Abstract
Moraxella catarrhalis is a common pathogen of the human respiratory tract. Multidrug efflux pumps play a major role in antibiotic resistance and virulence in many Gram-negative organisms. In the present study, the role of the AcrAB-OprM efflux pump in antibiotic resistance was investigated by constructing mutants that lack the acrA, acrB, and oprM genes in M. catarrhalis strain O35E. We observed a moderate (1.5-fold) decrease in the MICs of amoxicillin and cefotaxime and a marked (4.7-fold) decrease in the MICs of clarithromycin for acrA, acrB, and oprM mutants in comparison with the wild-type O35E strain. Exposure of the M. catarrhalis strains O35E and 300 to amoxicillin triggered an increased transcription of all AcrAB-OprM pump genes, and exposure of strains O35E, 300, and 415 to clarithromycin enhanced the expression of acrA and oprM mRNA. Inactivation of the AcrAB-OprM efflux pump genes demonstrated a decreased ability to invade epithelial cells compared to the parental strain, suggesting that acrA, acrB, and oprM are required for efficient invasion of human pharyngeal epithelial cells. Cold shock increases the expression of AcrAB-OprM efflux pump genes in all three M. catarrhalis strains tested. Increased expression of AcrAB-OprM pump genes after cold shock leads to a lower accumulation of Hoechst 33342 (H33342), a substrate of AcrAB-OprM efflux pumps, indicating that cold shock results in increased efflux activity. In conclusion, the AcrAB-OprM efflux pump appears to play a role in the antibiotic resistance and virulence of M. catarrhalis and is involved in the cold shock response.
INTRODUCTION
Moraxella catarrhalis colonizes the mucosal surface of the human nasopharynx and is a major cause of acute otitis media in children and of exacerbations of chronic obstructive pulmonary disease in adults (1–4). The proportion of cases of acute otitis media caused by M. catarrhalis varies between 5% and 20%, with recent studies showing a relative increase in M. catarrhalis-caused otitis media since the introduction of routine infant immunization with the pneumococcal conjugate vaccine (2, 4–6). Furthermore, clinical studies revealed that the prevalence of pharyngeal colonization and respiratory tract infections caused by M. catarrhalis displays seasonal variation and increases in winter (7–10). The human nasopharyngeal flora is recurrently exposed to rapid downshifts of environmental temperature. Breathing cold air (e.g., −1°C at 10 to 20 liters/min) reduces the nasopharyngeal temperature from 34°C at room temperature to about 26°C within several minutes and for extended periods (11). Such rapid variation of temperature induces adaptive events in the residential upper respiratory tract flora that may contribute to the transition from asymptomatic colonization to infection. Our previous in vitro studies demonstrated that a 26°C cold shock upregulates the expression of important virulence traits, such as adherence to epithelial cells, iron acquisition, complement resistance, and immune evasion (12–14).
Adaptive resistance also involves a temporary increase in the ability of a bacterium to survive exposure to antimicrobials due to alterations in gene/protein expression as a result of an environmental trigger, e.g., temperature, stress, nutrient conditions, or subinhibitory levels of the antibiotics themselves (15). One of the main antimicrobial resistance strategies of bacteria is altered porin expression to limit intracellular access of antibiotics. Recently, we showed that M. catarrhalis responds to exposure to aminopenicillins by reducing the expression level of the porin M35, thereby developing adaptive resistance to these antibiotics (16). Porin M35 is also regulated by temperature, being downregulated during growth at 26°C compared to growth at 37°C.
Bacterial efflux is another important mechanism of antimicrobial resistance, and bacterial efflux pumps of the resistance-nodulation-division (RND) family confer intrinsic resistance to multiple, structurally distinct, clinically relevant classes of antimicrobials, including the β-lactams, quinolones, and aminoglycosides (17). The AcrAB-OprM tripartite efflux system is the major RND efflux system found in M. catarrhalis (18) and other Gram-negative bacteria (17). The pump is composed of an inner membrane RND pump (AcrB), an outer membrane channel (OprM), and a periplasmic adaptor protein (AcrA). Some studies suggest that overexpression of AcrAB is a marker of multidrug resistance (19). The multidrug-resistant phenotype of carbenicillin-resistant clinical isolates of Pseudomonas aeruginosa can be explained as the consequence of the overexpression of multidrug efflux systems (20). However, the role, if any, that the AcrAB-OprM efflux pump plays in M. catarrhalis respiratory infections has not been investigated. Our recently performed transcriptome sequencing (RNA-seq) data analysis demonstrates that the expressions of genes encoding membrane fusion proteins of the RND family multidrug efflux pump (acrA and acrB) and the RND system membrane channel OprM (oprM) were considerably increased after exposure of M. catarrhalis to a 26°C cold shock (21). Therefore, consistent with these reports, it is possible that a 26°C cold shock may also influence the susceptibility of M. catarrhalis to several antimicrobial agents through the induction of the membrane multidrug efflux pump proteins AcrA, AcrB, and OprM.
The major aim of this study was to determine the mechanism by which the AcrAB-OprM efflux pump is involved in the susceptibility to antimicrobials. Consideration of the inducible expression of AcrAB-OprM by a cold shock led us to examine the implication of the efflux system in adaptive resistance.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. catarrhalis strain O35E and clinical isolates 300 and 415 were described elsewhere (12, 14). The collection of 16 M. catarrhalis strains (middle ear isolates from children with acute otitis media and nasopharyngeal isolates) was provided by R. Dagan, Israel, and G. A. Syrogiannopoulos, Greece. Bacteria were cultured at 37°C and 200 rpm in brain heart infusion (BHI) broth (Difco, Detroit, MI) or on BHI agar plates in an atmosphere containing 5% CO2. Cold shock experiments were performed as described previously (12). Bacteria were grown overnight at 37°C, resuspended in fresh medium, and grown to the mid-logarithmic phase (optical density at 600 nm [OD600] of 0.3). Subsequently, bacteria were exposed to 26°C or 37°C for 3 h.
For analysis of the effects of amoxicillin and clarithromycin, bacteria were cultured in BHI broth to an OD600 of 0.18. Afterward, 60 μg/ml of amoxicillin (Sigma, St. Louis, MO) or 0.01 μg/ml clarithromycin was added, and bacteria were cultured for an additional 4 h (16). To quantitate viable M. catarrhalis at various clarithromycin concentrations, bacteria were cultured at different concentrations (0 μg/ml, 0.01 μg/ml, 0.1 μg/ml, 0.5 μg/ml, and 1 μg/ml) for 4 h, and the OD600 and quantitation of CFU were determined at different time points.
DNA methods.
