Abstract
A novel New Delhi metallo-β-lactamase (NDM) variant, NDM-14, was identified in clinical isolate Acinetobacter lwoffii JN49-1, which was recovered from an intensive care unit patient at a local hospital in China. NDM-14, which differs from other existing enzymes by an amino acid substitution at position 130 (Asp130Gly), possesses enzymatic activity toward carbapenems that is greater than that of NDM-1. Kinetic data indicate that NDM-14 has a higher affinity for imipenem and meropenem.
TEXT
The emergence and global spread of carbapenem-resistant Enterobacteriaceae is of great concern. A novel metallo-β-lactamase (MBL), New Delhi MBL-1 (NDM-1), has attracted wide attention in recent years because it confers resistance to all classes of β-lactam antibiotics except for the monobactam aztreonam (1). NDM-1 is identified mainly in Escherichia coli, Acinetobacter spp., and Klebsiella pneumoniae. Ongoing research suggests that the gene conferring antibiotic resistance on these bacteria, blaNDM-1, is now widely spread throughout the world (2). Currently, there are 12 variants of NDM (NDM-1 to NDM-10, NDM-12, and NDM-13; NDM-11 is assigned without any information in GenBank) that differ by one, two, or five amino acid substitutions at 13 positions (see www.lahey.org/studies).
Acinetobacter lwoffii JN49-1 was isolated from the infected wound and feces of an intensive care unit (ICU) patient in Jinan, China. Identification of the isolate to the species level was carried out by using the Vitek 2 system (bioMérieux, France), 16S rRNA gene sequencing, and 16S-23S rRNA gene intergenic spacer sequencing (3). Antimicrobial susceptibility testing was performed by broth microdilution according to the Clinical and Laboratory Standards Institute (4). A. lwoffii JN49-1 was highly resistant to β-lactams, including imipenem and meropenem, and susceptible to tigecycline and colistin (Table 1). MBL detection with Etest MBL strips (bioMérieux, France) was positive. PCR screening for known β-lactamase genes and aminoglycoside resistance genes was also performed (5, 6). Interestingly, PCR product sequencing results revealed that JN49-1 carries the blaNDM and blaaac(6′)-Ib genes. Subsequent sequencing revealed that JN49-1 harbored a novel blaNDM gene with a point mutation at position 389 (A→G). Analysis of the predicted amino acid sequence showed an amino acid substitution (Asp130Gly), and it was designated NDM-14. Moreover, another NDM-1-positive A. lwoffii strain, JN247, that had a resistance pattern similar to that of JN49-1 was recovered from a different ICU patient at the same hospital (Table 1).
TABLE 1.
Antibiotic susceptibility profiles of NDM-carrying clinical isolates, transconjugants, and transformants
| Antibiotic | MIC (mg/liter) for NDM-carrying clinical isolate, transconjugant, and transformant: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| JN247 (NDM-1) | JN49 (NDM-14) | JN49-J53 | JN247-J53 | J53 | DH5α(pHSG398) | DH5α(pHSG398-NDM-1) | DH5α(pHSG398-NDM-14) | DH5α(pHSG398-NP-NDM-1) | DH5α(pHSG398-NP-NDM-14) | |
| Ampicillin | >256 | >256 | >256 | >256 | 4 | 2 | >256 | >256 | >256 | >256 |
| Ceftazidime | >256 | >256 | >256 | >256 | 0.25 | 0.25 | 32 | 32 | >256 | >256 |
| Cefotaxime | >256 | >256 | >256 | >256 | 2 | 1 | 128 | 128 | >256 | >256 |
| Meropenem | ≥32 | ≥32 | 4 | 2 | 0.023 | 0.023 | 0.094 | 0.094 | 4 | 16 |
| Imipenem | ≥32 | ≥32 | 2 | 1 | 0.19 | 0.19 | 0.25 | 0.38 | 6 | 16 |
| Aztreonam | 8 | >256 | 0.062 | 0.062 | 0.062 | 0.062 | 0.062 | 0.062 | 0.062 | 0.062 |
| Amikacin | 2 | 16 | 0.062 | 0.062 | 0.031 | 0.031 | 0.062 | 0.062 | 0.062 | 0.062 |
| Ciprofloxacin | 4 | 2 | 0.031 | 0.031 | 0.031 | 0.031 | 0.031 | 0.031 | 0.031 | 0.031 |
| Tigecycline | 0.125 | 0.125 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 |
| Colistin | 1 | 1 | 0.25 | 0.125 | 0.062 | 0.015 | 0.015 | 0.015 | 0.031 | 0.031 |
The horizontal-transfer capability of the blaNDM gene was assessed by broth and filter mating by using a standard E. coli J53 azide-resistant strain as the recipient. MacConkey agar containing 100 mg/liter sodium azide and 0.5 mg/liter meropenem was used to select for E. coli J53 transconjugants (7; see Materials and Methods in the supplemental material). Putative transconjugants were confirmed by blaNDM detection by PCR assay as described above.
