | >128 | 1 | 2 | Leu | |||||||
Amino acid position where an amino acid change is reported in quinolone-resistant mutants of U. parvum and/or U. urealyticum.
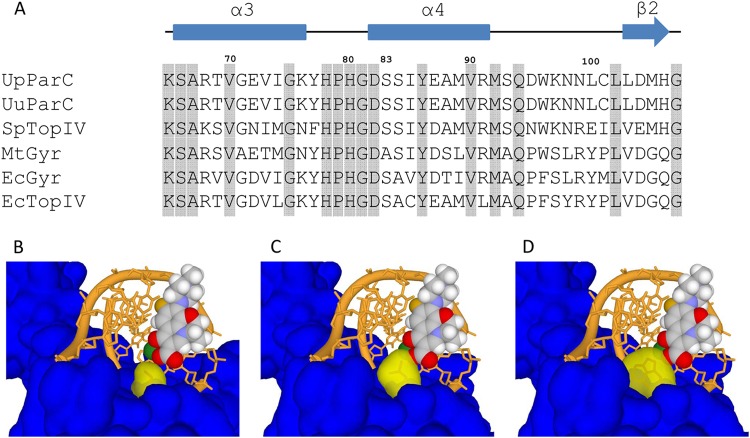
For Ureaplasma spp., the positions of mutations associated with quinolone resistance include GyrA 100 and 104 (84 and 88 for Escherichia coli), GyrB 462 (445 for E. coli) (13), ParC 82, 83, 87, and 88 (79, 80, 84, and 85 for E. coli), and ParE 457 (447 for E. coli) (9, 12, 13, 21). Nine strains (UP3, UP4, UP9, UP10, UP12, UP15, UP18, UU6, and UU8) out of the 28 (32.1%) showed a highly increased MICs of quinolones, including seven strains of U. parvum and two strains of U. urealyticum. The lowest MICs (≦4 μg/ml) in our clinical Ureaplasma strains were observed for GRFX and STFX, in contrast to those of the three other quinolones, which were characterized by increased MICs compared with those for the reference strain ATCC 700970. Among the strains showing high-level MICs of quinolones, we found identical mutations in the parC gene, corresponding to a Ser83-to-Leu (S83L) amino acid substitution (Table 2). No mutations associated with the quinolone-resistant alterations of the hot spots in gyrA, gyrB, and parE genes were detected in our strains. The ParC S83L substitution resulted in an increase in the MIC of up to 32-fold against tested quinolones.
We found the following amino acid substitutions in the QRDRs of 130 Ureaplasma culture-positive samples based on the DNA sequence: ParC S83L, serine (TCA) to leucine (TTA) (28 samples); ParC S83W, serine (TCA) to tryptophan (TGA) (TGA codes for tryptophan instead of being a termination codon in the Mycoplasmataceae [22]) (1 sample); ParC S84P, serine (TCA) to proline (CCA) (1 sample); GyrA D112H, aspartic acid (GAC) to histidine (CAC) (1 sample); and GyrB P462S, proline (CCA) to serine (TCA) (1 sample) (Table 3). The ParC mutation S83L was also detected in a DNA sample harboring GyrB P462S in the QRDR. Amino acid sequence alignment of QRDRs from type II topoisomerases is shown in Fig. 1A. Serines 83 and 84 are located at the second and third positions of the α4 helix, respectively. To evaluate the structural importance of the substituted amino acids of ParC S83L and S83W, homology modeling of U. parvum ParC was performed. The C-3 carboxylic acid and C-4 carbonyl of the quinolone together with a divalent magnesium cation are important for the binding interaction with the DNA-topoisomerase complex (Fig. 1B). From the model, the side chains of S83L (Fig. 1C) and S83W (Fig. 1D) occupied the quinolone binding pocket of the levofloxacin C-3 carboxylic acid. These S83L and S83W mutations in the side chains would interfere with the proper binding of quinolones via steric hindrance.
TABLE 3.
