Abstract
RX-P873 is a novel antibiotic from the pyrrolocytosine series which exhibits high binding affinity for the bacterial ribosome and broad-spectrum antibiotic properties. The pyrrolocytosines have shown in vitro activity against multidrug-resistant Gram-negative and Gram-positive strains of bacteria known to cause complicated urinary tract, skin, and lung infections, as well as sepsis. Enterobacteriaceae (657), Pseudomonas aeruginosa (200), and Acinetobacter baumannii (202) isolates from North America and Europe collected in 2012 as part of a worldwide surveillance program were tested in vitro by broth microdilution using Clinical and Laboratory Standards Institute (CLSI) methodology. RX-P873 (MIC90, 0.5 μg/ml) was >32-fold more active than ceftazidime and inhibited 97.1% and 99.5% of Enterobacteriaceae isolates at MIC values of ≤1 and ≤4 μg/ml, respectively. There were only three isolates with an MIC value of >4 μg/ml (all were indole-positive Protea). RX-P873 (MIC50/90, 2/4 μg/ml) was highly active against Pseudomonas aeruginosa isolates, including isolates which were nonsusceptible to ceftazidime or meropenem. RX-P873 was 2-fold less active against P. aeruginosa than tobramycin (MIC90, 2 μg/ml; 91.0% susceptible) and colistin (MIC90, 2 μg/ml; 99.5% susceptible) and 2-fold more potent than amikacin (MIC90, 8 μg/ml; 93.5% susceptible) and meropenem (MIC90, 8 μg/ml; 76.0% susceptible). RX-P873, the most active agent against Acinetobacter baumannii (MIC90, 1 μg/ml), was 2-fold more active than colistin (MIC90, 2 μg/ml; 97.0% susceptible) and 4-fold more active than tigecycline (MIC90, 4 μg/ml). This novel agent merits further exploration of its potential against multidrug-resistant Gram-negative bacteria.
INTRODUCTION
Multidrug-resistant (MDR) Gram-negative bacilli represent a serious public health problem (1–7). It is estimated that there are more than two million infections with antibiotic-resistant bacteria and at least 23,000 deaths annually (7). The CDC report Antibiotic Resistance Threats in the United States categorizes pathogens based on threat level categories (7). Among the Gram-negative bacilli, carbapenem-resistant Enterobacteriaceae (CRE) are listed as an urgent threat. Multidrug-resistant Acinetobacter spp., Pseudomonas aeruginosa, and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae are included in the serious threat category.
RX-P873 (Fig. 1) is a novel antibiotic from a series of compounds that target a biologically conserved region in the bacterial ribosome. RX-P873 is from the pyrrolocytosine series, one of three de novo-designed molecular scaffolds with high binding affinity and properties that have been rationally optimized for broad-spectrum activity (8–10). The pyrrolocytosines have shown in vitro activities and preclinical efficacies against multidrug-resistant (MDR) Gram-negative and Gram-positive strains of bacteria known to cause complicated urinary tract, skin, and lung infections, as well as sepsis (11–15).
FIG 1.
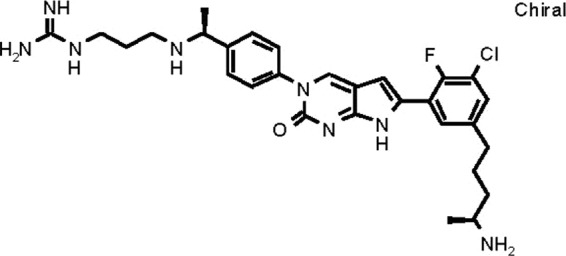
Chemical structure of RX-P873.
In this study, we evaluated the spectrum and activity of RX-P873 (a late preclinical stage compound) when tested against contemporary Enterobacteriaceae and nonfermentative Gram-negative bacilli collected in 2012 as part of a worldwide surveillance program.
(This work was presented in part at the 54th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 5 to 9 September 2014 [11].)
MATERIALS AND METHODS
Bacterial collection.
A total of 657 Enterobacteriaceae and 402 nonfermentative Gram-negative bacilli collected during 2012 as part of a global surveillance program were selected. Isolates were chosen from various medical institutions located in North America and Europe to represent the contemporary frequency distributions of antimicrobial susceptibility profiles within each organism species or genus group. We selected 202 Escherichia coli strains, 202 Klebsiella pneumoniae strains, 50 Enterobacter cloacae strains, 50 Enterobacter aerogenes strains, 51 Citrobacter freundii strains, 51 Proteus species strains, 51 Serratia marcescens strains, 200 Pseudomonas aeruginosa strains, and 202 Acinetobacter baumannii strains.
Susceptibility testing.
