Abstract
The development and maintenance of an arsenal of antibiotics is a major health care challenge. Ceftaroline is a new cephalosporin with activity against methicillin-resistant Staphylococcus aureus (MRSA); however, no reports concerning MRSA ceftaroline susceptibility have been reported in Switzerland. We tested the in vitro activity of ceftaroline against an archived set of 60 MRSA strains from the University Hospital of Geneva collected from 1994 to 2003. Our results surprisingly revealed ceftaroline-resistant strains (MIC, >1 μg/ml in 40/60 strains; EUCAST breakpoints, susceptible [S], ≤1 μg/ml; resistant [R], >1 μg/ml) were present from 1998 to 2003. The detected resistant strains predominantly belonged to sequence type 228 (ST228) (South German clonotype) but also to ST247 (Iberian clonotype). A sequence analysis of these strains revealed missense mutations in the penicillin-binding protein 2A (PBP2A) allosteric domain (N146K or E239K and N146K-E150K-G246E). The majority of our ST228 PBP2A mutations (N146K or E150K) were distinct from ST228 PBP2A allosteric domain mutations (primarily E239K) recently described for MRSA strains collected in Thailand and Spain during the 2010 Assessing Worldwide Antimicrobial Resistance Evaluation (AWARE) global surveillance program. We also found that similar allosteric domain PBP2A mutations (N146K) correlated with ceftaroline resistance in an independent external ST228 MRSA set obtained from the nearby University Hospital of Lausanne, Lausanne, Switzerland, collected from 2003 to 2008. Thus, ceftaroline resistance was observed in our archived strains (including two examples of an MIC of 4 µg/ml for the Iberian ST247 clonotype with the triple mutation N146K/E150K/G246E), at least as far back as 1998, considerably predating the commercial introduction of ceftaroline. Our results reinforce the notion that unknown parameters can potentially exert selective pressure on PBP2A that can subsequently modulate ceftaroline resistance.
INTRODUCTION
Staphylococcus aureus is a major pathogen worldwide and can provoke a range of diseases that range from relatively minor to life-threatening. It is the most common etiological agent of surgical site infections and ventilator-associated pneumonia. Of particular concern are infections arising from encounters with antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA). Methicillin resistance in S. aureus is dependent upon the acquisition of the mecA gene, which encodes penicillin-binding protein 2A (PBP2A), which is refractory to inhibition by methicillin and numerous other β-lactam antibiotics (1).
MRSA is widely recognized as a serious threat to public health, and new antibiotics are urgently needed (2). Penicillin-binding proteins (PBPs), which are bacterial enzymes catalyzing the last steps of cell wall biosynthesis, have long been used as lethal targets for β-lactam antibiotics. Ceftaroline, a novel β-lactam broad-spectrum cephalosporin, also binds to PBPs (3); however, an exceptional characteristic of ceftaroline is that it also binds to and inhibits PBP2A, effectively blocking the principal β-lactam resistance determinant of MRSA strains. Accordingly, numerous studies have attested that ceftaroline shows robust in vitro activity against MRSA strains (4–8).
Neither clinical nor in vitro studies have detected high percentages or rapid development of ceftaroline resistance (9). Nevertheless, some rare resistant strains have been reported (10, 11) (EUCAST MICs, >1 μg/ml or CLSI MIC, ≥4 μg/ml). To our knowledge, only a few studies have been conducted that identified PBP2A mutations as being correlated with a reduction in sensitivity to ceftaroline (11, 12). Some of these PBP2A mutations (N146K and E150K) correlated with decreased affinity to ceftaroline (13) and were found to be located in the recently discovered allosteric site that is considerably distant from the transpeptidase active site domain (13, 14). Interestingly, as shown by Otero and coworkers (14) using X-ray crystallographic analysis, ceftaroline is capable of binding both the PBP2A allosteric and the dd-transpeptidase domains. In the proposed model, noncovalent ceftaroline binding to the allosteric site might influence accessibility to the PBP2A active site by a second ceftaroline molecule over a considerable spatial distance by an intricate network of salt bridges and conformational changes. Consequently, mutations in the PBP2A allosteric site might conceivably potentiate β-lactam accessibility to and acylation of the PBP2A active site serine (Ser403), effectively modulating resistance to the antibiotic (14).
Experimental support for this model was recently obtained by biochemical and X-ray crystallographic analysis using mutant PBP2A variants harboring site-specific engineered single (N136K or E150K) or double (N136K-E150K) mutations within the allosteric domain (15). The results revealed that the mutations indeed disrupted the salt bridge network that underlies the allosteric response and gates the accessibility to the active site. Furthermore, the mutations clearly alter the electrostatic potential within the allosteric site and may further contribute to the resistance mechanism by altering the initial binding of the first ceftaroline molecule. The authors emphasized that this is an unprecedented mechanism of antibiotic resistance (15).
In our study reported here, we tested the ceftaroline susceptibility profiles of a collection of MRSA strains from the University Hospital of Geneva in Switzerland collected over the period of 1994 to 2003 (16). Our results revealed that a reduced susceptibility to ceftaroline (MIC, ≥2 μg/ml) strongly correlated with a known epidemic clonal type and altered PBP2A sequence. Importantly, strains archived as far back as 1998 and long before the commercial introduction of ceftaroline displayed altered PBP2A sequences within the allosteric domain, suggesting that some genetic selection, perhaps other β-lactams, can drive the appearance of PBP2A mutations.
(Portions of this work were presented orally and in poster format at the European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], 10 to 13 May 2014, Barcelona, Spain, ePoster 180 [17].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are primarily a collection of 60 clinical strains that includes hospital-acquired MRSA (HA-MRSA) strains derived from bacteremia episodes collected from 1994 to 2003, together with several well-studied laboratory-derived and clinical strains frequently used in our basic research program (18, 19). Notably, this particular HA-MRSA strain set was derived from a much larger HA-MRSA collection and was shown to harbor a large fraction of borderline glycopeptide-intermediate S. aureus (GISA) strains that were experimentally revealed by relatively minor modifications of standard MIC testing parameters (16). Quality control for antibiotic susceptibility testing was performed using S. aureus strain ATCC 29213. Standardized inocula for each bacterial isolate were prepared from overnight cultures at 37°C in cation-adjusted Mueller-Hinton broth (CAMHB) (ref. no. 212322; BBL), which were subsequently diluted 1:50 in fresh CAMHB and grown for 3 h at 37°C without shaking. Following adjustment to a 0.5 McFarland standard, each log-phase culture was diluted to deliver the final inoculum recommended, namely, 5 × 105 CFU/ml and 5 × 106 CFU/ml for the broth microdilution and disk diffusion methods, respectively.
