Abstract
Ribavirin, a guanosine analog, is a broad-spectrum antiviral agent. Ribavirin has been a fundamental component of the treatment of hepatitis C virus (HCV) infection for decades, but there is a very limited understanding of the clinical pharmacology of this drug. Furthermore, it is associated with a major dose-limiting toxicity, hemolytic anemia. Ribavirin undergoes intracellular phosphorylation by host enzymes to ribavirin monophosphate (RMP), ribavirin diphosphate (RDP), and ribavirin triphosphate (RTP). The intracellular forms have been associated with antiviral and toxic effects in vitro, but the kinetics of these phosphorylated moieties have not been fully elucidated in vivo. We developed a model to characterize the plasma pharmacokinetics of ribavirin and the difference between intracellular phosphorylation kinetics in red cells (nonnucleated) and in peripheral blood mononuclear cells (nucleated). A time-independent two-compartment model with first-order absorption described the plasma data well. The cellular phosphorylation kinetics was described by a one-compartment model for RMP, with the formation rate driven by plasma concentrations and the first-order degradation rate. RDP and RTP rapidly reached equilibrium with RMP. Concomitant telaprevir use, inosine triphosphatase genetics, creatinine clearance, weight, and sex were significant covariates. The terminal ribavirin half-life in plasma and phosphorylated anabolites in cells was approximately 224 h. We found no evidence of time-dependent kinetics. These data provide a foundation for uncovering concentration-effect associations for ribavirin and determining the optimal dose and duration of this drug for use in combination with newer direct-acting HCV agents. (This study has been registered at ClinicalTrials.gov under registration no. NCT01097395.)
INTRODUCTION
Ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide), a nucleoside analog first synthesized in 1972, exhibits broad-spectrum antiviral activity against several RNA and DNA viruses in vitro (1, 2). For several decades, ribavirin was combined with pegylated interferon alpha (Peg–IFN-α) as the standard of care for treating chronic hepatitis C virus (HCV) infections. Though several direct-acting antiviral agents have recently been approved for the treatment of HCV and dozens more are in various stages of clinical development, ribavirin remains an important component of several HCV treatment regimens (3–7).
Ribavirin's mechanism of antiviral action is not completely understood. In vitro, ribavirin has been shown to mimic the endogenous nucleoside guanosine, and its triphosphate anabolite (ribavirin triphosphate [RTP]) may be incorporated into replicating RNA strands by viral RNA polymerases. This erroneous incorporation inhibits chain elongation and viral replication. Other antiviral effects observed in vitro include immunologic modulation through switching the T-cell phenotype from phenotype 2 to phenotype 1, inhibition of host IMP dehydrogenase leading to depletion of intracellular GTP pools, induction of mutagenesis, and error catastrophes, as well as upregulation of genes involved in the interferon cascade or of IFN-stimulated genes and downregulation of IFN-inhibitory genes (8). It is unclear which mechanism(s) is responsible for the antiviral effects in vivo. While the therapeutic benefit of ribavirin in the treatment of HCV infection is undeniable, there is significant unexplained interpatient variability in the exposure-response relationship for this drug and it is associated with a major dose-limiting toxicity, hemolytic anemia (9, 10).
Following oral absorption, with a bioavailability of approximately 52% (11), ribavirin exhibits extensive distribution to various cells in the body via nucleoside transporters. Once inside the cell, it is phosphorylated to ribavirin monophosphate (RMP) by adenosine kinase or 5′ nucleotidase (12). Other cellular kinases further phosphorylate RMP to ribavirin diphosphate (RDP) and ribavirin triphosphate (RTP). Intracellular ribavirin accumulates in red blood cells (RBCs) because RBCs lack the 5′ nucleotidase and alkaline phosphatase needed to dephosphorylate RMP for transport out of the cells (13). The accumulation of RTP in RBCs interferes with ATP-dependent transport systems, causes RBC membrane destabilization, and induces premature RBC senescence and phagocytic removal by the reticuloendothelial system (14), which leads to hemolytic anemia. Depending on the severity of the anemia exhibited during treatment, a patient may require dose reduction or treatment discontinuation. It is difficult to predict which patients will develop hemolytic anemia from ribavirin treatment. Prior studies have found an association between hemoglobin decline and increased ribavirin concentrations in plasma (15, 16) and RBCs (17). Another study found that total intracellular ribavirin concentrations in RBCs, but not plasma levels, were significantly correlated with hemoglobin decline, which suggested that ribavirin levels in RBC would be a preferable parameter for assessing ribavirin-induced hemolytic anemia (18). In addition to drug exposure, host genetics may also play a role in the development of ribavirin-induced anemia. Polymorphisms in the gene encoding the inosine triphosphatase (ITPA) enzyme have been associated with protection against ribavirin-induced anemia (19). A quantitative understanding of the factors that influence the in vivo phosphorylation kinetics of ribavirin in RBCs and its relationship to hemolytic anemia may lead to improved treatment with ribavirin.
The overall goal of the study reported here was to quantify the relationship between the plasma concentrations of ribavirin and its phosphorylated metabolites in both RBCs and peripheral blood mononuclear cells (PBMCs) during the treatment of patients infected with HCV. To that end, we developed a population model for the plasma pharmacokinetics (PK) of ribavirin over the course of treatment which incorporated the effects of covariates, including coadministration of telaprevir as well as the ITPA phenotype, and evaluated the potential for time-dependent kinetics. This model was then linked to the RMP, RDP, and RTP concentrations measured in both RBCs and PBMCs to quantitatively evaluate and contrast ribavirin phosphorylation between these cell types. The developed complete plasma/intracellular models provide insights into the action of ribavirin, including the factors that affect its interpatient variability, and may guide improved dosing of ribavirin with direct-acting antiviral agents.
MATERIALS AND METHODS
Study design and patients.
This was an open-label, randomized study (ClinicalTrials.gov registration no. NCT01097395) of standard weight-based ribavirin dosing or concentration-guided dosing based on first-dose area under the concentration-time curve (AUC) data (0, 12). Thirty-six treatment-naive genotype 1 chronic HCV infection patients were enrolled between 2010 and 2013. Since telaprevir was approved in May 2011, the first 18 patients enrolled received Peg–IFN-α–2a at 180 μg weekly and ribavirin at 1,000 or 1,200 mg daily (dual-therapy group), whereas the other 18 patients enrolled after May 2011 received Peg–IFN-α–2a at 180 μg weekly, ribavirin at 1,000 or 1,200 mg daily, and telaprevir at 750 mg three times daily (triple-therapy group) for the first 12 weeks and then continued treatment with Peg–IFN-α–2a and ribavirin for an additional 12 or 24 weeks on the basis of clinical factors and antiviral response at week 4 of treatment. Patients who met the criteria for virologic futility discontinued treatment per the standard of care at the time. The study was conducted in accordance with the principles for human experimentation defined in the Declaration of Helsinki and was approved by the Colorado Multiple Institutional Review Board. All participants provided written informed consent.
Plasma ribavirin measurements.
Whole blood (4 ml) was obtained in EDTA tubes at predose and 1, 2, 3, 4, 6, 8, 10, and 12 h postdose during the 12-h intensive-PK visits (day 1 and the steady state at weeks 9 to 13) and at various times postdose (i.e., convenience sampling) at the other visits (weeks 1, 2, 4, 16, 24, and 48) for quantification of ribavirin in plasma using a validated high-performance liquid chromatography–UV (HPLC-UV) detection method. The assay was linear in the range of 0.05 to 10 μg/ml and had a minimum quantifiable limit of 0.05 μg/ml using 0.2 ml of human plasma. Interassay accuracy for the method was within ±8.2% at all concentrations, while precision was ≤12.3%.