MIC values were determined using Clinical and Laboratory Standards Institute (CLSI) broth microdilution methodology as described in CLSI document M07-A9 (16). The compound RX-P873 and comparator agents were tested in 96-well frozen-form panels produced by JMI Laboratories (North Liberty, IA) and consisted of one reference medium type, cation-adjusted Mueller-Hinton broth. RX-P873 powder was provided by Melinta Therapeutics, Inc. (Lincolnshire, IL). Quality control (QC) strains were tested daily, and inoculum densities were monitored by colony counts. QC ranges and interpretive criteria for the comparator compounds were as published in CLSI M100-S24 (17); we used Escherichia coli ATCC 25922 and ATCC 35218 and Pseudomonas aeruginosa ATCC 27853 as the QC strains.
RX-P873.
The chemical structure of RX-P873 is shown in Fig. 1.
RESULTS
Activity against Enterobacteriaceae.
The range of RX-P873 MIC values was 0.06 to ≥32 μg/ml, with MIC50 and MIC90 values of 0.25 and 0.5 μg/ml, respectively (Table 1). Only 4 of the 657 isolates (0.6%) had MIC values of >2 μg/ml for RX-P873; these isolates include one strain (Proteus mirabilis) with an MIC of 4 μg/ml, one strain (Providencia stuartii) with an MIC of 8 μg/ml, one strain (Providencia rettgeri) with an MIC of 16 μg/ml, and one strain (P. stuartii) with an MIC of >32 μg/ml. Against the majority of isolates, RX-P873 was the second most potent agent tested (MIC90, 0.5 μg/ml) after meropenem (MIC90, ≤0.06 μg/ml; Table 2).
TABLE 1.
Cumulative MIC distribution for RX-P873 when tested against 657 Enterobacteriaceae and 402 nonfermentative Gram-negative bacilli
| Organism group (no. of strains tested) | No. of strains (cumulative %) at an MIC (μg/ml) of: |
MIC (μg/ml) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ≥32 | MIC50 | MIC90 | |
| Enterobacteriaceae (657) | 0 (0.0) | 0 (0.0) | 2 (0.3) | 188 (28.9) | 287 (72.6) | 122 (91.2) | 39 (97.1) | 15 (99.4) | 1 (99.5) | 1 (99.7) | 1 (99.8) | 1 (100.0) | 0.25 | 0.5 |
| E. coli (202) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 111 (55.9) | 78 (94.6) | 8 (98.5) | 2 (99.5) | 1 (100.0) | 0.12 | 0.25 | ||||
| K. pneumoniae (202) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 52 (25.7) | 117 (83.7) | 28 (97.5) | 2 (98.5) | 3 (100.0) | 0.25 | 0.5 | ||||
| E. cloacae (50) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (6.0) | 21 (48.0) | 23 (94.0) | 3 (100.0) | 0.5 | 0.5 | |||||
| E. aerogenes (50) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 14 (28.0) | 30 (88.0) | 5 (98.0) | 1 (100.0) | 0.25 | 0.5 | |||||
| C. freundii (51) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (9.8) | 22 (52.9) | 19 (90.2) | 4 (98.0) | 1 (100.0) | 0.25 | 0.5 | ||||
| Proteus spp. (51)a | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.0) | 4 (9.8) | 9 (27.5) | 24 (74.5) | 9 (92.2) | 1 (94.1) | 1 (96.1) | 1 (98.0) | 1 (100.0) | 1 | 2 |
| S. marcescens (51) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (3.9) | 15 (33.3) | 30 (92.2) | 3 (98.0) | 1 (100.0) | 0.5 | 0.5 | ||||
| Nonfermentative bacilli (402) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 9 (2.2) | 41 (12.4) | 85 (33.6) | 136 (67.4) | 77 (86.6) | 45 (97.8) | 9 (100.0) | 1 | 4 | ||
| P. aeruginosa (200) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 5 (3.0) | 76 (41.0) | 70 (76.0) | 39 (95.5) | 9 (100.0) | 2 | 4 | ||
| A. baumannii (202) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 9 (4.5) | 40 (24.3) | 80 (63.9) | 60 (93.6) | 7 (97.0) | 6 (100.0) | 0.5 | 1 | |||
Includes the following species (no.): Proteus mirabilis (30), Morganella morganii (11), Proteus vulgaris (4), Providencia rettgeri (4), and Providencia stuartii (2).
TABLE 2.
