Abstract
Members of Fusarium solani species complex (FSSC) have been known as plant, animal, and human pathogens. Nevertheless, the taxonomic status of such an important group of fungi is still very confusing and many new species as well as lineages have been elucidated recently. Unfortunately, most of the new taxa came from temperate and subtropical regions. Therefore, the objectives of the present study were to identify strains of FSSC recovered from different sources in Malaysia. In the present study, 55 strains belonging to the FSSC were examined and phylogenetically analyzed on the basis of internal transcribed spacer (ITS) regions and partial translation elongation factor-1 (TEF-1α) sequences. Based on morphological features, a total of 55 strains were selected for molecular studies. Based on morphological features, the strains were classified into four described Fusarium species, namely Fusarium keratoplasticum, Fusarium falciforme, FSSC 5, and Fusarium cf. ensiforme, and one unknown phylogenetic species was introduced. Although the data obtained from morphological and molecular studies sufficiently supported each other, the phylogenetic trees based on ITS and TEF-1α dataset clearly distinguished closely related species and distinctly separated all morphological taxa. All members of FSSC in this research were reported for the first time for Malaysian mycoflora.
Introduction
Most members of the Fusarium solani species complex (FSSC) are frequently isolated from soils and act as decomposers, but some are putative parasites on plants, insects, humans, and animals [1–4]. Wollenweber and Reinking [5] divided members of F. solani into two sections of Ventricosum and Martiella. Snyder and Hansen [6] represented FSSC as a complex species in the Martiella section [1, 7, 8]. Seven mating populations (MPI–VII) were determined for F. solani [9–12]. Members of FSSC mating populations MPI, MPV, and MPVI were placed in distinct groups by phylogenetic analysis [13]. Molecular phylogenetic demonstrated that FSSC MPI and MPV as F. solani f. sp. cucurbitae races 1 and 2, respectively were polyphyletic [14, 15].
Because of the significant role of members of the FSSC in clinical infection and complication in their determination, molecular identification strategies have been emphasized for their identification in the last 20 years [2, 3]. Phylogenetic analysis by 28S ribosomal DNA, internal transcribed spacer (ITS) regions, and tef1 gene sequences revealed high variability within members of the FSSC, and all 55 diagnosable species were divided into three clades, termed clades 1, 2, and 3 [3, 15–17]. Members of clade 1 comprised two known species (Fusarium illudens and Nectria plagianthi) from New Zealand. Members of clade 2 included a number of important pathogens that cause sudden death syndrome (SDS) of soybean [18–20]. Nalim et al. [17] used molecular phylogeny to show that members of FSSC in clade 2 are paraphyletic.
Molecular phylogeny showed diverse phylogenetic affinities among members of clade 3. This group encompassed many species that are important in agricultural crops and medicine [15]. Members of clade 3 are the most common group of fusaria associated with plant diseases and human infections. Members of Fusarium falciforme (FSSC 3 + 4) and Fusarium keratoplasticum as most haplotype-diverse species were placed among clade 3 [2, 3, 15, 21, 22]. Several studies to date have revealed different phylogenetic species within this important evolutionary clade, though little work has been done to improve the taxonomy, and therefore correct identification of species as one of the prerequisites in any disease control program has become more challenging. Although the taxonomic status of FSSC from all over the world is being revised and a strong connection has been revealed among strains recovered from humans, insects, and plants [2, 17, 21–24], until today, no attempt has been made to classify members of the FSSC in tropical Southeast Asia, particularly Malaysia. Therefore, the objectives of this study were to identify strains of FSSC recovered from different substrates in Malaysia by using morphological characteristics and sequencing of ITS region and translation elongation factor-1α (TEF-1α) to determine genetic relationship among them.
Materials and Methods
Strains of Fusarium
A total of 55 strains were selected for the present study. Strains were obtained from Fusarium culture collection of the School of Biological Sciences, Universiti Sains Malaysia. A list of species names and culture collection numbers, geographical origins, original substrates, and GenBank accession numbers of the tef1 and ITS regions of the strains used in this study is in Table 1. To study the pigmentation and growth rates, all strains were transferred onto fresh potato dextrose agar (PDA) plates and incubated under 12 h alternating light (black/white) at 25 ± 2 °C for 1 week. For microscopic observations, all strains were transferred to carnation leaf-piece agar (CLA) [25] plates and incubated under 12 h alternating light (black/white) at 25 ± 2 °C for 1 week. Thirty randomly selected conidia of each septation class (macro- and microconidia) were measured and analyzed by two-sample t test using MINITAB® 15. For species determination, the descriptions by Summerbell and Schroers [26], Nalim et al. [17], and Short et al. [21] were adopted.