Phosphorylated ribavirin measurements.
Whole blood (8 ml) was obtained in a sodium heparin cell preparation tube at predose and 2 and 6 h postdose during the 12-h intensive-PK visits (day 1 and weeks 9 to 13) and at various times postdose at the other visits (weeks 1, 2, 4, 16, 24, and 48) for quantification of RMP, RDP, and RTP in PBMCs and RBCs. Isolation and counting procedures were reported previously (20). After separation of the RMP, RDP, and RTP metabolites, the components were dephosphorylated and analyzed using a validated LC-tandem mass spectrometry (LC-MS/MS) assay (21). The lower limit of quantification for the intracellular assay of phosphorylated ribavirin was 0.50 pmol/sample, when a range of 0.25 to 1.00 pmol/106 cells/sample was extracted. Interassay accuracy for the method was within ±8.9% at all concentrations, while precision was ≤7.9%.
ITPA and IL-28B genotyping.
DNA was extracted from whole blood using an automated system (Qiagen Autopure LS). Genotyping was performed using an ABI TaqMan allelic discrimination kit and an ABI Prism 7900HT sequence detection system (Applied Biosystems, Carlsbad, CA) in a 5-μl reaction volume using standard TaqMan Universal PCR conditions. Primer and probe sequences for the rs12979860 single nucleotide polymorphism (SNP) of interleukin-28B (IL-28B) were as follows: forward primer, 5′-GCCTGTCGTGTACTGAACCA-3′; reverse primer, 5′-GCGCGGAGTGCAATTCAAC-3′; probe, 5′-(VIC/6-carboxyfluorescein [FAM])-TGGTTC[G/A]CGCCTTC-3′. ITPA SNPs rs1127354 and rs7270101 were genotyped using custom genotyping assays manufactured by Applied Biosystems (Carlsbad, CA). Patients with the wild-type (WT) allele for both ITPA SNPs (C and C for the rs1127354 SNP and A and A for rs7270101) were categorized as the ITPA wild-type group, whereas patients with at least 1 variant allele were categorized as the ITPA variant group.
Plasma and intracellular pharmacokinetic modeling.
Population analyses of plasma and intracellular phosphorylated ribavirin were each performed using the maximum-likelihood, expectation maximization (MLEM) algorithm in ADAPT (Version 5) software (22). Model parameters were assumed to follow a multivariate lognormal distribution, and the residual error (defined as the difference between the observed and predicted concentrations) was described using proportional-error-variance terms. Plasma and intracellular metabolite measurements below their respective limits of quantification were incorporated into the likelihood calculation (using the M3 method reported in reference 23).
The schematic diagram of the final composite model is shown in Fig. 1. Plasma data were described using a two-compartment model with first-order absorption. The estimated parameters were apparent total clearance (CLt/F), apparent central compartment volume of distribution (Vc/F), apparent absorption rate constant (ka/F), apparent distributional clearance (CLd/F), and apparent peripheral compartment volume of distribution (Vp/F).
FIG 1.
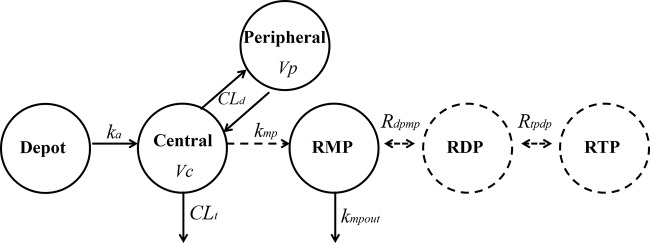
Composite model describing the plasma PK of ribavirin as well as its intracellular kinetics in both RBCs and PBMCs. CLt: total clearance. Vc: central compartment volume of distribution. CLd: distributional clearance. Vp: peripheral compartment volume of distribution. ka: absorption rate constant. kmp: transport/formation rate constant for ribavirin monophosphate (RMP). kmpout: rate constant for the disappearance of phosphorylated ribavirin. Rdpmp: rapid equilibrium partition coefficient between ribavirin diphosphate (RDP) and RMP. Rtpdp: rapid equilibrium partition coefficient between ribavirin triphosphate (RTP) and RDP.
The following covariates were examined for their influence on the plasma PK parameters: age, body weight, sex, race, creatinine clearance (CrCl), genotype 1 subtype, ITPA phenotype, degree of fibrosis, baseline HCV RNA level, baseline hemoglobin, and telaprevir coadministration. CrCl was calculated via the Cockroft-Gault equation using baseline serum creatinine level and total body weight unless the patient weight was ≥130% of the ideal, in which case an adjusted body weight was used. Covariate-parameter relationships were identified on the basis of exploratory graphics, scientific interest, and mechanistic plausibility. The final covariate model was selected based on model estimate precision and the Akaike information criterion (AIC) and backward elimination (P < 0.01).
A visual predictive check based on 500 simulation replicates for the intensive profiles on day 1 and at the steady state was performed as a model qualification method. Descriptive statistics of the simulated concentrations (median and 90% prediction intervals) were compared with those of the observed ribavirin plasma concentrations to assess model adequacy.
The individual subject parameter estimates from the PK modeling (conditional means of the MLEM algorithm) were then used to model the intracellular ribavirin metabolite kinetics (using RMP, RDP, and RTP simultaneously) in RBCs and also in PBMCs. In the final intracellular metabolism model depicted in Fig. 1 (for both RBCs and PBMCs), RDP rapidly reached equilibrium with RMP (Rdpmp = RDP/RMP) and RTP rapidly reached equilibrium with RDP (Rtpdp = RTP/RDP). Other models that included nonequilibrium phosphorylation and dephosphorylating rate constants were also examined but were rejected based on AIC. In the model, the transport of ribavirin into the cells and its initial phosphorylation to RMP (kmp) is driven by the plasma ribavirin concentration, while the disappearance of RMP follows a first-order process (kmpout). The parameter kmpout represents several mechanisms, including metabolism (dephosphorylation) of phosphorylated anabolites, degradation of PBMCs or RBCs, and potential efflux of RMP by nucleoside transporters.
The differential equations that describe the final model are given as follows:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Finally, the individual parameters from the intracellular models (RBCs and PBMCs) were used to simulate time profiles for each subject at each visit. The median and 5th to 95th percentile ranges of those predictions were plotted and overlaid with the observed RMP, RDP, and RTP data for PBMCs and RBCs, respectively, to assess the predictive ability of the model.
RESULTS
Forty subjects consented to participate in the study. Thirty-six were eligible for enrollment. Clinical, genetic, and pharmacokinetic data are available for all 36 subjects. The demographic and baseline characteristics of the 36 subjects are shown in Table 1. Based on the 2 ITPA SNPs, there were 26 subjects with a wild-type allele for both ITPA SNPs (C and C for rs1127354 SNP and A and A for rs7270101), associated with 100% ITPA enzyme activity. Of the remaining 10 subjects, 4 subjects (2 subjects with 10% activity and 2 subjects with 30% activity) received dual therapy and 6 subjects (4 subjects with 30% activity and 2 subjects with 60% activity) received triple therapy. The 10 subjects were pooled as representing an ITPA variant phenotype. The rate of sustained virological response was 33% (6 of 18 subjects) in the dual-therapy group and 78% (14 of 18 subjects) in the triple-therapy group. A total of 8 subjects (6 in the dual-therapy group and 2 in the triple-therapy group) had virologic failure, and 8 subjects (6 in the dual-therapy group and 2 in the triple-therapy group) dropped out or discontinued therapy due to an adverse event.
TABLE 1.