Susceptibility testing.
The MICs were determined by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) and EUCAST guidelines. Ceftaroline was supplied in the dephosphorylated active form of ceftaroline fosamil and prepared as recommended by the manufacturer (AstraZeneca). All MRSA isolates and quality control strains were tested at a final inoculum concentration of 5 × 105 CFU/ml and read after 24 h of incubation at 37°C. Three independent experiments were performed for each isolate. The disk diffusion assays were performed according to EUCAST recommendations (http://www.eucast.org). Briefly, inocula of exactly 5 × 106 CFU/ml each were spread on Mueller-Hinton agar plates. The plates were dried for ≤15 min and a ceftaroline disk provided by AstraZeneca (Oxoid) containing 5 μg of ceftaroline was immediately applied. The plates were immediately inverted and incubated at 37°C for 24 h. Three independent experiments were performed for each isolate.
mecA gene sequencing.
Genomic DNA (gDNA) was prepared from an overnight culture grown in MHB at 37°C, as previously described (20). Genomic DNA was isolated from a single colony of each isolate, and mecA was amplified by PCR (3 kb) using the primers seq_mec locus F (5′-TAAGGGAGAAGTAACAGCAC-3′) and seq_mec locus R (5′-ATCGCCCAAAGCTTCTTTAG-3′). The fragments were excised, purified, and directly sequenced using several appropriate nested primers within mecA. Sequence alignments were performed using ClustalW and mecA reference sequences, such as those of S. aureus N315 (SA0038) and COL (SACOL0033).
ST and SCCmec typing.
We determined the ST and staphylococcal cassette chromosome mec element (SCCmec) type for a subset of University Hospital of Geneva (HUG) isolates. Genomic DNA (gDNA) was isolated from a single colony of each isolate, as described above, and 7 genes (arcC, aroE, glpF, gmk, pta, tpi, and yqil) were amplified and sequenced with the appropriate primers. The sequence data were analyzed using the multilocus sequence typing (MLST) website (http://saureus.mlst.net/misc/info.asp). For SCCmec typing, gDNA was tested by quantitative PCR for the staphylococcal cassette chromosome mec element (SCCmec), as described previously (21).
RESULTS
Reduced ceftaroline susceptibility in a strain collection archived from 1994 to 2003.
The efficacy of ceftaroline was tested against a collection of bacteremic MRSA isolates (detailed in Materials and Methods) archived from 1994 to 2003 in our hospital (16). We tested ceftaroline efficacy using both disk diffusion and microdilution MIC assays, since both methods are recommended by EUCAST and are applicable to our hospital clinical laboratory. The MRSA strains showed inhibition zone diameters ranging from 18 to 26 mm (Table 1). The methicillin-susceptible S. aureus (MSSA) strains showed diameters between 30 and 33 mm. The majority of the strains tested showed diameters between 19 and 20 mm, which are considered borderline diameters according to the EUCAST breakpoints (susceptible [S], ≥20 mm; resistant [R], <20 mm), thus requiring additional analysis. As recommended by EUCAST for a diameter between 19 and 21 mm, a microdilution MIC determination was performed to confirm susceptibility (see http://www.eucast.org and reference 22). Accordingly, we performed ceftaroline microdilution assays for all the strains included in our study. The results revealed a ceftaroline microdilution MIC range of 0.5 to 4 μg/ml and a modal MIC of 2 μg/ml for this strain set (Table 1). The MICs for the susceptible control strain ATCC 29213 were within the EUCAST-approved values (microdilution MICs, 0.12 to 0.5 μg/ml; disk diameter, 29 mm). In accordance with EUCAST ceftaroline microdilution susceptibility breakpoints (S, ≤1 μg/ml; R, >1 μg/ml) we concluded that the majority of our strain collection showed resistance to ceftaroline. In Table 1, we report the corresponding final strain susceptibility phenotypes, taking into account the EUCAST recommendations, by using both the results from disk diffusion and microdilution MICs.
TABLE 1.