Amino acid substitutions and DNA mutations of QRDRs of Ureaplasma spp. among 130 DNA samples
| Protein | Position | Amino acid | DNA sequence |
U. urealyticum (n = 24) |
U. parvum (n = 106) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Amino acid substitution | DNA mutation | No. of mutant strains | Amino acid substitution | DNA mutation | No. of mutant strains | ||||
| GyrA | 100 | Gln | 0 | 0 | |||||
| 104 | Gln | 0 | 0 | ||||||
| 112 | Glu/Asp | GAA/GAC | 0 | His | CAC | 1 | |||
| GyrB | 462 | Pro | CCA | 0 | Ser | TCA | 1 | ||
| ParC | 82 | Asp | 0 | 0 | |||||
| 83 | Ser | TCA | Leu | TTA | 3 | Leu | TTA | 25 | |
| Trp | TGAa | 1 | |||||||
| 84 | Ser | TCA | 0 | Pro | CCA | 1 | |||
| 87 | Glu | 0 | 0 | ||||||
| 88 | Ala | 0 | 0 | ||||||
| ParE | 457 | Ala | 0 | 0 | |||||
The TGA codon encodes tryptophan in the Mycoplasmataceae (22).
FIG 1.
The QRDR model structures of U. parvum wild-type ParC and S83L and S83W mutants. (A) Sequence alignment of the QRDR domain from type II topoisomerases. The sequences are labeled as follows: UpParC, U. parvum ATCC 700970 ParC (GenBank no. AAF30879); UuParC, U. urealyticum ATCC 33699 ParC (GenBank no. ACI60320); SpTopIV, Streptococcus pneumoniae topoisomerase IV (PDB code 3RAE); MtGyr, Mycobacterium tuberculosis DNA gyrase (PDB code 3IFZ); EcGyr, Escherichia coli DNA gyrase (PDB code 1AB4); EcTopIV, E. coli topoisomerase IV (PDB code 1ZAU). Numbers above the sequences denote the amino acid positions according to U. parvum. The α3 and α4 helices (blue rectangles), the β2 strand (blue arrow), and identical amino acids (gray shading) are indicated. (B to D) Surface models of U. parvum wild-type ParC (by SWISS-MODEL server) (B) and the S83L (C) and S83W (D) mutants. Amino acid position 83 is in yellow; others are in blue. LVFX (ball), DNA (orange), and Mg2+ (green) are superimposed from PDB code 3RAE.
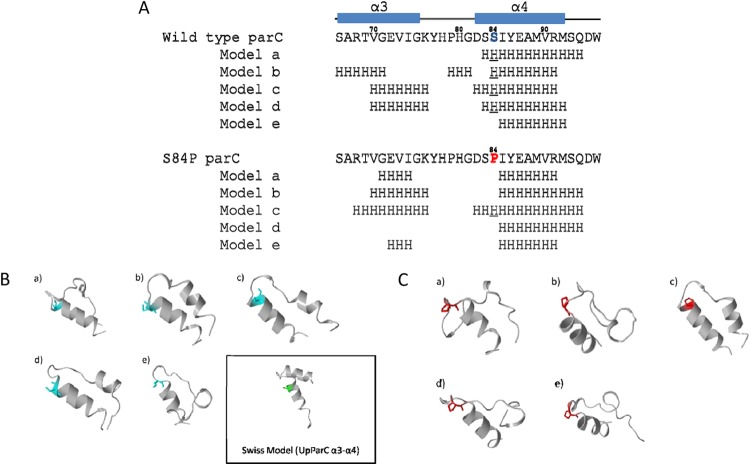
ParC serine 84 is located at the third position of the α4 helix based on the sequence alignment (Fig. 1A). Secondary structures of the predicted top 5 models are shown in Fig. 2A. Four of five models (80%) of wild-type QRDR formed α-helices (Fig. 2B), whereas the S84P mutant was predicted to form an α-helix in only one of five models (20%) (Fig. 2C). Our model suggested that the S84P mutant was less likely to form an α4 helix structure in QRDR than wild-type ParC.
FIG 2.
De novo prediction of U. parvum ParC peptide structures (amino acid positions 66 to 96) by the PEP-FOLD server. (A) The secondary structures of the top 5 predicted models are shown. Numbers above the sequences denote the amino acid positions according to U. parvum. Serine 84 is in blue, and proline 84 is in red. H refers to the α-helical structure, and underlining indicates helical serine 84 and helical proline 84. (B and C) Tertiary structures of the top 5 predicted peptide models of wild-type ParC (B) and ParC S84P (C). The outlined panel shows U. parvum ParC α3-α4 helix structures obtained by homology modeling.