Table 1.
Strains of members of the Fusarium solani species complex collected in Malaysia from soil and different substrate
| Culture no. | Species | Host/substratum | Plant part (symptom) | Location | tef1 a | ITSb |
|---|---|---|---|---|---|---|
| USM FSSC-C4383B | F. cf. solani | Bean | Root rot | Pahang | KC161396 | KC009602 |
| USM FSSC-R77O | F. cf. solani | Corn | Seed | Perlis | KC161404 | KC009610 |
| USM FSSC-R73O | F. cf. solani | Corn | Leaf (spot) | Perlis | KC161402 | KC009608 |
| USM FSSC-D8256C | F. cf. solani | Red chilli pepper | Leaf (spot) | Kelantan | KC161386 | KC009592 |
| USM FSSC-T8432C | F. cf. solani | Red chilli pepper | Stem (lesion) | Terengganu | KC161388 | KC009594 |
| USM FSSC-Q6002D | F. cf. solani | Pepper | Stem (lesion) | Sarawak | KC161414 | KC009620 |
| USM FSSC-Q729D | F. cf. solani | Pepper | Stem (lesion) | Sarawak | KC161418 | KC009624 |
| USM FSSC-Q1033Q | F. cf. solani | Sorghum | Root (rot) | Sarawak | KC161421 | KC009627 |
| USM FSSC-Q1017Q | F. cf. solani | Sorghum | Root (rot) | Sarawak | KC161399 | KC009605 |
| USM FSSC-P86S | F. cf. solani | Soil | – | Penang | KC161393 | KC009599 |
| USM FSSC-P104S | F. cf. solani | Soil | – | Penang | KC161391 | KC009597 |
| USM FSSC-P200S | F. cf. solani | Soil | – | Penang | KC161390 | KC009596 |
| USM FSSC-A2073S | F. cf. solani | Soil, Watermelon | – | Perak | KC161412 | KC009618 |
| USM FSSC-D310T | F. cf. solani | Tobacco | Stem (rot) | Kelantan | KC161394 | KC009600 |
| USM FSSC-J872T | F. cf. solani | Tobacco | Stem (rot) | Johor | KC161400 | KC009606 |
| USM FSSC-K600T | F. cf. solani | Tobacco | Stem (rot) | Kedah | KC161398 | KC009604 |
| USM FSSC-D216T | F. cf. solani | Tobacco | Stem (rot) | Kelantan | KC161392 | KC009598 |
| USM FSSC-J3516Gr | F. cf. solani | Grass | Root (rot) | Johor | KC161385 | KC009591 |
| USM FSSC-S2253Pu | F. cf. solani | Pumpkin | Fruit (canker) | Sabah | KC161410 | KC009616 |
| USM FSSC-D5281Pu | F. cf. solani | Pumpkin | Fruit (canker) | Kelantan | KC161409 | KC009615 |
| USM FSSC-A1969W | F. cf. solani | Watermelon | Root (rot) | Perak | KC161397 | KC009603 |
| USM FSSC-Q1094W | F. cf. solani | Watermelon | Root (rot) | Sarawak | KC161395 | KC009601 |
| USM FSSC-T617R | F. cf. solani | Rice | Root (rot) | Terengganu | KC161416 | KC009622 |
| USM FSSC-S2257W | F. cf. solani | Wood | Stem (lesion) | Sabah | KC161415 | KC009621 |
| USM FSSC-T921T | F. cf. solani | Tobacco | Stem (rot) | Terengganu | KC161403 | KC009609 |
| USM FSSC-A1881W | F. cf. solani | Watermelon | Root (rot) | Perak | KC161419 | KC009625 |
| USM FSSC-T2550W | F. cf. solani | Watermelon | Root (rot) | Terengganu | KC161422 | KC009628 |
| USM FSSC-B1770S | F. falciforme | Soil | Tree bark | Selangor | JX935592 | JX982559 |
| USM FSSC-A1444S | F. falciforme | Soil | Perak | JX935593 | JX982560 | |