Activities of RX-P873 and comparator antimicrobial agents when tested against 657 Enterobacteriaceae and 402 nonfermentative bacilli
| Organism group (no. tested) and antimicrobial agent | MIC (μg/ml) |
Susceptible/intermediate/resistant (%) according to: |
|||
|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | CLSIa | EUCASTa | |
| Enterobacteriaceae (657)b | |||||
| RX-P873 | 0.25 | 0.5 | 0.06–>32 | ||
| Piperacillin-tazobactam | 2 | 32 | ≤0.5–>64 | 87.5/6.3/6.2 | 83.9/3.6/12.5 |
| Ceftriaxone | 0.06 | >2 | ≤0.015–>2 | 79.6/0.3/20.1 | 79.6/0.3/20.1 |
| Ceftazidime | 0.25 | >16 | ≤0.12–>16 | 82.3/1.9/15.8 | 79.3/3.0/17.7 |
| Cefepime | ≤0.12 | 8 | ≤0.12–>16 | 87.1/3.9/9.0 | 85.5/4.1/10.4 |
| Ciprofloxacin | 0.03 | >8 | ≤0.004–>8 | 83.1/2.0/14.9 | 81.6/1.5/16.9 |
| Colistin | 0.5 | >8 | ≤0.12–>8 | 83.3/0.0/16.7 | |
| Gentamicin | 0.5 | 2 | ≤0.06–>8 | 92.4/0.3/7.3 | 91.0/1.4/7.6 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06–>8 | 98.0/0.2/1.8 | 98.2/0.1/1.7 |
| Tigecyclinec | 0.5 | 2 | 0.06–>4 | 97.1/2.6/0.3 | 88.9/8.2/2.9 |
| E. coli (202) | |||||
| RX-P873 | 0.12 | 0.25 | 0.06–2 | ||
| Piperacillin-tazobactam | 2 | 8 | ≤0.5–>64 | 93.1/3.4/3.5 | 90.6/2.5/6.9 |
| Ceftriaxone | 0.06 | >2 | ≤0.015–>2 | 88.6/0.0/11.4 | 88.6/0.0/11.4 |
| Ceftazidime | 0.25 | 2 | ≤0.12–>16 | 93.1/1.0/5.9 | 89.1/4.0/6.9 |
| Cefepime | ≤0.12 | 2 | ≤0.12–>16 | 91.1/1.5/7.4 | 89.1/3.0/7.9 |
| Ciprofloxacin | 0.015 | >8 | 0.008–>8 | 78.2/1.0/20.8 | 77.7/0.5/21.8 |
| Colistin | 0.25 | 0.5 | 0.25–0.5 | 100.0/0.0/0.0 | |
| Gentamicin | 0.5 | 1 | ≤0.06–>8 | 93.1/0.5/6.4 | 92.1/1.0/6.9 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Tigecyclinec | 0.12 | 0.25 | 0.06–1 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| K. pneumoniae (202) | |||||
| RX-P873 | 0.25 | 0.5 | 0.12–2 | ||
| Piperacillin-tazobactam | 2 | 32 | ≤0.5–>64 | 88.1/3.0/8.9 | 83.7/4.4/11.9 |
| Ceftriaxone | 0.06 | >2 | ≤0.015–>2 | 77.7/0.0/22.3 | 77.7/0.0/22.3 |
| Ceftazidime | 0.25 | >16 | ≤0.12–>16 | 76.7/3.5/19.8 | 75.2/1.5/23.3 |
| Cefepime | ≤0.12 | >16 | ≤0.12–>16 | 79.2/4.0/16.8 | 78.7/2.5/18.8 |
| Ciprofloxacin | 0.03 | >8 | 0.008–>8 | 80.2/2.5/17.3 | 79.7/0.5/19.8 |
| Colistin | 0.5 | 1 | 0.25–>8 | 97.5/0.0/2.5 | |
| Gentamicin | 0.25 | >8 | ≤0.06–>8 | 89.1/0.0/10.9 | 87.6/1.5/10.9 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06–>8 | 95.1/0.1/5.0 | 95.1/0.4/4.5 |
| Tigecyclinec | 0.5 | 2 | 0.12–4 | 96.5/3.5/0.0 | 88.6/7.9/3.5 |
| E. cloacae (50) | |||||
| RX-P873 | 0.5 | 0.5 | 0.12–1 | ||
| Piperacillin-tazobactam | 4 | >64 | 1–>64 | 70.0/12.0/18.0 | 64.0/6.0/30.0 |
| Ceftriaxone | 1 | >2 | 0.03–>2 | 54.0/0.0/46.0 | 54.0/0.0/46.0 |
| Ceftazidime | 0.5 | >16 | ≤0.12–>16 | 60.0/2.0/38.0 | 54.0/6.0/40.0 |
| Cefepime | ≤0.12 | 8 | ≤0.12–>16 | 76.0/16.0/8.0 | 72.0/14.0/14.0 |
| Ciprofloxacin | 0.015 | 8 | 0.008–>8 | 80.0/6.0/14.0 | 80.0/0.0/20.0 |
| Colistin | 0.5 | 8 | 0.25–>8 | 88.0/0.0/12.0 | |