DISCUSSION
Ureaplasma spp. are commonly found in the lower urogenital tracts of healthy women and have been implicated as the cause of acute and chronic urinary tract infections (21, 23–26). We recently reported an epidemiological relationship between Ureaplasma infection and pathological chorioamnionitis (1) and confirmed the lipoprotein multiple-banded antigen of Ureaplasma spp. as a virulence factor for preterm and intrauterine fetal death in pregnant mice (27). In Japan, Kamiya et al. detected a gene mutation associated with quinolone resistance in U. parvum and U. urealyticum by DNA sequencing of isolates from urine specimens from males with NGU (6). The eradication of ureaplasmas from the female genital tract is difficult, sometimes requiring prolonged courses of antimicrobial therapy (21, 28). Our data show that Ureaplasma spp. with a high MIC against quinolones are already widely distributed among females of reproductive age in Japan, which may complicate the eradication of ureaplasmas from the urogenital tract even in nonpregnant reproductive women, underscoring the importance of keeping quinolone treatment of adult NGU and respiratory infections under surveillance.
The S83L and S83W mutations were detected in the gyrA gene of E. coli and Staphylococcus pseudintermedius (S84W) (29, 30). A GyrA A87V/S88P (corresponding to positions 83 and 84, respectively, in Escherichia coli) double mutant was recently reported in multidrug-resistant Ochrobactrum intermedium (31). In Ureaplasma spp., the mutation corresponding to S83L is located in the parC gene. At present, a functional analysis of the ureaplasmal gyrA and/or parC gene product has not been provided. These two genes were assigned by in silico homology search against bacterial databases. Therefore, in accordance with previous reports, we report that the mutations corresponding to S83L, S83W, and also S84P were detected in the (putative) parC gene.
We found 9 strains from placental or vaginal swabs harboring the S83L mutation (32.1%; 9/28 strains) and showing high MICs of quinolones (Table 2). The S83L mutation in ParC was also detected in 28 of 130 ureaplasma-positive samples (21.5%) (Table 3). A few quinolone-resistant clinical Ureaplasma isolates have been described in China (32, 33), France (5, 10, 12), the United States (13, 21), Italy, South Korea (34), and Germany (35). Overall, the S83L mutant was identified in 37 of 158 samples (23.4%), indicating that quinolone-resistant Ureaplasma spp. are already widespread in the perinatal field in Japan. GRNX is one of the newer quinolones with strong activity against ureaplasmas (36). However, unfortunately, GRNX cannot be prescribed for patients with urogenital infections because it is permitted only for patients with respiratory infections in Japan. STFX is also one of the newer quinolones whose in vitro activity is higher than that of the older quinolones (37) or other antimicrobial agents (38). In our study, GRNX and STFX showed relatively high activity against ureaplasmas compared with CPFX, LVFX, and TFLX. Furthermore, GRNX and STFX had better activity against the S83L strains. UP6, UP14, and some other strains exhibited relatively high MICs of quinolones (CPFX, LVFX, and TFLX); however, we could not detect any known hot spot mutations associated with quinolone resistance. Resistance in these isolates could be linked to mutations outside the sequenced regions or alternative mechanisms, such as altered membrane permeability (5, 39).
A plasmid-mediated quinolone resistance gene was reported in Klebsiella pneumoniae (40) and Proteus mirabilis (41). To date, 20 human Ureaplasma sp. genomes have been fully sequenced. However, there is no evidence for the existence of a plasmid in 19 human Ureaplasma genomes (42), nor was any evidence uncovered in our previous study (43). Codon usage by the family Mycoplasmataceae, including ureaplasmas, differs from that of other bacteria, and the GC content of these species is lower than that of other bacteria (43). The plasmid gene transfer system does not appear to be a general evolution mechanism of ureaplasmas.
We found ParC S83W and S84P mutations in the QRDRs of Ureaplasma spp. Unfortunately, living bacteria harboring ParC S83W and S84P could not be recovered from the frozen LYO2 culture medium for MIC tests. To date, an efficient gene manipulation technology for Ureaplasma spp. has not yet been established. For these reasons, we performed in silico homology modeling of S83W and de novo predictions for S84P helix formation. In the analyses, S83W exhibited the potential to cause quinolone resistance via steric hindrance (Fig. 1D).
There is no available determined structure of S84P for homology modeling. Proline can be found at the N termini, but not in the middle, of the α-helices (44). Among all amino acids, proline has the lowest helix propensity (45) and the highest disorder propensity (46). For these reasons, proline is known to break or kink helical structures. This unique characteristic is associated with the backbone of proline, which cannot form hydrogen bonds and which exhibits rigid N-Cα rotation. Consequently, we performed de novo prediction using the S84P peptide structure instead of homology modeling using the wild-type serine 84 geometry (Fig. 2). As expected, the S84P protein exhibited a lower propensity for α4 helix formation, suggesting that this mutation may be an obstacle to the formation of the proper α3-α4 structure of QRDR. However, further research is needed, especially MIC tests for S83W and S84P. We also found a GyrA D112H mutation/polymorphism in one sample; however, we could not detect any evidence that this mutation is associated with quinolone resistance from the structural view (data not shown).
