Highlights
-
•
Serological and molecular assays exist for most economic important Theileria species.
-
•
Molecular assays are constantly being improved with regard to sensitivity and specificity.
-
•
The concept of what constitute a Theileria species impacts on accurate diagnostics.
-
•
Analytical specificity of molecular assays are >800 000 parasites/L blood.
-
•
Parasitemia ranges may determine practical limits of detection.
Keywords: Diagnostics, Serology, Parasitaemia, PCR, Species, Theileria
Graphical Abstract
Abstract
An extensive range of serological and molecular diagnostic assays exist for most of the economically important Theileira species such as T. annulata, T. equi, T. lestoquardi, T. parva, T. uilenbergi and other more benign species. Diagnostics of Theileria is considered with regard to sensitivity and specificity of current molecular and serological assays and their use in epidemiology. In the case of serological assays, cross-reactivity of genetically closely related species reduces the use of the gold standard indirect fluorescent antibody test (IFAT). Development of antigen-specific assays does not necessarily address this problem, since closely related species will potentially have similar antigens. Even so, serological assays remain an important line of enquiry in epidemiological surveys. Molecular based assays have exploded in the last decade with significant improvements in sensitivity and specificity. In this review, the current interpretation of what constitute a species in Theileria and its impact on accurate molecular diagnostics is considered. Most molecular assays based on conventional or real-time PCR technology have proven to be on standard with regard to analytical sensitivity. However, consideration of the limits of detection in regard to total blood volume of an animal indicates that most assays may only detect >400,000 parasites/L blood. Even so, natural parasitaemia distribution in carrier-state animals seems to be above this limit of detection, suggesting that most molecular assays should be able to detect the majority of infected individuals under endemic conditions. The potential for false-negative results can, however, only be assessed within the biological context of the parasite within its vertebrate host, i.e. parasitaemia range in the carrier-state that will support infection of the vector and subsequent transmission.
1. Introduction
The phylum Apicomplexa comprises a large group of complex eukaryotic organisms known to be obligate parasites of vertebrates and invertebrates. These organisms share a common characteristic of having an apical complex which contains secretory organelles considered to be involved in invasion and/or establishment of the parasite in the mammalian or invertebrate host (Bishop et al., 2004). The phylum is divided into four principal groups; the Coccidia, Gregarinasina (gregarines), Haemospororida (haemosporidians) and the Piroplasmorida (piroplasmids) (Adl et al., 2012). The Piroplasmorida comprises two main genera (Babesia and Theileria) responsible for the economic important diseases of domestic and wild animals. New species of the piroplasmids are still being discovered and their full biology is not completely documented yet. Many of this order's parasites were formerly classified based on morphology, host cells in which schizogony occurs, the observation of piroplasms in the red blood cells associated with disease manifestation and host-vector specificity (Barnett, 1977; Uilenberg, 2006). The genus Theileria is distinguished by infection of leukocytes by sporozoites, maturation of schizonts into merozoites and subsequent infection of red blood cells to form piroplasms (Uilenberg, 2006).
A generalised lifecycle for the Theileria genus include secretion of infective sporozoites during tick feeding into the feeding site (Fig. 1). Sporozoites then infect leukocytes and multiply by merogony, after which merozoites are released, which invade red blood cells thereby establishing the piroplasm stage. During a next feeding cycle, larval or nymphal vector ticks ingest piroplasms and the released parasites undergo syngamy in the tick gut, forming a zygote, the only diploid stage. The zygote divides into motile kinetes that infect the tick gut epithelial cells and migrate to the haemolymph and subsequently infect the salivary glands. After moulting and commencement of feeding by the tick, sporogony results in the multiplication of sporozoites in the salivary gland acini before injection into the feeding site by nymphs or adult ticks (McKeever, 2009).
Fig. 1.
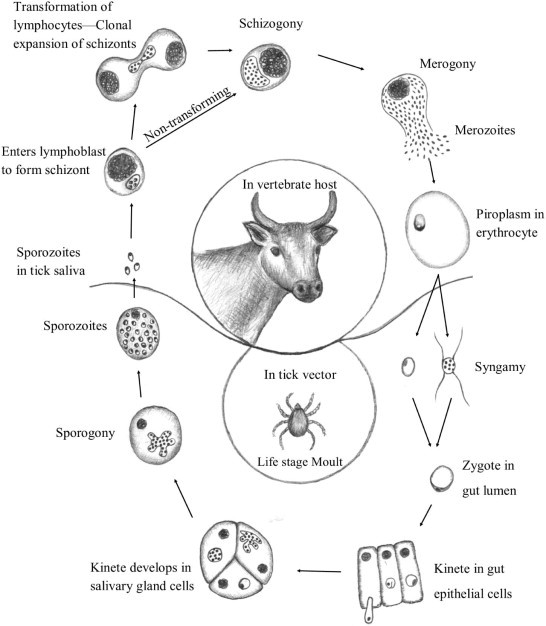
A generalised lifecycle for the Theileria using T. parva as example.
The Theileria species infect a wide range of both domestic and wild animals and are transmitted by ixodid ticks of the genera Amblyomma, Haemaphysalis, Hyalomma and Rhipicephalus. Most of these ticks are renowned for the large economic losses they cause to the agricultural industry due to disease outbreaks, mortalities, damage to hides and poor production in domestic animals (Bishop et al., 2004). The expansion of wildlife husbandry and conservation has also made Theileria of wildlife important subjects of study. The Theileria can be grouped into schizont “transforming” and “non-transforming” species (Sivakumar et al., 2014). Transforming parasites all group in the T. taurotragi clade (Fig. 2) (Sivakumar et al., 2014), and uncontrolled proliferation of schizonts results in the pathologies associated with Corridor disease (Theileria parva), East Coast fever (T. parva), Tropical theileriosis (T. annulata) in cattle and malignant theileriosis (T. lestoquardi) in goats and sheep (Bishop et al., 2004; McKeever, 2009). However, T. taurotragi, T. sp. (buffalo) and T. sp. (bougasvlei) do not cause schizont associated pathology (Young et al., 1977; Bishop et al., 2004; Pienaar et al., 2014). Theileria sp. (sable), that group within the antelope Theileria (Fig. 2), causes lymphoid hyperplasia typically associated with the transforming Theileria (Nijhof et al., 2005). This and the ability to culture schizonts (Zweygarth et al., 2009a) indicate that not all transforming parasites share a monophyletic origin (Sivakumar et al., 2014), which suggests that transformation of schizonts may occur more widely than expected. The non-transforming Theileria are regarded as being benign but still able to cause disease as a result of anaemia induced by the piroplasm stage (Sivakumar et al., 2014). The last decade has seen an increase in the discovery of new Theileria species and genotypes and the realisation that even domestic animals may harbour an extensive array of mixed infections (Criado-Fornelio et al., 2004; Mans et al., 2011a; Sivakumar et al., 2014). This is compounded by mixed Theileria infections in both mammalian hosts and tick vectors where wildlife and livestock share the same habitat and common tick species (Lawrence et al., 1983; Kariuki et al., 2012). The means to discriminate benign and virulent forms as well as species responsible for disease outbreaks is becoming more important, both for diagnostic and epidemiological purposes.
Fig. 2.


Phylogenetic analysis of Theileria genotypes. The 18S ribosomal RNA sequences were extracted from GenBank using text based queries and BLAST analysis. Final non-redundant dataset were obtained by phylogenetic analysis and manual curating. The dataset were trimmed to include the V4 hyper-variable region, aligned using MAFFT (auto, 200PAM/k = 2; Katoh and Standley, 2013) and analysed using neighbour-joining (bootstrap = 1000, matrix = number of differences) with Mega5 (Tamura et al., 2011). Host and geographic data for genotypes are indicated and were extracted from GenBank files or literature in the case of reverse line blot analysis.
2. Diagnostics
Diagnostic methods are used alone or in combination with other criteria to accurately diagnose disease. The use of assays as diagnostic tools needs to fulfil some pre-requisites albeit for human or veterinary use (Pfaffl, 2004; Bustin, 2010; Peeling et al., 2010). As such the basic performance characteristics of a test need to be determined against a reference standard. This would generate data on reproducibility, sensitivity and specificity of the particular assay, parasite prevalence, genetic variation and/or methodology used: type of antigen or antibody, automated or manual procedures, all contributing to the variables influencing any given assay's performance.
2.1. Microscopy
Historically most Theileria species were first described based on light microscopy examination after Koch's discovery of T. parva (Koch, 1898). It was usually performed on Giemsa stained blood smears, however, the limitation of light microscopy as a diagnostic tool was that the detection of carrier animals regularly went unnoticed and the discrimination between piroplasms of other Theileria species were difficult as they are morphologically very similar (Dschunkowsky and Luhs, 1904; Theiler, 1904; Lawrence, 1935, 1979; Uilenberg, 1981). Even so, discovery of novel species may still utilise morphology as part of their formal description (Nijhof et al., 2005; Clark and Spencer, 2007; Oosthuizen et al., 2009; Paparini et al., 2012), and remains a fast method to derive at an initial differential diagnosis of potential theileriosis in clinical cases (Nijhof et al., 2005; Izzo et al., 2010). In regard to routine diagnostics, microscopy is several orders of magnitude less sensitive than molecular methods (Criado-Fornelio, 2007), and generally unable to accurately detect all carrier-state animals (Lawrence, 1979; Zweygarth et al., 1997).
2.2. Xenodiagnosis
Xenodiagnosis (tick pickup and transmission linked with clinical disease) as demanded by Koch's postulate was used to confirm disease-causing species and tick vectors (Dschunkowsky and Luhs, 1904; Theiler, 1904; Young et al., 1977; Skilton et al., 2002). Direct injection of infected blood may also distinguish species amenable to proliferation in the piroplasm stage (Theiler, 1906). Xenodiagnosis remains a useful tool in basic parasite research and epidemiological investigations and may be the only means to confirm tick vector designation and clinical pathology of specific Theileria strains or as first step in parasite isolation (Ngumi et al., 1994; Steyl et al., 2012; Mbizeni et al., 2013). It is, however, not suitable for high-throughput or routine analysis and remains in the domain of specialised research groups.
2.3. Serological assays
Serological diagnosis by complement fixation (Lichtenheld, 1910) was followed by the more advanced indirect fluorescent antibody test (IFAT) (Schindler and Wokatsch, 1965; Burridge, 1971; Burridge and Kimber, 1972). Antigen may be prepared from schizont or piroplasm antigen, derived from infected animals or cell culture. Cell culture of Theileria parasites may be useful in diagnosis of carrier state animals (Zweygarth et al., 1997), to identify transforming parasites (Zweygarth et al., 2009a, 2009b), as means to generate IFAT antigen (Burridge and Kimber, 1972), and distinguish different species and investigate distinct parasite populations using monoclonal antibodies (Conrad et al., 1987; Bishop et al., 1994a; Alhassan et al., 2007a; Zweygarth et al., 2009b). The IFAT remains the gold standard assay recommended by the OIE for most economically important parasites (OIE, 2014). The IFAT has several drawbacks, namely, subjective operator-dependent interpretation of results, low throughput and difficulty in standardisation (Katende et al., 1998). However, the biggest problem with the IFAT is the significant cross-reactivity observed between closely related species. Cross-reactivity between T. parva and T. taurotragi antigen and anti-sera has been observed (de Vos and Roos, 1981; Jongejan et al., 1986). Similar observation has been made in regard to T. parva and T. sp. (buffalo) (Conrad et al., 1987; Pienaar et al., 2014). An IFAT assay for T. lestoquardi showed significant cross-reactivity with T. annulata and T. parva anti-sera and vice versa (Leemans et al., 1997). Detection by IFAT may therefore be a poor measure to assign parasites to species (Uilenberg, 1981; Uilenberg et al., 1985; Stewart et al., 1996), especially when closely related, such as the genotypes found in the T. buffeli, T. mutans and T. velifera clades (Fig. 2). Even so, IFAT may still be useful in epidemiological studies (Thompson et al., 2008; Mbizeni et al., 2013), or where certain species are absent in specific carrier hosts such as the case for T. taurotragi and African buffalo (T. parva carrier) (Mans et al., 2011a), or T. annulata in small ruminants (T. lestoquardi carrier) (Leemans et al., 1997).
The IFAT utilises whole-body antigen, but the realisation that a select number of antigens are responsible for the dominant immune response against most Theileria parasites has stimulated interest in enzyme-linked immunosorbent assays (ELISA) (Katende et al., 1998). These single molecule assays have the promise of more species specific antigens amenable to high throughput analysis. The polymorphic immunodominant molecule (PIM) of Theileria parva and the p32 antigen of T. mutans were used to develop ELISAs (Katende et al., 1990, 1998; Morzaria et al., 1999). Both were commercially marketed and shown to be more sensitive than the IFAT, but the tests were discontinued due to specificity issues (OIE, 2014). However, both are still being used in research (Swai et al., 2007; Bazarusanga et al., 2008; Kiara et al., 2014). The EMA-1 antigen for T. equi and a monoclonal antibody against this antigen were used to develop a competitive inhibition ELISA (Knowles et al., 1992) that is available as a commercial kit and considered as replacement for the IFAT (OIE, 2014). The EMA-2 antigen for T. equi was also investigated in an indirect ELISA and as rapid immune-chromatographic test (Huang et al., 2003, 2004). Other ELISAs developed include the T. annulata merozoite surface 1 antigen (Tams1) and the T. annulata macroschizont stage protein (TaSP) (Gubbels et al., 2000a; Seitzer et al., 2007; Renneker et al., 2008), a heat-shock 70 antigen for T. sp. (China) (Miranda et al., 2006) and T. uilenbergi immunodominant protein (TuIP) (Liu et al., 2010a). TaSP was also used to develop a rapid lateral flow device assay for T. annulata that compares well with other serodiagnostic assays (Abdo et al., 2010). One of the biggest advantages of ELISA is in its use as a high throughput, cheap and fast method to screen and diagnose large numbers of samples. In this regard, serodiagnosis remains the workhorse for laboratories where molecular infrastructure does not exist.