| USM FSSC-D436S | F. falciforme | Soil | Kelantan | JX935594 | JX982561 | |
| USM FSSC-S2216Ru | F. falciforme | Tree bark | Tree bark | Sabah | JX935586 | JX982553 |
| USM FSSC-S2228Tb | F. falciforme | Tree bark | Tree bark | Sabah | JX935588 | JX982555 |
| USM FSSC-S2237Ne | F. falciforme | Soil | – | Sabah | JX935587 | JX982554 |
| USM FSSC-S2238Ne | F. falciforme | Soil | – | Sabah | JX935585 | JX982552 |
| USM FSSC-S2236M | F. falciforme | Soil | – | Sabah | JX935584 | JX982551 |
| USM FSSC-K1418S | F. falciforme | Soil | – | Kedah | JX935589 | JX982556 |
| USM FSSC-P1531S | F. falciforme | Soil | – | Penang | JX935590 | JX982557 |
| USM FSSC-B1765S | F. falciforme | Soil | Tree bark | Selangor | JX935591 | JX982558 |
| USM FSSC-R43R | F. falciforme | Rice | Root (rot) | Perlis | KF836701 | KF836673 |
| USM FSSC-S2231Al | F. falciforme | Algae | – | Sabah | KF836700 | KF836672 |
| USM FSSC-P93S | F. falciforme | Soil | – | Penang | KF836702 | KF836674 |
| USM FSSC-P2108S | F. keratoplasticum | Soil | – | Penang | KF836695 | KF836667 |
| USM FSSC-S2138Se | F. keratoplasticum | Azadirachta excelsa | Root (rot) | Sabah | KF836699 | KF836671 |
| USM FSSC-S2126Se | F. keratoplasticum | Azadirachta excelsa | Root (rot) | Sabah | KF836698 | KF836670 |
| USM FSSC-Q1172Rh | F. keratoplasticum | Casuarina equisetifolia | Root (rot) | Sarawak | KF836696 | KF836668 |
| USM FSSC-Q4854D | F. keratoplasticum | Casuarina equisetifolia | Root (rot) | Sarawak | KF836697 | KF836669 |
| USM FSSC-Q1371W | FSSC 5 | Watermelon | Root (rot) | Sarawak | KF836691 | KF836676 |
| USM FSSC-C3496Gr | FSSC 5 | Echinochloa colonum | Root (rot) | Pahang | KF836692 | KF836677 |
| USM FSSC-Q1165Gr | FSSC 5 | Grass | Root (rot) | Sarawak | KF836693 | KF836678 |
| USM FSSC-B1409M | FSSC 5 | Mangifera indica | Fruit (spot) | Mango | KF836690 | KF836675 |
| USM FSSC-C1814An | FSSC 5 | Grass | Root (rot) | Pahang | KF836694 | KF836679 |
| USM FSSC-C4651Tb | F. cf. ensiforme | Tree bark | Wood (decay) | Sabah | KC161383 | KC009589 |
| USM FSSC-C4641Tb | F. cf. ensiforme | Tree bark | Wood (decay) | Sabah | KC161384 | KC009590 |
| USM FSSC-S2135Tb | F. cf. ensiforme | Tree bark | Wood (decay) | Sabah | KC161382 | KC009588 |
| USM FSSC-S2256Tb | F. cf. ensiforme | Tree bark | Wood (decay) | Sabah | KC161381 | KC009587 |
aGenBank numbers for translation elongation factor 1-alpha (tef1) partial sequences
bGenBank numbers for ITS regions
DNA Extraction
All FSSC strains were grown on PDA with sterile dialysis membranes [27] for 5 days. The mycelium grown over the membranes were harvested and grounded in a sterile mortar with liquid nitrogen to a fine powder. DNA extraction was done by using The DNeasy® Plant Mini Kit (Qiagen) according to the manufacturer’s instructions.