We report the first in vitro quinolone-resistant clinical strains of Ureaplasma spp. associated with an S83L mutation, and we also identified S83W and S84P mutations in the parC gene and a P462S mutation in the gyrB gene in samples obtained in the field of perinatal medicine in Japan. However, this study has some limitations. The total number of patients included in this study was relatively small. In addition, continuous surveillance of Ureaplasma antimicrobial resistance and appropriate treatments are required to prevent perinatal complications through Ureaplasma infections.
ACKNOWLEDGMENTS
This study was supported by research grants from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan (to Y.K. and I.Y.), and by SENTAN, JST, Japan (to I.Y.).
We thank K. Irimura (NHO, Saga) and N. Manno (Fujita Clinic) for technical help. We thank Daiichi Sankyo Co., Ltd., Tokyo, Japan, for providing sitafloxacin and levofloxacin and Toyama Chemical Co., Ltd., Tokyo, Japan, for providing garenoxacin and tosufloxacin.
REFERENCES
- 1.Namba F, Hasegawa T, Nakayama M, Hamanaka T, Yamashita T, Nakahira K, Kimoto A, Nozaki M, Nishihara M, Mimura K, Yamada M, Kitajima H, Suehara N, Yanagihara I. 2010. Placental features of chorioamnionitis colonized with Ureaplasma species in preterm delivery. Pediatr Res 67:166–172. doi: 10.1203/PDR.0b013e3181c6e58e. [DOI] [PubMed] [Google Scholar]
- 2.Waites KB, Schelonka RL, Xiao L, Grigsby PL, Novy MJ. 2009. Congenital and opportunistic infections: Ureaplasma species and Mycoplasma hominis. Semin Fetal Neonatal Med 14:190–199. doi: 10.1016/j.siny.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Taylor-Robinson D, McCormack WM. 1980. The genital mycoplasmas (first of two parts). N Engl J Med 302:1003–1010. doi: 10.1056/NEJM198005013021805. [DOI] [PubMed] [Google Scholar]
- 4.Schelonka RL, Waites KB. 2007. Ureaplasma infection and neonatal lung disease. Semin Perinatol 31:2–9. doi: 10.1053/j.semperi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Beeton ML, Chalker VJ, Kotecha S, Spiller OB. 2009. Comparison of full gyrA, gyrB, parC and parE gene sequences between all Ureaplasma parvum and Ureaplasma urealyticum serovars to separate true fluoroquinolone antibiotic resistance mutations from non-resistance polymorphism. J Antimicrob Chemother 64:529–538. doi: 10.1093/jac/dkp218. [DOI] [PubMed] [Google Scholar]
- 6.Kamiya Y, Shimada Y, Ito S, Kikuchi M, Yasuda M, Kawamura Y, Deguchi T. 2013. Analysis of the quinolone-resistance determining region of the gyrA gene and the analogous region of the parC gene in Ureaplasma parvum and Ureaplasma urealyticum detected in first-void urine of men with non-gonococcal urethritis. J Antimicrob Chemother 68:480–482. doi: 10.1093/jac/dks417. [DOI] [PubMed] [Google Scholar]
- 7.Waites KB, Crouse DT, Cassell GH. 1993. Therapeutic considerations for Ureaplasma urealyticum infections in neonates. Clin Infect Dis 17(Suppl 1):S208–S214. doi: 10.1093/clinids/17.Supplement_1.S208. [DOI] [PubMed] [Google Scholar]
- 8.Roberts MC, Kenny GE. 1986. Dissemination of the tetM tetracycline resistance determinant to Ureaplasma urealyticum. Antimicrob Agents Chemother 29:350–352. doi: 10.1128/AAC.29.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beeton ML, Chalker VJ, Maxwell NC, Kotecha S, Spiller OB. 2009. Concurrent titration and determination of antibiotic resistance in Ureaplasma species with identification of novel point mutations in genes associated with resistance. Antimicrob Agents Chemother 53:2020–2027. doi: 10.1128/AAC.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bebear CM, Renaudin H, Charron A, Gruson D, Lefrancois M, Bebear C. 2000. In vitro activity of trovafloxacin compared to those of five antimicrobials against mycoplasmas including Mycoplasma hominis and Ureaplasma urealyticum fluoroquinolone-resistant isolates that have been genetically characterized. Antimicrob Agents Chemother 44:2557–2560. doi: 10.1128/AAC.44.9.2557-2560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper DC. 1998. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin Infect Dis 27(Suppl 1):S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 12.Bebear CM, Renaudin H, Charron A, Clerc M, Pereyre S, Bebear C. 2003. DNA gyrase and topoisomerase IV mutations in clinical isolates of Ureaplasma spp. and Mycoplasma hominis resistant to fluoroquinolones. Antimicrob Agents Chemother 47:3323–3325. doi: 10.1128/AAC.47.10.3323-3325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao L, Crabb DM, Duffy LB, Paralanov V, Glass JI, Waites KB. 2012. Chromosomal mutations responsible for fluoroquinolone resistance in Ureaplasma species in the United States. Antimicrob Agents Chemother 56:2780–2783. doi: 10.1128/AAC.06342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govender S, Gqunta K, le Roux M, de Villiers B, Chalkley LJ. 2012. Antibiotic susceptibilities and resistance genes of Ureaplasma parvum isolated in South Africa. J Antimicrob Chemother 67:2821–2824. doi: 10.1093/jac/dks314. [DOI] [PubMed] [Google Scholar]
- 15.Kong F, Zhu X, Wang W, Zhou X, Gordon S, Gilbert GL. 1999. Comparative analysis and serovar-specific identification of multiple-banded antigen genes of Ureaplasma urealyticum biovar 1. J Clin Microbiol 37:538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallard K, Schopfer K, Bodmer T. 2005. Development of real-time PCR for the differential detection and quantification of Ureaplasma urealyticum and Ureaplasma parvum. J Microbiol Methods 60:13–19. doi: 10.1016/j.mimet.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Bebear CM KI. 2005. Antimicrobial therapy and antimicrobial resistance, p 535–568. In Blanchard A, Browning GF (ed), Mycoplasmas: molecular biology, pathogenicity and strategies for control. Horizon Bioscience, Norfolk, United Kingdom. [Google Scholar]
- 18.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 20.Thevenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tuffery P. 2012. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res 40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffy L, Glass J, Hall G, Avery R, Rackley R, Peterson S, Waites K. 2006. Fluoroquinolone resistance in Ureaplasma parvum in the United States. J Clin Microbiol 44:1590–1591. doi: 10.1128/JCM.44.4.1590-1591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamao F, Muto A, Kawauchi Y, Iwami M, Iwagami S, Azumi Y, Osawa S. 1985. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A 82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert GL, Garland SM, Fairley KF, McDowall DM. 1986. Bacteriuria due to ureaplasmas and other fastidious organisms during pregnancy: prevalence and significance. Pediatr Infect Dis 5:S239–243. doi: 10.1097/00006454-198611010-00007. [DOI] [PubMed] [Google Scholar]
- 24.McDonald MI, Lam MH, Birch DF, D'Arcy AF, Fairley KF, Pavillard ER. 1982. Ureaplasma urealyticum in patients with acute symptoms of urinary tract infection. J Urol 128:517–519. [DOI] [PubMed] [Google Scholar]
- 25.Potts JM, Ward AM, Rackley RR. 2000. Association of chronic urinary symptoms in women and Ureaplasma urealyticum. Urology 55:486–489. doi: 10.1016/S0090-4295(99)00555-5. [DOI] [PubMed] [Google Scholar]
- 26.Stamm WE, Running K, Hale J, Holmes KK. 1983. Etiologic role of Mycoplasma hominis and Ureaplasma urealyticum in women with the acute urethral syndrome. Sex Transm Dis 10:318–322. [PubMed] [Google Scholar]
- 27.Uchida K, Nakahira K, Mimura K, Shimizu T, De Seta F, Wakimoto T, Kawai Y, Nomiyama M, Kuwano K, Guaschino S, Yanagihara I. 2013. Effects of Ureaplasma parvum lipoprotein multiple-banded antigen on pregnancy outcome in mice. J Reprod Immunol 100:118–127. doi: 10.1016/j.jri.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Bowie WR, Willetts V. 1987. Suboptimal efficacy of erythromycin and tetracycline against vaginal Ureaplasma urealyticum. Sex Transm Dis 14:88–91. doi: 10.1097/00007435-198704000-00006. [DOI] [PubMed] [Google Scholar]