Demographic and baseline characteristics of the study patientsa
| Characteristic | No. | Mean ± SD | Minimum | Median | Maximum |
|---|---|---|---|---|---|
| Sex (male/female) | 22/14 | ||||
| Race (white/Hispanic/black/other) | 22/8/4/2 | ||||
| Age (yrs) | 50.4 ± 8.6 | 27 | 52 | 61 | |
| Wt (kg) | 82.5 ± 17.9 | 56 | 79 | 128 | |
| Height (cm) | 172 ± 10 | 154 | 172 | 191 | |
| Hemoglobin (g/dl) | 15.5 ± 1.4 | 12.9 | 15.7 | 17.7 | |
| Serum creatinine (mg/dl) | 0.85 ± 0.16 | 0.51 | 0.87 | 1.20 | |
| Creatinine clearance (ml/min) | 102 ± 27 | 52.1 | 97.7 | 176.8 | |
| Viral genotype (1A/1B/1) | 16/12/8 | ||||
| ITPA phenotype (wild type/variant) | 26/10 | ||||
| IL28B genotype (CC/CT/TT/unknown) | 12/10/12/2 |
Creatinine clearance was calculated based on total body weight using the Cockcroft-Gault equation. CC, both copies of the gene contain the “C” allele; CT, one copy of the gene contains the “C” allele and the other the “T” allele; TT, both copies of the gene contain the “T” allele.
Plasma pharmacokinetics.
Of the 36 subjects, steady-state (weeks 9 to 13) intensive plasma concentration profiles were available for 28 subjects (15 in the dual-therapy group and 13 in the triple-therapy group). At the steady state, the mean (standard deviation [SD]) plasma ribavirin trough (i.e., 12-h postdose) levels were 2.79 ± 0.80 μg/ml (2.46 ± 0.58 μg/ml in the dual-therapy group [n = 16] and 3.22 ± 0.88 μg/ml in the triple-therapy group [n = 12]) (P = 0.0177). The steady-state trough levels were higher in subjects with a variant ITPA phenotype (n = 10) than in subjects with a wild-type ITPA genotype (n = 18), but the differences were not statistically significant (3.06 ± 0.83 μg/ml versus 2.64 ± 0.77 μg/ml, respectively) (P = 0.209). The non-statistically significant difference was due to the higher creatinine clearance in the variant ITPA group than in the wild-type group (110 versus 99 ml/min), which attenuates the expected differences in CLt/F and the steady-state trough concentrations.
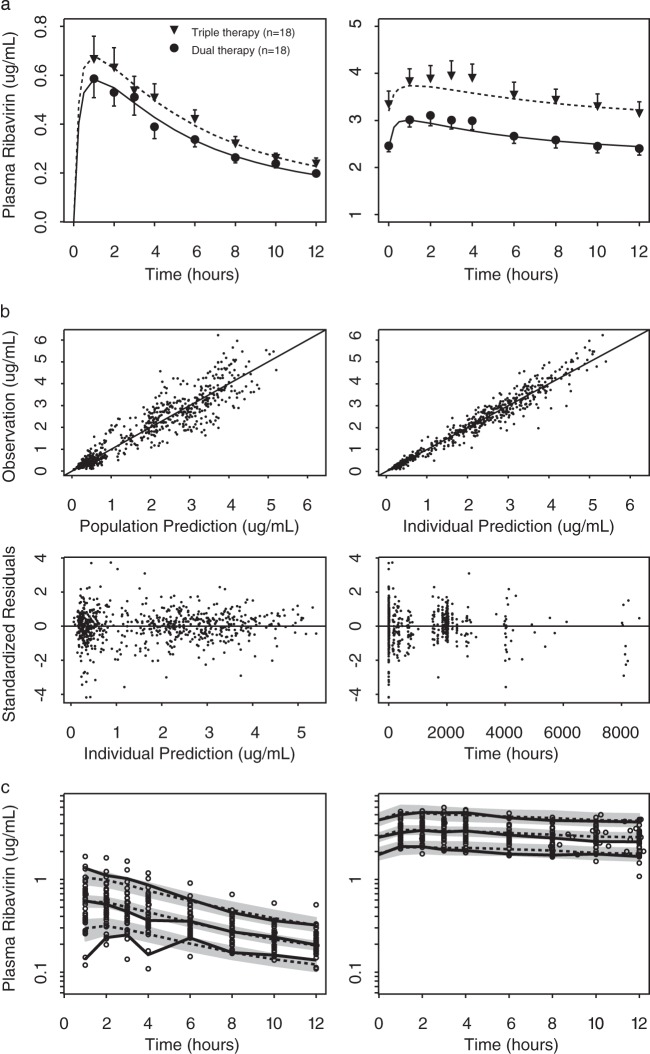
A two-compartment model with the following covariate relationships described the plasma ribavirin measurements: CLt/F = 17.0 · (CrCl/98)0.885 · 0.853T · 0.779ITPA (CrCl, creatinine clearance in ml/min; for T, dual-therapy group = 0 and triple-therapy group = 1; for ITPA, ITPA wild type = 0 and ITPA variant = 1); Vc/F = 756 · (WT/79)1.29 (WT, body weight in kg); and Vp/F = 3,460 · (WT/79)0.725 · 0.732SEX · 1.52ITPA (for SEX, males = 0 and females = 1). After incorporating these covariate relations, the values of interindividual variability of CLt/F, Vc/F, and Vp/F decreased from 33% to 23%, from 46% to 42%, and from 41% to 18%, respectively (Table 2). The population means were reliably estimated, with relative standard errors of less than 50%. However, the interindividual variability values had relative standard errors ranging from 23% to 98% (Table 2). The average individual predicted plasma concentration-time profiles for day 1 (left panel) and the steady state (right panel) are shown in Fig. 2a for the dual-therapy group (solid line) and for the triple-therapy group with telaprevir (dashed line), along with the means and standard errors of the respective measured plasma ribavirin concentrations. The plots show that the two-compartment model describes both the first-dose PK and the steady-state plasma PK and quantifies the role of telaprevir coadministration in ribavirin plasma PK. The goodness-of-fit plots in Fig. 2b (upper panels) indicate that the model describes the observations over the concentration range with no systematic prediction biases. The visual predictive check shown in Fig. 2c also confirms the model's predictive ability. The predicted 5th and 95th percentiles of the concentration-time profiles at day 1 and the steady state agree with the distribution in the observed data.
TABLE 2.
Population estimates for the parameters of the plasma ribavirin PK model without and with covariatesa
| Parameter (unit) | Estimate (relative SE) |
|
|---|---|---|
| Without covariates | With covariates | |
| AIC | 40.9969 | −20.5383 |
| CLt (liters/h) | 14.7 (7.98) | 17.0 (13.1) |
| Vc (liters) | 769 (15.7) | 756 (18.3) |
| ka (h−1) | 2.91 (32.4) | 2.58 (48.9) |
| CLd (liters/h) | 104 (11.4) | 103 (16.1) |
| Vp (liters) | 3,570 (11.1) | 3,460 (13.5) |
| Proportional error | 0.162 (2.16) | 0.162 (2.70) |
| IIV CLt | 33.2 (26.4) | 22.9 (23.1) |
| IIV Vc | 46.4 (30.8) | 41.7 (58.2) |
| IIV ka | 114 (58.7) | 95.7 (89.8) |
| IIV CLd | 42.0 (32.4) | 43.1 (43.5) |
| IIV Vp | 41.0 (33.3) | 17.8 (97.8) |
| Value representing effect of: | ||
| WT on Vc | 1.29 (50.6) | |
| WT on Vp | 0.725 (79.7) | |
| CrCL on CLt | 0.885 (30.4) | |
| Telaprevir on CLt | 0.853 (16.8) | |
| Sex on Vp | 0.732 (19.9) | |
| ITPA on CLt | 0.779 (16.9) | |
| ITPA on Vp | 1.52 (24.5) | |
IIV, interindividual.