Southern blot analysis was performed to locate the blaNDM genes by using specific blaNDM digoxigenin-labeled probes (Roche) (8). K. pneumoniae ATCC BAA-2146 was used as a positive control, and E. coli J53 was used as a negative control. Plasmid DNA was extracted and sequenced by the Ion Torrent sequencing platform (9).
To compare the relative contributions of NDM-1 and NDM-14 to carbapenem resistance, the entire open reading frame (ORF) (primers NDM-F [5′-CGGGATCCATGGAATTGCCCAATATTATG-3′] and NDM-R [5′-CCCAAGCTTTCAGCGCAGCTTGTCGGCCAT-3′]) and the complete gene with its native promoter (primers NP-NDM-F [5′-CGGGATCCCACCTCATGTTTGAATTCGC-3′] and NP-NDM-R [5′-CCCAAGCTTCTCTGTCACATCGAAATCGC-3′]) were amplified and cloned into the corresponding sites of pHSG398 (TaKaRa Bio). E. coli DH5α cells were transformed with pHSG398-NDM-1, pHSG398-NDM-14, pHSG398-NP-NDM-1, and pHSG398-NP-NDM-14 to determine β-lactam MICs (10, 11).
The ORFs of NDM-1 and NDM-14 without signal peptide regions were cloned into expression vector pET28a with primers BamHI-TEV-NDM-F (5′-CGGGATCCGAAAACCTGTATTTCCAAGGCCAGCAAATGGAAACTGGCGAC-3′) and XhoI-NDM-R (5′-CCGCTCGAGTCAGCGCAGCTTGTCGGCCATG-3′) (11).
E. coli BL21(DE3) was used to express the recombinant NDM proteins, which were purified by using nickel-nitrilotriacetic acid (Ni-NTA) agarose according to the manufacturer's instructions (Qiagen). His tags were cleaved with the TurboTEV protease (Accelagen, San Diego, CA), and the tags and protease were removed by an additional passage over Ni-NTA agarose. The purity of the recombinant NDM proteins was estimated up to 90% by SDS-PAGE. The protein concentration was measured with the Pierce bicinchoninic acid protein assay kit (Thermo Scientific). The hydrolysis rates were monitored in 50 mM phosphate buffer (pH 7.0) at 37°C with a SpectraMax 190 microplate reader (Molecular Devices). Km and kcat values and kcat/Km ratios were determined by using a Lineweaver-Burk plot. Wavelengths and extinction coefficients for β-lactam substrates have been reported previously (12–14).
The blaNDM-1 (in A. lwoffii JN247) and blaNDM-14 (in A. lwoffii JN49-1) genes were successfully transferred into E. coli J53 by filter mating with a low transfer frequency of approximately 1.0 × 10−8. No transfer was observed by broth mating. Transconjugation assays suggested that blaNDM might be located on a plasmid, and the E. coli J53 transconjugants carrying blaNDM from these two E. coli isolates were named JN49-J53 and JN247-J53. Both isolates exhibited resistance to ampicillin, ceftazidime, and cefotaxime and susceptibility to aztreonam. Two isolates showed different susceptibilities to meropenem and imipenem; JN49-J53 was resistant to meropenem and intermediately susceptible to imipenem, whereas JN247-J53 was intermediately susceptible to meropenem and susceptible to imipenem (Table 1).
A Southern blot assay showed that each isolate harbored multiple plasmids, yet only one plasmid in each isolate was positive for the blaNDM probe (Fig. 1). Further, these data clearly indicated that blaNDM was located on a plasmid of approximately 40 kb. The plasmid harboring blaNDM-14 from strain JN49-1 was named pNDM-JN01, and the plasmid harboring blaNDM-1 from strain JN247 was named pNDM-JN02.
FIG 1.
