Abstract
A direct assay of efflux by Escherichia coli AcrAB-TolC and related multidrug pumps would have great value in discovery of new Gram-negative antibiotics. The current understanding of how efflux is affected by the chemical structure and physical properties of molecules is extremely limited, derived from antibacterial data for compounds that inhibit growth of wild-type E. coli. We adapted a previously described fluorescent efflux assay to a 96-well microplate format that measured the ability of test compounds to compete for efflux with Nile Red (an environment-sensitive fluor), independent of antibacterial activity. We show that Nile Red and the lipid-sensitive probe DiBAC4-(3) [bis-(1,3-dibutylbarbituric acid)-trimethine oxonol] can quantify efflux competition in E. coli. We extend the previous findings that the tetracyclines compete with Nile Red and show that DiBAC4-(3) competes with macrolides. The extent of the competition shows a modest correlation with the effect of the acrB deletion on MICs within the compound sets for both dyes. Crystallographic studies identified at least two substrate binding sites in AcrB, the proximal and distal pockets. High-molecular-mass substrates bound the proximal pocket, while low-mass substrates occupied the distal pocket. As DiBAC4-(3) competes with macrolides but not with Nile Red, we propose that DiBAC4-(3) binds the proximal pocket and Nile Red likely binds the distal site. In conclusion, competition with fluorescent probes can be used to study the efflux process for diverse chemical structures and may provide information as to the site of binding and, in some cases, enable rank-ordering a series of related compounds by efflux.
INTRODUCTION
Tripartite multidrug efflux pumps are widely represented across Gram-negative bacteria. The two best studied examples are AcrAB-TolC of Escherichia coli and MexAB-OprM of Pseudomonas aeruginosa (1, 2). The normal physiological function for the E. coli AcrB pump is to protect bacterial cells from bile salts inside the human host (3). However, it appears that bacterial efflux pumps and their components may also serve other diverse functions (4–7). AcrB is a proton-driven antiporter that derives its energy from the proton motive force that exists across the bacterial cytoplasmic membrane (8, 9). AcrB is organized as a homotrimer and interacts with two other components, the outer membrane partner TolC and the small periplasmic adapter lipoprotein AcrA (10, 11). The broad substrate preference of AcrB includes bile salts and detergents as well as diverse structurally unrelated antibiotics and chemicals (12–14), making it a vital player in determining intrinsic antibiotic resistance (15).
AcrAB-TolC is the best-characterized bacterial efflux pump with regard to structure (16), with crystal structures solved for the individual components (17–22). The most recent high-resolution crystals show each monomer in a different configuration, lending support to a peristaltic mechanism for pump function (19). The three monomer forms, called Access (A), Binding (B), and Extrusion, show marked differences in their organization. Substrates have been visualized bound to the protein at two different sites (21, 22), called the proximal and distal pockets. Each monomer in the trimer has 12 transmembrane helices and a periplasmic headpiece, the latter of which has the pore (porter) domain and an upper region that interacts with TolC. The four subdomains in the pore region (PC1, PC2, PN1, and PN2) interact with or bind substrates in the crystals. Additionally, there is a deep periplasmic cleft in the pore region between PC1 and PC2 that ends in a hook-like bend (20). This loop, also referred to as the G-loop or switch or Phe-617 loop (21–23), physically separates the two binding pockets and plays an important role in substrate discrimination. Currently, substrate entry is thought to occur via three potential entrances: via an intermonomer vestibule (24), via the deep periplasmic cleft (25), or directly from the cytoplasmic membrane through a groove formed between transmembrane helices 7 and 8 (20). A functional rotation model that involves the three protomers cycling through the Access, Binding, and Extrusion states with accompanying conformational changes has been proposed (26, 27). The role of the central cavity remains to be elucidated (22, 28).
While structural studies have shed some light on the general mechanisms by which substrates enter the pump and pass through it, there is currently very little understanding of the molecular basis of substrate specificity. Efflux pump deletion mutants (ΔacrAB or ΔtolC) are more sensitive to most antibiotics than their wild-type parents, but the magnitude of the change in sensitivity varies greatly from one antibacterial compound to another. This suggests that although AcrB is able to extrude a wide range of structurally diverse chemicals, some compounds are much better efflux substrates than others. Similar observations have been made for MexAB-OprM, the major multidrug efflux pump of P. aeruginosa. The nature of substrate specificity is not only of great interest to the field of efflux pump biology, it is also of enormous practical importance for antibacterial drug discovery. Discovery of new Gram-negative antibiotics will require learning to design compounds with reduced susceptibility to efflux.
The differential susceptibility of wild-type and efflux-deficient strains to a given compound provides a rough estimate of that compound's sensitivity to efflux. One approach to quantify efflux is to determine the ratio of two MICs (wild type and efflux deficient) for each compound in a set in order to relate “efflux ratio” to chemical structure or to physical properties. This is not very useful because it is limited to compounds that have measurable antibacterial activity against wild-type strains and is dependent on potency. Antibacterial discovery programs have thus far yielded very few chemical scaffolds in which multiple compounds have measurable activity against wild-type strains. Although in vitro biochemical data are often useful for improving the potency of compounds toward the target, the antibacterial activity is often limited by poor outer membrane penetration and high levels of TolC-dependent efflux in E. coli as a consequence of their generally higher hydrophobic character (29). Without a direct assay of efflux, medicinal chemists have no rational basis for designing Gram-negative antibiotics other than the general observation that highly polar compounds are less subject to efflux.
An orthogonal approach to the study of efflux, not dependent on antibacterial assays, builds on previous work using dyes like N-phenyl-1-naphthylamine (NPN) (12) and trimethylammonium diphenylhexatriene (TMA-DPH) (30) to study the role of proton motive force on efflux by AcrAB-TolC and related pumps. These environment-sensitive dyes are highly fluorescent in a hydrophobic environment (such as membranes) and nonfluorescent in aqueous solvents. They are largely excluded from efflux-competent bacteria unless the cells are treated with a proton gradient-uncoupling agent such as carbonyl cyanide m-chlorophenylhydrazone (CCCP). Dye diffuses into CCCP-poisoned cells, making them fluorescent. Addition of glucose reverses the effect of CCCP, presumably by restoring the proton motive force across the cytoplasmic membrane, and restarts the pumps. The dye is then extruded out of the cells, leading to a concomitant drop in cell-associated fluorescence. A previous study used this system to study efflux of Nile Red in E. coli and showed that efflux of the dye was diminished in the presence of certain efflux pump substrates and efflux inhibitors (31). These data suggested that it might be possible to study the substrate specificity of AcrB by quantifying the effect of potential substrates on efflux of Nile Red.