| Gentamicin | 0.25 | >8 | 0.12–>8 | 88.0/0.0/12.0 | 84.0/4.0/12.0 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06–2 | 98.0/2.0/0.0 | 100.0/0.0/0.0 |
| Tigecyclinec | 0.5 | 2 | 0.25–>4 | 90.0/8.0/2.0 | 84.0/6.0/10.0 |
| E. aerogenes (50) | |||||
| RX-P873 | 0.25 | 0.5 | 0.12–1 | ||
| Piperacillin-tazobactam | 4 | 64 | ≤0.5–>64 | 64.0/32.0/4.0 | 56.0/8.0/36.0 |
| Ceftriaxone | 0.25 | >2 | ≤0.015–>2 | 54.0/0.0/46.0 | 54.0/0.0/46.0 |
| Ceftazidime | 1 | >16 | ≤0.12–>16 | 54.0/2.0/44.0 | 52.0/2.0/46.0 |
| Cefepime | ≤0.12 | 2 | ≤0.12–16 | 92.0/6.0/2.0 | 88.0/10.0/2.0 |
| Ciprofloxacin | 0.015 | 0.5 | 0.008–>8 | 94.0/0.0/6.0 | 90.0/4.0/6.0 |
| Colistin | 0.25 | 0.5 | 0.25–>8 | 96.0/0.0/4.0 | |
| Gentamicin | 0.25 | 1 | 0.12–>8 | 96.0/0.0/4.0 | 96.0/0.0/4.0 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06–0.25 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Tigecyclinec | 0.5 | 1 | 0.12–4 | 98.0/2.0/0.0 | 92.0/6.0/2.0 |
| C. freundii (51) | |||||
| RX-P873 | 0.25 | 0.5 | 0.12–2 | ||
| Piperacillin-tazobactam | 2 | 64 | ≤0.5–>64 | 82.4/7.8/9.8 | 78.4/4.0/17.6 |
| Ceftriaxone | 0.25 | >2 | 0.06–>2 | 76.5/0.0/23.5 | 76.5/0.0/23.5 |
| Ceftazidime | 0.5 | >16 | 0.25–>16 | 78.4/0.0/21.6 | 72.5/5.9/21.6 |
| Cefepime | ≤0.12 | 4 | ≤0.12–>16 | 88.2/4.0/7.8 | 86.3/5.9/7.8 |
| Ciprofloxacin | 0.06 | 2 | 0.008–>8 | 86.3/3.9/9.8 | 80.4/5.9/13.7 |
| Colistin | 0.5 | 1 | ≤0.12–1 | 100.0/0.0/0.0 | |
| Gentamicin | 0.5 | 1 | 0.12–>8 | 92.2/1.9/5.9 | 90.2/2.0/7.8 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06–>8 | 96.1/0.0/3.9 | 96.1/0.0/3.9 |
| Tigecyclinec | 0.25 | 1 | 0.12–2 | 100.0/0.0/0.0 | 92.2/7.8/0.0 |
| Proteus spp. (51)d | |||||
| RX-P873 | 1 | 2 | 0.12–>32 | ||
| Piperacillin-tazobactam | ≤0.5 | ≤0.5 | ≤0.5–4 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Ceftriaxone | ≤0.015 | 0.06 | ≤0.015–>2 | 96.1/0.0/3.9 | 96.1/0.0/3.9 |
| Ceftazidime | ≤0.12 | ≤0.12 | ≤0.12–8 | 98.0/2.0/0.0 | 96.1/1.9/2.0 |
| Cefepime | ≤0.12 | ≤0.12 | ≤0.12–8 | 96.1/3.9/0.0 | 96.1/1.9/2.0 |
| Ciprofloxacin | 0.03 | 2 | ≤0.004–>8 | 88.2/2.0/9.8 | 86.3/1.9/11.8 |
| Colistin | >8 | >8 | >8 | 0.0/0.0/100.0 | |
| Gentamicin | 0.5 | 2 | 0.12–>8 | 98.0/0.0/2.0 | 96.1/1.9/2.0 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06–0.12 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Tigecyclinec | 1 | 2 | 0.12–>4 | 92.2/5.8/2.0 | 52.9/39.3/7.8 |
| Serratia marcescens (51) | |||||
| RX-P873 | 0.5 | 0.5 | 0.12–2 | ||
| Piperacillin-tazobactam | 2 | 8 | ≤0.5–64 | 96.1/3.9/0.0 | 94.1/2.0/3.9 |
| Ceftriaxone | 0.25 | 2 | 0.03–>2 | 88.2/4.0/7.8 | 88.2/4.0/7.8 |
| Ceftazidime | 0.25 | 0.5 | ≤0.12–4 | 100.0/0.0/0.0 | 98.0/2.0/0.0 |
| Cefepime | ≤0.12 | 0.25 | ≤0.12–>16 | 98.0/0.0/2.0 | 98.0/0.0/2.0 |
| Ciprofloxacin | 0.06 | 0.12 | 0.015–>8 | 98.0/0.0/2.0 | 94.1/3.9/2.0 |
| Colistin | >8 | >8 | 1–>8 | 9.8/0.0/90.2 | |
| Gentamicin | 0.5 | 1 | 0.12–>8 | 98.0/0.0/2.0 | 98.0/0.0/2.0 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06–0.25 | 100.0/0.0/0.0 | 100.0/0.0/0.0 |