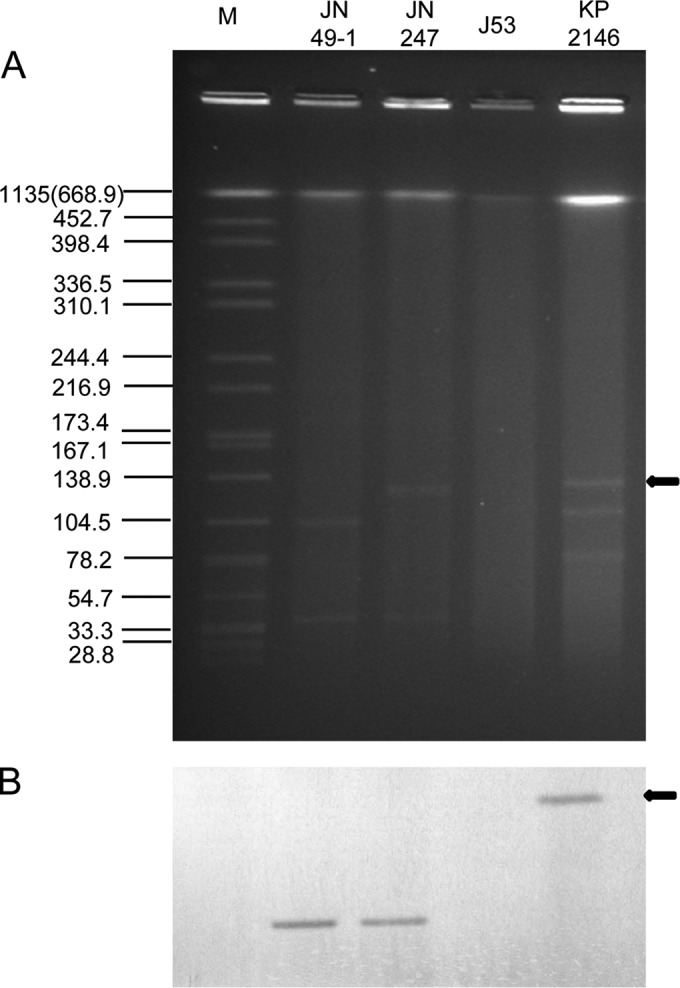
Identification of blaNDM-positive plasmids. (A) S1 nuclease plasmid pulsed-field gel electrophoresis profiles. (B) Southern blot hybridization for blaNDM. The arrows on the right indicate the positive control, K. pneumoniae ATCC BAA-2146 carrying an NDM-1-positive plasmid with a size of 140,825 bp. Lane M, reference standard strain H9812 restricted with XbaI. The molecular sizes on the left are in kilobases.
Plasmid sequencing revealed that pNDM-JN01 (harboring blaNDM-14) is 41,084 bp long with a GC content of 38%. Plasmid pNDM-JN02 (harboring blaNDM-1) was identical to pNDM-JN01, with the exception of a single nucleotide change in the ndm gene. A BLAST search showed that pNDM-JN01/pNDM-JN02 was similar to previously published plasmid pNDM-BJ01 (15). Sequence comparison revealed that a 6,187-bp region containing the genes groS, groEL, and insE and insertion sequence ISAba125 was absent from pNDM-JN01 and pNDM-JN02. We also found the same deletion in the blaNDM-1 downstream region in pNDM-44551 (GenBank accession no. KF208467.1) from A. pittii 445511 with a partial sequence in the NCBI database (Fig. 2). It was probable that pNDM-JN01, harboring the novel blaNDM-14 gene, evolved from pNDM-BJ01.
FIG 2.
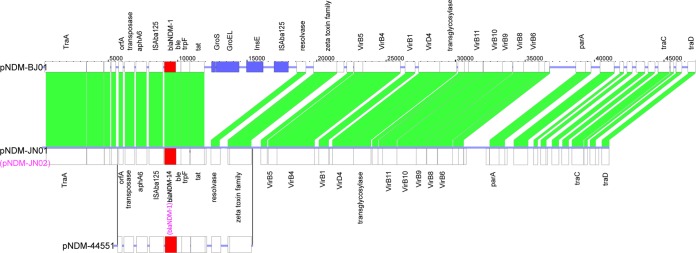
Sequence comparison of blaNDM-harboring plasmids pNDM-BJ01, pNDM-JN01/pNDM-JN02, and pNDM-44551 (only partial sequences of pNDM-44551 were accessible in the NCBI database). Boxes indicate ORFs identified by sequence analysis, and all regions are drawn to scale. Similar structures and high sequence homology are indicated by white boxes. Blue boxes indicate ORFs found only in pNDM-BJ01. Solid lines represent highly homologous sequences. The blaNDM genes are represented by red boxes.
Twelve NDM variants have been reported in several different countries (see www.lahey.org/studies). Upon sequence analysis of all of the variants, we determined that blaNDM-14 had a close relationship with blaNDM-8, as NDM-14 had only one amino acid difference (Asp130Gly) and NDM-8 had two differences (Asp130Gly and Met154Lys) from NDM-1.