2.4. Molecular assays
The molecular revolution saw an explosion of diagnostic assays (Criado-Fornelio, 2007), that is target specific genes and species (Table 1). This included conventional PCR followed by agarose gel electrophoretic analysis (Bishop et al., 1992; Pienaar et al., 2011a), PCR-RFLP methods (Bishop et al., 1992; Geysen et al., 1999; Heidarpour Bami et al., 2009; Zaeemi et al., 2011), nested-PCR (Odongo et al., 2010), PCR followed by dot blotting, capillary blotting or slot-blotting and hybridisation using radio-isotope labelled probes (Bishop et al., 1992; Allsopp et al., 1993; Collins et al., 2002; Skilton et al., 2002). The latter was improved by the non-radio-active reverse line blot method that used chemiluminescence (Gubbels et al., 1999; Schnittger et al., 2004), with recent variation on this technique using a DNA bead-based suspension array (Ros-García et al., 2012a, 2013). This was followed by probe based real-time PCR methods (Jeong et al., 2003; Kim et al., 2008; Sibeko et al., 2008; Bhoora et al., 2010a; Pienaar et al., 2011b, 2014; Ros-García et al., 2012b), SYBR green real-time PCR assays (Pienaar et al., 2013), loop-mediated isothermal amplification (LAMP) assays (Alhassan et al., 2007a, 2007b; Liu et al., 2008, 2012, 2013; Salih et al., 2008, 2012; Thekisoe et al., 2010; Wang et al., 2010a; Xie et al., 2013), pan-FRET based assays (Chaisi et al., 2013a; Perera et al., 2014; Yang et al., 2014a) and high-resolution melt analysis (Salim et al., 2013). In all cases, detection by molecular methods allow for direct confirmation of the presence of parasite genomic material, with the inference that live parasites are present in the animal at the moment of sampling. Developments from conventional to nested to real-time PCR has allowed improvement in sensitivity, quantification and speed of detection, while methods such as reverse line blot, bead arrays, pan-FRET assays and high-resolution melt analysis hold the promise of detection of multiple species or genotypes at the same time. Real-time melting profile based assays also hold the advantage that variation in probe regions may be detected by differences in melting profiles that may be related to genotypic or species differences. Whereas many of these assays require specialised equipment, LAMP assays hold the advantage of functioning at iso-thermal conditions, with possible application under field conditions.
Table 1.
A summary of diagnostic assays that exist for the Theileria. Indicated are Theileria species, tick vectors, hosts, diseases caused, serological and diagnostic assays developed.
| Theileria spp. | Tick vector | Hosts | Disease | Serology | Molecular |
|---|---|---|---|---|---|
| T. parva | Rhipicephalus appendiculatus; R. zambeziensis | Cattle, African buffalo | Fatal; East Coast fever, Corridor disease, Zimbabwean theileriosis (January disease) | 1, 2, 3 | 4, 5, 6, 7, 8 |
| T. mutans | Amblyomma sp. | African buffalo, cattle | Benign theileriosis | 1, 9, 10 | 4, 8, 11 |
| T. taurotragi | Rhipicephalus sp. | Cattle, eland (sheep) | Benign African theileriosis | 9, 12 | 4, 8 |
| T. velifera | Amblyomma sp. | African buffalo, cattle | Benign | None | 4, 8 |
|
T. buffeli/ T. sergenti T. orientalis T. sinesis |
Haemaphysalis sp. | African buffalo, cattle, Water buffalo | Oriental theileriosis | 13, 14, 15, 16 | 4, 8, 17, 18, 19, 20 |
| T. lestoquardi | Hyalomma sp. | Sheep, goats | Malignant sheep theileriosis | 21, 22 | 23, 24, 25 |
| T. ovis | Hyalomma sp. Rhipicephalus? | Sheep, goats | Benign | None | 23, 26, 27, 28, 29 |
| T. separata | Rhipicephalus sp., Hyalomma sp. | Sheep, antelope | Benign/pathogenic | None | 23, 24 |
| T. annulata | Hyalomma sp. | Cattle, water/domestic buffalo | Fata; Tropical or Mediterranean theileriosis | 30, 31, 32, 33 | 4, 17, 24, 26, 34, 35, 36, 37 |
| T. sp.(buffalo); T. sp. (bougasvlei) | Not known | African buffalo (South Africa); African buffalo, cattle (East Africa) | Benign | 38 | 8, 39 |
| T. sp. (sable) | R. evertsi/R. appendiculatus? | Sable | Malignant | None | 40 |
| T. equi | Hyalomma spp., Rhipicephalus spp. | Horses, donkeys, giraffes | Acute to chronic | 41, 42, 43, 44 | 45, 46, 47, 48 |
| T. annae | Ixodes sp., Rhipicephalus sp., Dermacentor sp. | Dogs, foxes | Severe, regenerative anaemia | None | 49, 50 |
| T. uilenbergi | Haemaphysalis sp. | Sheep, goats, sika, red deer | Cervine theileriosis | 51 | 52, 53 |
| T. luwenshuni | Haemaphysalis sp. | Sheep, goats, sika, red deer | Cervine theileriosis | 51 | 26, 52, 54 |
| T. bicornis | R. evertsi evertsi | Black, white and Indian rhinoceros | Benign | None | 55 |
| T. capreoli | Ixodes ricinus | Sika, red deer | Theileriosis? | None | 55, 56 |
| T. cervi | Amblyomma americanum | White-tailed deer, elk | Benign | 57 | 58 |
| T. sp. OT 3 | Haemaphysalis sp.? | ? | Theileriosis? | None | 26, 54, 59 |
| T. sp. MK | Not known | Goats, sheep | Benign | None | 60 |
1. Complement-fixation assay (Schindler and Mehlitz, 1969). 2. Piroplasm and schizont IFAT and ELISA (Burridge, 1971; Burridge and Kimber, 1972; Gray et al., 1980). 3. PIM indirect ELISA (Katende et al., 1998). 4. RLB (Gubbels et al., 1999). 5. Conventional and nested PCR for p67, p104, Tpr (Bishop et al., 1992; Skilton et al., 2002; Odongo et al., 2010; Pienaar et al., 2011a). 6. Real-time PCR targeting 18S (Sibeko et al., 2008; Papli et al., 2011; Pienaar et al., 2013). 7. LAMP targeting PIM and p150 (Thekisoe et al., 2010). 8. Pan-FRET real-time PCR for Cox III (Chaisi et al., 2013a). 9. Piroplasm IFAT (de Vos and Roos, 1981). 10. Indirect ELISA for p32 (Katende et al., 1990). 11. Conventional PCR based on non-coding region (Bishop et al., 1994b). 12. Piroplasm IFAT (Jongejan et al., 1986). 13. Piroplasm IFAT (Uilenberg et al., 1985; Papadopoulos et al., 1996), 14. Piroplasm extract ELISA (Shimizu et al., 1988). 15. Latex agglutination test for major piroplasm surface protein p33 (Jeong et al., 2005). 16. Indirect ELISA for p33 (Wang et al., 2010b). 17. Conventional PCR for β-tubulin, MPSP, p23, p32, p33, p34 (Tanaka et al., 1993; Kawazu et al., 1995; Kubota et al., 1996; Govaerts et al., 1998; Sarataphan et al., 1999; Cacciò et al., 2000; Liu et al., 2010b, 2011; Ota et al., 2010). 18. Semi-nested PCR for 18S rRNA (Ghaemi et al., 2012). 19. Multiplex-tandem real-time PCR for the major piroplasm surface protein, 23-kDa piroplasm membrane protein and ITS1 (Perera et al., 2014). 20. LAMP for p33 and ITS (Wang et al., 2010a; Liu et al., 2013). 21. Schizont IFAT (Leemans et al., 1997). 22. Clone 5 ELISA (Bakheit et al., 2006). 23. RLB (Schnittger et al., 2004). 24. PCR-RFLP of 18S rRNA (Heidarpour Bami et al., 2009). 25. LAMP of clone 5 (Salih et al., 2012). 26. Suspension arrays (Ros-García et al., 2012a). 27. Nested PCR of 18S rRNA (Altay et al., 2005). 28. PCR-RFLP of 18S RNA (Heidarpour Bami et al., 2009). 29. 5.8S RNA PCR (Zhang et al., 2014). 30. Piroplasm and schizont IFAT and ELISA (Gray et al., 1980; Kachani et al., 1992; Darghouth et al., 1996). 31. TaSP, Tams1 indirect ELISAs (Bakheit et al., 2004; Salih et al., 2005; Rajendran and Ray, 2014). 32. Competitive ELISA for TaSP (Renneker et al., 2008). 33. TaSP lateral flow device (Abdo et al., 2010). 34. Cytochrome b RLB (Bilgic et al., 2010). 35. Conventional PCR for cytochrome b, HSP70 and Tams1 (d'Oliveira et al., 1995; Shayan et al., 1998; Kirvar et al., 2000; Criado et al., 2006). 36. Conventional and real-time PCR for 18S rRNA gene (Ilhan et al., 1998; Ros-García et al., 2012b). 37. LAMP for the 18S rRNA, ITS and TA04795 (Salih et al., 2008; Liu et al., 2012). 38. Schizont IFAT (Conrad et al., 1987). 39. Real-time LNA probe assay (Pienaar et al., 2014). 40. Real-time PCR (Pienaar, Personal communication). 41. Piroplasm IFAT and complement fixation test (Ogunremi et al., 2007). 42. Competitive ELISA of EMA-1 (Knowles et al., 1992). 43. Indirect ELISA for EMA-2 (Huang et al., 2003). 44. Immuno-chromatographic test for EMA-2 (Huang et al., 2004). 45. RLB (Butler et al., 2008). 46. Conventional PCR of EMA-1, β-tubulin, 18S rRNA (Cacciò et al., 2000; Alhassan et al., 2005; Heim et al., 2007; Salim et al., 2008). 47. Real-time PCR and high resolution melt analysis of 18S rRNA (Kim et al., 2008; Bhoora et al., 2010a; Salim et al., 2013). 48. LAMP of EMA-1 and 18S rRNA (Alhassan et al., 2007a, 2007b; Xie et al., 2013). 49. RLB (Yisaschar-Mekuzas et al., 2013). 50. Conventional PCR-RFLP of 18S rRNA (Jefferies et al., 2007). 51. TlHSP70, rTulP ELISA (Miranda et al., 2006; Liu et al., 2014). 52. RLB (Niu et al., 2009). 53. Conventional PCR of 18S rRNA, RPS8 (Yin et al., 2008; Tian et al., 2013; Zhang et al., 2014). 54. RLB (Nagore et al., 2004). 55. RLB (Nijhof et al., 2003). 55. RLB (García-Sanmartín et al., 2007). 56. Conventional PCR of 18S rRNA (Li et al., 2014). 57. IFAT (Schaeffler, 1963). 58. PCR and sequencing of 18S rRNA (Chae et al., 1999a). 59. Conventional PCR of 18S rRNA (Tian et al., 2014). 60. RLB and conventional PCR of 18S rRNA (Altay et al., 2007, 2008).
3. Specificity of molecular assays and the species concept
Most molecular assays depend on primers and/or probes that target small regions of genes with the implicit assumption that these regions are conserved through all members of a species or genus. In the case of protein genes, the degenerate nature of the genetic code makes the design of specific primers or probes quite difficult, as exemplified by their reduced sensitivity compared to more conserved ribosomal genes (Pienaar et al., 2011a, 2013). Protein genes on the other hand may provide much greater specificity due to more distant orthologous relationships (Odongo et al., 2010; Pienaar et al., 2011a). The challenge in the development of specific assays therefore lies in the identification of these unique regions within genes or a genome, working with often limited information regarding the diversity of a gene or the availability of total genome information. For example, the reverse line blot (RLB) was developed well before extensive genomic information was available for the Theileria genus or estimates regarding 18S rRNA diversity in domestic animals and wildlife (Gubbels et al., 1999). The RLB primers for Theileria and Babesia target the 18S rRNA regions that flank the V4 hyper-variable region and to date have been found to be conserved in all members of these genera (Gubbels et al., 1999). The probes used in the reverse line blot were assumed to be species-specific and to be able to detect all members of a species, based on the assumption that the 18S hyper-variable region is conserved within species. It is therefore also useful to detect new species in the case where only a Theileria catch-all probe is detected (Oosthuizen et al., 2009; Chaisi et al., 2013b, 2014). More recently, some of these assumptions have been challenged.
Diagnostic screening of buffalo and cattle samples using the hybridisation real-time PCR for T. parva (Sibeko et al., 2008) detected samples with aberrant melting curves that suggested variation in the probe region of this assay that occurs within the 18S rRNA V4 hyper-variable region (Mans et al., 2011a). Sequencing of these samples indicated single nucleotide polymorphisms, suggesting that variation in the V4 hyper-variable region may occur in individuals within the T. parva population (Mans et al., 2011a). In geographically isolated populations these polymorphisms may become fixed, leading to cases where species identity may be questioned. It was as such proposed that organisms with 1–2 single nucleotide polymorphisms within the V4 18S rRNA hyper-variable region be considered the same species (Mans et al., 2011a). Conversely, the interesting phenomenon exists, that certain genotypes within the Theileria genus show extensive genotypic variation within the 18S rRNA hyper-variable region, but due to the fact that these groups within the same clade during phylogenetic analysis are considered to be members of the same species. As such, 12 main clades for Theileria can be distinguished (Fig. 2), comprising at least 91 unique genotypes which may or may not be unique species as will be discussed later. The main clades found include the T. buffeli, T. equi, T. mutans, T. taurotragi, T. velifera clades, three clades for antelope Theileria, a clade for marsupial Theileria and for Theileria annae.
The T. taurotragi clade comprises multiple well-recognised species, even though their inter-species genetic distances are very similar to other clades (Mans et al., 2011a). Theileria taurotragi has been found in antelope (bushbuck, eland) and cattle and occurs from East to Southern Africa (Martin and Brocklesby, 1960; Young et al., 1977; de Vos and Roos, 1981; Oura et al., 2011); T. annulata infects cattle and causes tropical theileriosis, while T. lestoquardi infects small ruminants such as sheep and goats and causes malignant theileriosis (Leemans et al., 1999); T. parva infects cattle and African buffalo (Norval et al., 1992); T. sp. (buffalo) infects African buffalo and has been detected in cattle (Allsopp et al., 1993; Zweygarth et al., 2009b; Mans et al., 2011a; Pienaar et al., 2011a; Githaka et al., 2014); T. sp. (bougasvlei) infects African buffalo (Zweygarth et al., 2009b; Mans et al., 2011a; Pienaar et al., 2011a, 2014). Theileria annulata and T. lestoquardi differ by 2 base pairs in the hyper-variable region, while differences in this region between T. parva, T. sp. (buffalo) and T. sp. (bougasvlei) range from 4 to 6 base pairs (Mans et al., 2011a). Even so, analyses using the nuclear S5 and mitochondrial COI genes distinguish these species quite well, while T. sp. (buffalo) and T. sp. (bougasvlei) also show disparate geographic distributions (Mans et al., 2011a; Pienaar et al., 2014). All members of this clade can transform lymphocytes (assumed for T. sp. (bougasvlei)), but this phenomenon is not associated with pathology for all members (Mans et al., 2011b; Sivakumar et al., 2014).