Plasmids were isolated using the Wizard Plus SV miniprep DNA purification system (Promega Corporation, Madison, WI). Escherichia coli DH5α was transformed as described previously (22). Restriction enzymes were purchased from New England BioLabs (Beverly, MA). Naturally competent M. catarrhalis was prepared and DNA was transformed as described previously (21, 23). The M. catarrhalis strains (middle ear and nasopharyngeal isolates) were analyzed for the presence of acrA, acrB, and oprM by PCR using forward and reverse primers (acrA_2F and acrA_2R, acrB_2F and acrB_2R, and oprM_2F and oprM_2R, respectively) and visualized by 1% agarose gel electrophoresis (Table 1).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Procedure | Sequence (5′ to 3′) |
|---|---|---|
| acrA_2F | PCR | CGTGTTGCCGCCATTGAGACTT |
| acrA_2R | PCR | GGGAACACAGCGCGTAAGGTCA |
| acrB_2F | PCR | GTACTTGCCATTGGCCTTTT |
| acrB_2R | PCR | AGGGCTTCATGACCTACACC |
| oprM_2F | PCR | CTGGATTTATGGGGCAAAGT |
| oprM_2R | PCR | GGCATTTTGAATGGCTTTTT |
| 16S RNA_F | PCR | AAGGTTTGATC(AC)TGG(CT)TCAG |
| 16S RNA_R | PCR | CTTTACGCCCA(AG)T(AG)A(AT)TCCG |
| acrA_F | Cloning | GCTGGTCATTGGGCTGAT |
| acrA_R | Cloning | GTTTCGTCAGCAGCGTTCAT |
| acrB_F | Cloning | ATCCTATCAGCCGTGTGGAG |
| acrB_R | Cloning | ACACCATCGAAGACACACCA |
| oprM_F | Cloning | TGCACGTATCTTGACGCTCT |
| oprM_R | Cloning | TGGGATTTTTCGTCATCCAT |
| acrA_F_SYBR green | SYBR green | CTCGAAACTGTGCCAGTGAT |
| acrA_R_SYBR green | SYBR green | ATCAATAATGCCCGTCACCT |
| acrB_F_SYBR green | SYBR green | TCGCTTGAGCAACAAAAATC |
| acrB_R_SYBR green | SYBR green | TACAGTGCTGCCAAACACAA |
| oprM_F_SYBR green | SYBR green | GCCAAGTCTACAAGCAGCAA |
| oprM_R_SYBR green | SYBR green | ACCAATCAGCAGCTGTAACG |
| 16S RNA _F_SYBR green | SYBR green | CAATGGGCGAAAGCCTGAT |
| 16S RNA _R_SYBR green | SYBR green | GTGCTTTACAACCAAAAGGCCT |
Construction of the isogenic mutants O35E.acrA, O35E.acrB, and O35E.oprM.
The AcrAB-OprM efflux system is encoded by the acrA acrB oprM operon. Parts of the acrA, acrB, and oprM genes of strain O35E were amplified using forward and reverse primers acrAF1 and acrAR1, acrBF1 and acrBR1, and oprMF1and oprMR1, respectively (Table 1). PCR products were ligated into the EcoRI restriction site of pGEM-T Easy (Promega). The kanamycin cassette from the pUC4K vector (Pharmacia, Sweden) was ligated into the HindIII restriction site of acrA, into the BamHI restriction site of acrB, and into the HpaI restriction site of oprM. The resulting constructs, ΔacrA::kan, ΔacrB::kan, and ΔoprM::kan, were used for natural transformation of the competent strain O35E. Transformants were selected on BHI agar plates containing 20 μg/ml of kanamycin. Insertional inactivation of each gene was confirmed by PCR analysis and by reverse transcriptase PCR (RT-PCR). We screened a number of kanamycin-resistant mutants in which we examined the expression of other genes by isolating the RNA and performing a PCR analysis for the appropriate gene. For subsequent experiments, we selected mutant strains without downstream effects of the kanamycin cassette.
RNA preparation and quantitative reverse transcriptase PCR (qRT-PCR) assays.
M. catarrhalis cultures were fixed with 2 volumes of RNA Protect (Qiagen, Valencia, CA) and harvested. RNA was isolated using the RNeasy kit (Qiagen) as described elsewhere (12). Residual DNA in the samples was removed using DNase I. The reverse transcription step was carried out using the Superscript II cDNA synthesis kit (Invitrogen, Basel, Switzerland) according to the manufacturer's instructions. Oligonucleotide primer pairs (Table 1) were designed for use in qRT-PCR with either PrimerExpress software (Applied Biosystems, Foster City, CA) or primer 3 (24). To assess DNA contamination, RNA samples were also run without RT. The constitutively expressed 16S rRNA gene was used as an internal control for relative quantification. Reactions for qRT-PCR were completed using Power SYBR green PCR master mix (Applied Biosystems) with a two-step reaction protocol consisting of 40 cycles of 94°C for 30 s and 60°C for 1 min, followed by a dissociation phase for quality control. The 25-μl qRT-PCR mixtures contained 0.2 μM specific primers, Power SYBR green (2×), and 2 μl of cDNA (10 ng/μl). Relative quantification of gene expression was performed using the comparative threshold method. The ratios obtained after normalization were expressed as folds of change compared with untreated samples or samples isolated from bacteria exposed to 37°C.
Antimicrobial resistance testing.
The MICs of amoxicillin, cefotaxime, clarithromycin, amikacin, tobramycin, gentamicin, tetracycline, chloramphenicol, ciprofloxacin, and trimethoprim-sulfamethoxazole were determined by Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. We tested the MIC of rifampin and erythromycin by a gradient test (Etest). However, the antimicrobial resistance of M. catarrhalis to these compounds was out of range for commercially available Etest strips. Therefore, the susceptibilities of M. catarrhalis to rifampin, erythromycin, and deoxycholate (all purchased from Sigma) were assayed using standard disk diffusion assays (25, 26). Bacteria were cultured in BHI broth to an OD600 of 0.2 and spread with a cotton swab onto BHI agar plates. Sterile blank paper disks (6 mm; Becton Dickinson, Franklin Lakes, NJ) were placed on the agar surface in duplicate and saturated with a solution of the antimicrobial at various concentrations (Table 2). After 18 h of incubation at 37°C, sensitivities were assessed by measuring the diameters of the zones of growth inhibition on two axes, and the mean values were calculated. The MICs and the susceptibilities of the tested agents for the mutant strains were determined in medium without kanamycin because of the synergistic effect between two substances.
TABLE 2.
Susceptibilities of M. catarrhalis wild-type strain O35E and its mutant strains to antimicrobial agents
| Measurement and antibiotic | Value for strain: |
|||
|---|---|---|---|---|
| O35E | O35E.acrA | O35E.acrB | O35E.oprM | |
| MIC (μg/ml)a | ||||
| Amoxicillin | 1.5 | 1.0 | 1.0 | 1.0 |
| Cefotaxime | 0.75 | 0.5 | 0.5 | 0.5 |
| Clarithromycin | 0.094 | 0.02 | 0.02 | 0.02 |
| Amikacin | 1.5 | 0.75 | 0.75 | 0.75 |
| Tobramycin | 0.5 | 0.38 | 0.25 | 0.25 |
| Zone of growth inhibition (mm)b | ||||
| Erythromycin (1.5 μg) | 23.8 ± 0.5 | 34 ± 0.0 | 35 ± 0.0 | 33.8 ± 0.5 |
| Rifampin (0.5 μg) | 26 ± 0.0 | 32 ± 0.0 | 32.3 ± 0.5 | 32.8 ± 0.5 |
| Deoxycholate (10 mg/ml) | 24 ± 0.0 | 32.25 ± 0.5 | 31.75 ± 0.5 | 31.75 ± 1.0 |
MICs of amoxicillin, cefotaxime, and clarithromycin were determined in two independent experiments with consistent results.
Sensitivities were assessed by measuring the diameters of the zones of growth inhibition on two axes, and the means ± 1 SD were calculated. A representative of two independent experiments performed in duplicate is shown.