In this work, we transformed the published method into a more quantitative assay in a 96-well format and also carried out a rigorous evaluation of the ability of the method to predict the effect of the acrB deletion on MICs in E. coli. We confirmed the previous observations that (i) members of the tetracycline class differentially compete with Nile Red and (ii) not all antibiotics known to be AcrB substrates are competitors of Nile Red. We identified another environmentally sensitive probe, DiBAC4-(3) [bis-(1,3-dibutylbarbituric acid)-trimethine oxonol], which is an AcrB substrate, and showed that it competes with macrolides, unlike Nile Red. We found that the extent of the competition for both dyes showed a modest correlation with the effect of the acrB deletion on efflux in MIC assays within the respective compound sets. Competition with the tetracyclines supports the conclusion that Nile Red binds to the “distal multisite pocket” on AcrB. Neither DiBAC4-(3) nor the macrolides competed with Nile Red, suggesting that the oxonol dye, DiBAC4-(3), was captured by AcrB at a site that was distinct from Nile Red and the tetracyclines, presumably at the “proximal multisite access pocket.” A third set of AcrB substrates studied were members of the azole antifungals. The azoles were able to inhibit efflux of both Nile Red and DiBAC4-(3), hinting at their ability to either interact with both sites on AcrB or function as efflux pump inhibitors. Our data are consistent with recent crystallographic studies indicating the presence of at least two major compound binding sites within AcrB.
MATERIALS AND METHODS
Strains and growth conditions.
E. coli K-12 strains AG100 and AG100A (ΔacrAB) (32) were kindly provided by H. Nikaido (University of California, Berkeley, CA) and S. Levy (Tufts University, Boston, MA). The tolC mutation was transferred into AG100 by P1 transduction using E. coli MC4100 tolC::Tn10 (Tetr) as a donor. Stationary-phase cultures were used in all experiments. Fifty-milliliter cultures were typically grown to stationary phase overnight in cation-adjusted Mueller-Hinton broth (MH2B) (Becton, Dickinson and Company, Franklin Lakes, NJ) prepared per the manufacturer's instructions.
Chemicals.
Nile Red, tetracyclines, macrolides, and azoles were from Sigma Chemical Company, St. Louis, MO; fluconazole was from Selleckchem, Houston, TX; tigecycline was from Sequoia Research Products, Pangbourne, United Kingdom. DiBAC4-(3) was from Invitrogen, Carlsbad, CA. Carbonyl cyanide m-chlorophenylhydrazone (CCCP), Nile Red, DiBAC4-(3), and the antibiotics were freshly solvated in dimethyl sulfoxide (DMSO) (Sigma Chemical Company, St. Louis, MO) prior to use. In the experiments with cells, the final DMSO concentration never exceeded 1.3%. Oxytetracycline and tetracycline (Sigma Chemical Company, St. Louis, MO) stocks were prepared in distilled water and distilled water-DMSO (1:1), respectively.
Measuring fluorescent probe efflux from cells.
We modified a previously described procedure, in which bacteria are treated sequentially with CCCP, an environment-sensitive fluorescent dye, and a potential competitor and are then reenergized with glucose to initiate efflux (31). The following method is similar in principle but has been modified extensively, simplifying the protocol and making it possible to assay multiple samples simultaneously.
A single colony of the desired E. coli strain was inoculated in 50 ml of prewarmed MH2B in a 250-ml glass Erlenmeyer flask and grown in a shaker (200 rpm, 35°C) to stationary phase (16 to 18 h). Bacteria in a 4-ml aliquot of stationary-phase culture were centrifuged, washed twice in phosphate-buffered saline (PBS), and resuspended in PBS containing 1 mM MgCl2 to a final optical density at 600 nm (OD600) of ∼1.0.
Efflux arrest.
The cells were transferred to a 125-ml glass Erlenmeyer flask, and CCCP (50 mM stock in DMSO) was added to a final concentration of 10 μM for AG100 and 5 μM for the efflux-compromised acrAB and tolC strains (31). The flask was incubated with shaking for 20 min (200 rpm, 35°C).
Competitor loading.
Five-hundred-microliter aliquots of the CCCP-treated cells were transferred to 96-well, glass-coated deep-well block microplates (Thermo Fisher Scientific, Waltham, MA; catalog no. 14-823-442) to which were added appropriate concentrations of the competitor (where applicable). Incubation with shaking was continued for an additional 20 min.
Probe loading.
The fluorescent probe being tested was added to the final desired concentration, and the block was reincubated with shaking for 20 min.
Reenergization.
Cells were spun down directly in the deep-well block at 4,500 rpm (3,800 × g; type S5700 rotor, Allegra 25R centrifuge) for 15 min at 18°C. Supernatants were carefully vacuum aspirated, and the pellets were gently resuspended in 500 μl PBS containing 1 mM MgCl2. Aliquots (144 μl) were transferred to wells of a 96-well clear-bottom plate (Corning Incorporated Life Sciences, Tewksbury, MA; catalog no. 3631), and efflux was initiated by automated injection of glucose (6 μl of a 25 mM stock; final concentration, 1 mM). All experiments were done in a BMG FluoStar Optima system (BMG Labtech, Cary, NC) equipped with a needle injector and a 96-well microplate reader. Measurements were recorded every 10 to 15 s for up to 300 s. In all cases, efflux of the fluorescent probe from cells began within 10 to 20 s of addition of glucose. Excitation (nm)/emission (nm) wavelengths were as follows: Nile Red, 544/650, and DiBAC4-(3), 490/520, respectively. Prior to running the experiments, we confirmed that the highest concentrations of the competitors tested did not affect the fluorescence of Nile Red or DiBAC4-(3) (tolerated changes were restricted to ±10% to 12% in either direction). We used this as a guideline in setting upper limits for competitor concentrations.
Curve fits of efflux data to calculate competitor concentration that inhibits probe efflux by 50% (EC50efflux).
Data from experiments for normalized dye fluorescence (y axis) versus time (x axis) were plotted using GraphPad Prism (GraphPad Software, La Jolla, CA). Both Nile Red and DiBAC4-(3) curves were fitted using a single exponential decay equation of the type
| (1) |
where X is time, Y starts at Y0 and then decays down to plateau in one phase, plateau is Y value at infinite times, span is the difference between Y0 and plateau and has the same units as Y, and K is rate constant.