Theileria annae is a parasite of dogs and other canines and is related to Babesia microti (Zahler et al., 2000; Camacho et al., 2001; Najm et al., 2014). It has been assigned to the genus Theileria based on the contention that the small canine Babesias are more closely related to Theileria than Babesia (Zahler et al., 2000). However, molecular analyses of the 18S rRNA gene indicate a close relationship to the Babesia (Matjila et al., 2008; Sivakumar et al., 2014). Other Theilerias found incidentally in dogs include T. annulata and T. equi (Criado-Fornelio et al., 2003; Criado et al., 2006; Beck et al., 2009). However, a genotype identical to T. sp. (sable) has been found extensively in dogs surveyed in KwaZulu-Natal, South Africa (Nijhof et al., 2005; Matjila et al., 2008). Theileria sp. (sable) groups within the clade of Antelope Theilerias have been found in an extensive range of other antelope as well (Nijhof et al., 2005). Since most of these identifications were done by RLB analysis these hosts remain to be confirmed by sequencing. The antelope clade is extensive and comprised of various genotypes found in different antelope species. In many cases, multiple genotypes have been found in single antelope species and are not distinguished by their genotype designation, even though their phylogenetic position would suggest that these may be unique species. This includes genotypes for T. sp. OT3 (Nagore et al., 2004; Tian et al., 2014), T. capreoli (García-Sanmartín et al., 2007; Yang et al., 2014b), T. cervi (Chae et al., 1998, 1999a), giraffe Theileria (Oosthuizen et al., 2009; Githaka et al., 2013), waterbuck Theileria (Githaka et al., 2014), white-tailed deer Theileria, water deer Theileria (Han et al., 2009). Many of these genotypes have been identified in multiple antelope species, making designation according to host specificity difficult. One genotype from this clade has been identified in cheetah and shows a close relationship to a genotype found in giraffes (Githaka et al., 2012).
The Theileria found in marsupials all cluster together and comprise 7 unique genotypes. Theileria fuliginosa is found in the western grey kangaroo (Macropus fuliginosus), T. penicillata in the woylie (Bettongia penicillata ogilbyi), T. gilberti in Gilbert's potoroo (Potorous gilbertii), T. sp. long-nosed potoroo in the long-nosed potoroo (Potorous tridactylus) and T. sp. K1 in the boodie (Bettongia lesueur) (Clark and Spencer, 2007; Lee et al., 2009; Paparini et al., 2012). Two distinct genotypes for T. brachyuri have been found in the quokka (Setonix brachyurus) (Clark and Spencer, 2007), suggesting that at least one is a different species. At least three other marsupial Theileria exist for which no molecular data are available; they include T. tachyglossi in the echidna, T. peramelis in the southern brown bandicoot (Isoodon obesulus), and T. ornithorhynchi in the platypus (Ornithorhynchus anatinus) (Lee et al., 2009; Paparini et al., 2012). The monophyletic relationship within this clade would suggest a single introduction into Australia, with subsequent host adaptations and speciation.
The T. buffeli clade possesses the most extensive number of genotypes (13) and occurs in all the major continents of the world, where they infect cattle, African buffalo, water buffalo and Yak (Chaisi et al., 2013a; Sivakumar et al., 2014). A number of different species names have historically been assigned to members of this clade including T. buffeli, T. orientalis and T. sergenti, but have been proposed to represent a single species designated as T. buffeli (Stewart et al., 1996; Gubbels et al., 2000b, 2002). This builds on a similar proposal by Uilenberg (1981) and Uilenberg et al. (1985) that used morphological and serological cross-reactivity to assign all members to T. orientalis. Both T. buffeli and T. orientalis continue to be used in literature depending on the historical background of different scientific groups (Chaisi et al., 2014; Sivakumar et al., 2014). Another “species” that belongs to this group is T. sinensis (Bai et al., 2002). The genotypes from this clade were postulated to be in the process of speciation that has not been completed yet (Schnittger et al., 2003), while others considered members to be distinct species (Kawazu et al., 1992; Fujisaki et al., 1994). Most members of this group are considered to be benign, but a recent outbreak of a virulent genotype (Ikeda type) in Australia readdressed the possibility of unique species in this group (Izzo et al., 2010; Kamau et al., 2011). More recently, related genotypes (T. buffeli C, T. sinensis-like) were found in African buffalo but not in cattle from the same geographic regions in southern Africa (Mans et al., 2011a; Chaisi et al., 2014). All these genotypes are generally considered to be genotypic variants of T. buffeli (Chae et al., 1999b; Gubbels et al., 2000b, 2002; Chaisi et al., 2014), even though host specificity, clinical presentation and specific geographic distributions are different. The use of other genes such as the major piroplasm surface protein (MPSP) also generally groups different genotypes into separate clades that correspond with those obtained using the 18S gene (Kamau et al., 2011; Sivakumar et al., 2014). Serological and molecular assays that can distinguish different genotypes have been developed (Kawazu et al., 1992, 1995; Liu et al., 2010b, 2013; Perera et al., 2014). It thus seem surprising that this clade does not have well recognised species, which complicates development of specific diagnostic assays and possible interpretation of diagnostic results. The relative caution in molecular species designation within this clade that occurred more than 14 years ago (Chae et al., 1999b; Gubbels et al., 2000b) has also permeated to other Theileria clades such as T. mutans, T. velifera, T. equi, the Theileria antelope clade and members of the T. taurotragi clade (discussed in their respective sections), irrespective of the fact that no recombination experiments have yet been attempted between different genotypes, that may warrant the conclusion of a single Theileria species per clade.
The T. mutans clade is comprised of five recognised genotypes, namely T. mutans that infects cattle and buffalo, T. mutans MSD that infects cattle and buffalo, T. mutans-like 1, T. mutans-like 2 and T. mutans-like 3 that has thus far been found only in African buffalo (Chae et al., 1999b; Mans et al., 2011a; Chaisi et al., 2013b). Of interest is that the T. mutans-like 1–3 genotypes were not detected by the T. mutans RLB probe, but is still considered to be genotypic variants of T. mutans (Chaisi et al., 2013b), even though they differ by 7–13 nucleotide as well as a gapped position (Mans et al., 2011a). However, to date T. mutans-like 1–3 have only been observed in African buffalo and occur at significantly higher prevalence and parasitaemia compared to T. mutans and T. mutans MSD, which occur extensively in cattle (B.J. Mans, personal observation). Host specificity would therefore suggest that at least two species exist in this clade.
The T. velifera clade is comprised of four genotypes, T. velifera, T. velifera A, T. velifera B and T. sp. RR-2012, an as yet undescribed genotype from Indian bovines. T. velifera and T. velifera A have been found in cattle and buffalo, while T. velifera B has only been found in buffalo (Mans et al., 2011a; Chaisi et al., 2013b). Significant similarity exists between the RLB probe for T. sp. (sable) and T. velifera, so that reports of the extensive occurrence of the former in cattle and African buffalo (Nijhof et al., 2005; Yusufmia et al., 2010) could be traced back to the presence of T. velifera (Mans et al., 2011a). This again underscores the pitfalls of the RLB approach, when used for screening of novel vertebrate species or animals from new geographic regions, for which the assay has not been validated.
The T. equi clade shows similar genetic diversity as the T. buffeli clade (twelve genotypes) and is also considered to be representative of a single species (Kim et al., 2008; Bhoora et al., 2009; Salim et al., 2010). This is supported by extensive cross-reactivity on IFAT (Bhoora et al., 2009). Some genotypes have thus far been confined to equids other than horses, such as donkeys and zebra, but have also been found in waterbuck (Bhoora et al., 2010b; Githaka et al., 2014). Whether these will infect horses remains to be determined, but the wide distribution observed for most genotypes suggests that none are host specific (Fig. 2). Multiple genotypes also generally occur in similar geographic localities which make the assessment of strain virulence difficult, with the general assumption that all strains present similar virulence and pathologies. A recently developed real-time PCR assay selected a region within the 18S ribosomal RNA gene, for which both primers and probes are conserved in the Theileria (Kim et al., 2008; Bhoora et al., 2010a). It will therefore detect all genotypes from T. equi when used to screen equids but will not be useful for screening other animals suspected to harbour T. equi such as waterbuck (Githaka et al., 2014). From an equid epidemiological control perspective this may be ideal if the clade is treated as a single species or virulent group. However, from a phylogeographic perspective where evolution of unique genotypes is considered, large-scale surveys that rely on diagnostic assays will be hampered.
Two genotypes that do not fit into any major clade include T. bicornis found in black, Indian and white rhinoceros (Nijhof et al., 2003; Otiende et al., 2014). The other was found in domestic cats in a Brazilian zoo and may be an incidental infection transmitted from another felid carrier (André et al., 2014).
The origin of the genotypic diversity observed should be considered. To date genomes for T. annulata, T. equi, T. parva and T. orientalis have been sequenced and all indicate the presence of two 18S ribosomal RNA genes (Gardner et al., 2005; Pain et al., 2005; Hayashida et al., 2012; Kappmeyer et al., 2012). In all cases these genes are highly conserved, most probably due to concerted evolution (Eickbush and Eickbush, 2007), with the implication that genotypic diversity observed within the 18S gene for the Theileria is not due to multiple gene copies within their genomes (Mans et al., 2011a). Genotypic diversity observed in the 18S genes will therefore be due to conventional divergent evolution, linked with mutation and insertion–deletion events.
The genotypic diversity observed within the Theileria has interesting implications from a diagnostic perspective. On the one hand, some species such as T. annulata, T. lestoquardi, T. parva, T. sp. (buffalo) and T. sp. (bougasvlei) seem to be highly conserved which allows for the development of specific probe-based assays that may be used to detect all individuals within the species (Schnittger et al., 2003; Pienaar et al., 2011b, 2014; Ros-García et al., 2012b). Conversely, if it is assumed that some clades do represent single species with genotypic diverse members, they are so diverse that no single assay may be able to detect all genotypes and therefore all individuals within the species. This necessitates the development of multiple assays or probes for geographically diverse populations (Gubbels et al., 2000b; Chaisi et al., 2013a; Perera et al., 2014).
The same generalities are found for all clades in the literature, i.e. when different genotypes are observed from a single host species and genotypes group within a monophyletic clade, it is assumed that these are the same Theileria species. Conversely, finding a single genotype associated with a single host species, even if the sampling was small and the genotype has not previously been observed, will lead to the conclusion that this is a unique species. From an evolutionary perspective this situation is extremely confusing. It suggests recent speciation of genotypes such as T. parva, T. sp. (buffalo) and T. sp. (bougasvlei) that allows conservation of the 18S V4 hyper-variable region (Mans et al., 2011a; Pienaar et al., 2014), even though evidence suggests a more ancient origin, since T. parva for example shows extensive genetic diversity in field populations (McKeever, 2009). It also implies that the 18S ribosomal genes in the T. buffeli, T. equi and T. mutans clades had undergone rapid evolution, contravening the ribosomal molecular clock (Lack et al., 2012), or that these clades have an ancient origin with diversification of lineages, while maintaining their species identity. The latter may occur via bottleneck events that result in clonal populations in which the 18S gene diverged (Tibayrenc and Ayala, 2002; Tibayrenc, 2006; McKeever, 2009). Why this would be more prone in certain clades is uncertain, while divergence of the whole genome may be expected. Designation of species then becomes arbitrary and retaining diverse genotypes in a single species may be a social and philosophic response to a complex problem (Tibayrenc, 2006). An alternative and more parsimonious scenario would posit dispersal and geographic isolation linked with speciation of vertebrate host species during the Middle–Late Miocene (~13–8 MYA) for the ancestral lineages to the Bubalina (African and Asiatic buffalo and cattle) (Hassanin et al., 2012) and the Pliocene (4–3 MYA) for the Bovina (cattle and bison) (Hassanin et al., 2012, 2013) and caballine and non-caballine equids (Vilstrup et al., 2013). This coincided with diversification of Theileria parasites within the different clades (Criado-Fornelio et al., 2003; Gou et al., 2013). Subsequent breakdown of geographic barriers could have resulted in co-infection of Bovini hosts by related genotypes unable to sexually recombine, but still able to infect similar vectors and hosts. In this case, the genotypic diversity observed within the respective clades should be interpreted as distinct lineages, i.e. species. With ~6000 named apicomplexan species, which comprise only 0.1% of the estimated number of apicomplexan species (1.2–10 million) (Morrison, 2009), it seems rather conservative that only 91 genotypes and ~40 species have been described thus far in the Theileria.
4. Sensitivity of molecular assays, carrier state and parasitaemia ranges
The theoretical limit of detection (LOD) of molecular PCR assays is three molecules per assay, which generally equates with a sensitivity cut-off of ~37 cycles (Bustin, 2010). In practice, however, this may range from 3 – > 100 molecules and a cut-off from 37 to 40 cycles (Burns and Valdivia, 2008; Bhoora et al., 2010a). This depends on a complex array of factors, including assay mixture, robustness of the polymerase enzyme, inhibitors in the sample, primer and probe design and thermodynamics, effective temperature cycling and detector sensitivity of PCR equipment. In the case of Theileria, most assays detect genomic DNA and sensitivity will therefore also be influenced by DNA extraction efficiency (i.e. yield), gene copy number in the genome, number of parasites (genomic copies) per cell sampled and the parasitaemia level.
Sensitive quantitative real-time PCR assays have been developed for T. parva and T. equi. In all cases the analytical LOD ranges from 3 to 10 copies, with efficiencies that fall within the acceptable levels for a PCR assay (Kim et al., 2008; Sibeko et al., 2008; Bhoora et al., 2010a; Pienaar et al., 2011b). It is, however, of interest to put this into practical perspective with regard to the detection limit in carrier animals. For the T. parva test, the LOD for whole blood was ~10 parasites from the equivalent of 5 µL whole blood (Sibeko et al., 2008). This calculates back to ~2000 parasites/mL of blood and 19 million parasites in a 500 kg buffalo. Similarly, nested PCR based on the p104 gene, conventional PCR based on the p104 gene and reverse line blot for T. parva had LOD of 400–2000 parasites/mL of blood (Skilton et al., 2002; Oura et al., 2004; Odongo et al., 2010). For the T. equi assay, the LOD was 190 parasites from the equivalent of 5 µL whole blood (Bhoora et al., 2010a). This calculates to a detection limit of 376–1520 million parasites in a 500 kg horse if the average number of parasites varies from 1 to 4 per red blood cell. Similarly, tests for T. annulata (d'Oliveira et al., 1995; Kirvar et al., 2000; Ros-García et al., 2012b) and T. buffeli (Jeong et al., 2003) gave LOD of 23–54 and 108 million parasites, respectively, in cattle weighing 500 kg. A sensitive PCR-RLB assay for T. annulata, based on the cytochrome b gene, gave a LOD of 100,000 parasites per liter (Bilgic et al., 2010). A nested PCR for T. ovis gave a LOD of 1200 parasites/mL of blood (Altay et al., 2005), which calculates to a detection limit of 5.7 million parasites in an 80 kg sheep. It would thus seem that in general, the LOD in animals is >400,000 parasites/L blood, implying that assays for most Theileria parasites, even if sensitive from an analytical perspective, may fail to detect carrier animals from a diagnostic perspective. The potential for false-negative results can, however, only be assessed within the biological context of the parasite within its vertebrate host, i.e. parasitaemia ranges and the carrier-state that impacts on the ability to infect the tick vector and subsequent transmission to a new host.