Cell lines and growth conditions.
Detroit 562 pharyngeal cells (ATCC CCL-138) were maintained in Eagle's minimal essential medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, 1 mM sodium pyruvate (Sigma), 1× nonessential amino acids (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2.
Adherence and invasion assay.
The ability of M. catarrhalis to adhere to and invade human Detroit 562 epithelial cells in vitro was measured as described previously (13, 27, 28). Each strain was analyzed in triplicate in each experiment.
Accumulation of Hoechst 33342.
To investigate the efflux activity of M. catarrhalis following a 26°C cold shock, we used an assay that measures the accumulation of the fluorescent DNA-binding dye Hoechst 33342 (H33342; Sigma), which is a substrate of the AcrAB pump in several Gram-negative pathogens, including E. coli and Salmonella enterica (29). When H33342 enters the cell, it binds to the DNA minor groove, becomes fluorescent, and can be detected by using a fluorescent plate reader. Efflux-competent cells extrude H33342 out of the cell and accumulate the dye sparingly, resulting in low levels of fluorescence. In contrast, efflux-defective cells (e.g., efflux pump mutants or heat-inactivated [hi] cells) accumulate H33342 at a higher rate, resulting in increased levels of fluorescence. Bacteria were exposed to 26°C or 37°C for 3 h, pelleted, and resuspended in NaCl (0.87%) to an OD600 of 0.2. H33342 was added to a final concentration of 2.5 μM. Fluorescence intensity was measured at excitation and emission wavelengths of 350 and 461 nm, respectively, using a Varioskan Flash (Thermo Scientific, Madison, WI).
Statistical analysis.
Data were expressed as means ± 1 standard deviation (SD). Differences between groups were analyzed by one-way analysis of variance with a Dunnett's correction using Prism software (GraphPad, version 5.01). A P value of <0.05 was defined as statistically significant.
RESULTS
In vitro growth of M. catarrhalis acrA, acrB, and oprM mutants and PCR and RT-PCR analyses of clinical isolates.
Standard growth curves of the O35E wild-type and mutant strains in BHI broth at 37°C revealed no significant differences in growth velocity, measured as broth density at OD600 (Fig. 1A). We studied 16 middle ear and nasopharyngeal isolates for the presence and transcription of acrA, acrB, and oprM genes of the AcrAB-OprM efflux pump. PCR and RT-PCR analyses resulted in positive products for all strains (data not shown).
FIG 1.
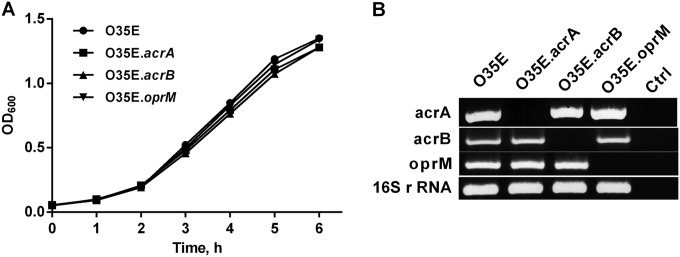
(A) Growth curves for wild-type O35E M. catarrhalis strain and its isogenic knockout strains in BHI medium at 37°C. (B) Expression of the mutated gene in each strain was determined by RT-PCR. For each strain, the product of RT-PCR using primers specific for acrA, acrB, oprM, or 16S rRNA, as a control gene, is shown. Ctrl indicates control reactions with no cDNA templates.
Genotypic and phenotypic characterization of the M. catarrhalis AcrAB-OprM operon knockout mutants.
To investigate the role of the AcrAB-OprM efflux pump in M. catarrhalis antimicrobial resistance and virulence, we constructed isogenic knockout mutants from the wild-type strain O35E. The periplasmic adaptor protein encoded by acrA is the first in an operon also encoding the cognate RND pump acrB and the outer membrane channel oprM. RT-PCR was used to check the inactivation of these genes by insertion of an antibiotic resistance cassette and the expression of the downstream pump genes (Fig. 1B). The inactivation of acrA by insertion of the kanamycin resistance gene was nonpolar; acrB and oprM were still transcribed at normal levels. The transcriptions of acrA and oprM were not affected by disruption of the acrB gene. The disruption of the oprM gene allowed the expression of upstream genes acrA and acrB.
Role of the AcrAB-OprM efflux pump in M. catarrhalis antimicrobial resistance.
The RND family multidrug efflux systems, which are common in Gram-negative bacteria, pump out a wide range of compounds, such as lipophilic and amphiphilic inhibitors, including antibiotics, and are responsible for intrinsic resistance to many antimicrobial agents (17, 30–32). To investigate this, Etests and susceptibility assays were performed with the mutants O35E.acrA, O35E.acrB, and O35E.oprM and the wild-type O35E strain. There were no differences in MICs for tetracycline, chloramphenicol, ciprofloxacin, and trimethoprim-sulfamethoxazole between wild-type and mutant strains. For amoxicillin and cefotaxime, there was a minor but consistent 1.5-fold decrease in the MICs of all mutants (1.5 μg/ml versus 1.0 μg/ml and 0.75 μg/ml versus 0.5 μg/ml, respectively) (Table 2). For clarithromycin, there was a 4.7-fold decrease in the MICs of all mutants tested in comparison with the wild-type strain (0.094 μg/ml versus 0.02 μg/ml) (Table 2). In a susceptibility test, insertional inactivation of acrA, acrB, and oprM also increased the antibacterial activity of many additional agents tested (erythromycin, rifampin, deoxycholic acid), implying that they are ordinarily substrates for the AcrAB-OprM efflux transporter (Table 2). Thus, M. catarrhalis strains lacking the efflux pump proteins display a MIC of amoxicillin, cefotaxime, and clarithromycin up to 1.5- and 4.7-fold, respectively, lower than that in the wild-type strain, indicating that the AcrAB-OprM efflux pump is involved in the susceptibility of these cells to the antimicrobials tested.
The antibiotic class of aminoglycosides is not a substrate for the E. coli AcrAB-TolC pump, although it is a substrate for the homologous Yersinia pestis, P. aeruginosa, and Klebsiella pneumoniae complexes (31, 32). The MICs of three aminoglycosides (amikacin, tobramycin, and gentamicin) were determined for M. catarrhalis O35E and its acrA, acrB, and oprM mutants. The acrA, acrB, and oprM mutant strains were more susceptible (2-fold) than the wild-type strain to amikacin (Table 2). The MICs of tobramycin were lower (2-fold) for the acrB and oprM knockout strains than for the wild-type strain, while there were only modest (1.3-fold) decreases in the MICs of this antibiotic in the case of O35E.acrA (Table 2). In contrast, gentamicin susceptibility was not affected in the acrA, acrB, and oprM strains. It was shown that the S. enterica acrB mutant strain was more susceptible to tobramycin (2-fold), whereas susceptibility to amikacin was not affected compared with that of the wild-type parental strain, reflecting substrate specificity of this pump (33).
Upregulation of acrA, acrB, and oprM mRNA expression after amoxicillin treatment.