It is interesting that while E. coli DH5α transformants without their native promoter (pHSG398-NDM-1 and pHSG398-NDM-14) exhibited resistance to ampicillin, ceftazidime, and cefotaxime but showed susceptibility to meropenem and imipenem, other transformants (pHSG398-NP-NDM-1 and pHSG398-NP-NDM-14) exhibited resistance to all β-lactams, including meropenem and imipenem, only when expressed under the control of their native promoters (Table 1). This is consistent with previous reports (10, 16, 17).
The most intriguing finding from our observations was that E. coli DH5α carrying pHSG398-NP-NDM-14 conferred meropenem and imipenem resistance higher than that of E. coli DH5α carrying pHSG398-NP-NDM-1 (Table 1). It was concluded that the differences in carbapenem MICs were caused by mutations outside the promoter region. As was the case for other MBLs, NDM-14 hydrolyzed all of the β-lactams tested except aztreonam (Table 2). Kinetic data showed that NDM-14 had an affinity for imipenem, meropenem, and ampicillin higher than that of NDM-1, with Km values reduced by 18 μM for imipenem, 16 μM for meropenem, and 74 μM for ampicillin, whereas slightly lower affinities of NDM-14 than of NDM-1 for cefotaxime, ceftazidime, and cefuroxime were observed. In addition, NDM-14 had an affinity for penicillin G significantly lower than that of NDM-1, with Km values of 186 and 58 μM for NDM-14 and NDM-1, respectively. Small differences (1- to 2-fold) between the kcat/Km values of NDM-1 and NDM-14 were observed (Table 2) (16, 18).
TABLE 2.
Kinetic parameters of NDM-14 and NDM-1 enzymesa
| β-Lactam | NDM-14 |
NDM-1 |
||||
|---|---|---|---|---|---|---|
| Km (μM)b | kcat (s−1)b | kcat/Km (μM−1s−1) ratio | Km (μM)b | kcat (s−1)b | kcat/Km (μM−1s−1) ratio | |
| Ampicillin | 80 ± 14 | 102 ± 7 | 1.28 | 154 ± 10 | 182 ± 5 | 1.18 |
| Penicillin G | 186 ± 9 | 230 ± 10 | 1.24 | 58 ± 5 | 142 ± 8 | 2.45 |
| Cefotaxime | 46 ± 5 | 63 ± 4 | 1.37 | 26 ± 4 | 49 ± 3 | 1.88 |
| Ceftazidime | 72 ± 4 | 16 ± 1 | 0.22 | 51 ± 3 | 22 ± 1 | 0.43 |
| Cefuroxime | 44 ± 2 | 15 ± 1 | 0.34 | 31 ± 1 | 20 ± 1 | 0.65 |
| Aztreonam | NHc | NH | NH | NH | NH | NH |
| Imipenem | 90 ± 4 | 42 ± 2 | 0.47 | 108 ± 5 | 58 ± 3 | 0.54 |
| Meropenem | 53 ± 2 | 60 ± 3 | 1.13 | 69 ± 7 | 72 ± 9 | 1.04 |
The proteins were initially modified by adding a His tag, which was removed after purification.
Values are means from three independent experiments ± standard deviations.
NH, no hydrolysis was detected under conditions with substrate concentrations of up to 1 mM and enzyme concentrations of up to 700 nM.
The amino acid substitution at position 130 (Asp130Gly) appears to confer higher carbapenemase activity, even though it is located outside the active center. This substitution is similar to those in NDM-7 (Asp130Asn and Met154Lys) and NDM-8 (Asp130Gly and Met154Lys) (10, 18). It remains unclear which sites play an important role in enzymatic activity. The crystal structure of NDM-1 shows that the active site of NDM-1 is decided at the bottom of a shallow groove enclosed by two important loops, L3 and L10 (19, 20). However, residue 130 is not located in these loops. We suggest that the residue may have an indirect effect on the formation of the active site. It has previously been proven that NDM-7 has increased carbapenemase activity (10). Thus, these data suggest that amino acid substitutions at position 130 contribute to the carbapenemase activity of NDM proteins.
In summary, we identified a novel NDM variant in A. lwoffii, NDM-14, possessing increased carbapenemase activity. In addition, another A. lwoffii isolate carrying blaNDM-1 was confirmed. Two blaNDM-positive plasmids, which were extracted from clinical isolates JN49-1 and JN247, harbored nearly identical sequences (one nucleotide difference between the blaNDM-1 genes). Taken together, these data suggest that the emergence of Acinetobacter spp. with similar NDM-positive plasmids promotes dissemination of the blaNDM gene, resulting in antibiotic resistance.