The K value or slope for each of these curves or the span in the presence or absence of the competitor was plotted against the external concentration of the competitor (Fig. 1A has an explanation of curve parameters). The apparent EC50efflux was calculated after fitting this dose response in GraphPad Prism using the log(inhibitor) versus response-variable slope equation of the general form:
| (2) |
where X is log of concentration; Y is response, decreasing as X increases; top and bottom are plateaus in the same units as Y; logEC50 is the same log units as X; and Hill slope is slope factor.
FIG 1.
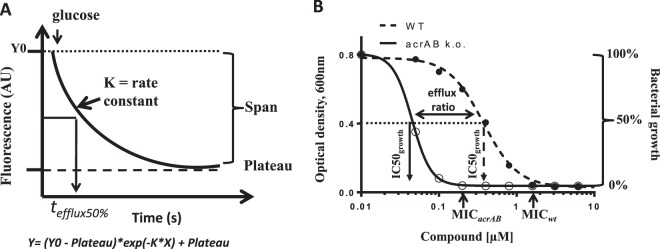
Schematic depicting parameters for efflux time course and bacterial growth inhibition curves. (A) Rate constant (K) and span from the single-phase exponential fits used to calculate EC50efflux are as indicated. Efflux is initiated at t = 0 by addition of glucose and is 50% complete at tefflux50%. (B) IC50growth values from inhibition of growth of the wild-type and acrAB strains are as shown and were used to calculate efflux ratio; MIC endpoints are also shown. k.o., knockout.
Assays of antibacterial activity and calculation of growth inhibition (IC50growth).
Antibacterial assays for the AG100 wild-type and AG100A ΔacrAB strains were run in 96-well microtiter plates (Corning Incorporated Life Sciences, Tewksbury, MA; catalog no. 3788) in Mueller-Hinton cation-adjusted broth, using 2-fold dilution series of the desired compound using CLSI guidelines. At the end of the standard 18-hour incubation at 35°C, the plates were scanned in an En Vision multilabel plate reader (PerkinElmer, Akron, OH) in multiwell mode equipped with a 600-nm filter. The standard readout of this assay is the MIC. We used the optical density data to generate growth inhibition curves and calculated 50% inhibitory concentration of growth (IC50growth) as the readout of antibacterial activity.
Absorbance at 600 nm was plotted using GraphPad Prism, and curves were empirically fitted using equation 2 above. The compound concentration causing a reduction of bacterial growth by 50% was determined from the curve fits for the wild-type and ΔacrAB strains and defined as IC50growth (Fig. 1B contains the details). For each antibiotic, we used IC50growth for the wild-type and ΔacrAB strains to calculate the efflux ratio. The ratio is interpreted to be an indirect measure of the extent of efflux by the AcrB pump. Also, being a ratio, the value is independent of the biochemical potency of the compound for the bacterial target and is unitless.
RESULTS
Modified assay to study efflux of Nile Red and DiBAC4-(3) by AcrAB-TolC.
The semiautomated microplate efflux assay described here differs in several respects from the previously described method for monitoring Nile Red efflux (31). We improved upon the previously reported method by using shorter incubation times for CCCP treatment and probe/competitor loading and an injector-enabled fluorimeter that enables more uniform start of efflux and makes it possible to assay multiple samples simultaneously. In addition, we used multiwell glass-coated blocks instead of single glass tubes to circumvent the problem of dye binding to plastic. To allow comparison of efflux data with standard assays of antibacterial activity, we grew bacteria in MH2B rather than LB. Our method, like the previous one, was developed for the K-12 strain AG100 and its derivatives. A serendipitous finding (data not shown) was that the closely related strain BW25113 was much less sensitive than AG100 to poisoning by CCCP, if grown in MH2B.
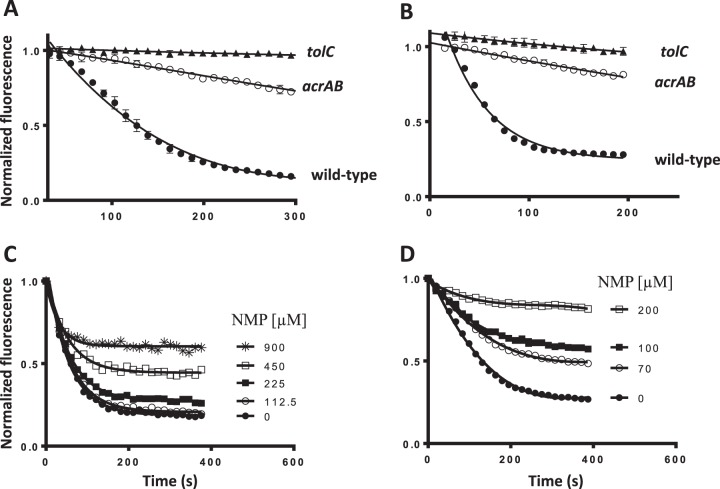
With these experimental modifications, the efflux of Nile Red from wild-type E. coli AG100 reached steady state within 5 min, reflecting the high efficiency of the AcrB pump (Fig. 2A). We used acrAB and tolC deletions, to confirm that most of this efflux depends on AcrB (Fig. 2A). The modest efflux of Nile Red observed from ΔacrAB cells was completely abolished by the presence of the tolC mutation (Fig. 2A), which suggested that at least one other TolC-dependent pump (in addition to AcrB) plays a role, albeit minor, in the export of Nile Red. As previously noted (31), efflux of Nile Red was also sensitive to the efflux pump inhibitor 1-(1-naphthylmethyl)-piperazine (NMP) (Fig. 2C).
FIG 2.
Nile Red and DiBAC4-(3) are primarily effluxed by AcrB in a TolC-dependent manner in E. coli, and efflux is inhibited by the pump blocker, 1-(1-naphthylmethyl)-piperazine (NMP). Real-time efflux curves for Nile Red (A) and DiBAC4-(3) (B) in wild-type, acrAB, and tolC E. coli cells with the corresponding strains indicated on the panels. NMP inhibits efflux of Nile Red (C) and DiBAC4-(3) (D) compared to its absence (●) in a dose-dependent manner in wild-type E. coli cells. Nile Red and DiBAC4-(3) concentrations were 1 μM and 10 μM, respectively. The concentrations of NMP in competition with Nile Red and DiBAC4-(3) are as shown in panels C and D for the two probes.