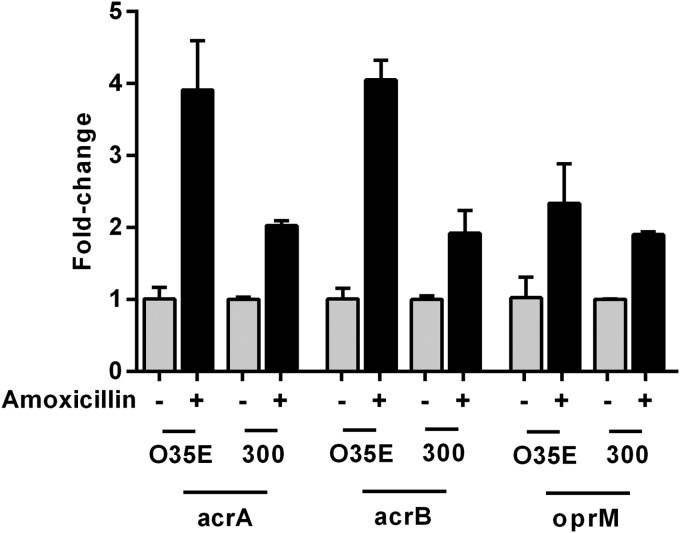
Reduced membrane permeability and increased active efflux—two of the main strategies used by bacteria for protection against antibiotics—are generally regulated by reduced porin expression and overexpression of efflux pumps (34, 35). Our previous findings of downregulation of m35 porin mRNA after amoxicillin (60 μg/ml) treatment (16) prompted us to investigate the expression of the AcrAB-OprM efflux pump genes of strains O35E and 300 during amoxicillin treatment by quantitative real-time PCR. Amoxicillin treatment of strain O35E for a period of 4 h induced the upregulation of acrA (4-fold), acrB (4-fold), and oprM (2.5-fold) mRNA expression compared with that of bacteria grown without amoxicillin (Fig. 2). Strain 300 demonstrated similar effects, with expression of acrA, acrB, and oprM mRNA induced (2-fold) after amoxicillin treatment. The expression of a control gene encoding type IV pilin A (pilA) was not altered by amoxicillin treatment, indicating that amoxicillin induces the expression of genes that are involved in the adaptive resistance (data not shown).
FIG 2.
Upregulation of acrA, acrB, and oprM mRNA expression of M. catarrhalis strains O35E and 300 during amoxicillin treatment. Quantitative real-time PCR was performed after 4 h of incubation with and without 60 μg/ml amoxicillin. Upregulation was normalized to 16S rRNA. The graph shows one of two representative experiments done in triplicate. Data are presented as means ± 1 SD.
Expression of acrA, acrB, and oprM mRNA after clarithromycin treatment.
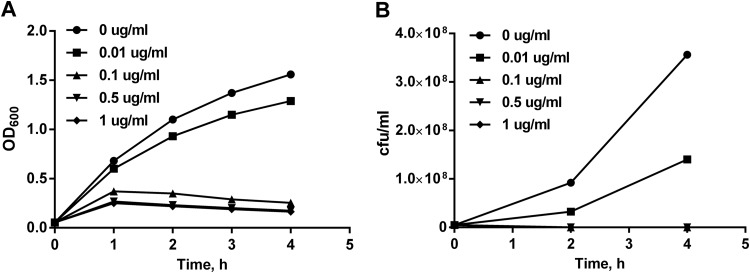
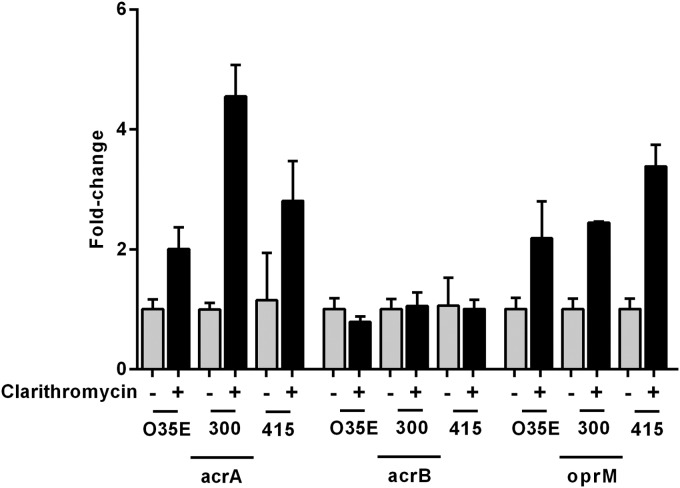
Because clarithromycin was found to be a substrate for the AcrAB-OprM pump, we investigated the expression of the acrA, acrB, and oprM genes after clarithromycin treatment by quantitative real-time PCR. The breakpoint of bacteriostatic clarithromycin concentration was evaluated by a time-kill curve assay (Fig. 3), and the subinhibitory concentration was estimated to be 0.01 μg/ml. Clarithromycin treatment of M. catarrhalis strain O35E, for a period of 4 h, induced the upregulation of acrA (2-fold) and oprM (2.2-fold) mRNA expression compared with that of bacteria grown without clarithromycin (Fig. 4). Clarithromycin treatment of M. catarrhalis strains 300 and 415 caused the induction of acrA (4.5- and 2.8-fold, respectively) and oprM (2.4- and 3.4-fold, respectively) mRNA expression. In contrast, the expression of acrB mRNA in these strains remained unaffected.
FIG 3.
Time-kill curves of strain O35E for different clarithromycin concentrations (0, 0.01, 0.1, 0.5, and 1 μg/ml).
FIG 4.
Expression of efflux pump AcrAB-OprM genes acrA, acrB, and oprM mRNA in M. catarrhalis strains O35E, 300, and 415 during clarithromycin treatment. Quantitative real-time PCR was performed after 4 h of incubation with and without 0.01 μg/ml clarithromycin. Upregulation was normalized to 16S rRNA. The graph shows one of three representative experiments done in triplicate. Data are presented as means ± 1 SD.
The expressions of acrA, acrB, and oprM genes are required for efficient invasion of Detroit 562 pharyngeal epithelial cells.
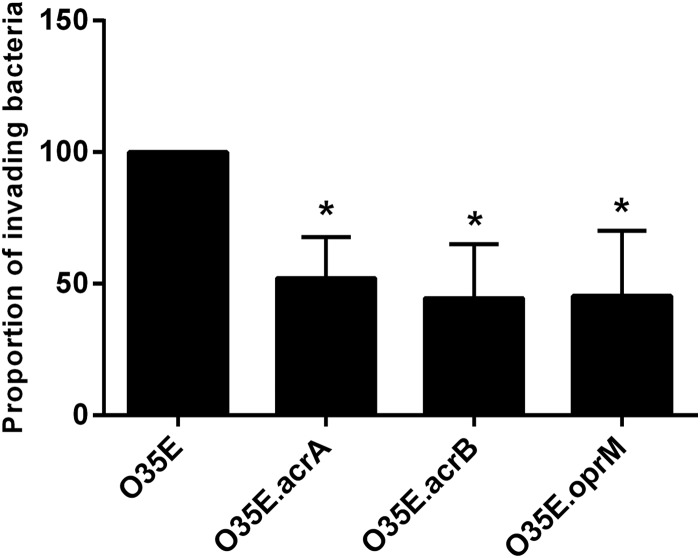
The abilities of a pathogen to adhere to and invade epithelial host cells are major virulence factors. Efflux pump systems have a role in mediating the adherence and uptake of bacteria into target host cells (29, 36, 37). To investigate if AcrAB-OprM efflux pump components mediate adherence and invasion, assays were performed on Detroit 562 pharyngeal epithelial cells. Adherence of O35E.acrA, O35E.acrB, and O35E.oprM mutants was as efficient as that of the O35E wild-type parent strain (data not shown). To determine if acrA, acrB, and oprM are required for efficient invasion, Detroit 562 cell monolayers were infected at a multiplicity of infection (MOI) of 30, and following a 3-h incubation, cells were washed, incubated with gentamicin for an additional 2 h, and then lysed, serially diluted, and plated to determine the proportion of invading bacteria. The O35E.acrA, O35E.acrB, and O35E.oprM strains demonstrated decreased (up to 50%) invasion compared to that of O35E (Fig. 5), indicating that acrA, acrB, and oprM are required for efficient invasion of human nasopharyngeal cells in vitro.