| Tigecyclinec | 1 | 2 | 0.5–4 | 96.1/3.9/0.0 | 80.4/15.7/3.9 |
| P. aeruginosa (200) | |||||
| RX-P873 | 2 | 4 | 0.25–8 | ||
| Piperacillin-tazobactam | 4 | 64 | ≤0.5–>64 | 80.5/11.0/8.5 | 80.5/0.0/19.5 |
| Ceftazidime | 2 | >16 | 0.5–>16 | 83.0/4.0/13.0 | 83.0/0.0/17.0 |
| Cefepime | 2 | 16 | 0.25–>16 | 84.5/10.5/5.0 | 84.5/0.0/15.5 |
| Ciprofloxacin | 0.12 | >8 | 0.008–>8 | 78.0/2.0/20.0 | 76.0/2.0/22.0 |
| Tobramycin | 0.5 | 2 | ≤0.25–>16 | 91.0/1.5/7.5 | 91.0/0.0/9.0 |
| Amikacin | 4 | 8 | ≤0.25–>32 | 93.5/2.0/4.5 | 91.0/2.5/6.5 |
| Meropenem | 0.5 | 8 | ≤0.06–>8 | 76.0/6.0/18.0 | 76.0/14.5/9.5 |
| Colistin | 1 | 2 | 0.25–4 | 99.5/0.5/0.0 | 100.0/0.0/0.0 |
| Acinetobacter baumannii (202) | |||||
| RX-P873 | 0.5 | 1 | 0.12–4 | ||
| Ampicillin-sulbactam | 16 | >32 | 0.5–>32 | 48.0/11.4/40.6 | |
| Ceftazidime | >16 | >16 | 0.5–>16 | 38.1/2.5/59.4 | |
| Cefepime | 16 | >16 | 0.25–>16 | 36.6/13.4/50.0 | |
| Ciprofloxacin | >8 | >8 | 0.06–>8 | 36.1/0.0/63.9 | 36.1/0.0/63.9 |
| Tobramycin | 4 | >16 | ≤0.25–>16 | 53.5/3.4/43.1 | 53.5/0.0/46.5 |
| Amikacin | 8 | >32 | ≤0.25–>32 | 58.4/6.0/35.6 | 53.5/4.9/41.6 |
| Meropenem | >8 | >8 | ≤0.06–>8 | 46.0/1.5/52.5 | 45.5/2.0/52.5 |
| Colistin | 1 | 2 | 0.25–>8 | 97.0/0.0/3.0 | 97.0/0.0/3.0 |
| Tigecycline | 1 | 4 | 0.12–>4 | ||
2013 CLSI and EUCAST criteria.
Includes Citrobacter freundii (51 strains), Enterobacter aerogenes (50 strains), Enterobacter cloacae (50 strains), Escherichia coli (202 strains), Klebsiella pneumoniae (202 strains), Morganella morganii (11 strains), Proteus mirabilis (30 strains), Proteus vulgaris (4 strains), Providencia rettgeri (4 strains), Providencia stuartii (2 strains), and Serratia marcescens (51 strains).
In the absence of a CLSI breakpoint, U.S. FDA breakpoints were applied when available (22).
Includes Morganella morganii (11 strains), Proteus mirabilis (30 strains), Proteus vulgaris (4 strains), Providencia rettgeri (4 strains), and Providencia stuartii (4 strains).
Against E. coli, the range of RX-P873 MIC values was 0.06 to 2 μg/ml, with MIC50 and MIC90 values of 0.12 and 0.25 μg/ml, respectively (Tables 1 and 2). RX-P873 had a potency identical to that of tigecycline (MIC50 and MIC90, 0.12 and 0.25 μg/ml, respectively, for each agent), with meropenem being the most potent (MIC90, ≤0.06 μg/ml; Table 2). Against K. pneumoniae, the range of RX-P873 MIC values was 0.12 to 2 μg/ml, with MIC50 and MIC90 values of 0.25 and 0.5 μg/ml, respectively (Table 1). Meropenem resistances were 5.0% and 4.5% according to CLSI and EUCAST criteria, respectively, and resistances to all other comparator agents (except tigecycline) ranged from 2.5% for colistin (EUCAST criteria) to 23.3% for ceftazidime (EUCAST criteria; Table 2). RX-P873 inhibited all isolates at an MIC value of ≤2 μg/ml, which is similar to the inhibition by tigecycline (all isolates inhibited at an MIC of ≤4 μg/ml).