Parasitaemia is generally expressed as percentage infected red blood cells. It would seem that all Theileria species have defined parasitaemia ranges (Young et al., 1986; Ueti et al., 2012). During clinical reactions, piroplasm parasitaemia may be as high as 1–45% depending on the species (Uilenberg and Schreuder, 1976; Young et al., 1978; de Waal, 1992; Latif et al., 2002; Schetters et al., 2010), but this generally drops well below 1% for the asymptomatic carrier state (Uilenberg and Schreuder, 1976; Young et al., 1986; Grootenhuis et al., 1987; Zaeemi et al., 2011; Ueti et al., 2012). Quantitative real-time PCR has allowed estimation of parasitaemia ranges in hosts and the first interesting observation was that parasitaemia ranges approximate normal distributions (Pienaar et al., 2011a, 2014). These ranges will be determined by the specific biology of the parasite in the carrier state, i.e. ability to maintain and propagate in the vertebrate host (will be discussed later). It also implies that the specific parasitaemia range of a species may determine its practical LOD. As such, >95% of the T. parva population in African buffalo have parasitaemias >0.00001%, which is well above the determined detection limit (Pienaar et al., 2011a). The same holds for T. sp. (buffalo) and T. sp. (bougasvlei) (Pienaar et al., 2011a, 2014). It is difficult to make similar estimations for other Theileria species since the numbers of animals sampled do not allow construction of representative frequency distribution plots of published Ct values (Jeong et al., 2003; Kim et al., 2008; Bhoora et al., 2010a). However, from work performed in our own laboratories we may conclude that the parasitaemia range for T. equi is above the LOD in horses from endemic areas (personal observation). Similarly, most benign Theileria species generally have higher parasitaemia ranges in the carrier state and it may be concluded that real-time PCR assays for most Theileria species should be able to detect the majority of infected animals in a herd.
Exceptions to this may occur where the sensitivity of the assay is affected by the presence of mixed infections. Sensitivity of the RLB is severely affected by the fact that universal primers are used and these are depleted by the predominant species present, thereby suppressing the signal from less abundant template (Pienaar et al., 2011a). The sensitivity of the hybridisation assay for T. parva (Sibeko et al., 2008) is similarly affected by the presence of T. sp. (buffalo)-like parasites, since the primers for this assay also amplify the latter parasite template (Pienaar et al., 2011a). Design of more specific primers and stringent conditions alleviated this suppression problem, at the cost of being slightly less sensitive in regard to analytical sensitivity, but with increased overall performance for field samples (Pienaar et al., 2011b). Other assays that may also be affected by sensitivity suppression include the cox III assay that uses universal primers and may be compounded by the implementation of a nested PCR strategy (Chaisi et al., 2013b). In regard to the latter, nested PCR may increase overall sensitivity (Schnittger et al., 2004; Odongo et al., 2010; Ueti et al., 2012), but may introduce contamination problems in the routine diagnostic laboratory, or if primers are not specific, still lead to PCR suppression by the most dominant genotypes. In addition, it is well accepted that parasitaemia can fluctuate and drop below detection limits, only to increase during relapse, the so-called sporadic carrier state (Norval et al., 1992). This generally occurs under conditions of stress, immune suppression or splenectomy (Hooshmand-Rad, 1976; Zobba et al., 2008). Fluctuation in parasitaemia from below the PCR LOD is, however, not extensively documented and may contribute little to herd surveillance, but may have a significant impact on disease epidemiology, precipitating in disease outbreaks. Multiple testing rounds are generally considered as solution for this eventuality (Ueti et al., 2012).
The carrier-state for piroplasmida may be defined as the long-term persistence of a parasite in its host, with the ability of transmission to other hosts via vector infection, to maintain a transmission cycle. Long-term persistence implies that the parasite can maintain itself and propagate in the vertebrate host while escaping the immune system. In most Theileria, it is accepted that the piroplasm stage is maintained in the host via asexual division and re-infection of red blood cells (Norval et al., 1992). In the case of T. parva, limited division of piroplasms has been observed (Conrad et al., 1986; Fawcett et al., 1987). However, it has been proposed that a reservoir of schizonts that maintains parasitaemia levels exists that escapes the immune system, especially since schizonts sampled 657 days after initial infection could be propagated in cell culture (Grootenhuis et al., 1987). Maintenance of parasitaemia levels is necessary, since red blood cells have a lifespan that range from 70 to 160 days in large mammals before being destroyed (Adili and Melizi, 2014). This would imply that parasites need to replicate and re-infect new red blood cells at least once in this period to maintain their parasitaemia levels. In this regard, remarkable stability in piroplasm parasitaemia levels has been observed in T. parva carrier buffalo kept under tick-free conditions where animals maintained parasitaemia levels for up to 20 years (Pienaar et al., 2014). It is therefore also accepted that once infected, animals may remain lifelong carriers of Theileria (de Waal, 1992; Norval et al., 1992). While this is generally true, fluctuations in parasitaemia and loss of carrier state has also been documented, especially in cases where true carrier-status cannot be established, such as the case for Corridor disease and East Coast Fever.
In the case of East Coast Fever, infection of animals using specific strains of Muguga resulted in fluctuating parasitaemia above and below the LOD of the assay, showing in principle that fluctuating parasitaemia was possible for T. parva. Animals infected with this strain will eventually clear it completely and will not be infective to ticks (Bishop et al., 1992; Skilton et al., 2002; Odongo et al., 2010). Whether this is representative for field strains is not known, since this strain has been propagated under laboratory conditions for extensive time periods. In contrast, the Marikebuni strain maintained the carrier-state indefinitely well above the LOD (Bishop et al., 1992; Skilton et al., 2002). Prevalence of T. parva in cattle from East Coast fever endemic regions range between 37 and 42% using nested PCR (Odongo et al., 2010), compared to ~80% in buffalo (Pienaar et al., 2011a), suggesting that the former is not compatible with an endemic stable situation. In East Coast fever, the carrier-state may therefore be strain dependent and intermittent. For Corridor disease, cattle infected with T. parva that survive may maintain the infection for up to 3 months, but with decreasing parasitaemia until the infection is cleared, with no relapse observed yet (even in splenectomised cattle) and unable to infect ticks (Mbizeni et al., 2013). The latter case may represent the dynamics of a parasite not fully adapted to its host and where the carrier-state may only be intermittent. In this case, the seasonality of brown ear ticks in South Africa preclude the maintenance of a transmission cycle (Norval et al., 1991; Mbizeni et al., 2013), and buffalo-adapted T. parva may therefore not have true carrier status in cattle. The latter becomes an important consideration in epidemiological surveillance, since the presence of parasite DNA or positive serology may not be indicative of true host carrier status, but only exposure to parasites during tick transmission.
5. Epidemiology in the context of species
Theileria epidemiology considers parasite and vector distribution, mortality and morbidity of disease outbreaks, disease outbreak risk assessment and disease control measures, socio-economic factors, climate change, host resistance and susceptibility (Gachohi et al., 2012). While many of these factors may be addressed by clinical differential diagnosis, vector mapping, vaccination strategies and control policy, serological and molecular diagnosis remain central to confirmation of parasite identity, distribution, disease surveillance, assessment of vaccine and as tool for informative control policies (Bakheit et al., 2007). From a disease control standpoint, accurate diagnostics and communication is hampered by loose treatment of species concepts and lumping of genotypes into large groups collectively designated as single species. Examples of this are the emergence of different virulent “strains or genotypes” from “T. buffeli” that cause outbreaks in geographic diverse regions (Izzo et al., 2010; Kamau et al., 2011; Sivakumar et al., 2014), or the presence of T. parva in South Africa that causes buffalo-derived Corridor disease, but not cattle-derived East Coast fever (Mbizeni et al., 2013). In contrast to the T. buffeli clade where ample genetic evidence exists for diverse genotypes, the case for separating the T. parva sub-types based on genetic evidence is not clear and the trinomial system used to distinguish Corridor disease (T. parva lawrencei), East Coast fever (T. parva parva) and January disease (T. parva bovis) was abolished for more than 20 years ago (Norval et al., 1992; Perry and Young, 1993). The biological differences between these “strains” are, however, well documented (Lawrence, 1979; Perry and Young, 1993) and treatment as one species disregards the epidemiology of these parasites. The search for markers to distinguish between these strains has continued, but with little success (Sibeko et al., 2010, 2011). Again, the probability exists that these parasites are related via an evolutionary continuum, i.e. recent divergence that will eventually result in speciation due to host specificity.
The study of parasite epidemiology is further complicated by treatment of distinct genotypes as discrete units (species), since this may mask additive or competitive effects between genotypic populations. Treatment of T. sp. (buffalo) and T. sp. (bougasvlei) as one species (Chaisi et al., 2011; Mans et al., 2011b) could have missed their skewed geographic distribution with regard to each other (Pienaar et al., 2014). Similarly, genotypes from the T. mutans, T. velifera and T. buffeli clades (Chaisi et al., 2013a, 2014) have skewed distributions in cattle and African buffalo in southern Africa based on screening data of ~1000 cattle and 1000 buffalo distributed across the region (B.J. Mans, personal observation). While these are anectodal in regard to all Theileria genotypes, similar disparities in genotypic distributions are likely to be prevalent in different hosts and geographic regions. To ultimately understand the phylogeographic history of the Theileria genus, all genotypes need to be accounted for. In this regard, lumping genotypes into species does not unify, but rather obscures lineage specific life histories.
6. Conclusion
Extensive resources exist for diagnostics of Theileria parasites. However, many of the available assays are restricted by limitations in specificity and sensitivity, and a positive diagnosis may in some cases only be made by multiple assays or xenodiagnosis. Critical examination of the measurements of uncertainty inherent in all assays will allow future improvements in diagnostic capability and may also contribute towards a better understanding of parasite biology. This in turn should have a positive impact on disease epidemiology.
Conflict of interest
The authors declared that there is no conflict of interest.
Acknowledgements
Funding for this review was provided by the Department of Agriculture, Forestry and Fisheries (DAFF) of South Africa (0V21/03/C148) and Theileria Diagnostics (15/08/1P01) of Parasites, Vectors and Vector-borne Diseases (PVVD), Onderstepoort Veterinary Institute, Agricultural Research Council, South Africa.