FIG 5.
Ability of M. catarrhalis wild-type strain O35E and its knockout mutants to invade human pharyngeal epithelial cells. Data are displayed as means of four separate experiments performed in triplicate ± 1 SD. *, P < 0.05 for wild-type versus respective mutant.
Serum resistance.
Resistance of M. catarrhalis to human complement, which is associated with disease-causing isolates (38), was not impaired by the lack of AcrAB-OprM efflux pump genes (data not shown).
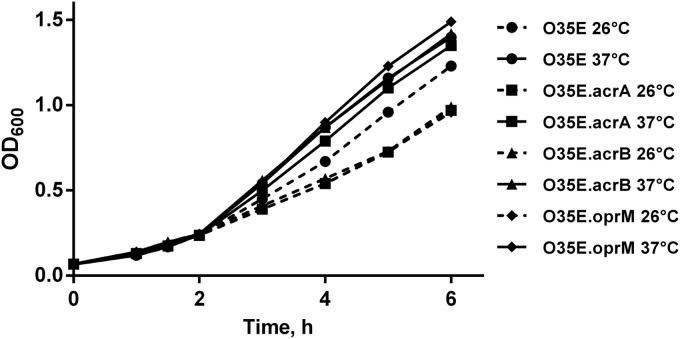
Growth of M. catarrhalis acrA, acrB, and oprM mutants after cold shock.
To investigate whether inactivation of the efflux pump genes affects the growth rate following cold shock conditions, growth curves were plotted for all strains studied. When the growth at 37°C was monitored, we observed that the parent strain O35E and its knockout mutants grew similarly (Fig. 6). The growth of M. catarrhalis acrA, acrB, and oprM mutants, subjected to a downshift to 26°C during the mid-logarithmic phase, was slightly diminished compared to the growth of parent strain O35E (Fig. 6), indicating that the AcrAB-OprM efflux pump might play a role in the adaptation to cold shock.
FIG 6.
Growth of M. catarrhalis O35E and its isogenic knockout strains at 26°C compared to that at 37°C. Bacteria were grown to mid-logarithmic phase (OD600 = 0.25) under standard conditions at 37°C. The cultures were then split, and equal aliquots were incubated further at 26°C and 37°C, respectively.
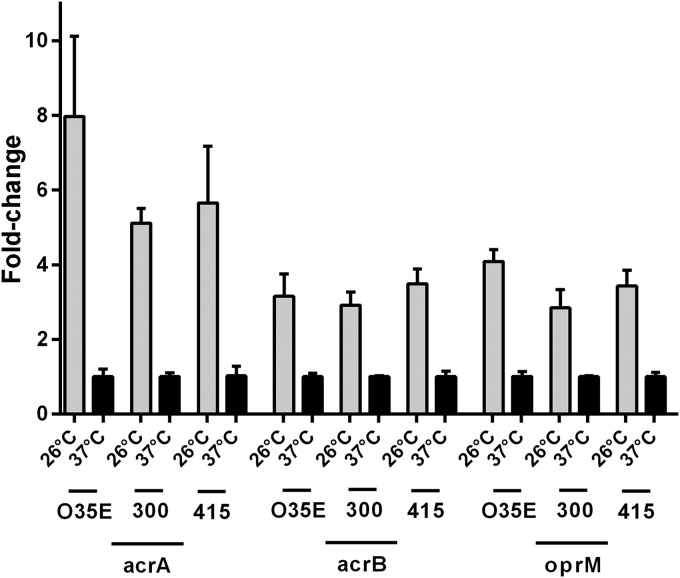
Cold shock induces upregulation of acrA, acrB, and oprM mRNA.
To confirm the RNA-seq data (21) and to investigate the contribution of the AcrAB-OprM efflux pump to the cold shock response, we assessed the acrA, acrB, and oprM mRNA expression levels of strain O35E exposed to either 26°C or 37°C. The expression levels of acrA, acrB, and oprM were significantly increased at 26°C (10.7-, 5.7-, and 6.5-fold, respectively) compared to expression at 37°C (Fig. 7). Similar expression patterns of acrA, acrB, and oprM were observed in M. catarrhalis clinical isolates 300 and 415, indicating that this effect is a general characteristic of M. catarrhalis (Fig. 7).
FIG 7.
Upregulation of acrA, acrB, and oprM mRNA expression of M. catarrhalis strains O35E, 300, and 415 after cold shock. Quantitative real-time PCR was performed after 3 h of incubation at 26°C or 37°C. Upregulation was normalized to 16S rRNA. The graph shows one of two representative experiments done in triplicate. Data are presented as means ± 1 SD.
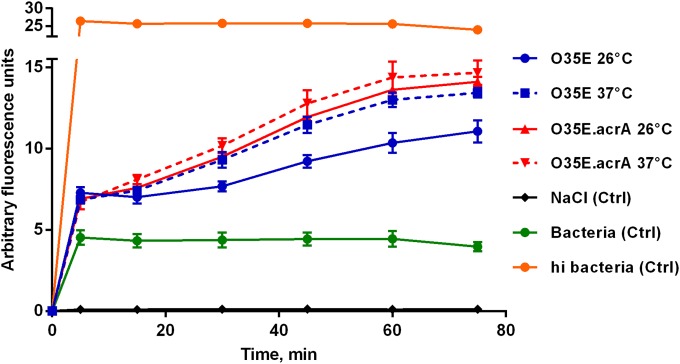
Cold shock results in the efflux upregulation of Hoechst 33342.
Since cold shock increases the expression of AcrAB-OprM efflux pump genes, we examined the effect of cold shock on active efflux. The Hoechst dye is a substrate of RND efflux pumps, and its accumulation can be used to infer the level of RND efflux (29). In order to compare the levels of active efflux in bacterial growth at 26°C, accumulation of H33342 was determined and compared with bacterial growth at 37°C. The results of the H33342 accumulation assay are shown in Fig. 8. Accumulation of H33342 was approximately 20% lower following cold shock at 26°C than after exposure to 37°C, indicating that cold shock resulted in increased efflux activity. Inactivation of acrA alone increased Hoechst 33342 accumulation, indicating that the inactivation of acrA resulted in reduced efflux activity. Furthermore, there was no cold shock effect on the accumulation of the dye, indicating that AcrA is required for active efflux of H33342 in cold shock-induced M. catarrhalis.
FIG 8.