Nucleotide sequence accession numbers.
Plasmid pNDM-JN01 and pNDM-JN02 sequences have been deposited in the GenBank database under accession numbers KM210086 and KM210088, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Mega-Projects of Science and Technology Research of China (grants 2011ZX10004-001 and 2013ZX10004-203), the National Natural Science Foundation of China (grants 31370093 and 81201320), and the National High Technology Research and Development Program of China (863 Program grant SS2014AA022210).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.05168-14.
REFERENCES
- 1.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D, Rolain J. 2014. New Delhi metallo-beta-lactamase around the world: an eReview using Google Maps. Euro Surveill 19:20809 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20809. [DOI] [PubMed] [Google Scholar]
- 3.Fu Y, Du X, Ji J, Chen Y, Jiang Y, Yu Y. 2012. Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J Antimicrob Chemother 67:2114–2122. doi: 10.1093/jac/dks192. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing: 24th informational supplement M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 5.Denisuik AJ, Lagacé-Wiens PR, Pitout JD, Mulvey MR, Simner PJ, Tailor F, Karlowsky JA, Hoban DJ, Adam HJ, Zhanel GG. 2013. Molecular epidemiology of extended-spectrum β-lactamase-, AmpC β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007-11. J Antimicrob Chemother 68:i57–i65. doi: 10.1093/jac/dkt027. [DOI] [PubMed] [Google Scholar]
- 6.Akers KS, Chaney C, Barsoumian A, Beckius M, Zera W, Yu X, Guymon C, Keen EF, Robinson BJ, Mende K. 2010. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J Clin Microbiol 48:1132–1138. doi: 10.1128/JCM.02006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 8.Borgia S, Lastovetska O, Richardson D, Eshaghi A, Xiong J, Chung C, Baqi M, McGeer A, Ricci G, Sawicki R, Pantelidis R, Low DE, Patel SN, Melano RG. 2012. Outbreak of carbapenem-resistant Enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin Infect Dis 55:e109–e117. doi: 10.1093/cid/cis737. [DOI] [PubMed] [Google Scholar]
- 9.Merriman B, Torrent I, Rothberg JM, Team D. 2012. Progress in Ion Torrent semiconductor chip based sequencing. Electrophoresis 33:3397–3417. doi: 10.1002/elps.201200424. [DOI] [PubMed] [Google Scholar]
- 10.Göttig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. 2013. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J Antimicrob Chemother 68:1737–1740. doi: 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 11.Tada T, Miyoshi-Akiyama T, Shimada K, Kirikae T. 2014. Biochemical analysis of metallo-beta-lactamase NDM-3 from a multidrug-resistant Escherichia coli strain isolated in Japan. Antimicrob Agents Chemother 58:3538–3540. doi: 10.1128/AAC.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boschi L, Mercuri PS, Riccio ML, Amicosante G, Galleni M, Frère J-M, Rossolini GM. 2000. The Legionella (Fluoribacter) gormanii metallo-β-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 β-lactamases. Antimicrob Agents Chemother 44:1538–1543. doi: 10.1128/AAC.44.6.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Queenan AM, Shang W, Flamm R, Bush K. 2010. Hydrolysis and inhibition profiles of β-lactamases from molecular classes A to D with doripenem, imipenem, and meropenem. Antimicrob Agents Chemother 54:565–569. doi: 10.1128/AAC.01004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. 1998. Overexpression, purification, and characterization of the cloned metallo-β-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 42:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makena A, Brem J, Pfeffer I, Geffen RE, Wilkins SE, Tarhonskaya H, Flashman E, Phee LM, Wareham DW, Schofield CJ. 2015. Biochemical characterization of New Delhi metallo-beta-lactamase variants reveals differences in protein stability. J Antimicrob Chemother 70:463–469. doi: 10.1093/jac/dku403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tada T, Miyoshi-Akiyama T, Dahal RK, Sah MK, Ohara H, Kirikae T, Pokhrel BM. 2013. NDM-8 metallo-beta-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob Agents Chemother 57:2394–2396. doi: 10.1128/AAC.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green VL, Verma A, Owens RJ, Phillips SE, Carr SB. 2011. Structure of New Delhi metallo-beta-lactamase 1 (NDM-1). Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1160–1164. doi: 10.1107/S1744309111029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Hao Q. 2011. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J 25:2574–2582. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