References
- Abdo J., Kristersson T., Seitzer U., Renneker S., Merza M., Ahmed J. Development and laboratory evaluation of a lateral flow device (LFD) for the serodiagnosis of Theileria annulata infection. Parasitol. Res. 2010;107:1241–1248. doi: 10.1007/s00436-010-1994-8. [DOI] [PubMed] [Google Scholar]
- Adili N., Melizi M. Preliminary study of the influence of red blood cells morphometry on the species determinism of domestic animals. Vet. World. 2014;7:219–223. [Google Scholar]
- Adl S.M., Simpson A.G., Lane C.E., Lukeš J., Bass D., Bowser S.S. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhassan A., Pumidonming W., Okamura M., Hirata H., Battsetseg B., Fujisaki K. Development of a single-round and multiplex PCR method for the simultaneous detection of Babesia caballi and Babesia equi in horse blood. Vet. Parasitol. 2005;129:43–49. doi: 10.1016/j.vetpar.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Alhassan A., Govind Y., Tam N.T., Thekisoe O.M.M., Yokoyama N., Inoue N. Comparative evaluation of the sensitivity of LAMP, PCR and in vitro culture methods for the diagnosis of equine piroplasmosis. Parasitol. Res. 2007;100:1165–1168. doi: 10.1007/s00436-006-0430-6. [DOI] [PubMed] [Google Scholar]
- Alhassan A., Thekisoe O.M., Yokoyama N., Inoue N., Motloang M.Y., Mbati P.A. Development of loop-mediated isothermal amplification (LAMP) method for diagnosis of equine piroplasmosis. Vet. Parasitol. 2007;143:155–160. doi: 10.1016/j.vetpar.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Allsopp B.A., Baylis H.A., Allsopp M.T.P.E., Cavalier-Smith T., Bishop R.P., Carrington D.M. Discrimination between six species of Theileria using oligonucleotide probes which detect small subunit ribosomal RNA sequences. Parasitology. 1993;107:157–165. doi: 10.1017/s0031182000067263. [DOI] [PubMed] [Google Scholar]
- Altay K., Dumanli N., Holman P.J., Aktas M. Detection of Theileria ovis in naturally infected sheep by nested PCR. Vet. Parasitol. 2005;127:99–104. doi: 10.1016/j.vetpar.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Altay K., Dumanli N., Aktas M. Molecular identification, genetic diversity and distribution of Theileria and Babesia species infecting small ruminants. Vet. Parasitol. 2007;147:161–165. doi: 10.1016/j.vetpar.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Altay K., Aktas M., Dumanli N., Aydin M.F. Evaluation of a PCR and comparison with RLB for detection and differentiation of Theileria sp. MK and other Theileria and Babesia species of small ruminants. Parasitol. Res. 2008;103:319–323. doi: 10.1007/s00436-008-0973-9. [DOI] [PubMed] [Google Scholar]
- André M.R., Baccarim Denardi N.C., Marques de Sousa K.C., Gonçalves L.R., Henrique P.C., Grosse Rossi Ontivero C.R. Arthropod-borne pathogens circulating in free-roaming domestic cats in a zoo environment in Brazil. Ticks Tick Borne Dis. 2014;5:545–551. doi: 10.1016/j.ttbdis.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Bai Q., Liu G.Y., Yin H., Zhao Q.Z., Liu D.K., Ren J.X. Theileria sinensis nov: study on molecular taxonomy. Xu Mu Shou Yi Xue Bao. 2002;33:185–190. [Google Scholar]
- Bakheit M.A., Schnittger L., Salih D.A., Boguslawski K., Beyer D., Fadl M. Application of the recombinant Theileria annulata surface protein in an indirect ELISA for the diagnosis of tropical theileriosis. Parasitol. Res. 2004;92:299–302. doi: 10.1007/s00436-003-1055-7. [DOI] [PubMed] [Google Scholar]
- Bakheit M.A., Seitzer U., Ahmed J.S. A new recombinant protein-based ELISA for the diagnosis of malignant theileriosis of sheep and goats. Parasitol. Res. 2006;98:145–149. doi: 10.1007/s00436-005-0034-6. [DOI] [PubMed] [Google Scholar]
- Bakheit M.A., Seitzer U., Mbati P.A., Ahmed J.S. Serological diagnostic tools for the major tick-borne protozoan diseases of livestock. Parassitologia. 2007;49:S53–S62. [PubMed] [Google Scholar]
- Barnett S.F. Theileria. In: Kreier J.P., editor. vol. IV. Academic Press; New York, USA: 1977. pp. 77–113. (Parasitic Protozoa). [Google Scholar]
- Bazarusanga T., Geysen D., Vercruysse J., Marcotty T. The sensitivity of PCR and serology in different Theileria parva epidemiological situations in Rwanda. Vet. Parasitol. 2008;154:21–31. doi: 10.1016/j.vetpar.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Beck R., Vojta L., Mrljak V., Marinculić A., Beck A., Zivicnjak T. Diversity of Babesia and Theileria species in symptomatic and asymptomatic dogs in Croatia. Int. J. Parasitol. 2009;39:843–848. doi: 10.1016/j.ijpara.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Bhoora R., Franssen L., Oosthuizen M.C., Guthrie A.J., Zweygarth E., Penzhorn B.L. Sequence heterogeneity in the 18S rRNA gene within Theileria equi and Babesia caballi from horses in South Africa. Vet. Parasitol. 2009;159:112–120. doi: 10.1016/j.vetpar.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Bhoora R., Quan M., Franssen L., Butler C.M., van der Kolk J.H., Guthrie A.J. Development and evaluation of real-time PCR assays for the quantitative detection of Babesia caballi and Theileria equi infections in horses from South Africa. Vet. Parasitol. 2010;168:201–211. doi: 10.1016/j.vetpar.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Bhoora R., Buss P., Guthrie A.J., Penzhorn B.L., Collins N.E. Genetic diversity of piroplasms in plains zebra (Equus quagga burchellii) and Cape mountain zebra (Equus zebra zebra) in South Africa. Vet. Parasitol. 2010;174:145–149. doi: 10.1016/j.vetpar.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Bilgic H.B., Karagenc T., Shiels B., Tait A., Eren H., Weir W. Evaluation of cytochrome b as a sensitive target for PCR based detection of T. annulata carrier animals. Vet. Parasitol. 2010;174:341–347. doi: 10.1016/j.vetpar.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Bishop R., Sohanpal B., Kariuki D.P., Young A.S., Nene V., Baylis H. Detection of a carrier state in Theileria parva-infected cattle by the polymerase chain reaction. Parasitology. 1992;104:215–232. doi: 10.1017/s0031182000061655. [DOI] [PubMed] [Google Scholar]
- Bishop R., Musoke A., Morzaria S., Gardner M., Nene V. Theileria: intracellular protozoan parasites of wild and domestic ruminants transmitted by ixodid ticks. Parasitology. 2004;129:S271–S283. doi: 10.1017/s0031182003004748. [DOI] [PubMed] [Google Scholar]
- Bishop R.P., Spooner P.R., Kanhai G.K., Kiarie J., Latif A.A., Hove T. Molecular characterization of Theileria parasites: application to the epidemiology of theileriosis in Zimbabwe. Parasitology. 1994;109:573–581. doi: 10.1017/s0031182000076459. [DOI] [PubMed] [Google Scholar]
- Bishop R.P., Sohanpal B.K., Morzaria S.P. Cloning and characterisation of a repetitive DNA sequence from Theileria mutans: application as a species-specific probe. Parasitol. Res. 1994;80:33–41. doi: 10.1007/BF00932621. [DOI] [PubMed] [Google Scholar]
- Burns M., Valdivia H. Modelling the limit of detection in real-time quantitative PCR. Eur. Food Res. Technol. 2008;226:1513–1524. [Google Scholar]
- Burridge M.J. Application of the indirect fluorescent antibody test in experimental East Coast fever (Theileria parva infection in cattle) Res. Vet. Sci. 1971;12:338–341. [PubMed] [Google Scholar]
- Burridge M.J., Kimber C.D. The indirect fluorescent antibody test for experimental East Coast fever (Theileria parva infection of cattle). Evaluation of a cell culture schizont antigen. Res. Vet. Sci. 1972;13:451–455. [PubMed] [Google Scholar]
- Bustin S.A. Why the need for qPCR publication guidelines? – the case for MIQE. Methods. 2010;50:217–226. doi: 10.1016/j.ymeth.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Butler C.M., Nijhof A.M., van der Kolk J.H., de Haseth O.B., Taoufik A., Jongejan F. Repeated high dose imidocarb dipropionate treatment did not eliminate Babesia caballi from naturally infected horses as determined by PCR-reverse line blot hybridization. Vet. Parasitol. 2008;151:320–322. doi: 10.1016/j.vetpar.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Cacciò S., Cammà C., Onuma M., Severini C. The beta-tubulin gene of Babesia and Theileria parasites is an informative marker for species discrimination. Int. J. Parasitol. 2000;30:1181–1185. doi: 10.1016/s0020-7519(00)00105-3. [DOI] [PubMed] [Google Scholar]
- Camacho A.T., Pallas E., Gestal J.J., Guitián F.J., Olmeda A.S., Goethert H.K. Infection of dogs in north-west Spain with a Babesia microti-like agent. Vet. Rec. 2001;149:552–555. doi: 10.1136/vr.149.18.552. [DOI] [PubMed] [Google Scholar]
- Chae J., Lee J., Kwon O., Holman P.J., Waghela S.D., Wagner G.G. Nucleotide sequence heterogeneity in the small subunit ribosomal RNA gene variable (V4) region among and within geographic isolates of Theileria from cattle, elk and white-tailed deer. Vet. Parasitol. 1998;75:41–52. doi: 10.1016/s0304-4017(97)00183-0. [DOI] [PubMed] [Google Scholar]
- Chae J.S., Waghela S.D., Craig T.M., Kocan A.A., Wagner G.G., Holman P.J. Two Theileria cervi SSU RRNA gene sequence types found in isolates from white-tailed deer and elk in North America. J. Wildl. Dis. 1999;35:458–465. doi: 10.7589/0090-3558-35.3.458. [DOI] [PubMed] [Google Scholar]
- Chae J.S., Allsopp B.A., Waghela S.D., Park J.H., Kakuda T., Sugimoto C. A study of the systematics of Theileria spp. based upon small-subunit ribosomal RNA gene sequences. Parasitol. Res. 1999;85:877–883. doi: 10.1007/s004360050651. [DOI] [PubMed] [Google Scholar]
- Chaisi M.E., Sibeko K.P., Collins N.E., Potgieter F.T., Oosthuizen M.C. Identification of Theileria parva and Theileria sp. (buffalo) 18S rRNA gene sequence variants in the African Buffalo (Syncerus caffer) in southern Africa. Vet. Parasitol. 2011;182:150–162. doi: 10.1016/j.vetpar.2011.05.041. [DOI] [PubMed] [Google Scholar]
- Chaisi M.E., Janssens M.E., Vermeiren L., Oosthuizen M.C., Collins N.E., Geysen D. Evaluation of a real-time PCR test for the detection and discrimination of Theileria species in the African buffalo (Syncerus caffer) PLoS ONE. 2013;8:e75827. doi: 10.1371/journal.pone.0075827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisi M.E., Collins N.E., Potgieter F.T., Oosthuizen M.C. Sequence variation identified in the 18S rRNA gene of Theileria mutans and Theileria velifera from the African buffalo (Syncerus caffer) Vet. Parasitol. 2013;191:132–137. doi: 10.1016/j.vetpar.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Chaisi M.E., Collins N.E., Oosthuizen M.C. Phylogeny of Theileria buffeli genotypes identified in the South African buffalo (Syncerus caffer) population. Vet. Parasitol. 2014;204:87–95. doi: 10.1016/j.vetpar.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Clark P., Spencer P.B.S. Description of three new species of Theileria Bettencourt, Franca Borges, 1907 from Macropodoidea in Western Australia. Trans. R. Soc. S. Aust. 2007;131:100–106. [Google Scholar]
- Collins N.E., Allsopp M.T., Allsopp B.A. Molecular diagnosis of theileriosis and heartwater in bovines in Africa. Trans. R. Soc. Trop. Med. Hyg. 2002;96:S217–S224. doi: 10.1016/s0035-9203(02)90079-9. [DOI] [PubMed] [Google Scholar]
- Conrad P.A., Denham D., Brown C.G.D. Intraerythrocytic multiplication of Theileria parva in vitro: an ultrastructural study. Int. J. Parasitol. 1986;16:223–229. doi: 10.1016/0020-7519(86)90047-0. [DOI] [PubMed] [Google Scholar]
- Conrad P.A., Stagg D.A., Grootenhuis J.G., Irvin A.D., Newson J., Njamunggeh R.E. Isolation of Theileria parasites from African buffalo (Syncerus caffer) and characterization with anti-schizont monoclonal antibodies. Parasitology. 1987;94:413–423. doi: 10.1017/s0031182000055761. [DOI] [PubMed] [Google Scholar]
- Criado A., Martinez J., Buling A., Barba J.C., Merino S., Jefferies R. New data on epizootiology and genetics of piroplasms based on sequences of small ribosomal subunit and cytochrome b genes. Vet. Parasitol. 2006;142:238–247. doi: 10.1016/j.vetpar.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Criado-Fornelio A. A review of nucleic acid-based diagnostic tests for Babesia and Theileria, with emphasis on bovine piroplasms. Parassitologia. 2007;49:S39–S44. [PubMed] [Google Scholar]
- Criado-Fornelio A., Martinez-Marcos A., Buling-Saraña A., Barba-Carretero J.C. Babesia, Theileria and Hepatozoon in southern Europe: part II. Phylogenetic analysis and evolutionary history. Vet. Parasitol. 2003;114:173–194. doi: 10.1016/s0304-4017(03)00141-9. [DOI] [PubMed] [Google Scholar]
- Criado-Fornelio A., Gónzalez-del-Río M.A., Buling-Saraña A., Barba-Carretero J.C. The “expanding universe” of piroplasms. Vet. Parasitol. 2004;119:337–345. doi: 10.1016/j.vetpar.2003.11.015. [DOI] [PubMed] [Google Scholar]
- de Vos A.J., Roos J.A. The isolation of Theileria taurotragi in South Africa. Onderstepoort J. Vet. Res. 1981;48:149–153. [PubMed] [Google Scholar]
- de Waal D.T. Equine piroplasmosis: a review. Br. Vet. J. 1992;148:6–14. doi: 10.1016/0007-1935(92)90061-5. [DOI] [PubMed] [Google Scholar]
- d'Oliveira C., van der Weide M., Habela M.A., Jacquiet P., Jongejan F. Detection of Theileria annulata in blood samples of carrier cattle by PCR. J. Clin. Microbiol. 1995;33:2665–2669. doi: 10.1128/jcm.33.10.2665-2669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darghouth M.E., Bouattour A., Ben Miled L., Sassi L. Diagnosis of Theileria annulata infection of cattle in Tunisia: comparison of serology and blood smears. Vet. Res. 1996;27:613–621. [PubMed] [Google Scholar]