The accumulation of H33342 in M. catarrhalis strain O35E and its isogenic mutant strain O35E.acrA over time. Cold shock results in the efflux upregulation of H33342, a substrate of AcrAB-OprM efflux pump. Bacteria were exposed to 26°C or 37°C for 3 h, pelleted, and resuspended in NaCl (0.87%) to an OD600 of 0.2. H33342 was added to a final concentration of 2.5 μM. The graph shows one of three representative experiments done in triplicate. Data are presented as means ± 1 SD. Ctrl indicates control reactions with no bacteria or heat-inactivated (hi; 60°C, 1 h) bacteria.
DISCUSSION
Antimicrobial resistance is based on three major strategies: detoxify enzymes to degrade or modify antibiotics, target protection to impair target recognition and thus antimicrobial activity, and target the membrane barrier to limit intracellular access of antimicrobials (35). For M. catarrhalis, two major mechanisms of bacterial resistance to antimicrobials have been described: the inactivation of antimicrobials by enzymes such as β-lactamases (39) and the reduced entry of the antibiotic into bacterial cells due to reducing the number of entry channels, such as porins (16). In our previous study, we demonstrated that the M35 porin was downregulated after amoxicillin treatment, leading to increased resistance and indicating a novel resistance mechanism against aminopenicillins in M. catarrhalis (16). Another important mechanism of antimicrobial resistance is enhancement of the active expulsion of antibiotics out of the cells—a process termed efflux. The expression of genes encoding porins and efflux pumps is carefully regulated in order to respond to certain signals, thereby altering the resistance of a bacterium depending on the growth conditions (15).
Sequence analysis of the genome of a variety of bacterial species has demonstrated that most of them encode several efflux pump systems. The multidrug efflux systems of M. catarrhalis have never before been characterized, to our knowledge, although the sequencing data for M. catarrhalis strain BBH18 indicate that only a few such pumps (Acr and Mtr efflux pumps) are potentially present (18). M. catarrhalis encodes an AcrAB-OprM efflux system homologous to the system described in E. coli. Most of the clinical studies have focused on the involvement of efflux pumps in antimicrobial resistance. In this study, the role of the AcrAB-OprM efflux pump in antibiotic resistance was investigated by constructing mutant strains containing acrA, acrB, and oprM genes in M. catarrhalis strain O35E. We observed a moderate (1.5-fold) decrease in the MICs of amoxicillin and cefotaxime and a marked (4.7-fold) decrease in the MICs of clarithromycin for all mutants in comparison with those for the wild-type strain. These results show that the AcrAB-OprM pump is important in clarithromycin resistance and is involved in macrolide efflux. Conversely, amoxicillin and cefotaxime resistances are only weakly mediated by the AcrAB-OprM efflux pump, suggesting that other resistance strategies, such as β-lactamases, porins (M35), or another efflux pump, may play a more important role. Furthermore, the insertional inactivation of acrA, acrB, and oprM increased the antibacterial activity of erythromycin, rifampin, and deoxycholic acid, indicating that this efflux pump may recognize multiple substrates, as already noted for the homologous proteins from E. coli (40).
Antimicrobials play a very important role in the induction of adaptive resistance by the overexpression of efflux systems (15). Several studies have shown increased bacterial efflux pump expression during antibiotic therapy, indicating that efflux pumps may play a role in survival within the host (41, 42). We found that exposure of M. catarrhalis strains O35E and 300 to amoxicillin for 4 h triggered the increased expression of all AcrAB-OprM pump genes (Fig. 2), and exposure of O35E and strains 300 and 415 to clarithromycin enhanced the expression of acrA and oprM (Fig. 4). We performed the expression analysis of the AcrAB-OprM efflux pump genes after amoxicillin treatment as an extension of the previous study by our group and used the already evaluated concentration of amoxicillin (16). Growth curves for treatment with 60 μg/ml of amoxicillin indicated that this concentration was not completely bactericidal but inhibited growth for a period of about 4 h before proliferation resumed (16). The breakpoint of the bacteriostatic clarithromycin concentration was evaluated by a time-kill curve assay (Fig. 3), and the subinhibitory concentration was estimated to be 0.01 μg/ml. The different correlations of amoxicillin and clarithromycin concentrations with the MICs obtained by Etest may be attributed to different resistance strategies of bacteria upon induction by specific antibiotics. It seems that the threshold concentrations for clarithromycin required to induce the antimicrobial resistance (e.g., increased expression of efflux genes) are lower than those of amoxicillin. As we already showed, exposure of M. catarrhalis to amoxicillin induces downregulation of the m35 porin to limit intracellular penetration of amoxicillin (16) and increases the expression of the AcrAB-OprM genes to enhance the active expulsion of this antibiotic.
The exposure of M. catarrhalis to clarithromycin increases the expression of particular components of the AcrAB-OprM efflux pump. Clarithromycin treatment of M. catarrhalis strains O35E, 300, and 415 caused the induction of acrA and oprM mRNA expression, while the expression of acrB mRNA in these strains remained unaffected. The regulatory mechanism controlling the transcription of the AcrAB-OprM genes after clarithromycin treatment is indistinct at this point and needs further investigation. However, it has been demonstrated that genes within an operon do not conform to the simple notion that they have equal levels of expression (43). Several studies revealed the existence of the internal promoters and read-through terminators in bacterial operons (43, 44). These additional promoters are often located downstream of the first gene so that only part of the operon is transcribed from the internal promoter. Similarly, some operons contain an internal read-through terminator at which partial continuation of transcription occurs (44). Additional antimicrobial strategies used by M. catarrhalis against clarithromycin (e.g., expression of the porin) are still unclear and need further investigation. Furthermore, it would be interesting to investigate antimicrobial resistance in M. catarrhalis mutant strains lacking the entire gene cluster of the AcrAB-OprM efflux pump in order to estimate the level of redundancy between potentially homologous efflux pump proteins. Our findings of the upregulation of AcrAB-OprM pump expression at the transcriptional level after treatment with amoxicillin and clarithromycin in all tested β-lactamase-producing strains combined with observations of the lower MICs for the AcrAB-OprM pump knockout mutants indicate that there is a potentially novel resistance mechanism against these antibiotics in M. catarrhalis.
Temperature is a critical environmental factor, and cold shock, as has been characterized for E. coli, affects the bacterial transcriptome in a concerted attempt to maintain essential cellular functions that favor survival under extreme and rapidly changing conditions (45). We have shown that the cold shock response is obviously an important mechanism for M. catarrhalis as an adaptation and survival mechanism in the nasopharyngeal habitat, as well as for the virulence and colonization abilities of M. catarrhalis (12–14, 16, 21). Our current results demonstrate that cold shock increases the expression of AcrAB-OprM efflux pump genes in all three M. catarrhalis strains tested (Fig. 7), suggesting that this effect is a general characteristic of M. catarrhalis. We also show that increased expression of AcrAB-OprM pump genes following incubation of M. catarrhalis at 26°C leads to a lower accumulation of Hoechst 33342 (Fig. 8), a substrate of the AcrAB-OprM efflux pump, indicating that cold shock results in increased efflux activity. Our findings of the upregulation of AcrAB-OprM pump expression following cold shock and pretreatment with subinhibitory concentrations of clarithromycin together with an observation of higher efflux activity upon cold shock indicate that cold shock could contribute to increased resistance of M. catarrhalis to this antibiotic. Since both cold shock and treatment with amoxicillin and clarithromycin induce expression of the efflux pump genes, it would be interesting to investigate whether an additive effect exists with both cold shock and antibiotic treatment. M. catarrhalis exposed to a 26°C cold shock might transiently increase its ability to withstand the presence of subinhibitory concentrations of an antibiotic by limiting its entry (e.g., M35 porin) into the cell and/or expelling it more efficiently, leading to increased survival of bacteria. The relative and synergistic effect of these two antimicrobial resistance mechanisms (porin loss/active efflux) in M. catarrhalis is still unclear and needs further investigation by constructing mutant strains lacking both components (porin/efflux pump). Since it has been shown that one stress response might help bacteria to contend with other stress conditions (46, 47), it is possible that cold shock could improve the ability of M. catarrhalis to survive subinhibitory concentrations of antibiotics because several pathways of stress responses are activated. Clinical studies in children demonstrated that the density of M. catarrhalis in the nasopharynx is positively associated with prolonged respiratory tract symptoms and a greater likelihood of purulent otitis media (48, 49).