One of the objectives in our study was to find additional environment-sensitive fluorescent dyes that could be used to quantify AcrB-dependent compound efflux by competition, because Nile Red does not compete with all classes of antibiotics that are AcrB efflux substrates (31). We did a brief survey of other commonly used environment-sensitive probes like N-phenyl-1-naphthylamine (NPN) and trimethylammonium diphenylhexatriene (TMA-DPH) (data not shown) that have been used as reporters of AcrB-dependent efflux (12, 30). One of the probes that we tested was the anionic oxonol DiBAC4-(3), which we found to be exported by wild-type E. coli cells (Fig. 2B) and is commonly used to measure membrane potential changes in biological systems (33). We confirmed that the fluorescence time course seen in wild-type cells was indeed due to efflux pump activity and not due to changes in the membrane potential. First, efflux of DiBAC4-(3) was dependent on the presence of AcrAB and TolC, as strains missing either set of genes no longer showed the rapid time-dependent decay in fluorescence upon reenergization with glucose (Fig. 2B). Second, export of DiBAC4-(3) by wild-type cells was blocked by 1-(1-naphthylmethyl)-piperazine (NMP) (Fig. 2D). Export of DiBAC4-(3) reached steady state by 5 min and appeared to be much more sensitive to inhibition by NMP than Nile Red (Fig. 2D). DiBAC4-(3) is known to partition into the cytoplasm of bacterial cells (34). It is possible that the higher residual fluorescence at the end of efflux is due to dye that is still bound to cytoplasmic components during the time frame of our experiments and, hence, unavailable to the pumps. We concluded that the two fluorescent probes, Nile Red and DiBAC4-(3), can be used as probes to monitor the efflux activity of AcrAB-TolC in E. coli.
Use of Nile Red competition to quantify efflux of substrates by AcrB.
To confirm the relevance of the fluorescence-based readout, we compared efflux rates for a group of related compounds for which we could also measure growth-based efflux ratios from MIC assays. It has been reported previously that minocycline and other tetracyclines inhibit efflux of Nile Red (31). The same authors quantified inhibition of the rate of Nile Red efflux in the presence or absence of the competitor using a parameter called tefflux50%. This was the time required for cells to extrude half of the preloaded probe molecule (Fig. 1A). Compounds that increased tefflux50% were interpreted as being competitors of Nile Red efflux. A limitation in that study was that data at only two concentrations were available, for most compounds. For example, minocycline was indeed very potent, but fluorescence of Nile Red was fully inhibited and did not reach 50% at either concentration tested, so an exact value could not be determined (31). Their analysis was, therefore, unable to rank-order the competition by the extent of their efflux within the set.
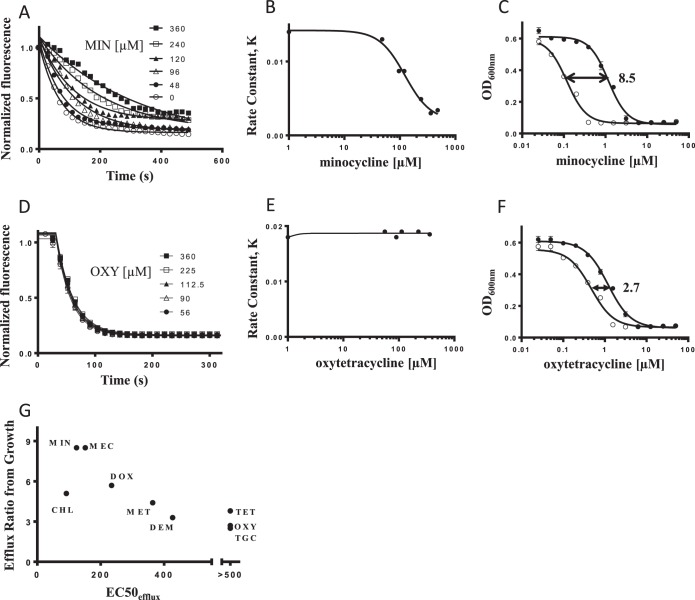
We greatly improved upon this finding by testing a larger set of tetracyclines, performed full dose responses to quantify the extent of competition, and showed that the extent of Nile Red competition paralleled the effect of the acrAB deletion on MICs (see below). The Nile Red efflux curves in the presence or absence of the competitor were fitted using a single-phase exponential decay function, from which we extracted an apparent rate constant (K). Graphing this parameter, K, for Nile Red efflux at each concentration of competitor compound versus the total competitor concentration generated a new term that we call EC50efflux. This parameter corresponded to the competitor concentration that inhibits the efflux of Nile Red by 50% (see Materials and Methods). We found that one of the best competitors was indeed minocycline, which slowed the efflux of Nile Red in a dose-dependent manner (Fig. 3A). Using this type of analysis, we calculated the minocycline EC50efflux value to be 124 μM (Fig. 3B). In contrast to minocycline, oxytetracycline was unable to slow the efflux of Nile Red at the highest concentration tested (Fig. 3D and E). We used this method to determine EC50efflux values for 10 tetracycline-class antibiotics (Table 1).
FIG 3.
EC50efflux values correlate with the impact of AcrB-mediated efflux on bacterial growth assays. (A and D) Effect of increasing concentrations of minocycline (A) and oxytetracycline (D) on the efflux of Nile Red. The concentrations of competing compounds are shown in each panel. (B and E) Each Nile Red efflux competition curve was fitted to obtain an apparent rate constant, and these rate constants were plotted versus minocycline (B) or oxytetracycline (E) concentrations. EC50efflux values for the tetracyclines were then calculated from these plots (see Materials and Methods). (C and F) Growth inhibition plots for E. coli wild-type (●) and ΔacrAB (○) strains grown in the presence of minocycline (C) and oxytetracycline (F). The concentration of the drug producing 50% growth inhibition for each strain was used to calculate growth efflux ratios (Table 1). The mean efflux ratios for minocycline and oxytetracycline appear on each panel for reference. (G) Correlation between growth efflux ratios and apparent EC50efflux values for tetracycline-class antibiotics. Tetracycline, tigecycline, and oxytetracycline did not compete for efflux with Nile Red up to the highest concentration tested (500 μM). MIN, minocycline; MEC, meclocycline; CHL, chlortetracycline; DOX, doxycycline; MET, methacycline; DEM, demeclocycline; TET, tetracycline; OXY, oxytetracycline; TGC, tigecycline.