- Dschunkowsky E., Luhs J. Die piroplasmosen der rinder. Zentralbl. Bakteriol. Mikrobiol. Hyg. 1904;35:486–493. [Google Scholar]
- Eickbush T.H., Eickbush D.G. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D.W., Conrad P.A., Grootenhuis J.G., Morzaria S.P. Ultrastructure of the intra-erythrocytic stage of Theileria species from cattle and waterbuck. Tissue Cell. 1987;19:643–655. doi: 10.1016/0040-8166(87)90071-1. [DOI] [PubMed] [Google Scholar]
- Fujisaki K., Kawazu S., Kamio T. The taxonomy of the bovine Theileria spp. Parasitol. Today. 1994;10:31–33. doi: 10.1016/0169-4758(94)90355-7. [DOI] [PubMed] [Google Scholar]
- Gachohi J., Skilton R., Hansen F., Ngumi P., Kitala P. Epidemiology of East Coast fever (Theileria parva infection) in Kenya: past, present and the future. Parasit. Vectors. 2012;5:194. doi: 10.1186/1756-3305-5-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sanmartín J., Aurtenetxe O., Barral M., Marco I., Lavin S., García-Pérez A.L. Molecular detection and characterization of piroplasms infecting cervids and chamois in Northern Spain. Parasitology. 2007;134:391–398. doi: 10.1017/S0031182006001569. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Bishop R., Shah T., de Villiers E.P., Carlton J.M., Hall N. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science. 2005;309:134–137. doi: 10.1126/science.1110439. [DOI] [PubMed] [Google Scholar]
- Geysen D., Bishop R., Skilton R., Dolan T.T., Morzaria S. Molecular epidemiology of Theileria parva in the field. Trop. Med. Int. Health. 1999;4:A21–A27. doi: 10.1046/j.1365-3156.1999.00447.x. [DOI] [PubMed] [Google Scholar]
- Ghaemi P., Hoghooghi-Rad N., Shayan P., Eckert B. Detection of Theileria orientalis in Iran by semi-nested PCR. Parasitol. Res. 2012;110:527–531. doi: 10.1007/s00436-011-2517-y. [DOI] [PubMed] [Google Scholar]
- Githaka N., Konnai S., Kariuki E., Kanduma E., Murata S., Ohashi K. Molecular detection and characterization of potentially new Babesia and Theileria species/variants in wild felids from Kenya. Acta Trop. 2012;124:71–78. doi: 10.1016/j.actatropica.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Githaka N., Konnai S., Skilton R., Kariuki E., Kanduma E., Murata S. Genotypic variations in field isolates of Theileria species infecting giraffes (Giraffa camelopardalis tippelskirchi and Giraffa camelopardalis reticulata) in Kenya. Parasitol. Int. 2013;62:448–453. doi: 10.1016/j.parint.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Githaka N., Konnai S., Bishop R., Odongo D., Lekolool I., Kariuki E. Identification and sequence characterization of novel Theileria genotypes from the waterbuck (Kobus defassa) in a Theileria parva-endemic area in Kenya. Vet. Parasitol. 2014;202:180–193. doi: 10.1016/j.vetpar.2014.02.056. [DOI] [PubMed] [Google Scholar]
- Gou H., Guan G., Liu A., Ma M., Chen Z., Liu Z. Coevolutionary analyses of the relationships between piroplasmids and their hard tick hosts. Ecol. Evol. 2013;3:2985–2993. doi: 10.1002/ece3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts M.M., Voet M., Volckaert G., Goddeeris B.M. PCR amplification and sequence of the p33 piroplasm surface antigen gene of a Theileria species isolated from cattle in west Java, Indonesia. Ann. N. Y. Acad. Sci. 1998;849:126–136. doi: 10.1111/j.1749-6632.1998.tb11041.x. [DOI] [PubMed] [Google Scholar]
- Gray M.A., Luckins A.G., Rae P.F., Brown C.G. Evaluation of an enzyme immunoassay for serodiagnosis of infections with Theileria parva and T. annulata. Res. Vet. Sci. 1980;29:360–366. [PubMed] [Google Scholar]
- Grootenhuis J.G., Leitch B.L., Stagg D.A., Dolan T.T., Young A.S. Experimental induction of Theileria parva lawrencei carrier state in an African buffalo (Syncerus caffer) Parasitology. 1987;94:425–431. doi: 10.1017/s0031182000055773. [DOI] [PubMed] [Google Scholar]
- Gubbels J.M., de Vos A.P., van der Weide M., Viseras J., Schouls L.M., de Vries E. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 1999;37:1782–1789. doi: 10.1128/jcm.37.6.1782-1789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels M.J., d'Oliveira C., Jongejan F. Development of an indirect Tams1 enzyme-linked immunosorbent assay for diagnosis of Theileria annulata infection in cattle. Clin. Diagn. Lab. Immunol. 2000;7:404–411. doi: 10.1128/cdli.7.3.404-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels M.J., Hong Y., van der Weide M., Qi B., Nijman I.J., Guangyuan L. Molecular characterisation of the Theileria buffeli/orientalis group. Int. J. Parasitol. 2000;30:943–952. doi: 10.1016/s0020-7519(00)00074-6. [DOI] [PubMed] [Google Scholar]
- Gubbels M.J., Yin H., Bai Q., Liu G., Nijman I.J., Jongejan F. The phylogenetic position of the Theileria buffeli group in relation to other Theileria species. Parasitol. Res. 2002;88:S28–S32. doi: 10.1007/s00436-001-0566-3. [DOI] [PubMed] [Google Scholar]
- Han J.I., Jang H.J., Lee S.J., Na K.J. High prevalence of Theileria sp. in wild Chinese Water Deer (Hydropotes inermis argyropus) in South Korea. Vet. Parasitol. 2009;164:311–314. doi: 10.1016/j.vetpar.2009.05.027. [DOI] [PubMed] [Google Scholar]
- Hassanin A., Delsuc F., Ropiquet A., Hammere C., Jansen van Vuuren B., Matthee C. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C. R. Biol. 2012;335:32–50. doi: 10.1016/j.crvi.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Hassanin A., An J., Ropiquet A., Nguyen T.T., Couloux A. Combining multiple autosomal introns for studying shallow phylogeny and taxonomy of Laurasiatherian mammals: application to the tribe Bovini (Cetartiodactyla, Bovidae) Mol. Phylogenet. Evol. 2013;66:766–775. doi: 10.1016/j.ympev.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Hayashida K., Hara Y., Abe T., Yamasaki C., Toyoda A., Kosuge T. Comparative genome analysis of three eukaryotic parasites with differing abilities to transform leukocytes reveals key mediators of Theileria-induced leukocyte transformation. mBio. 2012;3:e204–e212. doi: 10.1128/mBio.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidarpour Bami M., Haddadzadeh H.R., Kazemi B., Khazraiinia P., Bandehpour M., Aktas M. Molecular identification of ovine Theileria species by a new PCR-RFLP method. Vet. Parasitol. 2009;161:171–177. doi: 10.1016/j.vetpar.2009.01.035. [DOI] [PubMed] [Google Scholar]
- Heim A., Passos L.M., Ribeiro M.F., Costa-Júnior L.M., Bastos C.V., Cabral D.D. Detection and molecular characterization of Babesia caballi and Theileria equi isolates from endemic areas of Brazil. Parasitol. Res. 2007;102:63–68. doi: 10.1007/s00436-007-0726-1. [DOI] [PubMed] [Google Scholar]
- Hooshmand-Rad P. The pathogenesis of anaemia in Theileria annulata infection. Res. Vet. Sci. 1976;20:324–329. [PubMed] [Google Scholar]
- Huang X., Xuan X., Yokoyama N., Xu L., Suzuki H., Sugimoto C. High-level expression and purification of a truncated merozoite antigen-2 of Babesia equi in Escherichia coli and its potential for immunodiagnosis. J. Clin. Microbiol. 2003;41:1147–1151. doi: 10.1128/JCM.41.3.1147-1151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Xuan X., Xu L., Zhang S., Yokoyama N., Suzuki N. Development of an immunochromatographic test with recombinant EMA-2 for the rapid detection of antibodies against Babesia equi in horses. J. Clin. Microbiol. 2004;42:359–361. doi: 10.1128/JCM.42.1.359-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilhan T., Williamson S., Kirvar E., Shiels B., Brown C.G. Theileria annulata: carrier state and immunity. Ann. N. Y. Acad. Sci. 1998;849:109–125. doi: 10.1111/j.1749-6632.1998.tb11040.x. [DOI] [PubMed] [Google Scholar]
- Izzo M.M., Poe I., Horadagoda N., De Vos A.J., House J.K. Haemolytic anaemia in cattle in NSW associated with Theileria infections. Aust. Vet. J. 2010;88:45–51. doi: 10.1111/j.1751-0813.2009.00540.x. [DOI] [PubMed] [Google Scholar]
- Jefferies R., Ryan U.M., Irwin P.J. PCR-RFLP for the detection and differentiation of the canine piroplasm species and its use with filter paper-based technologies. Vet. Parasitol. 2007;144:20–27. doi: 10.1016/j.vetpar.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Jeong W., Kweon C.H., Kang S.W., Paik S.G. Diagnosis and quantification of Theileria sergenti using TaqMan PCR. Vet. Parasitol. 2003;111:287–295. doi: 10.1016/s0304-4017(02)00388-6. [DOI] [PubMed] [Google Scholar]
- Jeong W., Kweon C.H., Kim J.M., Jang H., Paik S.G. Serological investigation of Theileria sergenti using latex agglutination test in South Korea. J. Parasitol. 2005;91:164–169. doi: 10.1645/GE-3294. [DOI] [PubMed] [Google Scholar]
- Jongejan F., Musisi F.L., Moorhouse P.D.S., Snacken M., Uilenberg G. Theileria taurotragi in Zambia. Vet. Q. 1986;8:261–263. doi: 10.1080/01652176.1986.9694051. [DOI] [PubMed] [Google Scholar]
- Kachani M., Spooner R.L., Rae P., Bell-Sakyi L., Brown C.G. Stage-specific responses following infection with Theileria annulata as evaluated using ELISA. Parasitol. Res. 1992;78:43–47. doi: 10.1007/BF00936180. [DOI] [PubMed] [Google Scholar]
- Kamau J., de Vos A.J., Playford M., Salim B., Kinyanjui P., Sugimoto C. Emergence of new types of Theileria orientalis in Australian cattle and possible cause of theileriosis outbreaks. Parasit. Vectors. 2011;4:22. doi: 10.1186/1756-3305-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappmeyer L.S., Thiagarajan M., Herndon D.R., Ramsay J.D., Caler E., Djikeng A. Comparative genomic analysis and phylogenetic position of Theileria equi. BMC Genomics. 2012;13:603. doi: 10.1186/1471-2164-13-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki E.K., Penzhorn B.L., Horak I.G. Ticks (Acari: Ixodidae) infesting cattle and African buffaloes in the Tsavo conservation area, Kenya. Onderstepoort J. Vet. Res. 2012;79:437. doi: 10.4102/ojvr.v79i1.437. [DOI] [PubMed] [Google Scholar]
- Katende J., Morzaria S., Toye P., Skilton R., Nene V., Nkonge C. An enzyme-linked immunosorbent assay for detection of Theileria parva antibodies in cattle using a recombinant polymorphic immunodominant molecule. Parasitol. Res. 1998;84:408–416. doi: 10.1007/s004360050419. [DOI] [PubMed] [Google Scholar]
- Katende J.M., Goddeeris B.M., Morzaria S.P., Nkonge C.G., Musoke A.J. Identification of a Theileria mutans-specific antigen for use in an antibody and antigen detection ELISA. Parasite Immunol. 1990;12:419–433. doi: 10.1111/j.1365-3024.1990.tb00978.x. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu S., Sugimoto C., Kamio T., Fujisaki K. Antigenic differences between Japanese Theileria sergenti and other benign Theileria species of cattle from Australia (T. buffeli) and Britain (T. orientalis) Parasitol. Res. 1992;78:130–135. doi: 10.1007/BF00931654. [DOI] [PubMed] [Google Scholar]
- Kawazu S., Kamio T., Sekizaki T., Fujisaki K. Theileria sergenti and T. buffeli: polymerase chain reaction-based marker system for differentiating the parasite species from infected cattle blood and infected tick salivary gland. Exp. Parasitol. 1995;81:430–435. doi: 10.1006/expr.1995.1135. [DOI] [PubMed] [Google Scholar]
- Kiara H., Jennings A., Bronsvoort B.M., Handel I.G., Mwangi S.T., Mbole-Kariuki M. A longitudinal assessment of the serological response to Theileria parva and other tick-borne parasites from birth to one year in a cohort of indigenous calves in western Kenya. Parasitology. 2014;141:1289–1298. doi: 10.1017/S003118201400050X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.M., Blanco L.B., Alhassan A., Iseki H., Yokoyama N., Xuan X. Diagnostic real-time PCR assay for the quantitative detection of Theileria equi from equine blood samples. Vet. Parasitol. 2008;151:158–163. doi: 10.1016/j.vetpar.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Kirvar E., Ilhan T., Katzer F., Hooshmand-Rad P., Zweygarth E., Gerstenberg C. Detection of Theileria annulata in cattle and vector ticks by PCR using the Tams1 gene sequences. Parasitology. 2000;120:245–254. doi: 10.1017/s0031182099005466. [DOI] [PubMed] [Google Scholar]
- Knowles D.P., Lowell J.R., Kappmeyer S., Stiller D., Hennager S.G., Perryman L.E. Antibody to a recombinant merozoite protein epitope identifies horses infected with Babesia equi. J. Clin. Microbiol. 1992;30:3122–3126. doi: 10.1128/jcm.30.12.3122-3126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch R. Springer; Berlin: 1898. Reiseberichte über Rinderpest, Bubonenpest in Indien und Africa, Tsetse und Surrakrankhet, Texasfieber, Tropische Malaria, Schwarzwasserfieber. [Google Scholar]
- Kubota S., Sugimoto C., Kakuda T., Onuma M. Analysis of immunodominant piroplasm surface antigen alleles in mixed populations of Theileria sergenti and T. buffeli. Int. J. Parasitol. 1996;26:741–747. doi: 10.1016/0020-7519(96)00047-1. [DOI] [PubMed] [Google Scholar]
- Lack J.B., Reichard M.V., Van Den Bussche R.A. Phylogeny and evolution of the Piroplasmida as inferred from 18S rRNA sequences. Int. J. Parasitol. 2012;42:353–363. doi: 10.1016/j.ijpara.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Latif A.A., Hove T., Kanhai G.K., Masaka S. Buffalo-associated Theileria parva: the risk to cattle of buffalo translocation into the Highveld of Zimbabwe. Ann. N. Y. Acad. Sci. 2002;969:275–279. doi: 10.1111/j.1749-6632.2002.tb04392.x. [DOI] [PubMed] [Google Scholar]