FIG 2.
(a) Means and standard errors of ribavirin concentration-time profiles at day 1 and the steady state with and without telaprevir coadministration, overlaid with the mean of the model predictions. Black circles: dual therapy (Peg–IFN-α–2a and ribavirin). Black triangles: triple therapy (Peg–IFN-α–2a, ribavirin, and telaprevir). Solid and dashed lines: mean individual predictions for dual therapy and triple therapy, respectively. (b) Goodness-of-fit plots of the final plasma PK model. Top left: observed plasma ribavirin versus population prediction. Top right: observed plasma ribavirin versus individual prediction. Bottom left: standardized residuals versus individual prediction. Bottom right: standardized residuals versus time. (c) Visual predictive check overlaying plasma ribavirin concentration-time profiles at day 1 and the steady state (weeks 9 to 13) with simulation. Left: day 1. Right: steady state (weeks 9 to 13). Gray shaded areas and dashed lines: median and 90% prediction intervals of the median, 5th and 95th percentiles of the simulated profiles. Solid lines: median and 5th and 95th percentiles of the observed concentrations. Open circles: observed data points.
Intracellular PBMC and RBC kinetics.
Over the time course of the study in the 36 subjects, 266, 266, and 312 measurements of RMP, RDP, and RTP, respectively, were obtained in both RBCs and PBMCs. Gradual accumulation of RMP, RDP, and RTP was observed (see Fig. 3a). In PBMCs, RDP levels were lower than RMP levels, and the RTP levels were the highest. In RBCs, RDP levels were higher than RMP levels, and the RTP levels were also the highest among the three metabolites. These relative differences between PBMCs and RBCs in the abundances of RMP, RDP, and RTP were in agreement with the in vitro findings of Page and Connor (13). Our data showed that accumulation in RBCs was much more extensive than in PBMCs, which was also consistent with the in vitro results (13).
FIG 3.
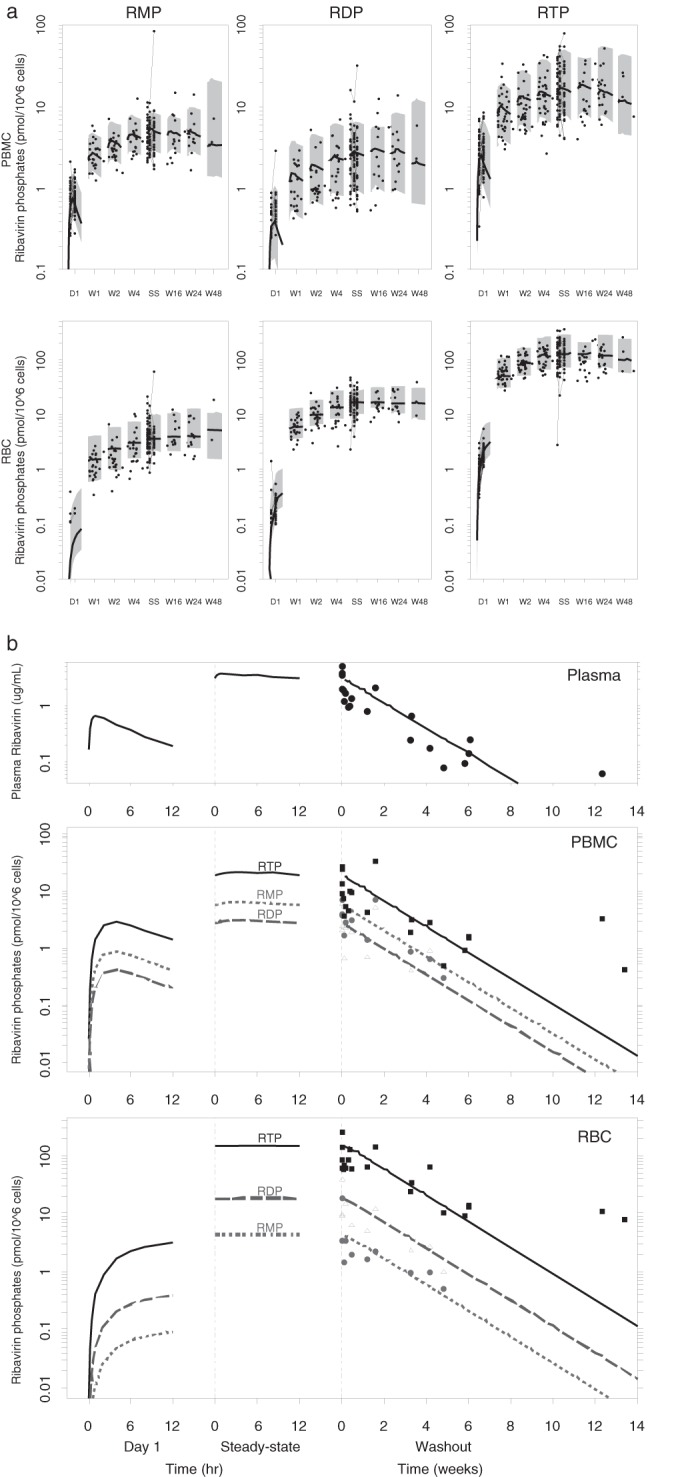
(a) Observed intracellular data overlaid with the medians (solid lines) and 90% prediction intervals (gray-shaded area) for RMP (left), RDP (middle), and RTP (right) concentration-time profiles in PBMCs (top panels) and RBCs (bottom panels). Gray circles represent the data observed within 12 h postdose at different visits. D1: day 1. W1: week 1. W2: week 2. W4: week 4. SS: steady state (weeks 9 to 13). W16: week 16. W24: week 24. W48: week 48. (b) Simulation of ribavirin plasma and intracellular concentrations after a single dose, at the steady state, and during washout, based on the typical population parameter values. Data points represent the concentrations observed after the last administered ribavirin dose (solid circles = RMP, open triangles = RDP, solid squares = RTP).
The average steady-state concentrations for RMP, RDP, and RTP were 5.70 ± 9.09, 3.21 ± 4.04, and 19.8 ± 14.3 pmol/106 cells in PBMCs and 5.00 ± 7.03, 17.2 ± 8.31, and 128 ± 71.5 pmol/106 cells in RBCs. The steady-state RTP concentrations were higher in the triple-therapy group than in the dual-therapy group: 24.0 ± 15.0 versus 16.2 ± 12.8 pmol/106 cells and 161 ± 81.0 versus 100 ± 47.0 pmol/106 cells in PBMCs and RBCs, respectively. Also, a >2-fold difference was observed in the mean steady-state RTP concentrations in subjects with the ITPA variant versus the wild type: 31.3 ± 17.0 versus 13.5 ± 7.1 pmol/106 cells and 196 ± 73 versus 91.2 ± 33.8 pmol/106 cells in PBMCs and RBCs, respectively.
Using the individual estimates of the plasma PK parameters, the intracellular RMP, RDP, and RTP measurements were fitted simultaneously, but were fitted separately for PBMCs and RBCs. The model described the accumulation of phosphorylated ribavirin over the complete duration of treatment, as well as during washout (Fig. 3). The population means were reliably estimated, with standard errors of less than 40% (Table 3). Interindividual variability ranged between 22% and 85%, with standard errors ranging from 40% to 77%. The residual errors of 31% to 50% observed for the intracellular data were higher than the 16% residual error observed for plasma ribavirin concentrations.
TABLE 3.