Ceftaroline MICs and susceptibility data for S. aureus strains in this study
| Strain group and no. | Archived date | Strain characteristic | Ceftaroline microdilution MIC (μg/ml)a | Ceftaroline 5-μg disk diam (mm)b | Susceptibility phenotypec |
|---|---|---|---|---|---|
| HUG strain collectiond | |||||
| 2 | 1995 | Clinical MRSA | 0.5–1 | 22 | S |
| 3 | 1995 | Clinical MRSA | 0.5 | 25 | S |
| 4 | 1996 | Clinical MRSA | 1 | 21 | S |
| 5 | 1996 | Clinical MRSA | 0.5 | 24 | S |
| 6 | 1996 | Clinical MRSA | 0.5 | 22 | S |
| 7 | 1997 | Clinical MRSA | 0.5 | 24 | S |
| 8 | 1997 | Clinical MRSA | 0.5 | 24 | S |
| 9 | 1997 | Clinical MRSA | 1 | 21 | S |
| 10 | 1998 | Clinical MRSA | 1 | 21 | S |
| 11 | 1998 | Clinical MRSA | 1 | 22 | S |
| 12 | 1998 | Clinical MRSA | 2 | 19 | R |
| 13 | 1998 | Clinical MRSA | 4 | 18 | R |
| 14 | 1998 | Clinical MRSA | 2 | 21 | R |
| 15 | 1998 | Clinical MRSA | 0.5 | 24 | S |
| 16 | 1998 | Clinical MRSA | 4 | 18 | R |
| 17 | 1999 | Clinical MRSA | 2 | 22 | R |
| 18 | 1999 | Clinical MRSA | 0.5 | 24 | S |
| 19 | 1999 | Clinical MRSA | 2 | 19 | R |
| 20 | 1999 | Clinical MRSA | 2 | 19 | R |
| 21 | 1999 | Clinical MRSA | 2 | 19 | R |
| 22 | 1999 | Clinical MRSA | 2 | 19 | R |
| 23 | 1999 | Clinical MRSA | 2 | 19 | R |
| 24 | 2000 | Clinical MRSA | 2 | 19 | R |
| 25 | 2000 | Clinical MRSA | 2 | 19 | R |
| 26 | 2000 | Clinical MRSA | 2 | 19 | R |
| 27 | 2000 | Clinical MRSA | 2 | 19 | R |
| 28 | 2000 | Clinical MRSA | 2 | 18 | R |
| 29 | 2000 | Clinical MRSA | 2 | 19 | R |
| 30 | 2000 | Clinical MRSA | 4 | 18 | R |
| 31 | 2000 | Clinical MRSA | 2 | 19 | R |
| 32 | 2000 | Clinical MRSA | 2 | 19 | R |
| 33 | 2000 | Clinical MRSA | 2 | 20 | R |
| 34 | 2000 | Clinical MRSA | 2 | 20 | R |
| 35 | 2001 | Clinical MRSA | 1 | 25 | S |
| 36 | 2001 | Clinical MRSA | 1 | 26 | S |
| 37 | 2002 | Clinical MRSA | 2 | 20 | R |
| 38 | 2002 | Clinical MRSA | 2 | 19 | R |
| 39 | 2002 | Clinical MRSA | 2 | 20 | R |
| 40 | 2002 | Clinical MRSA | 2 | 20 | R |
| 41 | 2002 | Clinical MRSA | 2 | 20 | R |
| 42 | 2002 | Clinical MRSA | 2 | 20 | R |
| 43 | 2002 | Clinical MRSA | 2 | 20 | R |
| 44 | 2002 | Clinical MRSA | 2 | 20 | R |
| 45 | 2002 | Clinical MRSA | 2 | 20 | R |
| 46 | 2002 | Clinical MRSA | 2 | 20 | R |
| 47 | 2003 | Clinical MRSA | 2 | 21 | R |
| 48 | 2003 | Clinical MRSA | 2 | 20 | R |
| 49 | 2003 | Clinical MRSA | 2 | 19 | R |
| 50 | 2003 | Clinical MRSA | 2 | 20 | R |
| 51 | 2003 | Clinical MRSA | 2 | 20 | R |
| 52 | 2003 | Clinical MRSA | 2 | 20 | R |
| 53 | 2003 | Clinical MRSA | 0.5 | 22 | S |
| 54 | 2003 | Clinical MRSA | 2 | 20 | R |
| 55 | 1994 | Clinical MRSA | 0.5 | 23 | S |
| 56 | 1999 | Clinical MRSA | 2 | 20 | R |
| 57 | 2000 | Clinical MRSA | 2 | 19 | R |
| 58 | 1997 | Clinical MRSA | 1 | 23 | S |
| 59 | 1979 | Clinical MRSA | 1 | 23 | S |
| 60 | 2000 | Clinical MRSA | 1 | 21 | S |
| 61 | 2000 | Clinical MRSA | 0.5 | 26 | S |
| 62 | 1983 | Laboratory MSSA | 0.25 | 33 | S |
| 63 | 2009 | Laboratory MSSA | 0.25 | 33 | S |
| 64 | 2010 | Laboratory MSSA | 0.25 | 30 | S |
| Control strainse | |||||
| NRS3 | HIP 5827 | 1 | 24 | S | |
| Mu50 | NRS1, ATCC 700699 | 1 | 26 | S | |
| ATCC 29213 | ATCC 29213 | 0.25 | 29 | S |
EUCAST microdilution ceftaroline susceptibility breakpoint: susceptible (S), ≥1 μg/ml; resistant (R), >1 μg/ml.
EUCAST ceftaroline disk susceptibility breakpoints: S, ≥20 mm; R, <20 mm.
Susceptibility phenotype based on EUCAST recommendation. If the diameter was between 19 and 21 mm, the MIC determination was used to confirm susceptibility.
Network on Antimicrobial Resistance in S. aureus (NARSA) collection. The MICs for ATCC 29213 are within the EUCAST-approved ranges for ceftaroline.
Remarkably, our strain collection showed a high percentage of strains with reduced ceftaroline susceptibility (MIC, >1 μg/ml). This result is, to our knowledge, unprecedented, but with the caveat that this result was based on a monocentric retrospective strain set initially chosen for study because of its particular borderline GISA characteristics (see Materials and Methods). Numerous multicentric studies have observed ceftaroline-resistant strains (MIC, 2 μg/ml by EUCAST breakpoints) but at significantly lower percentages (3 to 19.4%) (5, 6, 10, 23). The modal values reported in these MRSA strains for ceftaroline susceptibility were reported in the range of 0.5 to 1 μg/ml. For clarity in the interpretation of our findings, it is worthwhile to mention that by both current CLSI and EUCAST breakpoints, an MIC of 2 μg/ml does not indicate susceptibility (see http://www.eucast.org, CLSI document M100-S23 [24], and reference 11). According to the CLSI, an MIC of 2 μg/ml is considered in a separate group labeled intermediate, whereas EUCAST considers an MIC of 2 μg/ml to be resistant.
PBP2A allosteric site mutations detected in strains archived since 1998 correlate with reduced ceftaroline susceptibility.