In addition to antibiotic sensitivity, efflux pumps are used in bacteria to excrete cellular metabolites that are toxic and/or have a signaling role (50). This could potentially explain the reduced growth of the M. catarrhalis efflux pump mutant strains after cold shock conditions (Fig. 6). Cold shock induces the expression of AcrAB-OprM pump genes (Fig. 7), suggesting that the AcrAB-OprM efflux pump might play an important role in adaptation to cold shock. Inactivation of the AcrAB-OprM efflux pump would lead to accumulation of potentially toxic metabolites and impair growth at 26°C. On the other hand, the parent strain O35E and its knockout mutants grew similarly at 37°C, indicating that at this temperature, other efflux pumps, such as Mtr, would accomplish this function. Our RNA-seq data indicate that some components of the Mtr efflux pump (mtrF) are induced (1.8-fold) after exposure to 37°C (21). The contribution of other environmental triggers like heat shock or nutrient conditions to the expression of AcrAB-OprM efflux pump genes is still unclear and needs further scrutiny.
In addition to an established role in antimicrobial resistance, efflux pumps have been shown to have a role in pathogenicity in a variety of clinically relevant bacterial species, including Salmonella, P. aeruginosa, Neisseria gonorrhoeae, and Campylobacter jejuni (36). In Salmonella, disruption of acrA or acrB led not only to susceptibility to a range of antimicrobial compounds but also to a reduced ability to adhere to and invade human intestinal epithelial cells (29, 37). This effect can be partially explained by the finding that many efflux pumps are also responsible for exporting host-derived substrates, such as bile salts and fatty acids. Inactivation of the AcrAB-OprM efflux pump genes of M. catarrhalis strain O35E demonstrated a decreased invasion ability compared with the parental strain (Fig. 5), suggesting that acrA, acrB, and oprM are required for efficient invasion of human nasopharyngeal cells.
In summary, we describe here the use of specific knockout mutants to investigate the effect of the AcrAB-OprM efflux system on antimicrobial resistance and virulence in M. catarrhalis. We demonstrate that the AcrAB-OprM efflux pump is involved in resistance to amoxicillin, cefotaxime, and clarithromycin and is required for efficient invasion of pharyngeal epithelial cells. A physiologic cold shock, as occurs when humans breathe cold air for prolonged periods, increases the expression of AcrAB-OprM pump genes in vitro, resulting in increased efflux activity of M. catarrhalis. These data support the notion that M. catarrhalis uses physiologic exposure to cold air to upregulate pivotal survival systems in the human pharynx that may contribute to bacterial virulence.
ACKNOWLEDGMENT
This work was supported by Swiss National Science Foundation (SNF) grants 3100A0-102246 and 3100A0-116053 (to C.A.).
REFERENCES
- 1.Faden H, Duffy R, Wasielewski R, Wolf J, Krystofik D, Tung Y. 1997. Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis 175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 2.Palmu A, Herva E, Savolainen H, Karma P, Mäkelä PH, Kilpi T. 2004. Association of clinical signs and symptoms with bacterial findings in acute otitis media. Clin Infect Dis 38:234–242. doi: 10.1086/380642. [DOI] [PubMed] [Google Scholar]
- 3.Murphy TF, Brauer AL, Grant BJ, Sethi S. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am J Respir Crit Care Med 172:195–199. doi: 10.1164/rccm.200412-1747OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy TF, Parameswaran GI. 2009. Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis 49:124–131. doi: 10.1086/599375. [DOI] [PubMed] [Google Scholar]
- 5.Eskola J, Kilpi T, Palmu A, Jokinen J, Haapakoski J, Herva E, Takala A, Kayhty H, Karma P, Kohberger R, Siber G, Makela PH. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N Engl J Med 344:403–409. doi: 10.1056/NEJM200102083440602. [DOI] [PubMed] [Google Scholar]
- 6.Revai K, McCormick DP, Patel J, Grady JJ, Saeed K, Chonmaitree T. 2006. Effect of pneumococcal conjugate vaccine on nasopharyngeal bacterial colonization during acute otitis media. Pediatrics 117:1823–1829. doi: 10.1542/peds.2005-1983. [DOI] [PubMed] [Google Scholar]
- 7.Van Hare GF, Shurin PA. 1987. The increasing importance of Branhamella catarrhalis in respiratory infections. Pediatr Infect Dis J 6:92–94. doi: 10.1097/00006454-198701000-00047. [DOI] [PubMed] [Google Scholar]
- 8.Mbaki N, Rikitomi N, Nagatake T, Matsumoto K. 1987. Correlation between Branhamella catarrhalis adherence to oropharyngeal cells and seasonal incidence of lower respiratory tract infections. Tohoku J Exp Med 153:111–121. doi: 10.1620/tjem.153.111. [DOI] [PubMed] [Google Scholar]
- 9.Sarubbi FA, Myers JW, Williams JJ, Shell CG. 1990. Respiratory infections caused by Branhamella catarrhalis. Selected epidemiologic features. Am J Med 88:9S–14S. [DOI] [PubMed] [Google Scholar]
- 10.Hendley JO, Hayden FG, Winther B. 2005. Weekly point prevalence of Streptococcus pneumoniae, Hemophilus influenzae and Moraxella catarrhalis in the upper airways of normal young children: effect of respiratory illness and season. APMIS 113:213–220. doi: 10.1111/j.1600-0463.2005.apm1130310.x. [DOI] [PubMed] [Google Scholar]
- 11.Rouadi P, Baroody FM, Abbott D, Naureckas E, Solway J, Naclerio RM. 1999. A technique to measure the ability of the human nose to warm and humidify air. J Appl Physiol 87:400–406. [DOI] [PubMed] [Google Scholar]
- 12.Heiniger N, Troller R, Meier PS, Aebi C. 2005. Cold shock response of the UspA1 outer membrane adhesin of Moraxella catarrhalis. Infect Immun 73:8247–8255. doi: 10.1128/IAI.73.12.8247-8255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spaniol V, Troller R, Aebi C. 2009. Physiologic cold shock increases adherence of Moraxella catarrhalis to and secretion of interleukin 8 in human upper respiratory tract epithelial cells. J Infect Dis 200:1593–1601. doi: 10.1086/644640. [DOI] [PubMed] [Google Scholar]