TABLE 1.
Nile Red EC50efflux and growth efflux ratios for tetracyclinesa
| Compound | Mean ± SD (n) |
|
|---|---|---|
| EC50efflux (μM) | Efflux ratio | |
| Minocycline | 124 ± 7 (4) | 8.5 ± 1.6 (6) |
| Meclocycline | 152 (2) | 8.5 ± 0.7 (6) |
| Doxycycline | 235 (2) | 5.7 (2) |
| Chlortetracycline | 92 (2) | 5.1 ± 1.6 (6) |
| Methacycline | 364 ± 28 (3) | 4.4 ± 0.4 (6) |
| Demeclocycline | 427 (2) | 3.3 ± 0.3 (6) |
| Tetracycline | >500 (2) | 3.8 ± 1.3 (5) |
| Oxytetracycline | >500 (2) | 2.7 ± 0.5 (6) |
| Tigecycline | >500 (2) | 2.5 (5) |
See Materials and Methods for explanations of EC50efflux and efflux ratios.
Tetracycline-class EC50efflux values correlated with AcrB-mediated efflux determined from bacterial growth.
The 10 members of the tetracycline class of antibiotics for which we measured Nile Red competition were chosen because they all have measurable activity versus wild-type E. coli. Deletion of acrAB affected susceptibility to various extents, producing a range of efflux ratio values (Table 1). For example, the IC50growth of minocycline for E. coli was reduced 8- to 11-fold by deletion of acrAB (Fig. 3C), while that of oxytetracycline was reduced only 2- to 3-fold (Fig. 3F). This is consistent with the observation that minocycline is a much stronger competitor of Nile Red efflux than oxytetracycline (Fig. 3B and E). For the 10 tetracyclines, there was indeed a modest relationship between the efflux ratio and EC50efflux parameters, confirming that EC50efflux from Nile Red competition was a measure of the ability of the different tetracyclines to serve as AcrB substrates (Fig. 3G). Thus, at least for this class of antibiotics, EC50efflux seems to accurately reflect the role of AcrB-mediated efflux on antibacterial activity resulting from an overnight incubation.
DiBAC4-(3) competed with macrolides for efflux by AcrB and might occupy the proximal AcrB site.
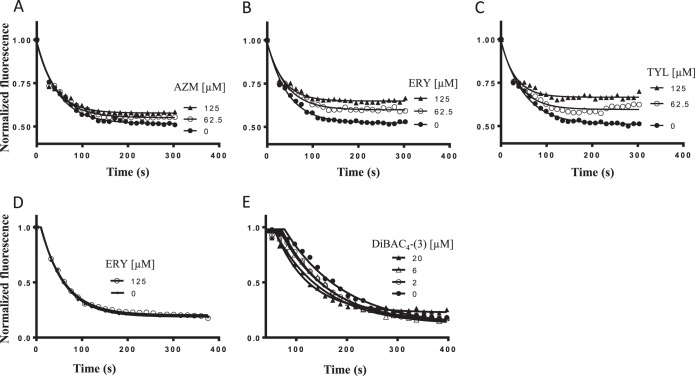
Several macrolide antibiotics, known to be AcrB substrates in E. coli (13), had efflux ratios of 4- to 16-fold or more in E. coli (Table 2) and did not compete with Nile Red (Fig. 4D) (31). However, we found that the macrolides competed to differing extents with the AcrB-mediated efflux of the probe DiBAC4-(3). Azithromycin, with the lowest growth efflux ratio among the macrolide set, competed the least or not at all with efflux of DiBAC4-(3), compared with the other more-effluxed macrolides (Fig. 4A to C; Table 2). Free DiBAC4-(3) fluorescence in DMSO (data not shown) was not affected by macrolide concentrations as high as 500 μM. However, the initial cell-associated DiBAC4-(3) fluorescence values (prior to energization) were substantially higher at macrolide concentrations of 250 μM or greater depending on the specific macrolide. We do not understand the underlying molecular basis for the increased cell-associated DiBAC4-(3) fluorescence at these higher macrolide concentrations. Macrolides like azithromycin and erythromycin have been suggested as having the ability to interact with the lipopolysaccharide (LPS) layer in E. coli, as their MICs were affected by the presence of Mg2+ ions in the medium (35); we speculate that the increase in DiBAC4-(3) fluorescence at these higher macrolide concentrations is because of this interaction with the bacterial membrane compartment. Regardless, this observation prevented us from running more-detailed dose responses to quantify the extent of the competition with the different macrolides (Table 2). X-ray crystallographic studies of AcrB showed that the macrolide erythromycin was bound to the access monomer at a second site called the proximal pocket (22), distinct from the distal pocket where tetracycline and doxorubicin are reported to bind (17, 22, 36). In the context of the competition with the macrolides, we propose that DiBAC4-(3) most likely binds at this second site in AcrB and does not overlap Nile Red or the tetracyclines. Two lines of evidence support our proposal: (i) the macrolide antibiotics did not compete with Nile Red (Fig. 4D) and (ii) DiBAC4-(3) did not compete with the AcrB-mediated efflux of Nile Red (Fig. 4E).
TABLE 2.
Growth efflux ratios for a set of macrolide antibioticsa
| Compound | Efflux ratio, mean ± SD (n) |
|---|---|
| Azithromycin | 4.0 ± 0.8 (3) |
| Clarithromycin | 10.8 ± 1.2 (3) |
| Spiramycin | 11.8 ± 1.7 (3) |
| Erythromycin | 13.8 ± 1.9 (3) |
| Tylosin | 16.5 (2) |
| Josamycin | >19 (2) |
See Materials and Methods for an explanation of efflux ratios. EC50efflux values were not determined for any compound.
FIG 4.
The macrolide class of antibiotics competes with DiBAC4-(3). Effect of increasing concentrations of azithromycin (AZM) (A), erythromycin (ERY) (B), and tylosin (TYL) (C) on the efflux of DiBAC4-(3). Macrolides do not compete with the efflux of Nile Red (erythromycin competition is shown at the concentration indicated in panel D). DiBAC4-(3) does not inhibit efflux of Nile Red (E). The concentrations of the competing compounds are shown in each panel. The concentrations of Nile Red and DiBAC4-(3) in the macrolide competition experiments were 1 μM and 10 μM, respectively. The DiBAC4-(3) and Nile Red efflux competition curves are fitted using the single exponential decay model (see Materials and Methods).