Population estimates for the parameters of the intracellular kinetic RBC and PMBC models without and with covariatesa
| Parameter (unit) | Estimate (relative SE) |
|||
|---|---|---|---|---|
| PBMC | PBMC with covariate | RBC | RBC with covariate | |
| AIC | 3,275.09 | 3,256.32 | 4,523.26 | 4,503.86 |
| kmp (pmol/106 cells)/(μg/mlh) | 1.11 (14.8) | 1.10 (16.2) | 0.0231 (36.6) | 0.0238 (39.3) |
| kmpout (h−1) | 0.617 (14.7) | 0.755 (18.6) | 0.0184 (28.7) | 0.0188 (29.1) |
| Rdpmp | 0.483 (14.9) | 0.379 (14.7) | 4.26 (26.8) | 3.59 (25.7) |
| Rtpdp | 6.85 (12.7) | 7.94 (20.2) | 8.18 (8.78) | 8.23 (9.08) |
| IIV kmp | 49.4 (43.9) | 49.5 (55.3) | 85.8 (39.9) | 85.1 (46.8) |
| IIV kmpout | 55.7 (27.8) | 49.4 (54.1) | 69.9 (39.7) | 70.1 (40.7) |
| IIV Rdpmp | 35.8 (40.5) | 27.6 (58.4) | 46.2 (46.0) | 37.3 (49.1) |
| IIV Rtpdp | 30.8 (34.9) | 22.2 (77.6) | 27.0 (25.7) | 27.3 (47.2) |
| Res RMP | 0.382 (9.00) | 0.390 (11.7) | 0.506 (14.6) | 0.503 (14.6) |
| Res RDP | 0.471 (9.46) | 0.465 (10.7) | 0.395 (10.2) | 0.395 (11.3) |
| Res RTP | 0.389 (5.71) | 0.395 (6.66) | 0.313 (7.23) | 0.315 (6.99) |
| Value representing effect of: | ||||
| Telaprevir on Rdpmp | 1.51 (20.2) | |||
| Telaprevir on Rtpdp | 0.669 (17.3) | |||
| ITPA on kmpout | 0.697 (31.7) | |||
| ITPA on Rdpmp | 1.28 (29.6) | 1.55 (15.7) | ||
Res, residual.
The transport/formation rate constant (kmp) was greater in PBMCs than in RBCs, reflecting the higher RMP concentrations at day 1 in PBMCs (0.82 ± 0.36) versus the mostly undetectable concentrations at day 1 in RBC samples. The kmpout parameter estimates suggested that the aggregate elimination of cellular ribavirin phosphates was more rapid in PBMCs than in RBCs, reflecting the higher degree of accumulation of ribavirin phosphates in RBCs than in PBMCs. For example, average RTP concentrations in PBMCs were 3.04 ± 1.72 at day 1 and 19.8 ± 14.3 pmol/106 cells at the steady state (6.5-fold accumulation), compared to the increase in the average RTP concentrations in RBCs from day 1 to the steady state of 1.52 ± 0.91 to 128 ± 71.5 pmol/106 cells (84-fold accumulation). Regardless of the differences between the rates of elimination of cellular ribavirin phosphates from PBMCs and RBCs determined on the basis of kmpout, the values seen with both were much higher than the typical subject's terminal elimination half-life in plasma, which was 224 h. Therefore, the intracellular kinetics of ribavirin exhibits a flip-flop phenomenon, and as a result, we observed clinically that the phosphorylated ribavirin moieties have the same terminal elimination half-life as that of ribavirin in plasma (Fig. 3b). As shown in the figure, the plasma concentrations and the RTP levels in PBMCs and RBCs measured 13 weeks postdose during washout appear to be higher than those predicted for a typical subject. These data are from two subjects who received telaprevir and who were of the ITPA variant phenotype—both factors contributed to the correspondingly longer half-lives of 350 to 400 h. Since RTP accumulation is highest in RBCs, the model predicts that it takes, on average, 14 weeks to reach an undetectable level (<0.1 pmol/106 cells). Considering the interindividual variability in the parameters, the validity of the warning in the package insert to avoid pregnancy for 6 months after the discontinuation of therapy due to the teratogenic effect associated with ribavirin is confirmed.
Similarly, because the plasma pharmacokinetics is the rate-limiting step, the times to near the steady state are also the same for plasma ribavirin and RMP, RDP, and RTP in PBMCs and RBCs. Judging on the basis of the typical population parameters, it takes about 6 weeks to reach 95% of the concentration at the steady state.
Telaprevir was found to be a significant covariate and to be associated with 1.53-times-higher Rdpmp and 0.673-times-lower Rtpdp in the PBMCs. ITPA genetics was also associated with the PBMC concentrations. The kmpout rate was 0.693 times lower and the Rdpmp was 1.31 times higher in subjects with ITPA variant phenotypes (i.e., with more ITP) than in those with a 100% functional phenotype. In RBCs, the ITPA variant phenotype was associated with a 1.57-times-higher Rdpmp. Upon inclusion of the covariates in the PBMC model, the interindividual variability of kmpout, Rdpmp, and Rtpdp decreased from 59% to 52%, from 36% to 28%, and from 31% to 22%, respectively, while in the RBC model, the effect of the inclusion of the ITPA phenotype on Rdpmp lowered the interindividual variability from 46% to 37%.
DISCUSSION
Therapy with ribavirin remains challenging due in part to high interindividual variability and the dose-limiting toxicity of hemolytic anemia that is related to the accumulation of RTP in the RBCs. To better understand the relation between plasma ribavirin and intracellular RTP, including the factors that may contribute to the interindividual variability observed clinically, we used population modeling to quantify the relationship between ribavirin plasma concentration and its phosphorylated metabolites in both RBCs and PBMCs during chronic treatment of subjects infected with genotype 1 HCV. In this first clinical study to measure and model the intracellular RMP, RDP, and RTP in PBMCs and RBCs following treatment in HCV-infected subjects, we also quantified the role of coadministration of telaprevir and the ITPA phenotype in interindividual variability. This study and the resulting model provide a framework for integrating cumulative knowledge of relationships between ribavirin dosing, systemic and intracellular kinetics, ribavirin-induced anemia, and HCV kinetics to achieve higher sustained virological response rates with less-hemolytic anemia.
Consistent with previous reports (10, 24, 25), we found that a linear two-compartment pharmacokinetic model with first-order absorption described ribavirin plasma concentration-time profiles following oral administration in the study population. The estimates for the apparent volumes of distribution for Vc/F of 756 liters and Vp/F of 3,460 liters correspond to an apparent volume of distribution at the steady state of 4,216 liters, which is within the range of reported values of 2,000 to 5,000 liters (10, 26).
Some previous studies, however, reported a difference between single-dose and multiple-dose studies in the ribavirin plasma half-life values. Glue (26) published the first report comparing a ribavirin plasma half-life value of 79 h after a single dose (27) to values of 274 to 298 h after multiple doses (28). Lertora et al. also reported ribavirin half-life values of 30 h following a single dose and 151 h after multiple doses (29). The population model from the current study described single-dose and multiple-dose data over the entire time frame of the study using the time-invariant, two-compartment model given in Table 2. Several factors may explain these differences. The half-life on day 1 in those prior studies may have been underestimated due to a limited duration of the sampling which captured distribution versus elimination kinetics and/or due to insufficient bioanalytical sensitivity to quantify samples taken during the terminal elimination phase. In our study, availability of plasma concentration accumulation values from day 1 and from weeks 1, 2, and 4 to the steady state (weeks 9 to 13) and washout after treatment cessation allowed accurate estimation of time-independent CLt/F values even though we did not have single-dose washout data. Based on the two-compartment typical-population PK parameters, ribavirin terminal half-life was calculated to be around 224 h. This is similar to the 274-to 298-h multiple-dose half-life values reported by Khakoo et al. (28) and the 288-h multiple-dose half-life stated in the package insert (30).