Against Enterobacter cloacae, the range of RX-P873 MIC values was 0.12 to 1 μg/ml, with MIC50 and MIC90 values of 0.5 and 0.5 μg/ml, respectively (Table 2). All the isolates were inhibited by meropenem at an MIC value of ≤2 μg/ml (98.0% and 100.0% susceptible according to CLSI and EUCAST interpretive criteria, respectively). Resistances (CLSI criteria) for the other agents ranged from 2.0% for tigecycline to 46.0% for ceftriaxone. For colistin, 12.0% of the isolates were found to be resistant by EUCAST interpretive criteria (Table 2). RX-P873 retained activity against all E. cloacae resistance phenotypes, inhibiting all 50 strains at an MIC value of ≤1 μg/ml. The range of RX-P873 MIC values against the 50 E. aerogenes strains was 0.12 to 1 μg/ml, with MIC50 and MIC90 values of 0.25 and 0.5 μg/ml, respectively (Table 1). All the isolates were inhibited by meropenem at an MIC value of ≤0.25 μg/ml (100.0% susceptible according to CLSI and EUCAST interpretive criteria). Resistances (CLSI criteria) for other agents ranged from 0% for tigecycline to 46.0% for ceftriaxone. For colistin, 4.0% of the isolates were resistant according to EUCAST interpretive criteria (Table 2). RX-P873 retained activity against all resistance phenotypes, inhibiting all 50 strains at an MIC value of ≤1 μg/ml.
The range of RX-P873 MIC values against 51 C. freundii strains was 0.12 to 2 μg/ml, with MIC50 and MIC90 values of 0.25 and 0.5 μg/ml, respectively (Table 1). Meropenem resistance was 3.9% according to CLSI and EUCAST criteria, and resistances (CLSI criteria) to all other comparator agents (except tigecycline) ranged from 5.9% for gentamicin to 23.5% for ceftriaxone (Table 2). RX-P873 and tigecycline inhibited all the isolates at an MIC value of ≤2 μg/ml (U.S. FDA susceptibility breakpoint). All the isolates were susceptible to colistin (EUCAST criteria). RX-P873 inhibited 47/51 (92.2%) isolates of Proteus spp. at an MIC value of ≤2 μg/ml, with MIC50 and MIC90 values of 1 and 2 μg/ml, respectively, which represent the highest MIC parameters observed for all seven species/groups tested (Table 1).The range of RX-P873 MIC values against 51 S. marcescens strains was 0.12 to 2 μg/ml, with MIC50 and MIC90 values of 0.5 and 0.5 μg/ml, respectively (Table 1).
Activities against P. aeruginosa and Acinetobacter baumannii.
Against all 402 strains of P. aeruginosa and Acinetobacter baumannii tested, the range of RX-P873 MIC values was 0.12 to 8 μg/ml, with MIC50 and MIC90 values of 1 and 4 μg/ml, respectively (Table 1).The range of RX-P873 MIC values against the 200 P. aeruginosa strains was 0.25 to 8 μg/ml, with MIC50 and MIC90 values of 2 and 4 μg/ml, respectively (Table 1). For β-lactam agents, nonsusceptibilities for the agents against P. aeruginosa ranged from 15.5% for cefepime to 24.0% for meropenem. For non-β-lactam agents, nonsusceptibilities ranged from 0.5% for colistin to 22.0% (CLSI criteria) for ciprofloxacin (Table 2). RX-P873 and colistin were the most potent agents tested. Against A. baumannii, the range of RX-P873 MIC values was 0.12 to 4 μg/ml, with MIC50 and MIC90 values of 0.5 and 1 μg/ml, respectively (Table 1). For β-lactam agents, nonsusceptibilities ranged from 52.0% for ampicillin-sulbactam to 63.4% for cefepime. Meropenem resistance was 52.5%. For non-β-lactam agents, nonsusceptibilities ranged from 3.0% for colistin to 63.9% for ciprofloxacin (Table 2). RX-P873 was the most potent agent tested.
DISCUSSION
RX-P873 was identified in a preclinical program focused on the de novo design of new classes of antibiotics that bind to an unexploited site in the large subunit (50S) of the bacterial ribosome. The program was designed to optimize the classes to treat multidrug- and extremely drug-resistant Gram-positive and Gram-negative pathogens. The research approach uses crystal structures of the bacterial ribosome to identify antibiotics that bind to the ribosome and inhibit its function. Three novel families of antibiotics with broad-spectrum activities were designed, optimized, and evaluated (fewer than 900 total compounds) to deliver broad-spectrum compounds (18).
The lead chemical series of the program, the pyrrolocytosine class, consists of broad-spectrum antibacterial agents that are bactericidal and have activities against multidrug-resistant Enterococcus spp., Staphylococcus aureus (including methicillin-resistant S. aureus [MRSA]), K. pneumoniae, A. baumannii, P. aeruginosa, and E. coli (ESKAPE pathogens) and Bacillus anthracis, Yersinia pestis, Francisella tularensis, Burkholderia mallei, and Burkholderia pseudomallei (11, 19, 20). There is a lack of cross-resistance to classes used for the treatment of Gram-negative infections and to other 50S ribosome-binding antibiotics. Efficacies for the pyrrolocytosine class have been shown in multiple murine models of infection, including skin, kidney, and lung infections, and in sepsis. RX-P873 is a lead compound in the pyrrolocytosine class (12–15, 21).