To explain the unexpectedly high percentage of strains with reduced susceptibility to ceftaroline, we next performed a detailed examination of our susceptibility data. The analysis revealed a remarkable change in the distribution of ceftaroline susceptibility when listing our strain collection in chronological order by the archived dates. We observed that the strains from the period before 1998 (strains 2 to 11) showed lower ceftaroline MICs than those of the strains from the period 1998 to 2003 (strains 12 to 54). The University Hospital of Geneva, Geneva, Switzerland, and the University Hospital of Lausanne, Lausanne, Switzerland, experienced a documented period around 1998 in which an aggressive nosocomial MRSA strain type (named the South German SCCmec I ST228) became established and predominant (25). Given that our strain collection is a subset of a larger nosocomial strain collection, we hypothesized that a large fraction of this collection contained SCCmec I ST228 with features that led to reduced ceftaroline susceptibility. Particularly relevant to this hypothesis was the study conducted by Mendes et al. (13), which showed that one important feature that correlated with reduced ceftaroline susceptibility was the reduced affinity of ceftaroline to its PBP2A target, which is attributed to amino acid alterations within the PBP2A protein (Fig. 1).
FIG 1.
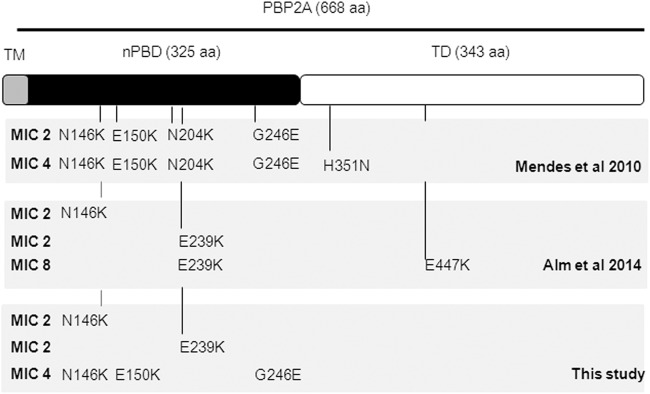
Schematic representation of PBP2A showing mutations correlated with increased ceftaroline MICs. TM, transmembrane domain; nPBD, non-penicillin-binding domain; TD, transpeptidase domain. The locations of the different detected PBP2A mutations in the different studies (Mendes et al. [13], Alm et al. [11], and this study) are indicated.
We thus determined the SCCmec, ST, and deduced PBP2A sequence for each strain in a subset of our strain collection, including strains with different resistance profiles (MICs, 0.25 to 4 μg/ml) (Table 2). An analysis of the PBP2A sequences showed the presence of strains with PBP2A missense mutations (N146K, E150K, E239K, and G246E) in the allosteric non-penicillin-binding domain (Table 2 and Fig. 1). Interestingly, in a comparison of the deduced PBP2A sequences from our strain collection and the previously published Mendes et al. (13) study that analyzed the Greek strains 4977 and 13101, we observed a striking overlap of PBP2A mutations (Fig. 1). Furthermore, the existence of a PBP2A mutation(s) strongly correlated with our observed reduced ceftaroline susceptibility. In contrast, the majority of the strains in our collection showing sensitivity to ceftaroline did not show mutations in PBP2A, with the exception of strains 35, 36, and 53.
TABLE 2.
PBP2A mutations in strains by collection
| Strain collection | Archived date | ST, SCCmec type | S. aureus strain no. orGenBank accession no. | MIC (μg/ml) | PBP2A mutation |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N146K | E150K | N204K | E239K | G246E | H351N | E447K | |||||
| Greecea | ST5, SCCmec II | 4981 | 1 | ||||||||
| Greecea | ST239, SCCmec III | 4977 | 2 | K | K | K | E | ||||
| Greecea | 2008 | ST239, SCCmec III | 13101 | 4 | K | K | K | E | N | ||
| HUG | 1994 | ST30, SCCmec III | 55 | 0.5 | |||||||
| HUG | 1995 | ST30, SCCmec III | 3 | 0.5 | |||||||
| HUG | 1996 | ST45, SCCmec IV | 5 | 0.5 | |||||||
| HUG | 1996 | ST572, SCCmec I | 4 | 1 | |||||||
| HUG | 1997 | ST45, SCCmec IV | 7 | 0.5 | |||||||
| HUG | 1997 | ST45, SCCmec IV | 58 | 1 | |||||||
| HUG | 1998 | ST30, SCCmec III | 10 | 1 | |||||||
| HUG | 1998 | ST228, SCCmec I | 12 | 2 | K | ||||||
| HUG | 1998 | ST228, SCCmec I | 14 | 2 | K | ||||||
| HUG | 1998 | ST247, SCCmec I | 13 | 4 | K | K | E | ||||
| HUG | 1998 | ST247, SCCmec I | 16 | 4 | K | K | E | ||||
| HUG | 1999 | ST45, SCCmec IV | 18 | 0.5 | |||||||
| HUG | 1999 | ST228, SCCmec I | 56 | 2 | K | ||||||
| HUG | 1999 | ST228, SCCmec I | 17 | 2 | K | ||||||
| HUG | 1999 | ST228, SCCmec I | 21 | 2 | K | ||||||
| HUG | 2000 | ST228, SCCmec I | 57 | 2 | K | ||||||
| HUG | 2000 | ST228, SCCmec I | 25 | 2 | K | ||||||
| HUG | 2000 | ST228, SCCmec I | 28 | 2 | K | ||||||
| HUG | 2000 | ST228, SCCmec I | 30 | 4 | K | ||||||
| HUG | 2001 | ST228, SCCmec I | 35 | 1 | K | ||||||
| HUG | 2001 | ST228, SCCmec I | 36 | 1 | K | ||||||
| Lausanneb | 2001 | ST228, SCCmec I | HE579059 | 2 | K | ||||||
| Lausanneb | 2001 | ST228, SCCmec I | HE579061 | 2 | K | ||||||
| HUG | 2002 | ST228, SCCmec I | 42 | 2 | K | ||||||
| HUG | 2003 | ST8, SCCmec IV | 53 | 0.5 | K | ||||||
| HUG | 2003 | ST228, SCCmec I | 48 | 2 | K | ||||||
| HUG | 2003 | ST228, SCCmec I | 52 | 2 | K | ||||||
| Lausanneb | 2006 | ST228, SCCmec I | HE579063 | 2 | K | ||||||
| Lausanneb | 2006 | ST228, SCCmec I | HE579065 | 2–4 | K | ||||||
| Lausanneb | 2006 | ST228, SCCmec I | HE579067 | 2 | K | ||||||
| Lausanneb | 2008 | ST228, SCCmec I | HE579069 | 2 | K | ||||||
| Lausanneb | 2008 | ST228, SCCmec I | HE579071 | 2 | K | ||||||
| Lausanneb | 2008 | ST228, SCCmec I | HE579073 | 2 | K | ||||||
| HUG | ST250 | COL | 0.25 | E | |||||||
| HUG | ST5, SCCmec II | N315 | 1 | ||||||||
| Spainc | 2010 | ST228 | ARC3824 | 8 | K | K | |||||
| Thailandc | 2010 | ST228 | ARC3827 | 2 | K | ||||||
| Thailandc | 2010 | ST228 | ARC3828 | 8 | K | K | |||||
| Thailandc | 2010 | ST228 | ARC3829 | 8 | K | K | |||||
| Thailandc | 2010 | ST228 | ARC3830 | 8 | K | K | |||||
| Spainc | 2010 | ST228 | ARC5493 | 2 | K | ||||||
| Spainc | 2010 | ST228 | ARC5495 | 2 | K | ||||||