- Lawrence D.A. Rhodesian Printing and Publishing Co; Salisbury, Rhodesia: 1935. Report of the Director of Veterinary Research, Southern Rhodesia, for the Year 1934. [Google Scholar]
- Lawrence J.A. The differential diagnosis of the bovine Theilerias of Southern Africa. J. S. Afr. Vet. Assoc. 1979;50:311–313. [PubMed] [Google Scholar]
- Lawrence J.A., Norval R.A.I., Uilenberg G. Rhipicephalus zambeziensis as a vector of bovine Theileriae. Trop. Anim. Health Prod. 1983;15:39–42. doi: 10.1007/BF02250760. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Ryan U.M., Jefferies R., McInnes L.M., Forshaw D., Friend J.A. Theileria gilberti n. sp. (Apicomplexa: Theileriidae) in the Gilbert's potoroo (Potorous gilbertii) J. Eukaryot. Microbiol. 2009;56:290–295. doi: 10.1111/j.1550-7408.2009.00398.x. [DOI] [PubMed] [Google Scholar]
- Leemans I., Hooshmand-Rad P., Uggla A. The indirect fluorescent antibody test based on schizont antigen for study of the sheep parasite Theileria lestoquardi. Vet. Parasitol. 1997;69:9–18. doi: 10.1016/s0304-4017(96)01098-9. [DOI] [PubMed] [Google Scholar]
- Leemans I., Brown D., Hooshmand-Rad P., Kirvar E., Uggla A. Infectivity and cross-immunity studies of Theileria lestoquardi and Theileria annulata in sheep and cattle: I. In vivo responses. Vet. Parasitol. 1999;82:179–192. doi: 10.1016/S0304-4017(99)00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen Z., Liu Z., Liu J., Yang J., Li Q. Molecular identification of Theileria parasites of northwestern Chinese Cervidae. Parasit. Vectors. 2014;7:225. doi: 10.1186/1756-3305-7-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenheld G. Preliminary communication on the fixing of complement in horse-sickness and East Coast fever. Z. Immun. Forsch. 1910;8:232–238. [Google Scholar]
- Liu A., Guan G., Liu Z., Liu J., Leblanc N., Li Y. Detecting and differentiating Theileria sergenti and Theileria sinensis in cattle and yaks by PCR based on major piroplasm surface protein (MPSP) Exp. Parasitol. 2010;126:476–481. doi: 10.1016/j.exppara.2010.05.024. [DOI] [PubMed] [Google Scholar]
- Liu A., Guan G., Du P., Liu Z., Gou H., Liu J. Loop-mediated isothermal amplification (LAMP) assays for the detection of Theileria annulata infection in China targeting the 18S rRNA and ITS sequences. Exp. Parasitol. 2012;131:125–129. doi: 10.1016/j.exppara.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Liu A., Guan G., Du P., Gou H., Zhang J., Liu Z. Rapid identification and differentiation of Theileria sergenti and Theileria sinensis using a loop-mediated isothermal amplification (LAMP) assay. Vet. Parasitol. 2013;191:15–22. doi: 10.1016/j.vetpar.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Liu A.H., Guan G.Q., Liu J.L., Liu Z.J., Leblanc N., Li Y.Q. Polymorphism analysis of Chinese Theileria sergenti using allele-specific polymerase chain reaction of the major piroplasm surface protein gene. J. Parasitol. 2011;97:116–121. doi: 10.1645/GE-2444.1. [DOI] [PubMed] [Google Scholar]
- Liu Z., Hou J., Bakheit M.A., Salih D.A., Luo J., Yin H. Development of loop-mediated isothermal amplification (LAMP) assay for rapid diagnosis of ovine theileriosis in China. Parasitol. Res. 2008;103:1407–1412. doi: 10.1007/s00436-008-1149-3. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wang Z., Yin H., Luo J., Zhang B., Kullmann B. Identification of Theileria uilenbergi immunodominant protein for development of an indirect ELISA for diagnosis of ovine theileriosis. Int. J. Parasitol. 2010;40:591–598. doi: 10.1016/j.ijpara.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Liu Z., Li Y., Salih D.E., Luo J., Ahmed J.S., Seitzer U. Validation of a recombinant protein indirect ELISA for the detection of specific antibodies against Theileria uilenbergi and Theileria luwenshuni in small ruminants. Vet. Parasitol. 2014;204:139–145. doi: 10.1016/j.vetpar.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Mans B.J., Pienaar R., Latif A.A., Potgieter F.T. Diversity in the 18S SSU rRNA V4 hyper-variable region of Theileria in bovines and African buffalo (Syncerus caffer) from southern Africa. Parasitology. 2011;138:766–779. doi: 10.1017/S0031182011000187. [DOI] [PubMed] [Google Scholar]
- Mans B.J., Pienaar R., Potgieter F.T., Latif A.A. Theileria parva, T. sp. (buffalo) and T. sp. (bougasvlei) 18S variants. Vet. Parasitol. 2011;182:382–383. doi: 10.1016/j.vetpar.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Martin H., Brocklesby D.W. A new parasite of Eland. Vet. Rec. 1960;72:331–332. [Google Scholar]
- Matjila P.T., Leisewitz A.L., Oosthuizen M.C., Jongejan F., Penzhorn B.L. Detection of a Theileria species in dogs in South Africa. Vet. Parasitol. 2008;157:34–40. doi: 10.1016/j.vetpar.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Mbizeni S., Potgieter F.T., Troskie C., Mans B.J., Penzhorn B.L., Latif A.A. Field and laboratory studies on Corridor disease (Theileria parva infection) in cattle population at the livestock/game interface of uPhongolo-Mkuze area, South Africa. Ticks Tick Borne Dis. 2013;4:227–234. doi: 10.1016/j.ttbdis.2012.11.005. [DOI] [PubMed] [Google Scholar]
- McKeever D.J. Bovine immunity – a driver for diversity in Theileria parasites? Trends Parasitol. 2009;25:269–276. doi: 10.1016/j.pt.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Miranda J., Bakheit M.A., Liu Z., Yin H., Mu Y., Guo S. Development of a recombinant indirect ELISA for the diagnosis of Theileria sp. (China) infection in small ruminants. Parasitol. Res. 2006;98:561–567. doi: 10.1007/s00436-005-0105-8. [DOI] [PubMed] [Google Scholar]
- Morrison D.A. Evolution of the Apicomplexa: where are we now? Trends Parasitol. 2009;25:375–382. doi: 10.1016/j.pt.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Morzaria S.P., Katende J., Musoke A., Nene V., Skilton R., Bishop R. Development of sero-diagnostic and molecular tools for the control of important tick-borne pathogens of cattle in Africa. Parassitologia. 1999;41:S73–S80. [PubMed] [Google Scholar]
- Nagore D., García-Sanmartín J., García-Pérez A.L., Juste R.A., Hurtado A. Identification, genetic diversity and prevalence of Theileria and Babesia species in a sheep population from Northern Spain. Int. J. Parasitol. 2004;34:1059–1067. doi: 10.1016/j.ijpara.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Najm N.A., Meyer-Kayser E., Hoffmann L., Herb I., Fensterer V., Pfister K. A molecular survey of Babesia spp. and Theileria spp. in red foxes (Vulpes vulpes) and their ticks from Thuringia, Germany. Ticks Tick Borne Dis. 2014;5:386–391. doi: 10.1016/j.ttbdis.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Ngumi P.N., Lesan A.C., Williamson S.M., Awich J.R. Isolation and preliminary characterisation of a previously unidentified Theileria parasite of cattle in Kenya. Res. Vet. Sci. 1994;57:1–9. doi: 10.1016/0034-5288(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Nijhof A.M., Penzhorn B.L., Lynen G., Mollel J.O., Morkel P., Bekker C.P. Babesia bicornis sp. nov. and Theileria bicornis sp. nov.: tick-borne parasites associated with mortality in the black rhinoceros (Diceros bicornis) J. Clin. Microbiol. 2003;41:2249–2254. doi: 10.1128/JCM.41.5.2249-2254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhof A.M., Pillay V., Steyl J., Prozesky L., Stoltsz W.H., Lawrence J.A. Molecular characterization of Theileria species associated with mortality in four species of African antelopes. J. Clin. Microbiol. 2005;43:5907–5911. doi: 10.1128/JCM.43.12.5907-5911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q., Luo J., Guan G., Ma M., Liu Z., Liu A. Detection and differentiation of ovine Theileria and Babesia by reverse line blotting in China. Parasitol. Res. 2009;104:1417–1423. doi: 10.1007/s00436-009-1344-x. [DOI] [PubMed] [Google Scholar]
- Norval R.A., Lawrence J.A., Young A.S., Perry B.D., Dolan T.T., Scott J. Theileria parva: influence of vector, parasite and host relationships on the epidemiology of theileriosis in southern Africa. Parasitology. 1991;102:347–356. doi: 10.1017/s0031182000064295. [DOI] [PubMed] [Google Scholar]
- Norval R.A., Perry B.D., Young A.S. Academic Press Inc; London, UK: 1992. The Epidemiology of Theileriosis in Africa; pp. 136–154. [Google Scholar]
- Odongo D.O., Sunter J.D., Kiara H.K., Skilton R.A., Bishop R.P. A nested PCR assay exhibits enhanced sensitivity for detection of Theileria parva infections in bovine blood samples from carrier animals. Parasitol. Res. 2010;106:357–365. doi: 10.1007/s00436-009-1670-z. [DOI] [PubMed] [Google Scholar]
- Ogunremi O., Georgiadis M.P., Halbert G., Benjamin J., Pfister K., Lopez-Rebollar L. Validation of the indirect fluorescent antibody and the complement fixation tests for the diagnosis of Theileria equi. Vet. Parasitol. 2007;148:102–108. doi: 10.1016/j.vetpar.2007.06.006. [DOI] [PubMed] [Google Scholar]
- OIE . seventh ed. vol. 1–2. Office International Des Epizooties; Paris: 2014. (Manual of Diagnostic Tests and Vaccines for Terrestrial Animals). [Google Scholar]
- Oosthuizen M.C., Allsopp B.A., Troskie M., Collins N.E., Penzhorn B.L. Identification of novel Babesia and Theileria species in South African giraffe (Giraffa camelopardalis, Linnaeus, 1758) and roan antelope (Hippotragus equinus, Desmarest 1804) Vet. Parasitol. 2009;163:39–46. doi: 10.1016/j.vetpar.2009.03.045. [DOI] [PubMed] [Google Scholar]
- Ota N., Mizuno D., Kuboki N., Igarashi I., Nakamura Y., Yamashina H. Epidemiological survey of Theileria orientalis infection in grazing cattle in the eastern part of Hokkaido, Japan. J. Vet. Med. Sci. 2010;71:937–944. doi: 10.1292/jvms.71.937. [DOI] [PubMed] [Google Scholar]
- Otiende M.Y., Kivata M.W., Jowers M.J., Makumi J.N., Runo S., Obanda V. Three novel haplotypes of Theileria bicornis in black and white Rhinoceros in Kenya. Transbound. Emerg. Dis. 2014 doi: 10.1111/tbed.12242. [DOI] [PubMed] [Google Scholar]
- Oura C.A., Bishop R., Wampande E.M., Lubega G.W., Tait A. The persistence of component Theileria parva stocks in cattle immunized with the ‘Muguga cocktail’ live vaccine against East Coast fever in Uganda. Parasitology. 2004;129:27–42. doi: 10.1017/s003118200400513x. [DOI] [PubMed] [Google Scholar]
- Oura C.A., Tait A., Asiimwe B., Lubega G.W., Weir W. Theileria parva genetic diversity and haemoparasite prevalence in cattle and wildlife in and around Lake Mburo National Park in Uganda. Parasitol. Res. 2011;108:1365–1374. doi: 10.1007/s00436-010-2030-8. [DOI] [PubMed] [Google Scholar]
- Pain A., Renauld H., Berriman M., Murphy L., Yeats C.A., Weir W. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- Papadopoulos B., Perié N.M., Uilenberg G. Piroplasms of domestic animals in the Macedonia region of Greece. 1. Serological cross-reactions. Vet. Parasitol. 1996;63:41–56. doi: 10.1016/0304-4017(95)00878-0. [DOI] [PubMed] [Google Scholar]
- Paparini A., Ryan U.M., Warren K., McInnes L.M., de Tores P., Irwin P.J. Identification of novel Babesia and Theileria genotypes in the endangered marsupials, the woylie (Bettongia penicillata ogilbyi) and boodie (Bettongia lesueur) Exp. Parasitol. 2012;131:25–30. doi: 10.1016/j.exppara.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Papli N.E., Landt O., Fleischer C., Koekemoer J.O., Mans B.J., Pienaar R. Evaluation of a TaqMan real-time PCR for the detection of Theileria parva in buffalo and cattle. Vet. Parasitol. 2011;175:356–359. doi: 10.1016/j.vetpar.2010.10.038. [DOI] [PubMed] [Google Scholar]
- Peeling R.W., Smith P.G., Bossuyt P.M.M. A guide for diagnostic evaluations. Nat. Rev. Microbiol. 2010;8:S2–S6. [PubMed] [Google Scholar]
- Perera P.K., Gasser R.B., Firestone S.M., Smith L., Roeber F., Jabbar A. Semi-quantitative multiplexed-tandem PCR for the detection and differentiation of four Theileria orientalis genotypes in cattle. J. Clin. Microbiol. 2014 doi: 10.1128/JCM.02536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry B.D., Young A.S. The naming game: the changing fortunes of East Coast fever and Theileria parva. Vet. Rec. 1993;133:613–616. [PubMed] [Google Scholar]
- Pfaffl M.W. Quantification strategies in real-time PCR. In: Bustin S.A., editor. A–Z of Quantitative PCR. International University Line (IUL); La Jolla, CA, USA: 2004. pp. 87–112. [Google Scholar]
- Pienaar R., Potgieter F.T., Latif A.A., Thekisoe O.M.M., Mans B.J. Mixed Theileria infections in free-ranging buffalo herds: implications for diagnosing Theileria parva infections in Cape buffalo (Syncerus caffer) Parasitology. 2011;138:884–895. doi: 10.1017/S0031182011000503. [DOI] [PubMed] [Google Scholar]
- Pienaar R., Potgieter F.T., Latif A.A., Thekisoe O.M.M., Mans B.J. The HybridII assay: a sensitive and specific real-time hybridization assay for the diagnosis of Theileria parva infection in Cape buffalo (Syncerus caffer) and cattle. Parasitology. 2011;138:1935–1944. doi: 10.1017/S0031182011001454. [DOI] [PubMed] [Google Scholar]
- Pienaar, R., Latif, A.A., Thekisoe, O.M.M., Mans, B.J., 2013. Protein gene candidates for the qualitative molecular detection of Theileria parva in African buffalo (Syncerus caffer) using the real-time SYBR green polymerase chain reaction. Proceedings of the 1st Annual Conference on Advances in Veterinary Science Research, Singapore.