- 14.Spaniol V, Troller R, Schaller A, Aebi C. 2011. Physiologic cold shock of Moraxella catarrhalis affects the expression of genes involved in the iron acquisition, serum resistance and immune evasion. BMC Microbiol 11:182. doi: 10.1186/1471-2180-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez L, Hancock RE. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jetter M, Spaniol V, Troller R, Aebi C. 2010. Down-regulation of porin M35 in Moraxella catarrhalis by aminopenicillins and environmental factors and its potential contribution to the mechanism of resistance to aminopenicillins. J Antimicrob Chemother 65:2089–2096. doi: 10.1093/jac/dkq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol 178:5853–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries SP, van Hijum SA, Schueler W, Riesbeck K, Hays JP, Hermans PW, Bootsma HJ. 2010. Genome analysis of Moraxella catarrhalis strain BBH18, [corrected] a human respiratory tract pathogen. J Bacteriol 192:3574–3583. doi: 10.1128/JB.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. 2011. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother 55:921–924. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XZ, Nikaido H, Poole K. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother 39:1948–1953. doi: 10.1128/AAC.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spaniol V, Wyder S, Aebi C. 2013. RNA-Seq-based analysis of the physiologic cold shock-induced changes in Moraxella catarrhalis gene expression. PLoS One 8:e68298. doi: 10.1371/journal.pone.0068298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Meier PS, Troller R, Heiniger N, Hays JP, van Belkum A, Aebi C. 2006. Unveiling electrotransformation of Moraxella catarrhalis as a process of natural transformation. FEMS Microbiol Lett 262:72–76. doi: 10.1111/j.1574-6968.2006.00365.x. [DOI] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 25.Peng D, Hong W, Choudhury BP, Carlson RW, Gu XX. 2005. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect Immun 73:7569–7577. doi: 10.1128/IAI.73.11.7569-7577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duval BD, Mathew A, Satola SW, Shafer WM. 2010. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob Agents Chemother 54:2283–2290. doi: 10.1128/AAC.01786-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spaniol V, Heiniger N, Troller R, Aebi C. 2008. Outer membrane protein UspA1 and lipooligosaccharide are involved in invasion of human epithelial cells by Moraxella catarrhalis. Microbes Infect 10:3–11. doi: 10.1016/j.micinf.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Murphy TF, Brauer AL, Kirkham C, Johnson A, Koszelak-Rosenblum M, Malkowski MG. 2013. Role of the zinc uptake ABC transporter of Moraxella catarrhalis in persistence in the respiratory tract. Infect Immun 81:3406–3413. doi: 10.1128/IAI.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blair JM, La Ragione RM, Woodward MJ, Piddock LJ. 2009. Periplasmic adaptor protein AcrA has a distinct role in the antibiotic resistance and virulence of Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 64:965–972. doi: 10.1093/jac/dkp311. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido H, Pages JM. 2012. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lister IM, Raftery C, Mecsas J, Levy SB. 2012. Yersinia pestis AcrAB-TolC in antibiotic resistance and virulence. Antimicrob Agents Chemother 56:1120–1123. doi: 10.1128/AAC.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padilla E, Llobet E, Domenech-Sanchez A, Martinez-Martinez L, Bengoechea JA, Alberti S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blair JM, Smith HE, Ricci V, Lawler AJ, Thompson LJ, Piddock LJ. 2015. Expression of homologous RND efflux pump genes is dependent upon AcrB expression: implications for efflux and virulence inhibitor design. J Antimicrob Chemother 70:424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pages JM, James CE, Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 35.Davin-Regli A, Bolla JM, James CE, Lavigne JP, Chevalier J, Garnotel E, Molitor A, Pages JM. 2008. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets 9:750–759. doi: 10.2174/138945008785747824. [DOI] [PubMed] [Google Scholar]
- 36.Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith HE, Blair JM. 2014. Redundancy in the periplasmic adaptor proteins AcrA and AcrE provides resilience and an ability to export substrates of multidrug efflux. J Antimicrob Chemother 69:982–987. doi: 10.1093/jac/dkt481. [DOI] [PubMed] [Google Scholar]
- 38.Murphy S, Fitzgerald M, Mulcahy R, Keane C, Coakley D, Scott T. 1997. Studies on haemagglutination and serum resistance status of strains of Moraxella catarrhalis isolated from the elderly. Gerontology 43:277–282. doi: 10.1159/000213863. [DOI] [PubMed] [Google Scholar]
- 39.Khan MA, Northwood JB, Levy F, Verhaegh SJ, Farrell DJ, Van Belkum A, Hays JP. 2010. bro β-lactamase and antibiotic resistances in a global cross-sectional study of Moraxella catarrhalis from children and adults. J Antimicrob Chemother 65:91–97. doi: 10.1093/jac/dkp401. [DOI] [PubMed] [Google Scholar]
- 40.Nikaido H, Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hocquet D, Vogne C, El Garch F, Vejux A, Gotoh N, Lee A, Lomovskaya O, Plesiat P. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 47:1371–1375. doi: 10.1128/AAC.47.4.1371-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Garch F, Lismond A, Piddock LJ, Courvalin P, Tulkens PM, Van Bambeke F. 2010. Fluoroquinolones induce the expression of patA and patB, which encode ABC efflux pumps in Streptococcus pneumoniae. J Antimicrob Chemother 65:2076–2082. doi: 10.1093/jac/dkq287. [DOI] [PubMed] [Google Scholar]
- 43.Laing E, Mersinias V, Smith CP, Hubbard SJ. 2006. Analysis of gene expression in operons of Streptomyces coelicolor. Genome Biol 7:R46. doi: 10.1186/gb-2006-7-6-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Hoon MJ, Makita Y, Nakai K, Miyano S. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput Biol 1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber MH, Marahiel MA. 2003. Bacterial cold shock responses. Sci Prog 86:9–75. doi: 10.3184/003685003783238707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H, Lee HY, Ahn J. 2008. Cross-protective effect of acid-adapted Salmonella enterica on resistance to lethal acid and cold stress conditions. Lett Appl Microbiol 47:290–297. doi: 10.1111/j.1472-765X.2008.02429.x. [DOI] [PubMed] [Google Scholar]
- 47.Gunasekera TS, Csonka LN, Paliy O. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190:3712–3720. doi: 10.1128/JB.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kristo A, Uhari M, Kontiokari T, Glumoff V, Kaijalainen T, Leinonen M, Luotonen J, Koivunen P, Kujala T, Pokka T, Alho OP. 2006. Nasal middle meatal specimen bacteriology as a predictor of the course of acute respiratory infection in children. Pediatr Infect Dis J 25:108–112. doi: 10.1097/01.inf.0000201048.65828.b5. [DOI] [PubMed] [Google Scholar]
- 49.Smith-Vaughan H, Byun R, Nadkarni M, Jacques NA, Hunter N, Halpin S, Morris PS, Leach AJ. 2006. Measuring nasal bacterial load and its association with otitis media. BMC Ear Nose Throat Disord 6:10. doi: 10.1186/1472-6815-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruiz C, Levy SB. 2014. Regulation of acrAB expression by cellular metabolites in Escherichia coli. J Antimicrob Chemother 69:390–399. doi: 10.1093/jac/dkt352. [DOI] [PMC free article] [PubMed] [Google Scholar]