| Spainc | 2010 | ST228 | ARC5496 | 2 | K | ||||||
| Spainc | 2010 | ST228 | ARC5497 | 2 | K | ||||||
| Spainc | 2010 | ST228 | ARC5498 | 2 | K | ||||||
| Thailandc | 2010 | ST228 | ARC5500 | 2 | K | ||||||
| Thailandc | 2010 | ST228 | ARC5503 | 2 | K | ||||||
The strains showing an MIC of >1 μg/ml were predominantly of South German clonotype SCCmec I and ST228 containing mutations in the PBP2A allosteric site. However, in this study, we also detected two Iberian clones of SCCmec I ST247 (strains 13 and 16) that displayed a ceftaroline MIC of 4 μg/ml and contained strikingly similar PBP2A mutations as those of Greek strains 4977 and 13101 (13). Notably, strains 4977 and 13101 (both ST239) are of SCCmec III, whereas our strains of the South German ST228 and Iberian ST247 are of SCCmec I, suggesting that remarkably similar PBP2A mutations affecting ceftaroline susceptibility can arise independently in variant MRSA type genetic backgrounds.
Additional evidence for reduced ceftaroline susceptibility correlating with missense PBP2A mutations was obtained by analyzing an independent set of South German SCCmec I ST228 strains external to our collection. The University Hospital of Lausanne (CHUV) recently published the complete genome sequence of eight South German clonotype ST228 strains collected over the period of 2001 to 2008 (26). An analysis of PBP2A sequences extracted from the public database repository showed amino acid changes (N146K) identical to those of the majority of our ST228 strain isolates. The subsequent ceftaroline MICs from all eight CHUV ST228 strains showed a ceftaroline MIC of 2 μg/ml, thus also defining these ST228 strains as ceftaroline resistant according to the EUCAST breakpoints.
Collectively, we conclude that the most likely explanation for our strain collection results arises from the high fractional representation of strains carrying a missense mutation in PBP2A that renders these strains resistant to ceftaroline. This finding is most likely linked to the local epidemiology of resident HA-MRSA strains that have colonized western Switzerland since 1998.
DISCUSSION
In this work, we report the existence of an HA-MRSA collection that spans the period of 1994 to 2003 and contains a high percentage of strains from 1998 to 2003 showing ceftaroline MICs of >1 μg/ml. Reduced ceftaroline susceptibility predominantly correlated with South German ST228 SCCmec I MRSA strains carrying allosteric site mutations in PBP2A. Sequenced ST228 strains from Lausanne collected in the period of 2003 to 2008 also showed PBP2A allosteric site mutations and reduced susceptibilities to ceftaroline. In addition, two ST247 (Iberian) strains from our HA-MRSA collection archived in 1998 showed an MIC of 4 μg/ml and contained three mutations within the PBP2A allosteric site at positions identical to those previously described for ST239 SCCmec III isolates (4977 and 13101) from Greece (10, 13). The high-level resistance to ceftaroline (MIC = 4 μg/ml, undisputed by either EUCAST or CLSI breakpoints) arising in distinct SCCmec types with identical mutations over an interval of 12 years is intriguing.
While this work was in progress, PBP2A mutations showing reduced ceftaroline susceptibility with MICs of 2 and 8 μg/ml were identified independently in other ST228 strains isolated from Spain and Thailand during the Assessing Worldwide Antimicrobial Resistance Evaluation (AWARE) global surveillance study conducted in 2010 (13). However, these mutations were different from those found in our study (Fig. 1) (11). These mutations were also located in both PBP2A allosteric and/or transpeptidase domains (particularly in the cases of strains with an elevated MIC of 8 μg/ml). The whole-genome sequence-based epidemiologic analysis performed by Alm and coworkers (11) showed that the ST228 strains identified in their study do not cluster with previously sequenced Swiss ST228 strains (University Hospital of Lausanne), suggesting that they belong to distinct lineages.
We have considered several possible causes for our observations. Our hospital in western Switzerland has experienced a documented establishment of ST228 as a prevalent HA-MRSA local endemic clonotype since 1998. While the existence of variation in local geographic epidemic MRSA strains is well known (27, 28), the existence of particular regional geographic concentrations of ST228 strains showing PBP2A mutations may have gone unreported despite large-scale sampling in other geographical regions where other strain variants predominated during the course of global susceptibility studies. Second, our strain set was chosen in particular for its characteristics as an HA-MRSA set showing a large fraction of borderline GISA phenotypes; thus, the set per se was not likely a representative sample of all circulating MRSA strains in our institution or geographic region during the study period. It is presently not known whether genetic mechanisms underlying the GISA phenotype affect ceftaroline resistance. Nevertheless, reports independent of ours have turned up PBP2A mutations within the allosteric domain in ST228 and other strains without prior consideration for the underlying GISA phenotype. Finally, testing methodology variations or reagents were excluded as potential causes for reduced ceftaroline susceptibility, since our study employed methods established by EUCAST, and similar methods applied by other laboratories have revealed strains in other geographically diverse sites with reduced susceptibilities to ceftaroline.