In this study, RX-P873 demonstrated a high level of activity and consistent potency against the contemporary collection of Enterobacteriaceae bacilli selected from various medical institutions located in North America and Europe. The isolates were selected to represent the frequency distributions of antimicrobial susceptibility profiles for uncomplicated and complicated urinary tract infections and complicated intra-abdominal infections. Only three of the 657 Enterobacteriaceae isolates tested exhibited an RX-P873 MIC value of >4 μg/ml (all three were indole-positive Protea). The overall MIC90 for RX-P873 (0.5 μg/ml) was >32-fold higher than that of ceftazidime (MIC90, >16 μg/ml; 82.3% susceptible). A total of 97.1% of all Enterobacteriaceae isolates exhibited an RX-P873 MIC value of ≤1 μg/ml. RX-P873 was shown to be very active against Enterobacteriaceae strains with acquired and intrinsic resistance to piperacillin-tazobactam, ceftriaxone, ceftazidime, cefepime, ciprofloxacin, colistin, gentamicin, meropenem, and tigecycline.
RX-P873 was also shown to be highly active (MIC50/90, 2/4 μg/ml) against P. aeruginosa, including strains which were not susceptible to ceftazidime or meropenem. RX-P873 was 2-fold less active than tobramycin (MIC90, 2 μg/ml; 91.0% susceptible) and colistin (MIC90, 2 μg/ml; 99.5% susceptible) and 2-fold more potent than amikacin (MIC90, 8 μg/ml; 93.5% susceptible) and meropenem (MIC90, 8 μg/ml; 76.0% susceptible). RX-P873 was the most active agent tested against Acinetobacter spp. (MIC50/90, 0.5/1 μg/ml). Its activity was 2-fold greater than that of colistin (MIC90, 2 μg/ml; 97.0% susceptible). Susceptibilities to other agents were compromised. Meropenem, tobramycin, and amikacin susceptibilities ranged from 45.5% to 53.5%. RX-P873 demonstrated activities against isolates of P. aeruginosa and A. baumannii with acquired and intrinsic resistances to piperacillin-tazobactam, ampicillin-sulbactam, ceftazidime, cefepime, meropenem, ciprofloxacin, colistin, tobramycin, amikacin, and tigecycline.
Overall, RX-P873, a novel agent targeting protein synthesis, exhibited potent activities against contemporary Gram-negative bacteria collected from patients in North American and European medical centers. These activities against Enterobacteriaceae, P. aeruginosa, and Acinetobacter baumannii, including isolates resistant to many commonly used antibacterial agents, indicate that RX-P873 merits further exploration of its potential use against multidrug-resistant Gram-negative bacteria.
ACKNOWLEDGMENTS
We thank K. Hass and M. Janechek for the preparation of the manuscript and the JMI staff members for scientific assistance in performing this study.
This study was funded under a service agreement with Melinta Therapeutics, Inc.
JMI Laboratories, Inc., received research and educational grants from 2012 to 2014 from Achaogen, Actelion, Affinium, American Proficiency Institute (API), AmpliPhi BioSciences, Anacor, Astellas, AstraZeneca, Basilea, BioVersys, Cardeas, Cempra, Cerexa, Cubist, Daiichi, Dipexium, Durata, Exela, Fedora, Forest Research Institute, Furiex, Genentech, GlaxoSmithKline, Janssen, Johnson & Johnson, Medpace, Meiji Seika Kaisha, Melinta, Merck, MethylGene, Nabriva, Nanosphere, Novartis, Pfizer, Polyphor, Rempex, Roche, Seachaid, Shionogi, Synthes, The Medicines Co., Theravance, Thermo Fisher, VenatoRx, Vertex, Waterloo, Wockhardt, and some other corporations. Some JMI employees are advisors/consultants for Astellas, Cubist, Pfizer, Cempra, Cerexa-Forest, and Theravance. In regard to speakers' bureaus and stock options, we have none to declare.