The current study found that CrCl had a significant effect on CLt/F, which is consistent with the conclusions reported by Bruchfeld et al. (10) and Kamar et al. (24) but not with those reported by Jin et al. (25). Since ribavirin is eliminated in part by renal excretion, with approximately 24% to 29% of the dose excreted unchanged in urine (11, 31, 32), it is expected that CrCl would be found to influence ribavirin clearance in studying a sample of patients exhibiting a range of CrCl values such as those observed in the current study. In contrast to Bruchfeld et al. (10), however, we did not find that adding body weight as a covariate for ribavirin clearance, in addition to CrCl, explained any additional interindividual variability.
The effects of telaprevir on ribavirin pharmacokinetics have not been studied previously via a formal population analysis. A prior review suggested that telaprevir has no effect on ribavirin pharmacokinetics (33). However, several studies have reported that telaprevir impaired renal function, leading to increased serum creatinine levels and thus resulting in increased ribavirin concentrations (34–36). Mauss et al. also reported that treatment with telaprevir or boceprevir was associated with a reduction in the estimation of the glomerular filtration rate (eGFR) to <60 ml/min (37). The population analysis in the current study found that telaprevir therapy was associated with a 15% decrease in plasma ribavirin clearance (see Table 2). This effect of telaprevir on ribavirin clearance is independent of its effect on CrCl. In the current study, CrCl rates decreased to similar extents in the dual-therapy and triple-therapy groups: from 112 ± 26 ml/min to 100 ± 29 ml/min at week 12 for the dual-therapy group versus from 100 ± 29 ml/min to 88 ± 21 ml/min at week 12 for the triple-therapy group. In the triple-therapy group, when telaprevir therapy was discontinued at week 12, the mean CrCl rate returned to 99 ± 22 ml/min at week 24. Telaprevir is known to inhibit the efflux transporters P-glycoprotein and breast cancer resistance protein (38, 39), and it is possible that these transporters are involved in the renal and hepatic clearance of ribavirin. Since telaprevir itself is associated with a 15% decrease in ribavirin clearance, in addition to the CrCl effect and ITPA phenotype effect on CLt/F in the model, this may explain the more frequent and severe anemia observed in patients receiving telaprevir (40).
The ITPA variant phenotype has been shown to protect against ribavirin-induced anemia, and its effect on ribavirin plasma PK has not previously been studied via a formal population analysis. Our study found that the ITPA variant phenotype was associated with 0.779-times-lower CLt/F and a 1.52-times-higher Vp/F, resulting in higher plasma concentrations in these subjects. This finding suggests that ITPA deficiency does not protect against ribavirin-induced anemia by reducing the systemic exposure of ribavirin but rather operates through some other mechanism(s).
In vitro studies have found that ribavirin nucleotide formation is dependent on extracellular ribavirin concentrations (41) and therefore, in the model, plasma concentration was assumed to drive the formation of RMP in both cell types. Also, on the basis of previous in vitro studies, uptake of ribavirin by RBC via equilibrative nucleoside transporter 1 (ENT1) was found to be saturable, reaching equilibrium rapidly (<10 min) (42). In addition, the intestinal absorption processes operating through nucleoside transporters CNT2, CNT3, ENT1, and ENT2 (43, 44), as well as the enzymatic reactions of adenosine kinase and 5′ nucleotidase, which phosphorylate ribavirin to RMP (12, 41, 45), are all saturable processes. The data in the current study, however, are not sufficient to fully characterize these nonlinear processes. Therefore, the first-order rate constant kmp in the model is used to describe the aggregate processes of cellular uptake and phosphorylation to RMP. Similarly, the parameter kmpout, which represents the rate constant for the disappearance of phosphorylated ribavirin, also represents multiple processes, including the dephosphorylation of RMP to ribavirin and transport out of the cell, the further phosphorylation to RDP and RTP, and cell lysis. In the final model for the cellular metabolism of ribavirin, it was determined that RDP and RTP rapidly reach equilibrium with RMP, which is consistent with the phosphorylation kinetic models of other nucleoside analogs following similarly sparse sampling designs (46).
In the population model for the cellular kinetics, telaprevir was found to increase Rdpmp and to reduce Rtpdp in PBMCs (see Table 3), as determined from our joint plasma PBMC population analysis, indicating a potential direct effect of telaprevir on nucleoside kinases that alters the phosphorylation/dephosphorylation equilibrium.
The modeling results also indicate that the ITPA variant phenotype is associated with a higher Rdpmp in both PBMCs and RBCs, and with a lower kmpout in PBMCs, relative to wild-type ITPA patients. Hitomi et al. proposed that decreased activity of the ITPA enzyme leads to higher levels of ITP, which can substitute for GTP in generation of AMP in a reaction catalyzed by adenylsuccinate synthase (47). Subsequently, elevated AMP levels allow increased ATP synthesis in these subjects compared to subjects with wild-type ITPA. It is possible that higher levels of ATP lead to more-extensive phosphorylation of RMP to RDP; therefore, higher values for Rdpmp were estimated in both PBMCs and RBCs of patients with the ITPA variant phenotype. Thus, the data and subsequent modeling results from the current study suggest that the associated protective attribute of the ITPA variant phenotype against ribavirin-induced anemia is not mediated through limiting the RTP accumulation. We hypothesize that this may result instead from restored ATP levels in subjects with ITPA variants.
The composite population modeling analysis of the plasma and cellular ribavirin kinetics of the patients from the current study resulted in an estimated phosphorylated ribavirin turnover half-life in RBCs of 1.5 days. The half-life of ribavirin elimination from RBC was originally reported to be approximately 40 days (48). The latter estimate, however, was based on an analysis that did not account for the contribution of plasma kinetics to the elimination time course of the cellular ribavirin measurements that were used to estimate the reported ribavirin elimination half-life. Inspection of Fig. 4 in reference 48 shows that the slope of the terminal mean ribavirin concentrations in RBCs is parallel to the terminal slope of the plasma ribavirin data in that figure. Another more recent study (49) using a modeling analysis that incorporated plasma kinetics together with total phosphorylated ribavirin concentrations in RBCs of HCV infection patients (the data came from reference 18) reported a loss rate constant of 0.5 day−1 (0.0208 h−1) (half-life = 33 h), which is consistent with our kmpout in RBCs of 0.019 h−1 (half-life = 36 h). The half-life in nucleated cells was estimated to be 1 h (13), which is shorter than in RBCs, because RBCs appear to lack the enzymes that dephosphorylate RMP. In addition, a report from an ex vivo study performed with uninfected and respiratory syncytial virus-infected cells indicated the half-lives of RTP catabolism to be in the range of 70 to 100 min (41), which agrees with our estimate of kmpout in PBMCs (65 min).
Ribavirin has been used for decades to treat HCV and other viruses despite an incomplete understanding of its clinical pharmacology. This fundamental knowledge gap has prevented a more informed use of this agent. The wide interindividual variability results in inconsistent antiviral response, and hemolytic anemia remains a challenge in many patient populations. Our report represents the first comprehensive assessment of both plasma and intracellular ribavirin kinetics in treatment-naive genotype 1 chronically HCV-infected patients. We measured RMP, RDP, and RTP concentrations in vivo in RBCs and PBMCs and, via population modeling, found that CrCl, sex, telaprevir use, and ITPA genetics were important predictors of ribavirin plasma kinetics and that use of telaprevir together with ITPA explained interindividual differences in cellular metabolism of ribavirin in PBMCs and RBCs. Integration of these pharmacokinetic findings with clinical outcomes in larger trials may ultimately provide critical guidance on the dose and duration of ribavirin in combination with direct-acting antiviral agents.
ACKNOWLEDGMENTS
We gratefully acknowledge the patients who participated in the study and Genentech and Vertex for supplying the study medications.