One of the most important observations of our study was the detection of ceftaroline resistance in strains archived prior to the commercial introduction of ceftaroline in 2010 (Fig. 2). In agreement with the conclusion of Alm and coworkers (11), we speculate that biological pressure other than exposure to ceftaroline allowed the selection of these PBP2A mutations. The existence of strains with an MIC of 2 μg/ml with different single amino acid changes in the same or different MRSA STs may be explained by independent mutational events generated under unidentified, but perhaps similar, biological selective pressures. The findings from single or multistep resistance selection suggest a low potential of ceftaroline itself to select for resistance (29). We speculate that encounters with other therapeutic agents, such as other β-lactams, may lead to the selection of mutations within PBP2A. Indeed, such a proof of principle has been shown in vitro by the demonstration that β-lactam antibiotic pressure can result in PBP2A mutations showing some overlap at or near the PBP2A mutations detected in our collection (11, 30, 31).
FIG 2.
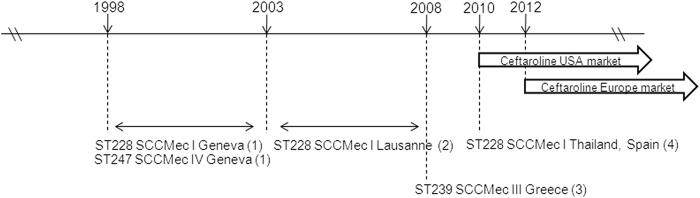
Time interval between detection of ceftaroline-resistant strains and arrival of ceftaroline drug in the U.S. and European markets. Only bacterial strains showing ceftaroline resistance with concomitant PBP2A sequencing data in the literature are indicated (1 and 2, this study; 3, Mendes et al. [13]; 4, Alm et al. [11]).
Nearly all of the mutations described in published studies and our work here map in the PBP2A allosteric domain. Sequence database searches that we performed for allosteric mutations turned up additional evidence in strains from various diverse geographical regions; however, these strains have not been tested for reduced susceptibility to ceftaroline. Some examples (GenBank accession no.) are: an N146K single mutation in strains IS-157 (EHS72684.1), DBM17 (AEQ76893.1), and M600 (ADB44841); an E150K single mutation in strains CH10 (ABS12108 [32]) and BK16691 (ADC53314 [33]); an E239K single mutation in strains DAR116 (EXM51016) and TN/CN/1/2 (AGC51118); and an N146K-E150K double mutation in strains SN1M89 (AEV66267), M16 (ADB44843), and DARS883 (EUJ99152).
While we have primarily discussed mutations mapping within the allosteric domain of PBP2A, mutations within the dd-transpeptidase domain have also been described, such as E447K (11), H351N (13), and Y446N-E447K (12). The latter mutations are particularly important and found in an ST5 strain named 3125 isolated from a cystic fibrosis patient in 2013 and yielding high-level ceftaroline resistance (MIC, >32 μg/ml).
Taking into account all the studies concerning PBP2A sequencing analysis and ceftaroline susceptibility, no clear conclusions can be drawn concerning a particular pattern of PBP2A mutations and predictable ceftaroline MICs. Conceivably, mutations within the transpeptidase domain might be associated with higher ceftaroline MICs, while mutations within the allosteric domain might result in more moderate changes in ceftaroline MICs. All strains in our collection with an MIC of 2 μg/ml show predominantly single amino acid substitutions at position 146 (N146K), while strains with an MIC of 2 μg/ml collected from Spain and Thailand show single amino acid substitutions predominantly at position 239 (E239K) (Fig. 1). Nevertheless, more than one PBP2A amino acid change, regardless of the amino acid position, appears to be associated with decreased ceftaroline susceptibility. The mechanistic insights provided by crystallographic and biochemical methods hint that allosteric site mutations result in complex alterations of salt bridge networks that comprise the allosteric trigger. Their individual contributions to phenotype may not simply be additive (15). Alternative mechanisms cannot be excluded either; for example, mutations that impact protein-protein interactions perhaps occur on the outer leaflet of the membrane surface (11).
In our study, we note that our laboratory strain COL, an ancestral MRSA strain SCCmec I ST250 (34), harbored the PBP2A missense mutation G246E but without an experimentally detectable effect upon ceftaroline sensitivity (MIC = 0.5 μg/ml; Table 2). Interestingly, the mutation G246E is shared by both of our ST247 strains and both ST239 Greek strains 4977 and 13101, raising the question of whether this mutation contributes at all to reduced ceftaroline susceptibility. ST247 is thought to be derived from ST250 (34), and it remains to be determined whether other COL isolates and their derivatives harbor G246E PBP2A variants. Notably, this mutation is also present in the sequenced COL reference strain (NCBI accession no. CP000049). Refined studies must examine the contribution of individual and multiple mutations in more detail, to determine whether they are additive or synergistic, and to identify mutations that do not contribute to ceftaroline resistance.
Another comparison of the genetic loci known to affect β-lactam resistance between fully sequenced genomes from reference and ceftaroline-resistant strains indicated that changes in the mecA gene encoding PBP2A were the only changes correlated with ceftaroline MICs (11). However, our results suggest that unidentified chromosomal genes must also affect ceftaroline susceptibility. This hypothesis is supported by an analysis of our strain panel, in which we observed the three susceptible strains 35, 36, and 53 (MIC assays tested repeatedly) that nevertheless carried identical mutations as those of other strains with an MIC of 2 μg/ml. Moreover, it is also supported by the work of Fernandez et al. (35), in which they found that strains with an MIC of 2 μg/ml for ceftaroline do not show PBP2A sequence mutations but instead show penicillin-binding protein 2 mutations; this was further supported by the observation that no PBP2A sequence mutation was found in the heterogeneous vancomycin-intermediate S. aureus (hVISA) strain Mu3, but ceftaroline MIC analysis classified this strain as resistant (MIC = 2 μg/ml) (7).