PCR Amplification
Amplification of the TEF-1α gene and ITS regions was conducted using primer pair ef1 and ef2 for TEF-1α [16] and ITS1 and ITS4 for the ITS regions [28]. Polymerase chain reaction (PCR) was performed in a Peltier Thermal Cycler, PTC-100® (MJ Research, Inc. USA) in a total volume of 25 μl for each strain. The PCR mixture contained 4 μl 5× buffer (Promega, Madison, WI, USA), 4 mM MgCl2, 0.2 mM deoxynucleotide triphosphate (dNTP; Promega), 0.8 μM each primer, 0.75 units of Taq DNA polymerase (Promega), and 6 ng of template DNA. DNA amplification of TEF-1α was performed with an initial denaturation of 1 min at 94 °C followed by 35 cycles of 30 s at 95 °C, 55 s at 59 °C, and 90 s at 72 °C and a final extension of 10 min at 72 °C. The PCR for ITS regions was performed at 95 °C (2 min) for a hot start, followed by 35 cycles of 94 °C (1 min), 56 °C (30 s), 72 °C (2 min), and a final extension of 72 °C (10 min). PCR products were purified using Qiagen columns according to the manufacturer’s protocol and sent for sequencing to a service provider.
Sequencing Alignment and Phylogenetic Analysis
The program Molecular Evolutionary Genetic Analysis software, ver. 4.0 (MEGA4.0; http://www.megasoftware.net) was performed in order to edit and align the sequence files [29], which were manually adjusted. In order to assess the relationships between the major taxa, ambiguous parts of the ITS regions and TEF-1α were removed from further analysis and more conserved and alignable parts of the region and all genes were used to generate phylogenetic trees containing representative taxa from major groups. The aligned sequences were BLAST in two genome databases, GenBank and Fusarium database, to identify all the 55 strains. In this study, phylogenetic tree was generated using maximum parsimony (MP) in MEGA4.0. Bootstrap values for the maximum parsimony tree (MPT) were calculated for 1,000 replicates. The edited ITS and TEF-α sequences were compared with other available Fusarium species sequences in the GenBank. Furthermore, the sequences of some known species of FSSC were downloaded from GenBank and used to reconstruct a combined ITS region and TEF-α phylogenetic trees. For phylogenetic analysis, 91 taxa were included in the combined dataset and Fusarium staphyleae (NRRL 22316) was used as an outgroup.
Results
Morphological Analysis
The 55 strains were again investigated for their macro- and microscopic characteristics as shown in Table 2. All possible morphological dissimilarities were therefore taken into account and represented by the 55 strains selected. For species determination, the descriptions by Summerbell and Schroers [26], Nalim et al. [17], and Short et al. [21] were adopted. No sexual structures were observed in this study. Based on ITS and TEF-1α sequence data and morphological characteristics (Table 2), five species were identified. Of these, F. keratoplasticum (5 strains) and F. falciforme (14 strains) were known species. Features showed (Table 2) five isolates belonging to undescribed FSSC 5 (USM FSSC-C3496Gr) as the known phylogenetic species among FSSC and four isolates were identified as F. cf. ensiforme (USM FSSC-C4651Tb) that were described by Nalim et al. [17]. Also, 27 strains were identified as F. cf. solani (USM FSSC-K600T). Macro- and microscopic characteristics including means and ranges of spore dimensions of individual isolates of FSSC are summarized in Table 2.
Table 2.