We gratefully acknowledge our funding support from the National Institutes of Health (K23 DK 082621 [J.J.K.], R03 DK 09612 [J.J.K.], and P41 EB 001978 [D.Z.D.]) and the Colorado CTSI (UL1 TR000154).
We declare that we have no conflicts of interest.
REFERENCES
- 1.Witkowski JT, Robins RK, Sidwell RW, Simon LN. 1972. Design, synthesis, and broad spectrum antiviral activity of 1-d-ribofuranosyl-1,2,4-triazole-3-carboxamide and related nucleosides. J Med Chem 15:1150–1154. doi: 10.1021/jm00281a014. [DOI] [PubMed] [Google Scholar]
- 2.Sidwell RW, Huffman JH, Khare GP, Allen LB, Witkowski JT, Robins RK. 1972. Broad-spectrum antiviral activity of Virazole: 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 3.Osinusi A, Meissner EG, Lee YJ, Bon D, Heytens L, Nelson A, Sneller M, Kohli A, Barrett L, Proschan M, Herrmann E, Shivakumar B, Gu W, Kwan R, Teferi G, Talwani R, Silk R, Kotb C, Wroblewski S, Fishbein D, Dewar R, Highbarger H, Zhang X, Kleiner D, Wood BJ, Chavez J, Symonds WT, Subramanian M, McHutchison J, Polis MA, Fauci AS, Masur H, Kottilil S. 2013. Sofosbuvir and ribavirin for hepatitis C genotype 1 in patients with unfavorable treatment characteristics: a randomized clinical trial. JAMA 310:804–811. doi: 10.1001/jama.2013.109309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, Weiland O, Aguilar H, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 5.Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, Svarovskaia E, Brainard DM, Symonds WT, Subramanian GM, McHutchison JG, Weiland O, Reesink HW, Ferenci P, Hezode C, Esteban R, VALENCE Investigators . 2014. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med 370:1993–2001. doi: 10.1056/NEJMoa1316145. [DOI] [PubMed] [Google Scholar]
- 6.Ferenci P, Bernstein D, Lalezari J, Cohen D, Luo Y, Cooper C, Tam E, Marinho RT, Tsai N, Nyberg A, Box TD, Younes Z, Enayati P, Green S, Baruch Y, Bhandari BR, Caruntu FA, Sepe T, Chulanov V, Janczewska E, Rizzardini G, Gervain J, Planas R, Moreno C, Hassanein T, Xie W, King M, Podsadecki T, Reddy KR, PEARL-III Study, PEARL-IV Study . 2014. ABT-450/r-ombitasvir and dasabuvir with or without ribavirin for HCV. N Engl J Med 370:1983–1992. doi: 10.1056/NEJMoa1402338. [DOI] [PubMed] [Google Scholar]
- 7.Gane EJ, Hyland RH, An D, Pang PS, Symonds WT, Hutchison JG, Stedman CA. 2014. ELECTRON-2 (ledipasvir + sofosbuvir ± ribavirin). Abstr 49th Annu Meet Eur Assoc Study Liver (EASL), abstract O46. [Google Scholar]
- 8.Hofmann WP, Herrmann E, Sarrazin C, Zeuzem S. 2008. Ribavirin mode of action in chronic hepatitis C: from clinical use back to molecular mechanisms. Liver Int 28:1332–1343. doi: 10.1111/j.1478-3231.2008.01896.x. [DOI] [PubMed] [Google Scholar]
- 9.Jen JF, Glue P, Gupta S, Zambas D, Hajian G. 2000. Population pharmacokinetic and pharmacodynamic analysis of ribavirin in patients with chronic hepatitis C. Ther Drug Monit 22:555–565. [DOI] [PubMed] [Google Scholar]
- 10.Bruchfeld A, Lindahl K, Schvarcz R, Stahle L. 2002. Dosage of ribavirin in patients with hepatitis C should be based on renal function: a population pharmacokinetic analysis. Ther Drug Monit 24:701–708. doi: 10.1097/00007691-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Preston SL, Drusano GL, Glue P, Nash J, Gupta SK, McNamara P. 1999. Pharmacokinetics and absolute bioavailability of ribavirin in healthy volunteers as determined by stable-isotope methodology. Antimicrob Agents Chemother 43:2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu JZ, Larson G, Walker H, Shim JH, Hong Z. 2005. Phosphorylation of ribavirin and viramidine by adenosine kinase and cytosolic 5′-nucleotidase II: implications for ribavirin metabolism in erythrocytes. Antimicrob Agents Chemother 49:2164–2171. doi: 10.1128/AAC.49.6.2164-2171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page T, Connor JD. 1990. The metabolism of ribavirin in erythrocytes and nucleated cells. Int J Biochem 22:379–383. doi: 10.1016/0020-711X(90)90140-X. [DOI] [PubMed] [Google Scholar]
- 14.De Franceschi L, Fattovich G, Turrini F, Ayi K, Brugnara C, Manzato F, Noventa F, Stanzial AM, Solero P, Corrocher R. 2000. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology 31:997–1004. doi: 10.1053/he.2000.5789. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl K, Schvarcz R, Bruchfeld A, Stahle L. 2004. Evidence that plasma concentration rather than dose per kilogram body weight predicts ribavirin-induced anaemia. J Viral Hepat 11:84–87. doi: 10.1046/j.1365-2893.2003.00475.x. [DOI] [PubMed] [Google Scholar]
- 16.Rendón AL, Núñez M, Romero M, Barreiro P, Martín-Carbonero L, García-Samaniego J, Jiménez-Nácher I, González-Lahoz J, Soriano V. 2005. Early monitoring of ribavirin plasma concentrations may predict anemia and early virologic response in HIV/hepatitis C virus-coinfected patients. J Acquir Immune Defic Syndr 39:401–405. doi: 10.1097/01.qai.0000170034.90438.68. [DOI] [PubMed] [Google Scholar]
- 17.Homma M, Matsuzaki Y, Inoue Y, Shibata M, Mitamura K, Tanaka N, Kohda Y. 2004. Marked elevation of erythrocyte ribavirin levels in interferon and ribavirin-induced anemia. Clin Gastroenterol Hepatol 2:337–339. doi: 10.1016/S1542-3565(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 18.Inoue Y, Homma M, Matsuzaki Y, Shibata M, Matsumura T, Ito T, Kohda Y. 2006. Erythrocyte ribavirin concentration for assessing hemoglobin reduction in interferon and ribavirin combination therapy. Hepatol Res 34:23–27. doi: 10.1016/j.hepres.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, Little LD, Qiu P, Bertelsen AH, Watson M, Warner A, Muir AJ, Brass C, Albrecht J, Sulkowski M, McHutchison JG, Goldstein DB. 2010. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature 464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 20.Bushman LR, Kiser JJ, Rower JE, Klein B, Zheng JH, Ray ML, Anderson PL. 2011. Determination of nucleoside analog mono-, di-, and tri-phosphates in cellular matrix by solid phase extraction and ultra-sensitive LC-MS/MS detection. J Pharma Biomed Anal 56:390–401. doi: 10.1016/j.jpba.2011.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimmerson LC, Ray ML, Bushman LR, Anderson PL, Klein B, Rower JE, Zheng JH, Kiser JJ. 17 December 2014, posting date Measurement of intracellular ribavirin mono-, di- and triphosphate using solid phase extraction and LC-MS/MS quantification. J Chromatogr B Analyt Technol Biomed Life Sci doi: 10.1016/j.jchromb.2014.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Argenio DZ, Schumitzky A, Wang X. 2009. ADAPT 5 user's guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA. [Google Scholar]
- 23.Ahn JE, Karlsson MO, Dunne A, Ludden TM. 2008. Likelihood based approaches to handling data below the quantification limit using NONMEM VI. J Pharmacokin Pharmacodyn 35:401–421. doi: 10.1007/s10928-008-9094-4. [DOI] [PubMed] [Google Scholar]