- Pienaar R., Potgieter F.T., Latif A.A., Thekisoe O.M.M., Mans B.J. Geographic distribution of Theileria sp. (buffalo) and Theileria sp. (bougasvlei) in Cape buffalo (Syncerus caffer) in southern Africa: implications for speciation. Parasitology. 2014;141:411–424. doi: 10.1017/S0031182013001728. [DOI] [PubMed] [Google Scholar]
- Rajendran C., Ray D.D. Diagnosis of tropical bovine theileriosis by ELISA with recombinant merozoite surface protein of Theileria annulata (Tams1) J. Parasit. Dis. 2014;38:41–45. doi: 10.1007/s12639-012-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renneker S., Kullmann B., Gerber S., Dobschanski J., Bakheit M.A., Geysen D. Development of a competitive ELISA for detection of Theileria annulata infection. Transbound. Emerg. Dis. 2008;55:249–256. doi: 10.1111/j.1865-1682.2008.01036.x. [DOI] [PubMed] [Google Scholar]
- Ros-García A., Juste R.A., Hurtado A. A highly sensitive DNA bead-based suspension array for the detection and species identification of bovine piroplasms. Int. J. Parasitol. 2012;42:207–214. doi: 10.1016/j.ijpara.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Ros-García A., Nicolás A., García-Pérez A.L., Juste R.A., Hurtado A. Development and evaluation of a real-time PCR assay for the quantitative detection of Theileria annulata in cattle. Parasit. Vectors. 2012;5:171. doi: 10.1186/1756-3305-5-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros-García A., Barandika J.F., García-Pérez A.L., Juste R.A., Hurtado A. Assessment of exposure to piroplasms in sheep grazing in communal mountain pastures by using a multiplex DNA bead-based suspension array. Parasit. Vectors. 2013;6:277. doi: 10.1186/1756-3305-6-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih D.A., Liu Z., Bakheit M.A., Ali A.M., El Hussein A.M., Unger H. Development and evaluation of a loop-mediated isothermal amplification method for diagnosis of tropical theileriosis. Transbound. Emerg. Dis. 2008;55:238–243. doi: 10.1111/j.1865-1682.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- Salih D.A., Ali A.M., Liu Z., Bakheit M.A., Taha K.M., El Imam A.H. Development of a loop-mediated isothermal amplification method for detection of Theileria lestoquardi. Parasitol. Res. 2012;110:533–538. doi: 10.1007/s00436-011-2518-x. [DOI] [PubMed] [Google Scholar]
- Salih D.E., Ahmed J.S., Bakheit M.A., Ali E.B., El Hussein A.M., Hassan S.M. Validation of the indirect TaSP enzyme-linked immunosorbent assay for diagnosis of Theileria annulata infection in cattle. Parasitol. Res. 2005;97:302–308. doi: 10.1007/s00436-005-1431-6. [DOI] [PubMed] [Google Scholar]
- Salim B., Bakheit M., Kamau J., Nakamura I., Sugimoto C. Nucleotide sequence heterogeneity in the small subunit ribosomal RNA gene within Theileria equi from horses in Sudan. Parasitol. Res. 2010;106:493–498. doi: 10.1007/s00436-009-1691-7. [DOI] [PubMed] [Google Scholar]
- Salim B., Bakheit M.A., Sugimoto C. Rapid detection and identification of Theileria equi and Babesia caballi by high-resolution melting (HRM) analysis. Parasitol. Res. 2013;112:3883–3886. doi: 10.1007/s00436-013-3581-2. [DOI] [PubMed] [Google Scholar]
- Salim B.O., Hassan S.M., Bakheit M.A., Alhassan A., Igarashi I., Karanis P. Diagnosis of Babesia caballi and Theileria equi infections in horses in Sudan using ELISA and PCR. Parasitol. Res. 2008;103:1145–1150. doi: 10.1007/s00436-008-1108-z. [DOI] [PubMed] [Google Scholar]
- Sarataphan N., Nilwarangkoon S., Tananyutthawongese C., Kakuda T., Onuma M., Chansiri K. Genetic diversity of major piroplasm surface protein genes and their allelic variants of Theileria parasites in Thai cattle. J. Vet. Med. Sci. 1999;61:991–994. doi: 10.1292/jvms.61.991. [DOI] [PubMed] [Google Scholar]
- Schaeffler W.F. Serologic tests for Theileria cervi in white-tailed deer and for other species of Theileria in cattle and sheep. Am. J. Vet. Res. 1963;24:784–791. [PubMed] [Google Scholar]
- Schetters T.P., Arts G., Niessen R., Schaap D. Development of a new score to estimate clinical East Coast Fever in experimentally infected cattle. Vet. Parasitol. 2010;167:255–259. doi: 10.1016/j.vetpar.2009.09.027. [DOI] [PubMed] [Google Scholar]
- Schindler R., Mehlitz D. Serologic studies on Theileria mutans infection in cattle. Z. Tropenmed. Parasitol. 1969;20:459–473. [PubMed] [Google Scholar]
- Schindler R., Wokatsch R. Versuche zur differenzierung der Theilerien spezies des rindes durch serologische unterschungen. Tropenmed. Parasitol. 1965;16:85–87. [PubMed] [Google Scholar]
- Schnittger L., Yin H., Gubbels M.J., Beyer D., Niemann S., Jongejan F. Phylogeny of sheep and goat Theileria and Babesia parasites. Parasitol. Res. 2003;91:398–406. doi: 10.1007/s00436-003-0979-2. [DOI] [PubMed] [Google Scholar]
- Schnittger L., Yin H., Qi B., Gubbels M.J., Beyer D., Niemann S. Simultaneous detection and differentiation of Theileria and Babesia parasites infecting small ruminants by reverse line blotting. Parasitol. Res. 2004;92:189–196. doi: 10.1007/s00436-003-0980-9. [DOI] [PubMed] [Google Scholar]
- Seitzer U., Bakheit M.A., Salih D.E., Ali A., Haller D., Yin H. From molecule to diagnostic tool: Theileria annulata surface protein TaSP. Parasitol. Res. 2007;101:S217–S223. doi: 10.1007/s00436-007-0685-6. [DOI] [PubMed] [Google Scholar]
- Shayan P., Biermann R., Schein E., Gerdes J., Ahmed J.S. Detection and differentiation of Theileria annulata and Theileria parva using macroschizont-derived DNA probes. Ann. N. Y. Acad. Sci. 1998;849:88–95. doi: 10.1111/j.1749-6632.1998.tb11038.x. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Suzuki K., Nakamura K., Kadota K., Fujisaki K., Ito S. Isolation of Theileria sergenti piroplasms from infected erythrocytes and development of an enzyme-linked immunosorbent assay for serodiagnosis of T. sergenti infections. Res. Vet. Sci. 1988;45:206–212. [PubMed] [Google Scholar]
- Sibeko K.P., Oosthuizen M.C., Collins N.E., Geysen D., Rambritch N.E., Latif A.A. Development and evaluation of a real-time polymerase chain reaction test for the detection of Theileria parva infections in Cape buffalo (Syncerus caffer) and cattle. Vet. Parasitol. 2008;155:37–48. doi: 10.1016/j.vetpar.2008.03.033. [DOI] [PubMed] [Google Scholar]
- Sibeko K.P., Geysen D., Oosthuizen M.C., Matthee C.A., Troskie M., Potgieter F.T. Four p67 alleles identified in South African Theileria parva field samples. Vet. Parasitol. 2010;167:244–254. doi: 10.1016/j.vetpar.2009.09.026. [DOI] [PubMed] [Google Scholar]
- Sibeko K.P., Collins N.E., Oosthuizen M.C., Troskie M., Potgieter F.T., Coetzer J.A. Analyses of genes encoding Theileria parva p104 and polymorphic immunodominant molecule (PIM) reveal evidence of the presence of cattle-type alleles in the South African T. parva population. Vet. Parasitol. 2011;181:120–130. doi: 10.1016/j.vetpar.2011.04.035. [DOI] [PubMed] [Google Scholar]
- Sivakumar T., Hayashida K., Sugimoto C., Yokoyama N. Evolution and genetic diversity of Theileria. Infect. Genet. Evol. 2014;27:250–263. doi: 10.1016/j.meegid.2014.07.013. [DOI] [PubMed] [Google Scholar]
- Skilton R.A., Bishop R.P., Katende J.M., Mwaura S., Morzaria S.P. The persistence of Theileria parva infection in cattle immunized using two stocks which differ in their ability to induce a carrier state: analysis using a novel blood spot PCR assay. Parasitology. 2002;124:265–276. doi: 10.1017/s0031182001001196. [DOI] [PubMed] [Google Scholar]
- Stewart N.P., Uilenberg G., de Vos A.J. Review of the Australian species of Theileria, with special reference to Theileria buffeli of cattle. Trop. Anim. Health Prod. 1996;28:81–90. doi: 10.1007/BF02250731. [DOI] [PubMed] [Google Scholar]
- Steyl J.C.A., Prozesky L., Stoltsz W.H., Lawrence J.A. Theileriosis (Cytauxzoonosis) in Roan antelope (Hippotragus equinus): field exposure to infection and identification of potential vectors. Onderstepoort J. Vet. Res. 2012;79:367. doi: 10.4102/ojvr.v79i1.367. [DOI] [PubMed] [Google Scholar]
- Swai E.S., Karimuribo E.D., Kambarage D.M., Moshy W.E., Mbise A.N. A comparison of seroprevalence and risk factors for Theileria parva and T. mutans in smallholder dairy cattle in the Tanga and Iringa regions of Tanzania. Vet. J. 2007;174:390–396. doi: 10.1016/j.tvjl.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Onoe S., Matsuba T., Katayama S., Yamanaka M., Yonemichi H. Detection of Theileria sergenti infection in cattle by polymerase chain reaction amplification of parasite-specific DNA. J. Clin. Microbiol. 1993;31:2565–2569. doi: 10.1128/jcm.31.10.2565-2569.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler A. Rhodesian tick fever. Transvaal Agr. J. 1904;2:421–438. [Google Scholar]
- Theiler A. Piroplasma mutans (n. spec) of South African cattle. J. Comp. Path. Ther. 1906;19:292–300. [Google Scholar]
- Thekisoe O.M., Rambritch N.E., Nakao R., Bazie R.S., Mbati P., Namangala B. Loop-mediated isothermal amplification (LAMP) assays for detection of Theileria parva infections targeting the PIM and p150 genes. Int. J. Parasitol. 2010;40:55–61. doi: 10.1016/j.ijpara.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Thompson B.E., Latif A.A., Oosthuizen M.C., Troskie M., Penzhorn B.L. Occurrence of Theileria parva infection in cattle on a farm in the Ladysmith district, KwaZulu-Natal, South Africa. J. S. Afr. Vet. Assoc. 2008;79:31–35. doi: 10.4102/jsava.v79i1.237. [DOI] [PubMed] [Google Scholar]
- Tian Z., Liu G., Yin H., Luo J., Guan G., Luo J. Discrimination between ovine Babesia and Theileria species in China based on the ribosomal protein S8 (RPS8) gene. Vet. Parasitol. 2013;197:354–359. doi: 10.1016/j.vetpar.2013.04.033. [DOI] [PubMed] [Google Scholar]
- Tian Z., Liu G., Yin H., Xie J., Wang S., Yuan X. First report on the occurrence of Theileria sp. OT3 in China. Parasitol. Int. 2014;63:403–407. doi: 10.1016/j.parint.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M. The species concept in parasites and other pathogens: a pragmatic approach? Trends Parasitol. 2006;22:66–70. doi: 10.1016/j.pt.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- Ueti M.W., Mealey R.H., Kappmeyer L.S., White S.N., Kumpula-McWhirter N., Pelzel A.M. Re-emergence of the apicomplexan Theileria equi in the United States: elimination of persistent infection and transmission risk. PLoS ONE. 2012;7:e44713. doi: 10.1371/journal.pone.0044713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uilenberg G. Theilerial species of domestic livestock. In: Irving A.D., Cunningham M.P., Young A.S., editors. Advances in the Control of Theileriosis. Martinus Nijhoff Publishers; The Hague, Boston, London: 1981. pp. 4–37. [Google Scholar]
- Uilenberg G. Babesia – a historical perspective. Vet. Parasitol. 2006;138:3–10. doi: 10.1016/j.vetpar.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Uilenberg G., Schreuder B.E.C. Studies on Theileriidae (Sporozoa) in Tanzania. I. Tick transmission of Haematoxenus veliferus. Tropenmed. Parasitol. 1976;27:106–111. [PubMed] [Google Scholar]
- Uilenberg G., Perié N.M., Spanjer A.A., Franssen F.F. Theileria orientalis, a cosmopolitan blood parasite of cattle: demonstration of the schizont stage. Res. Vet. Sci. 1985;38:352–360. [PubMed] [Google Scholar]
- Vilstrup J.T., Seguin-Orlando A., Stiller M., Ginolhac A., Raghavan M., Nielsen S.C. Mitochondrial phylogenomics of modern and ancient equids. PLoS ONE. 2013;8:e55950. doi: 10.1371/journal.pone.0055950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.X., He L., Fang R., Song Q.Q., Tu P., Jenkins A. Loop-mediated isothermal amplification (LAMP) assay for detection of Theileria sergenti infection targeting the p33 gene. Vet. Parasitol. 2010;171:159–162. doi: 10.1016/j.vetpar.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Wang L.X., Zhao J.H., He L., Liu Q., Zhou D.N., Zhou Y.Q. An indirect ELISA for detection of Theileria sergenti antibodies in water buffalo using a recombinant major piroplasm surface protein. Vet. Parasitol. 2010;170:323–326. doi: 10.1016/j.vetpar.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Xie J., Liu G., Tian Z., Luo J. Development of loop-mediated isothermal amplification (LAMP) for detection of Theileria equi. Acta Trop. 2013;127:245–250. doi: 10.1016/j.actatropica.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Yang J.F., Li Y.Q., Liu Z.J., Liu J.L., Guan G.Q., Chen Z. Molecular evidence for piroplasms in wild Reeves' muntjac (Muntiacus reevesi) in China. Parasitol. Int. 2014;63:713–716. doi: 10.1016/j.parint.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Yang Y., Mao Y., Kelly P., Yang Z., Luan L., Zhang J. A pan-Theileria FRET-qPCR survey for Theileria spp. in ruminants from nine provinces of China. Parasit. Vectors. 2014;7:413. doi: 10.1186/1756-3305-7-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Liu Z., Guan G., Liu A., Ma M., Ren Q. Detection and differentiation of Theileria luwenshuni and T. uilenbergi infection in small ruminants by PCR. Transbound. Emerg. Dis. 2008;55:233–237. doi: 10.1111/j.1865-1682.2008.01031.x. [DOI] [PubMed] [Google Scholar]
- Yisaschar-Mekuzas Y., Jaffe C.L., Pastor J., Cardoso L., Baneth G. Identification of Babesia species infecting dogs using reverse line blot hybridization for six canine piroplasms, and evaluation of co-infection by other vector-borne pathogens. Vet. Parasitol. 2013;191:367–373. doi: 10.1016/j.vetpar.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Young A.S., Grootenhuis J.G., Kimber C.D., Kanhai G.K., Stagg D.A. Isolation of a Theileria species from Eland (Taurotragus oryx) infective for cattle. Tropenmed. Parasitol. 1977;27:185–194. [PubMed] [Google Scholar]
- Young A.S., Purnell R.E., Payne R.C., Brown C.G., Kanhai G.K. Studies on the transmission and course of infection of a Kenyan strain of Theileria mutans. Parasitology. 1978;76:99–115. doi: 10.1017/s0031182000047430. [DOI] [PubMed] [Google Scholar]
- Young A.S., Leitch B.L., Newson R.M., Cunningham M.P. Maintenance of Theileria parva parva infection in an endemic area of Kenya. Parasitology. 1986;93:9–16. doi: 10.1017/s0031182000049787. [DOI] [PubMed] [Google Scholar]
- Yusufmia S.B.A.S., Collins N.E., Nkuna R., Troskie M., Van den Bossche P., Penzhorn B.L. Occurrence of Theileria parva and other haemoprotozoa in cattle at the edge of Hluhluwe-iMfolozi Park, KwaZulu-Natal, South Africa. J. S. Afr. Vet. Assoc. 2010;81:45–49. doi: 10.4102/jsava.v81i1.95. [DOI] [PubMed] [Google Scholar]
- Zaeemi M., Haddadzadeh H., Khazraiinia P., Kazemi B., Bandehpour M. Identification of different Theileria species (Theileria lestoquardi, Theileria ovis, and Theileria annulata) in naturally infected sheep using nested PCR-RFLP. Parasitol. Res. 2011;108:837–843. doi: 10.1007/s00436-010-2119-0. [DOI] [PubMed] [Google Scholar]
- Zahler M., Rinder H., Schein E., Gothe R. Detection of a new pathogenic Babesia microti-like species in dogs. Vet. Parasitol. 2000;89:241–248. doi: 10.1016/s0304-4017(00)00202-8. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu Z., Yang J., Chen Z., Guan G., Ren Q. Multiplex PCR for diagnosis of Theileria uilenbergi, Theileria luwenshuni, and Theileria ovis in small ruminants. Parasitol. Res. 2014;113:527–531. doi: 10.1007/s00436-013-3684-9. [DOI] [PubMed] [Google Scholar]
- Zobba R., Ardu M., Niccolini S., Chessa B., Manna L., Cocco R. Clinical and laboratory findings in equine piroplasmosis. J. Equine Vet. Sci. 2008;28:301–308. [Google Scholar]
- Zweygarth E., Just M.C., De Waal D.T. In vitro cultivation of Babesia equi: detection of carrier animals and isolation of parasites. Onderstepoort J. Vet. Res. 1997;64:51–56. [PubMed] [Google Scholar]
- Zweygarth E., Benade J., Steyl J., Prozesky L., Koekemoer O., Josemans A.I. In vitro cultivation of a Theileria species from a roan antelope (Hippotragus equinus) Parasitol. Res. 2009;105:1755–1757. doi: 10.1007/s00436-009-1589-4. [DOI] [PubMed] [Google Scholar]
- Zweygarth E., Koekemoer O., Josemans A.I., Rambritch N., Pienaar R., Putterill J. Theileria-infected cell line from an African buffalo (Syncerus caffer) Parasitol. Res. 2009;105:579–581. doi: 10.1007/s00436-009-1467-0. [DOI] [PubMed] [Google Scholar]