Azole antifungal compounds competed with Nile Red and DiBAC4-(3) for AcrB-dependent efflux.
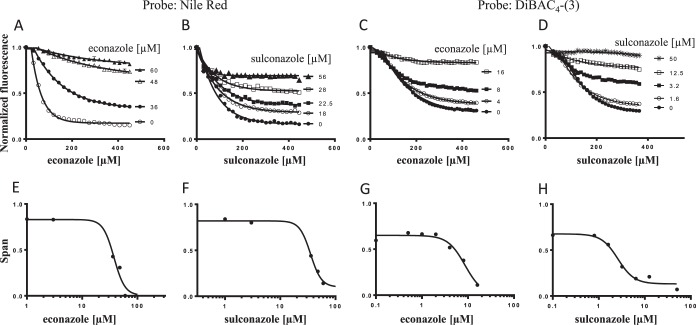
The azoles are commercially marketed antifungal drugs (37, 38) that have been reported to have antibacterial activity versus Mycobacterium and Streptomyces spp. via inhibition of essential P450 enzymes (39). Although E. coli lacks a P450 enzyme (40), we found that several azoles are active versus the acrAB deletion mutant of E. coli but not wild-type E. coli (Table 3). We used these compounds to further evaluate fluorescent dyes as reporters of AcrB-dependent efflux. We found that the azoles inhibited the AcrB-dependent efflux of both Nile Red (Fig. 5A and B) and DiBAC4-(3) (Fig. 5C and D) in a dose-dependent manner. Unlike the tetracyclines, we observed that the efflux of both DiBAC4-(3) and Nile Red in the presence of the azoles tended to plateau at different values that depended on the external competitor concentration. The difference between the plateau values in the presence or absence of the competitor and the starting fluorescence value, also called the span (Fig. 1A), was a robust, reproducible, and distinctive feature of azole competition with either dye. For this reason, span values from the single-phase exponential fits of the efflux curves were extracted and plotted against the corresponding external competitor concentration to generate EC50efflux values (Fig. 5E to H). It was curious that the azole competition efflux curves resembled those observed with the efflux pump inhibitor NMP (Fig. 2C and D). Specifically, miconazole, econazole, clotrimazole, and sulconazole competed best, with both Nile Red and DiBAC4-(3) giving the best EC50efflux values among the set (Table 3); ketoconazole and fluconazole competed either less well or not at all with either probe. Because of the lack of antibacterial activity versus the wild type and the acrAB deletion mutant and the lack of competition at the highest concentration tested, no conclusion can be made for fluconazole. Also, the azoles had consistently lower EC50efflux values with DiBAC4-(3) than with Nile Red (Table 3).
TABLE 3.
EC50efflux values for the azole-class compoundsa
| Compound | Mean ± SD (n) |
||||
|---|---|---|---|---|---|
| EC50efflux, μM |
IC50growth, μM |
Efflux ratio | |||
| Nile Red | DiBAC4-(3) | Wild type | acrAB | ||
| Miconazole | 20.0 (2) | 2.5 ± 2.1 (3) | >100 | 29.7 (2) | >3 (2) |
| Econazole | 34.9 ± 5.9 (3) | 7.2 ± 1.6 (3) | >100 | 14.3 (2) | >7 (2) |
| Clotrimazole | 42.7 ± 11.7 (3) | 20.6 ± 16.1 (3) | >100 | 12.0 (2) | >8 (2) |
| Sulconazole | 29.0 ± 6.0 (3) | 3.4 (2) | >100 | 7.8 (2) | >12 (2) |
| Ketoconazole | >90 (2) | >90 (2) | >100 | 28 | >3 |
| Fluconazole | >100 (2) | >100 (2) | >100 | >100 (2) | Not active |
See Materials and Methods for explanation of EC50efflux and efflux ratios.
FIG 5.
The azole class of antimicrobials competes with Nile Red and DiBAC4-(3). Effect of increasing concentrations of econazole (A and C) and sulconazole (B and D) on the efflux of Nile Red and DiBAC4-(3) as indicated. The concentrations of competing compounds are shown in each panel. The Nile Red and DiBAC4-(3) efflux competition curves are fitted to obtain the span value (where the individual curves plateau), and these are plotted versus econazole (E and G) or sulconazole (F and H) concentrations for the two dyes, respectively. EC50efflux values for the azoles are then calculated from these plots (see Materials and Methods).
DISCUSSION
One impetus for this study was to evaluate the possibility of using Nile Red competition as a routine assay of efflux. This would be most useful if (i) all or nearly all AcrB substrates were found to be competitors of Nile Red and (ii) Nile Red competition could be used as a quantitative measure of AcrB-dependent efflux. Such an assay would allow a set of compounds to be rank-ordered by efflux even if they had no antibacterial activity. Analyzing efflux data for a large set of diverse chemical compounds could provide the basis for proposing guidelines for efflux avoidance. Ideally, in a discovery program, the assay would be used during hit evaluation, offering the possibility of choosing a low-efflux series as a starting point even if it had little antibacterial activity. During structure-activity relationship (SAR) development, the assay would provide a rational basis for designing compounds that have lower efflux. It should be noted that design of compounds with improved general antibacterial activity would also require optimization of entry across the bacterial outer membrane, an aspect that is not inherent in the assay presented here.
The tetracyclines, by virtue of having antibacterial activity versus both the wild-type and efflux null strains, made a good initial set of related compounds to evaluate if there was a relationship between EC50efflux and MIC-based efflux ratios. The relationship between EC50efflux and AcrB-dependent MIC efflux ratios for the tetracyclines and the macrolides (to a limited extent) enabled us to validate the fluorescent approach. It provided confidence that the fluorescent assay was a good surrogate measure of AcrB-mediated efflux, notwithstanding the differences in both the methodology and measurement time scales, i.e., several minutes versus an overnight incubation. Our data provide encouragement that differences in EC50efflux values could be employed to sort compounds into high- or low-efflux categories, especially as antibacterial activity or even potency toward a target is not a prerequisite to observe competition in the assay.