An intriguing question concerns why PBP2A mutations have been discovered predominantly in ST228 and, to a lesser extent, a few other scattered sequence types, such as ST247, ST239, ST5, ST8, and ST764, which collectively fall into clonal complex categories 5 and 8 (11–13, 22). Do certain clonotypes more easily acquire and support potential fitness costs associated with PBP2A variation? The evolutionary history of MRSA strains, including those of ST247, ST228, and ST239, has been well documented (34, 36–40). Recent epidemiological studies of MRSA revealed that ST228 and ST247 still circulate in Europe and Switzerland (27, 28, 34, 41) in widely dispersed geographical regions, providing evidence for local epidemiological clusters. The important question arises as to whether strains, such as those belonging to ST228 with PBP2A mutations, represent sporadic isolated events. The performance of additional studies using isolates from multiple sources seems imperative in order to answer this question. To this end, a follow-up study (A. Renzoni, unpublished data) examined MRSA strains collected in 2013 and 2014 in the University Hospital of Geneva and indeed detected contemporary examples of ST228 carrying the N146K mutation.
ACKNOWLEDGMENTS
This work was supported in part by an investigator-initiated ISSSP grant of AstraZeneca Pharmaceuticals and by the Swiss National Science Foundation grants AR 310030-149762 and WLK 10030-146540.
We acknowledge Dominique Blanc and Jacques Bille from the Laboratoire de Typage Microbien of Lausanne University Hospital for kindly providing their sequenced strains.
REFERENCES
- 1.Llarrull LI, Fisher JF, Mobashery S. 2009. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge. Antimicrob Agents Chemother 53:4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosowska-Shick K, McGhee PL, Appelbaum PC. 2010. Affinity of ceftaroline and other β-lactams for penicillin-binding proteins from Staphylococcus aureus and Streptococcus pneumoniae. Antimicrob Agents Chemother 54:1670–1677. doi: 10.1128/AAC.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell DJ, Flamm RK, Sader HS, Jones RN. 2013. Spectrum and potency of ceftaroline tested against leading pathogens causing skin and soft-tissue infections in Europe (2010). Int J Antimicrob Agents 41:337–342. doi: 10.1016/j.ijantimicag.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Farrell DJ, Castanheira M, Mendes RE, Sader HS, Jones RN. 2012. In vitro activity of ceftaroline against multidrug-resistant Staphylococcus aureus and Streptococcus pneumoniae: a review of published studies and the AWARE Surveillance Program (2008–2010). Clin Infect Dis 55(Suppl 3):S206–S214. doi: 10.1093/cid/cis563. [DOI] [PubMed] [Google Scholar]
- 6.Sader HS, Flamm RK, Jones RN. 2013. Antimicrobial activity of ceftaroline tested against staphylococci with reduced susceptibility to linezolid, daptomycin, or vancomycin from U.S. hospitals, 2008 to 2011. Antimicrob Agents Chemother 57:3178–3181. doi: 10.1128/AAC.00484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werth BJ, Steed ME, Kaatz GW, Rybak MJ. 2013. Evaluation of ceftaroline activity against heteroresistant vancomycin-intermediate Staphylococcus aureus and vancomycin-intermediate methicillin-resistant S. aureus strains in an in vitro pharmacokinetic/pharmacodynamic model: exploring the “seesaw effect.” Antimicrob Agents Chemother 57:2664–2668. doi: 10.1128/AAC.02308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saravolatz LD, Stein GE, Johnson LB. 2011. Ceftaroline: a novel cephalosporin with activity against methicillin-resistant Staphylococcus aureus. Clin Infect Dis 52:1156–1163. doi: 10.1093/cid/cir147. [DOI] [PubMed] [Google Scholar]
- 9.Frampton JE. 2013. Ceftaroline fosamil: a review of its use in the treatment of complicated skin and soft tissue infections and community-acquired pneumonia. Drugs 73:1067–1094. doi: 10.1007/s40265-013-0075-6. [DOI] [PubMed] [Google Scholar]
- 10.Jones RN, Mendes RE, Sader HS. 2010. Ceftaroline activity against pathogens associated with complicated skin and skin structure infections: results from an international surveillance study. J Antimicrob Chemother 65(Suppl 4):iv17–iv31. doi: 10.1093/jac/dkq252. [DOI] [PubMed] [Google Scholar]
- 11.Alm RA, McLaughlin RE, Kos VN, Sader HS, Iaconis JP, Lahiri SD. 2014. Analysis of Staphylococcus aureus clinical isolates with reduced susceptibility to ceftaroline: an epidemiological and structural perspective. J Antimicrob Chemother 69:2065–2075. doi: 10.1093/jac/dku114. [DOI] [PubMed] [Google Scholar]
- 12.Long SW, Olsen RJ, Mehta SC, Palzkill TG, Cernoch PL, Perez KK, Musick WL, Rosato AE, Musser JM. 2014. PBP2a mutations causing high-level ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 58:6668–6674. doi: 10.1128/AAC.03622-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendes RE, Tsakris A, Sader HS, Jones RN, Biek D, McGhee P, Appelbaum PC, Kosowska-Shick K. 2012. Characterization of methicillin-resistant Staphylococcus aureus displaying increased MICs of ceftaroline. J Antimicrob Chemother 67:1321–1324. doi: 10.1093/jac/dks069. [DOI] [PubMed] [Google Scholar]
- 14.Otero LH, Rojas-Altuve A, Llarrull LI, Carrasco-Lopez C, Kumarasiri M, Lastochkin E, Fishovitz J, Dawley M, Hesek D, Lee M, Johnson JW, Fisher JF, Chang M, Mobashery S, Hermoso JA. 2013. How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc Natl Acad Sci U S A 110:16808–16813. doi: 10.1073/pnas.1300118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fishovitz J, Rojas-Altuve A, Otero LH, Dawley M, Carrasco-Lopez C, Chang M, Hermoso JA, Mobashery S. 2014. Disruption of allosteric response as an unprecedented mechanism of resistance to antibiotics. J Am Chem Soc 136:9814–9817. doi: 10.1021/ja5030657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaudaux P, Huggler E, Bernard L, Ferry T, Renzoni A, Lew DP. 2010. Underestimation of vancomycin and teicoplanin MICs by broth microdilution leads to underdetection of glycopeptide-intermediate isolates of Staphylococcus aureus. Antimicrob Agents Chemother 54:3861–3870. doi: 10.1128/AAC.00269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley WL, Jousselin A, Barras C, Lelong E, Renzoni A. 2014. Allosteric site missense mutations in PBP2A affecting ceftaroline susceptibility detected in epidemic HA-MRSA clonotypes ST228 and ST247 in western Switzerland, e-poster 180 European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Barcelona, Spain, 10 to 13 May 2014. [Google Scholar]