REFERENCES
- 1.Cohen J. 2013. Confronting the threat of multidrug-resistant Gram-negative bacteria in critically ill patients. J Antimicrob Chemother 68:490–491. doi: 10.1093/jac/dks460. [DOI] [PubMed] [Google Scholar]
- 2.de Kraker ME, Wolkewitz M, Davey PG, Koller W, Berger J, Nagler J, Icket C, Kalenic S, Horvatic J, Seifert H, Kaasch A, Paniara O, Argyropoulou A, Bompola M, Smyth E, Skally M, Raglio A, Dumpis U, Melbarde Kelmere A, Borg M, Xuereb D, Ghita MC, Noble M, Kolman J, Grabljevec S, Turner D, Lansbury L, Grundmann H. 2011. Burden of antimicrobial resistance in European hospitals: excess mortality and length of hospital stay associated with bloodstream infections due to Escherichia coli resistant to third-generation cephalosporins. J Antimicrob Chemother 66:398–407. doi: 10.1093/jac/dkq412. [DOI] [PubMed] [Google Scholar]
- 3.Kallen AJ, Hidron AI, Patel J, Srinivasan A. 2010. Multidrug resistance among gram-negative pathogens that caused healthcare-associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect Control Hosp Epidemiol 31:528–531. doi: 10.1086/652152. [DOI] [PubMed] [Google Scholar]
- 4.Fauci AS, Morens DM. 2012. The perpetual challenge of infectious diseases. N Engl J Med 366:454–461. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- 5.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 6.Boucher HW, Talbot GH, Benjamin DK Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Diseases Society of America . 2013. 10 × '20 progress—development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/ Accessed 24 September 2014. [Google Scholar]
- 8.Scheideman M, Bhattacharjee A, Chen S, Collin F, Dalton J, Devito J, Devivo M, Kanyo Z, Leggio M, Lou R, O'Dowd H, Paik I, Remy J, Sinishtaj S, Wimberly B, Duffy E. 2011. Completely novel antibiotics for treating multidrug-resistant Gram-negative infections: the phenoxazinocytosines, abstr F1-1844 Abstr 51st Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 9.Sinishtaj S, Bhattacharjee A, Chen S, Collin F, Dalton J, Devito J, Devivo MD, Ippolito YJ, Kanyo Z, Leggio M, Lou R, O'Dowd H, Paik I, Remy J, Scheideman M, Tang Y, Wimberly B, Duffy E. 2011. Completely novel antibiotics for treating multidrug-resistant Gram-negative infections: the isocytosines, abstr F1-1845 Abstr 51st Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 10.Bhattacharjee A, Chen S, Collin F, Dalton J, Devito J, Devivo M, Kanyo Z, Leggio M, Lou R, O'Dowd H, Paik I, Remy J, Scheideman M, Sinishtaj S, Tang Y, Wimberly B, Duffy E. 2011. Completely novel antibiotics for treating multidrug-resistant Gram-negative infections: the pyrrolocytosines, abstr F1-1846 Abstr 51st Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 11.Flamm RK, Rhomberg PR, Jones RN, Farrell DJ, Duffy E. 2014. In vitro activity of RX-P873 tested against Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter spp., abstr 1555b Abstr 54th Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 12.Marra A, Bortolon E, Molstad D, Wu Y, Jing H, Duffy E. 2011. Novel ribosome inhibitors are efficacious in a murine skin and soft tissue infection model caused by Klebsiella pneumoniae, abstr F1-1853 Abstr 51st Intersci Conf Antimicrob Agents Chemother American Society for Microbiology, Washington, DC. [Google Scholar]
- 13.Marra A, Bortolon E, Molstad D, Wu Y, Jing H, Duffy E. 2011. Novel ribosome inhibitors are efficacious in a murine kidney infection model caused by Staphylococcus aureus MRSA USA300, abstr F1-1852 Abstr 51st Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 14.Marra A, Bortolon E, Molstad D, Wu Y, Jing H, Duffy E. 2011. Novel ribosome inhibitors are efficacious in a murine peritonitis model caused by different bacterial pathogens, abstr F1-1854 Abstr 51st Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 15.Marra A, Bortolon E, Molstad D, Wu Y, Jing H, Duffy E. 2012. Novel ribosome inhibitors are efficacious in a murine respiratory tract infection model caused by Streptococcus pneumoniae, abstr F-1523 Abstr 52nd Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—9th ed CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Duffy E, Devivo M, Kanyo Z, Bhattacharjee A. 2011. The molecular tuning of RX-04, a novel broad spectrum antibacterial class, for coverage of MDR Gram-negative pathogens, abstr F1-1843 Abstr 51st Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 19.Hershfield J, Miller L, Halasohoris S, Marchand C, Remy J, Leggio M, Devito J, Bhattacharjee A, Marra A, Kanyo Z, Duffy E. 2012. Antibacterial activity of novel RX-04 compounds against biodefense pathogens, abstr F-1522 Abstr 52nd Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 20.Hershfield J, Miller L, Halasohoris S, Miller J, Schmadel H, Poinsette J, Wasbrough E, Leggio M, Kanyo Z, Duffy E. 2013. Optimization of novel RX-04 compounds against biodefense pathogens, abstr F-635 Abstr 53rd Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 21.Lamb LM, Crandon JL, Nicolau DP. 2013. Pharmacokinetic and pharmacodynamic evaluation of P-873 versus Klebsiella pneumoniae in a neutropenic murine thigh infection model. Antimicrob Agents Chemother 57:1971–1973. doi: 10.1128/AAC.02170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfizer. 2013. Tygacil product insert. Pfizer, New York, NY. [Google Scholar]