- 29.Cullen ME, Wyke AW, Kuroda R, Fisher LM. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother 33:886–894. doi: 10.1128/AAC.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruger T, Nitiss JL, Maxwell A, Zechiedrich EL, Heisig P, Seeber S, Pommier Y, Strumberg D. 2004. A mutation in Escherichia coli DNA gyrase conferring quinolone resistance results in sensitivity to drugs targeting eukaryotic topoisomerase II. Antimicrob Agents Chemother 48:4495–4504. doi: 10.1128/AAC.48.12.4495-4504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnning A, Moore ER, Svensson-Stadler L, Shouche YS, Larsson DG, Kristiansson E. 2013. Acquired genetic mechanisms of a multiresistant bacterium isolated from a treatment plant receiving wastewater from antibiotic production. Appl Environ Microbiol 79:7256–7263. doi: 10.1128/AEM.02141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Wu Y, Yin W, Yu M. 2002. Study of isolation of fluoroquinolone-resistant Ureaplasma urealyticum and identification of mutant sites. Chin Med J 115:1573–1575. [PubMed] [Google Scholar]
- 33.Xie X, Zhang J. 2006. Trends in the rates of resistance of Ureaplasma urealyticum to antibiotics and identification of the mutation site in the quinolone resistance-determining region in Chinese patients. FEMS Microbiol Lett 259:181–186. doi: 10.1111/j.1574-6968.2006.00239.x. [DOI] [PubMed] [Google Scholar]
- 34.Choi SJ, Park SD, Jang IH, Uh Y, Lee A. 2012. The prevalence of vaginal microorganisms in pregnant women with preterm labor and preterm birth. Annals Lab Med 32:194–200. doi: 10.3343/alm.2012.32.3.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krausse R, Schubert S. 2010. In-vitro activities of tetracyclines, macrolides, fluoroquinolones and clindamycin against Mycoplasma hominis and Ureaplasma ssp. isolated in Germany over 20 years. Clin Microbiol Infect 16:1649–1655. doi: 10.1111/j.1469-0691.2010.03155.x. [DOI] [PubMed] [Google Scholar]
- 36.Waites KB, Crabb DM, Bing X, Duffy LB. 2003. In vitro susceptibilities to and bactericidal activities of garenoxacin (BMS-284756) and other antimicrobial agents against human mycoplasmas and ureaplasmas. Antimicrob Agents Chemother 47:161–165. doi: 10.1128/AAC.47.1.161-165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milatovic D, Schmitz FJ, Brisse S, Verhoef J, Fluit AC. 2000. In vitro activities of sitafloxacin (DU-6859a) and six other fluoroquinolones against 8,796 clinical bacterial isolates. Antimicrob Agents Chemother 44:1102–1107. doi: 10.1128/AAC.44.4.1102-1107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamasuna R, Jensen JS, Osada Y. 2009. Antimicrobial susceptibilities of Mycoplasma genitalium strains examined by broth dilution and quantitative PCR. Agents Chemother 53:4938–4939. doi: 10.1128/AAC.00724-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hooper DC. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist Updates 2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Martinez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 41.Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Hooper DC, Wang M. 2009. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother 53:1892–1897. doi: 10.1128/AAC.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paralanov V, Lu J, Duffy LB, Crabb DM, Shrivastava S, Methe BA, Inman J, Yooseph S, Xiao L, Cassell GH, Waites KB, Glass JI. 2012. Comparative genome analysis of 19 Ureaplasma urealyticum and Ureaplasma parvum strains. BMC Microbiol 12:88. doi: 10.1186/1471-2180-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu HN, Nakura Y, Motooka D, Nakamura S, Nishiumi F, Ishino S, Kawai Y, Tanaka T, Takeuchi M, Nakayama M, Fujita T, Yanagihara I. 2014. Complete genome sequence of Ureaplasma parvum serovar 3 strain SV3F4, isolated in Japan. Genome Announc 2:e00256-14. doi: 10.1128/genomeA.00256-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim MK, Kang YK. 1999. Positional preference of proline in alpha-helices. Protein Sci 8:1492–1499. doi: 10.1110/ps.8.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pace CN, Scholtz JM. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J 75:422–427. doi: 10.1016/S0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theillet FX, Han K-H, Selenko P, Dunker AK, Daughdrill GW, Uversky VN. 2013. The alphabet of intrinsic disorder I. Act like a Pro: on the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disordered Proteins 1:e24360. doi: 10.4161/idp.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]