- 18.Vaudaux P, Huggler E, Arhin FF, Moeck G, Renzoni A, Lew DP. 2009. Comparative activity of oritavancin against meticillin-resistant Staphylococcus aureus (MRSA) bloodstream isolates from Geneva University Hospital. Int J Antimicrob Agents 34:540–543. doi: 10.1016/j.ijantimicag.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Uçkay I, Bernard L, Buzzi M, Harbarth S, François P, Huggler E, Ferry T, Schrenzel J, Renzoni A, Vaudaux P, Lew PD. 2012. High prevalence of isolates with reduced glycopeptide susceptibility in persistent or recurrent bloodstream infections due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 56:1258–1264. doi: 10.1128/AAC.05808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renzoni A, Kelley WL, Barras C, Monod A, Huggler E, Francois P, Schrenzel J, Studer R, Vaudaux P, Lew DP. 2009. Identification by genomic and genetic analysis of two new genes playing a key role in intermediate glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 53:903–911. doi: 10.1128/AAC.01287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.François P, Renzi G, Pittet D, Bento M, Lew D, Harbarth S, Vaudaux P, Schrenzel J. 2004. A novel multiplex real-time PCR assay for rapid typing of major staphylococcal cassette chromosome mec elements. J Clin Microbiol 42:3309–3312. doi: 10.1128/JCM.42.7.3309-3312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koeth LM, Matuschek E, Kahlmeter G, Alm RA, Ambler JE. 2014. Development of EUCAST zone diameter breakpoints and quality control range for Staphylococcus aureus with ceftaroline 5-μg disk. Eur J Clin Microbiol Infect Dis 33:1511–1517. doi: 10.1007/s10096-014-2089-8. [DOI] [PubMed] [Google Scholar]
- 23.Sader HS, Flamm RK, Jones RN. 2013. Antimicrobial activity of ceftaroline and comparator agents tested against bacterial isolates causing skin and soft tissue infections and community-acquired respiratory tract infections isolated from the Asia-Pacific region and South Africa (2010). Diagn Microbiol Infect Dis 76:61–68. doi: 10.1016/j.diagmicrobio.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd information supplement CLSI document M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.François P, Harbarth S, Huyghe A, Renzi G, Bento M, Gervaix A, Pittet D, Schrenzel J. 2008. Methicillin-resistant Staphylococcus aureus, Geneva, Switzerland, 1993–2005. Emerg Infect Dis 14:304–307. doi: 10.3201/eid1402.070229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel V, Falquet L, Calderon-Copete SP, Basset P, Blanc DS. 2012. Short term evolution of a highly transmissible methicillin-resistant Staphylococcus aureus clone (ST228) in a tertiary care hospital. PLoS One 7:e38969. doi: 10.1371/journal.pone.0038969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, European Staphylococcal Reference Laboratory Working Group . 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med 7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark C, McGhee P, Appelbaum PC, Kosowska-Shick K. 2011. Multistep resistance development studies of ceftaroline in Gram-positive and -negative bacteria. Antimicrob Agents Chemother 55:2344–2351. doi: 10.1128/AAC.01602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama Y, Zhang HZ, Chambers HF. 2004. PBP 2a mutations producing very-high-level resistance to beta-lactams. Antimicrob Agents Chemother 48:453–459. doi: 10.1128/AAC.48.2.453-459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee R, Gretes M, Basuino L, Strynadka N, Chambers HF. 2008. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 52:2089–2096. doi: 10.1128/AAC.01403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens AJ, Huygens F, Giffard PM. 2007. Systematic derivation of marker sets for staphylococcal cassette chromosome mec typing. Antimicrob Agents Chemother 51:2954–2964. doi: 10.1128/AAC.01323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Mediavilla JR, Smyth DS, Chavda KD, Ionescu R, Roberts RB, Robinson DA, Kreiswirth BN. 2010. Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob Agents Chemother 54:3347–3354. doi: 10.1128/AAC.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez R, Paz LI, Rosato RR, Rosato AE. 2014. Ceftaroline is active against heteroresistant methicillin-resistant Staphylococcus aureus clinical strains despite associated mutational mechanisms and intermediate levels of resistance. Antimicrob Agents Chemother 58:5736–5746. doi: 10.1128/AAC.03019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crisostomo MI, Westh H, Tomasz A, Chung M, Oliveira DC, de Lencastre H. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc Natl Acad Sci U S A 98:9865–9870. doi: 10.1073/pnas.161272898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth DS, McDougal LK, Gran FW, Manoharan A, Enright MC, Song JH, de Lencastre H, Robinson DA. 2010. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS One 5:e8582. doi: 10.1371/journal.pone.0008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira DC, Tomasz A, de Lencastre H. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist 7:349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 39.Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanc DS, Petignat C, Wenger A, Kuhn G, Vallet Y, Fracheboud D, Trachsel S, Reymond M, Troillet N, Siegrist HH, Oeuvray S, Bes M, Etienne J, Bille J, Francioli P, Zanetti G. 2007. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small geographic area over an eight-year period. J Clin Microbiol 45:3729–3736. doi: 10.1128/JCM.00511-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senn L, Basset P, Greub G, Prod'hom G, Frei R, Zbinden R, Gaia V, Balmelli C, Pfyffer GE, Muhlemann K, Zanetti G, Blanc DS. 2013. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Switzerland: sampling only invasive isolates does not allow a representative description of the local diversity of clones. Clin Microbiol Infect 19:E288–E290. doi: 10.1111/1469-0691.12185. [DOI] [PubMed] [Google Scholar]