- 24.Kamar N, Chatelut E, Manolis E, Lafont T, Izopet J, Rostaing L. 2004. Ribavirin pharmacokinetics in renal and liver transplant patients: evidence that it depends on renal function. Am J Kidney Dis 43:140–146. doi: 10.1053/j.ajkd.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Jin R, Fossler MJ, McHutchison JG, Howell CD, Dowling TC. 2012. Population pharmacokinetics and pharmacodynamics of ribavirin in patients with chronic hepatitis C genotype 1 infection. AAPS J 14:571–580. doi: 10.1208/s12248-012-9368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glue P. 1999. The clinical pharmacology of ribavirin. Semin Liver Dis 19(Suppl 1):17–24. [PubMed] [Google Scholar]
- 27.Schering-Plough Corporation. 1998. Ribavirin. Schering-Plough Corporation, Kenilworth, NJ. [Google Scholar]
- 28.Khakoo S, Glue P, Grellier L, Wells B, Bell A, Dash C, Murray-Lyon I, Lypnyj D, Flannery B, Walters K, Dusheiko GM. 1998. Ribavirin and interferon alfa-2b in chronic hepatitis C: assessment of possible pharmacokinetic and pharmacodynamic interactions. Br J Clin Pharmacol 46:563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lertora JJ, Rege AB, Lacour JT, Ferencz N, George WJ, VanDyke RB, Agrawal KC, Hyslop NE Jr. 1991. Pharmacokinetics and long-term tolerance to ribavirin in asymptomatic patients infected with human immunodeficiency virus. Clin Pharmacol Ther 50:442–449. doi: 10.1038/clpt.1991.162. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann-La Roche Inc. 2013. Copegus package insert. Hoffmann-La Roche Inc., Nutley, NJ. [Google Scholar]
- 31.Laskin OL, Longstreth JA, Hart CC, Scavuzzo D, Kalman CM, Connor JD, Roberts RB. 1987. Ribavirin disposition in high-risk patients for acquired immunodeficiency syndrome. Clin Pharmacol Ther 41:546–555. doi: 10.1038/clpt.1987.70. [DOI] [PubMed] [Google Scholar]
- 32.Paroni R, Del Puppo M, Borghi C, Sirtori CR, Galli Kienle M. 1989. Pharmacokinetics of ribavirin and urinary excretion of the major metabolite 1,2,4-triazole-3-carboxamide in normal volunteers. Int J Clin Pharmacol Ther Toxicol 27:302–307. [PubMed] [Google Scholar]
- 33.Garg V, Kauffman RS, Beaumont M, van Heeswijk RP. 2012. Telaprevir: pharmacokinetics and drug interactions. Antivir Ther 17:1211–1221. doi: 10.3851/IMP2356. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda K, Imai Y, Hiramatsu N, Irishio K, Igura T, Sawai Y, Kogita S, Makino Y, Mizumoto R, Matsumoto Y, Nakahara M, Zushi S, Kajiwara N, Oze T, Kawata S, Hayashi N, Takehara T. 7 January 2014, posting date Renal impairment during the treatment of telaprevir with peginterferon and ribavirin in patients with chronic hepatitis C. Hepatol Res doi: 10.1111/hepr.12229. [DOI] [PubMed] [Google Scholar]
- 35.Hara T, Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S, Kumada H. 2013. A pilot study of triple therapy with telaprevir, peginterferon and ribavirin for elderly patients with genotype 1 chronic hepatitis C. J Med Virol 85:1746–1753. [DOI] [PubMed] [Google Scholar]
- 36.Karino T, Ozeki I, Hige S, Kimura M, Arakawa T, Nakajima T, Kuwata Y, Sato T, Ohmura T, Toyota J. 2014. Telaprevir impairs renal function and increases blood ribavirin concentration during telaprevir/pegylated interferon/ribavirin therapy for chronic hepatitis C. J Viral Hepat 21:341–347. doi: 10.1111/jvh.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauss S, Hueppe D, Alshuth U. 2014. Renal impairment is frequent in chronic hepatitis C patients under triple therapy with telaprevir or boceprevir. Hepatology 59:46–48. doi: 10.1002/hep.26602. [DOI] [PubMed] [Google Scholar]
- 38.Kunze A, Huwyler J, Camenisch G, Gutmann H. 2012. Interaction of the antiviral drug telaprevir with renal and hepatic drug transporters. Biochem Pharmacol 84:1096–1102. doi: 10.1016/j.bcp.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 39.Fujita Y, Noguchi K, Suzuki T, Katayama K, Sugimoto Y. 2013. Biochemical interaction of anti-HCV telaprevir with the ABC transporters P-glycoprotein and breast cancer resistance protein. BMC Res Notes 6:445. doi: 10.1186/1756-0500-6-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S, ADVANCE Study Team . 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med 364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 41.Smee DF, Matthews TR. 1986. Metabolism of ribavirin in respiratory syncytial virus-infected and uninfected cells. Antimicrob Agents Chemother 30:117–121. doi: 10.1128/AAC.30.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarvis SM, Thorn JA, Glue P. 1998. Ribavirin uptake by human erythrocytes and the involvement of nitrobenzylthioinosine-sensitive (es)-nucleoside transporters. Br J Pharmacol 123:1587–1592. doi: 10.1038/sj.bjp.0701775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patil SD, Ngo LY, Glue P, Unadkat JD. 1998. Intestinal absorption of ribavirin is preferentially mediated by the Na+-nucleoside purine (N1) transporter. Pharm Res 15:950–952. doi: 10.1023/A:1011945103455. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto T, Kuniki K, Takekuma Y, Hirano T, Iseki K, Sugawara M. 2007. Ribavirin uptake by cultured human choriocarcinoma (BeWo) cells and Xenopus laevis oocytes expressing recombinant plasma membrane human nucleoside transporters. Eur J Pharmacol 557:1–8. doi: 10.1016/j.ejphar.2006.10.062. [DOI] [PubMed] [Google Scholar]
- 45.Willis RC, Carson DA, Seegmiller JE. 1978. Adenosine kinase initiates the major route of ribavirin activation in a cultured human cell line. Proc Natl Acad Sci U S A 75:3042–3044. doi: 10.1073/pnas.75.7.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z, Rodman JH, Flynn PM, Robbins BL, Wilcox CK, D'Argenio DZ. 2006. Model for intracellular Lamivudine metabolism in peripheral blood mononuclear cells ex vivo and in human immunodeficiency virus type 1-infected adolescents. Antimicrob Agents Chemother 50:2686–2694. doi: 10.1128/AAC.01637-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hitomi Y, Cirulli ET, Fellay J, McHutchison JG, Thompson AJ, Gumbs CE, Shianna KV, Urban TJ, Goldstein DB. 2011. Inosine triphosphate protects against ribavirin-induced adenosine triphosphate loss by adenylosuccinate synthase function. Gastroenterology 140:1314–1321. doi: 10.1053/j.gastro.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 48.Catlin DH, Smith RA, Samuels AI. 1980. 14C-Ribavirin: distribution and pharmacokinetic studies in rats, baboons and man, p 83–98. In Smith RA, Kirkpatrick W (ed), Ribavirin: a broad spectrum antiviral agent. Academic Press, New York, NY. [Google Scholar]
- 49.Krishnan SM, Dixit NM. 2011. Ribavirin-induced anemia in hepatitis C virus patients undergoing combination therapy. PLoS Comput Biol 7:e1001072. doi: 10.1371/journal.pcbi.1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]