We then tested the fluorescent competition approach to see if we could quantify efflux for a set of antimicrobials (the azoles), which, in spite of being antifungal compounds, actually had some measurable antibacterial activity versus the acrAB deletion mutant but not the wild type—a theme similar to antibacterial screening hits. For the azoles that we tested, we found that miconazole, econazole, clotrimazole, and sulconazole had better EC50efflux values than did ketoconazole for both probes. A simple interpretation of this result was that the former are better AcrB substrates than the latter. An alternative use of the dye competition assay is the study of potential efflux pump inhibitors. We noted that the azoles produced fluorescence curves with shapes similar to those of NMP (Fig. 2C and D) and phenylalanine-arginine β-naphthylamide (PAβN) (31). These are known inhibitors of efflux pumps that may physically jam the functional rotation of the monomers (41).
The proton-driven AcrB antiporter has been proposed to extrude bile salts, detergents, and host defense molecules (3, 42, 43). The pump accepts a wide range of unrelated substrates, from organic solvents (44) to many antibiotics (45, 46), and is responsible for the intrinsic resistance of E. coli (15) to antibiotics. Observations comparing efflux among different members of the β-lactam class of antibiotics led to the conclusion that more hydrophobic compounds or ones with amphipathic character tend to be better substrates for the pump (47). Consistent with this view is the observation that the aminoglycosides, which as a class are more hydrophilic in nature, are not recognized by AcrB (28, 48). A clearer picture of the underlying substrate discrimination mechanism would help in understanding pump biology and aid in identifying structural features that are associated with strong AcrB substrates.
Minocycline and doxorubicin have been shown to bind to an aromatic, phenylalanine-rich region designated the distal pocket, formed between PN2 and PC1 in the Binding protomer (17). A later publication from the same research group described a second binding site, called the proximal or access pocket. This site, in the cleft region of the Access (A) monomer, accommodates high-molecular-mass drugs, specifically rifampin and the macrolide erythromycin (22). The findings for the distal pocket were confirmed by a second group who reported, in addition, that two stacked doxorubicin molecules were seen in the access pocket of the A monomer (21). The G-loop or Phe-617 loop forms a partition between the proximal and distal pocket, sitting above a narrow substrate channel connecting these two sites of interaction (22). The flexibility of this loop has been shown to be important for the efflux of higher-molecular-weight substrates like the macrolides and rifampin, as mutations around Phe-617 increased the sensitivity of cells toward these antibiotics (23). More hydrophilic substrates are thought to enter from the periplasm via the deep cleft, while amphipathic or hydrophobic ones from the membrane enter via the vestibule to gain access to the region containing the proximal high-volume site. As the Access monomer switches to the Binding state, substrates presumably move toward the deep, aromatic, distal multisite pocket and then are thought to be pushed out of the binding pocket toward the TolC tunnel as the Binding monomer switches to the Extrusion form (28).
Our data from the competition studies with DiBAC4-(3) and Nile Red were generally consistent with the presence of at least two functional and distinct sites in AcrB supporting Nile Red and the tetracycline interaction with the distal pocket and DiBAC4-(3) (and the macrolides) at the proximal pocket in AcrB. Several pieces of evidence potentially supported interaction of DiBAC4-(3) with the proximal pocket: (i) DiBAC4-(3) did not compete with Nile Red, (ii) Nile Red did not compete with any of the macrolides that we tested, and, (iii) DiBAC4-(3) did not compete with the tetracyclines (data not shown). In addition, we found that the efflux of DiBAC4-(3) was more sensitive than Nile Red to inhibition by NMP. This is consistent with the report that NMP straddles the G-loop and occludes the substrate extrusion channel closer to the proximal pocket (41). Recently, NMP-resistant mutations in AcrB, located away from the distal pocket near the outer face, have been described (49). Finally, the higher molecular mass of DiBAC4-(3) (∼520 Da) than of Nile Red (∼320 Da) fitted in the range of molecular weights for antibiotics that were likely to be proximal pocket substrates (22, 23).
The lack of competition observed for some antibiotic classes versus others (31) limits the utility of using single fluorescent probes as universal reporters of AcrB-mediated efflux, as a single probe may not be able to capture all aspects of the process. Several antibiotics that are subject to AcrB-dependent efflux were tested at concentrations comparable to those of the antibiotics used in this study and did not compete with either probe; these included penicillin G, chloramphenicol, linezolid, rifampin, and trimethoprim (data not shown). For a compound lacking antibacterial activity, lack of competition with a given fluorescent probe would be difficult to interpret as the compound could be captured by residues in either of the two high-volume binding sites not privy to the probe. In practice, it is rare to find compounds with antibacterial activities that are not substrates for AcrB or one of the other TolC-dependent efflux pumps. It would therefore not be reasonable to use these or any probes to screen a library with the hope of finding compounds not subject to efflux. A more practical approach would be to use Nile Red and DiBAC4-(3) to survey compounds of interest (for example, a set of screening hits) in order to identify those that are strong competitors of either probe and might provide guidance for optimization away from this liability. Finally, as this is a cell-based approach, we cannot exclude the possibility that some degree of competition could also be occurring at the site of the membrane rather than at a discrete AcrB binding pocket. Site-directed mutagenesis of specific residues in AcrB that have been described as being involved in compound transit (24, 25, 28) would be essential to confirming the molecular basis for the observed competition.
In summary, we further optimized a previously published assay that measures competition with Nile Red for AcrB-dependent efflux. The semiautomated, microplate format of the current assay allowed us to extend the previous study considerably. Key findings are (i) the identification of a second fluorescent probe that we believe binds at a different site within AcrB and (ii) validation of the assay with a small compound set for potentially quantifying efflux among members of a chemical class of compounds. We recognize that only a limited set of compounds were used to validate this approach as presented in this study, and as such, this limits the general applicability of this assay. It would be desirable to conduct further validation studies, using additional sets of related compounds for which several members are active versus wild-type E. coli and for which appropriate probes can be identified. This work has implications for the general study of multidrug efflux pumps and for antibacterial drug discovery.
ACKNOWLEDGMENTS
All authors were employed at Vertex Pharmaceuticals Incorporated when these studies were performed. R.I. is still an employee of the company. At the time of the writing of this paper, both R.I. and R.R. are shareholders of Vertex Pharmaceuticals Incorporated.
We thank Ute Müh, Len Duncan, Nagraj Mani, and Joshua Leeman for reading the manuscript and providing suggestions during discussions related to completion of this effort and Sam Pazhanisamy for advice with fitting efflux rate data.
REFERENCES
- 1.Poole K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotechnol 3:255–264. [PubMed] [Google Scholar]
- 2.Nikaido H, Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deininger KN, Horikawa A, Kitko RD, Tatsumi R, Rosner JL, Wachi M, Slonczewski JL. 2011. A requirement of TolC and MDR efflux pumps for acid adaptation and GadAB induction in Escherichia coli. PLoS One 6:e18960. doi: 10.1371/journal.pone.0018960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horiyama T, Nishino K. 2014. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One 9:e108642. doi: 10.1371/journal.pone.0108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamarche MG, Deziel E. 2011. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310. doi: 10.1371/journal.pone.0024310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJ. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 8.Zgurskaya HI, Nikaido H. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A 96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zgurskaya HI, Nikaido H. 1999. AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol 285:409–420. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]
- 10.Nikaido H, Zgurskaya HI. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J Mol Microbiol Biotechnol 3:215–218. [PubMed] [Google Scholar]
- 11.Lomovskaya O, Zgurskaya HI, Nikaido H. 2002. It takes three to tango. Nat Biotechnol 20:1210–1212. doi: 10.1038/nbt1202-1210. [DOI] [PubMed] [Google Scholar]
- 12.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chollet R, Chevalier J, Bryskier A, Pages JM. 2004. The AcrAB-TolC pump is involved in macrolide resistance but not in telithromycin efflux in Enterobacter aerogenes and Escherichia coli. Antimicrob Agents Chemother 48:3621–3624. doi: 10.1128/AAC.48.9.3621-3624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikaido H, Pages JM. 2012. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikaido H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol 12:215–223. doi: 10.1006/scdb.2000.0247. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido H. 2011. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol 77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 18.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, Pos KM. 2008. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol 15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 20.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 21.Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grutter MG, Diederichs K, Pos KM. 2012. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A 109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. 2011. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 23.Cha HJ, Muller RT, Pos KM. 2014. Switch-loop flexibility affects transport of large drugs by the promiscuous AcrB multidrug efflux transporter. Antimicrob Agents Chemother 58:4767–4772. doi: 10.1128/AAC.02733-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husain F, Bikhchandani M, Nikaido H. 2011. Vestibules are part of the substrate path in the multidrug efflux transporter AcrB of Escherichia coli. J Bacteriol 193:5847–5849. doi: 10.1128/JB.05759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husain F, Nikaido H. 2010. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol 78:320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 27.Seeger MA, Diederichs K, Eicher T, Brandstatter L, Schiefner A, Verrey F, Pos KM. 2008. The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance. Curr Drug Targets 9:729–749. doi: 10.2174/138945008785747789. [DOI] [PubMed] [Google Scholar]
- 28.Ruggerone P, Murakami S, Pos KM, Vargiu AV. 2013. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr Top Med Chem 13:3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- 29.Brown DG, May-Dracka TL, Gagnon MM, Tommasi R. 2014. Trends and exceptions of physical properties on antibacterial activity for Gram positive and Gram negative pathogens. J Med Chem 57:10144–10161. doi: 10.1021/jm501552x. [DOI] [PubMed] [Google Scholar]
- 30.Ocaktan A, Yoneyama H, Nakae T. 1997. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa. J Biol Chem 272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 31.Bohnert JA, Karamian B, Nikaido H. 2010. Optimized Nile Red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob Agents Chemother 54:3770–3775. doi: 10.1128/AAC.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hachler H, Cohen SP, Levy SB. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol 173:5532–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waggoner A. 1976. Optical probes of membrane potential. J Membr Biol 27:317–334. doi: 10.1007/BF01869143. [DOI] [PubMed] [Google Scholar]
- 34.Plasek J, Sigler K. 1996. Slow fluorescent indicators of membrane potential: a survey of different approaches to probe response analysis. J Photochem Photobiol B 33:101–124. doi: 10.1016/1011-1344(96)07283-1. [DOI] [PubMed] [Google Scholar]
- 35.Farmer S, Li ZS, Hancock RE. 1992. Influence of outer membrane mutations on susceptibility of Escherichia coli to the dibasic macrolide azithromycin. J Antimicrob Chemother 29:27–33. doi: 10.1093/jac/29.1.27. [DOI] [PubMed] [Google Scholar]
- 36.Pos KM. 2009. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta 1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 37.Kathiravan MK, Salake AB, Chothe AS, Dudhe PB, Watode RP, Mukta MS, Gadhwe S. 2012. The biology and chemistry of antifungal agents: a review. Bioorg Med Chem 20:5678–5698. doi: 10.1016/j.bmc.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 38.Rani N, Sharma A, Gupta GK, Singh R. 2013. Imidazoles as potential antifungal agents: a review. Mini Rev Med Chem 13:1626–1655. [DOI] [PubMed] [Google Scholar]
- 39.McLean KJ, Marshall KR, Richmond A, Hunter IS, Fowler K, Kieser T, Gurcha SS, Besra GS, Munro AW. 2002. Azole antifungals are potent inhibitors of cytochrome P450 mono-oxygenases and bacterial growth in mycobacteria and streptomycetes. Microbiology 148:2937–2949. [DOI] [PubMed] [Google Scholar]
- 40.Guengerich FP, Cheng Q. 2011. Orphans in the human cytochrome P450 superfamily: approaches to discovering functions and relevance in pharmacology. Pharmacol Rev 63:684–699. doi: 10.1124/pr.110.003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. 2014. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother 58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elkins CA, Mullis LB. 2006. Mammalian steroid hormones are substrates for the major RND- and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J Bacteriol 188:1191–1195. doi: 10.1128/JB.188.3.1191-1195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 44.White DG, Goldman JD, Demple B, Levy SB. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol 179:6122–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol 16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 46.Okusu H, Ma D, Nikaido H. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol 178:306–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikaido H, Basina M, Nguyen V, Rosenberg EY. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J Bacteriol 180:4686–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elkins CA, Nikaido H. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J Bacteriol 184:6490–6498. doi: 10.1128/JB.184.23.6490-6499.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuster S, Kohler S, Buck A, Dambacher C, Konig A, Bohnert JA, Kern WV. 2014. Random mutagenesis of the multidrug transporter AcrB from Escherichia coli for identification of putative target residues of efflux pump inhibitors. Antimicrob Agents Chemother 58:6870–6878. doi: 10.1128/AAC.03775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]