Morphological characteristics of individual strains of FSSC collected from different places in Malaysia
| Culture no. | Species identified | Shape of Microconidia | Shape of basal cell and apical cell | Length × width of macroconidia (μm)a | |
|---|---|---|---|---|---|
| 3- and 4-septate | 5-septate | ||||
| FSSC-R73O | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 50.2 ± 2.5 × 5.7 ± 0.5 | 54.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-K600T | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 47.5 ± 2.5 × 5.8 ± 0.5 | 52.5 ± 2.5 × 5.6 ± 0.5 |
| FSSC-J872T | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 48.3 ± 2.5 × 5.6 ± 0.5 | 54.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-R77O | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 47.5 ± 2.5 × 5.9 ± 0.5 | 54.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-Q6002D | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 47.0 ± 2.5 × 5.9 ± 0.5 | 54.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-Q1033Q | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 51.2 ± 2.5 × 5.8 ± 0.5 | 52.5 ± 2.5 × 6 ± 0.5 |
| FSSC-Q1017Q | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 49.2 ± 2.5 × 5.8 ± 0.5 | 54.5 ± 2.5 × 6 ± 0.5 |
| FSSC-P200S | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 48.5 ± 2.5 × 5.5 ± 0.5 | 54.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-C4383B | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 52.2 ± 2.5 × 5.8 ± 0.5 | 56.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-Q729D | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 49.3 ± 2.5 × 5.8 ± 0.5 | 55.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-A2073S.W | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 48.2 ± 2.5 × 5.8 ± 0.5 | 56.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-P104S | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 51.2 ± 2.5 × 5.8 ± 0.5 | 56.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-Q1094W | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 49.2 ± 2.5 × 5.8 ± 0.5 | 56.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-J3516& | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 50.2 ± 2.5 × 5.8 ± 0.5 | 56.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-A1969W | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 48.6 ± 2.5 × 5.6 ± 0.5 | 57.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-D8256C | F. cf. solani | Oval, reniform, and elongated oval | Foot cell and tapered, curved | 48.0 ± 2.5 × 5. 5 ± 0.5 | 54.5 ± 2.5 × 5.9 ± 0.5 |
| FSSC-S2236Mo | F. falciforme | Elongated oval and obovoid with a truncate | Barely notched and pointed, curved | 43.5 ± 2.5 × 5.1 ± 0.5 | – |
| FSSC-S2238Ne | F. falciforme | Elongated oval and obovoid with a truncate | Barely notched and pointed, curved | 42.5 ± 2.5 × 5.0 ± 0.5 | – |
| FSSC-S2216Ru | F. falciforme | Elongated oval and obovoid with a truncate | Barely notched and pointed, curved | 40.5 ± 1.5 × 5.1 ± 0.5 | – |
| FSSC-K1418S | F. falciforme | Elongated oval and obovoid with a truncate | Barely notched and pointed, curved | 41.0 ± 1.0 × 5.2 ± 0.5 | – |
| USM FSSC-P2106S | F. keratoplasticum | Oval and elongated and clavate | Notched and blunt | 33.0 ± 2.3 × 4.9 ± 0.3 | – |
| USM FSSC-P2108S | F. keratoplasticum | Oval and elongated and clavate | Notched and blunt | 33.0 ± 1.5 × 4.9 ± 0.3 | – |
| USM FSSC-S2138Se | F. keratoplasticum | Oval and elongated and clavate | Notched and blunt | 34.0 ± 2.3 × 4.8 ± 0.3 | – |
| USM FSSC-S2126Se | F. keratoplasticum | Oval and elongated and clavate | Notched and blunt | 35.0 ± 2.3 × 4.9 ± 0.3 | – |
| USM FSSC-C4651Tb | F. cf. ensiforme | Elongated oval | Foot cell and papillate curved | 52.5 ± 2.3 × 6.1 ± 0.2 | 60.5 ± 2 × 6.3 ± 0.3 |
| USM FSSC-C4641Tb | F. cf. ensiforme | Elongated oval | Foot cell and papillate curved | 53.5 ± 2.3 × 6.1 ± 0.2 | 60.5 ± 2 × 6.3 ± 0.3 |
| USM FSSC-S2135Tb | F. cf. ensiforme | Elongated oval | Foot cell and papillate curved | 52.5 ± 2.3 × 6.1 ± 0.2 | 61.5 ± 2 × 6.3 ± 0.3 |
| USM FSSC-C3496Gr | FSSC 5 | Oval, elongated oval, clavate, and reniform | Barely notched and papillate curved | 38.5 ± 1.5 × 5.3 ± 0.2 | 45.5 ± 2.5 × 5.7 ± 0.2 |
| USM FSSC-Q1165Gr | FSSC 5 | Oval, elongated oval, clavate, and reniform | Barely notched and papillate curved | 40.5 ± 1.5 × 5.4 ± 0.2 | 45.5 ± 2.5 × 5.8 ± 0.2 |
| USM FSSC-B1409M | FSSC 5 | Oval, elongated oval, clavate, and reniform | Barely notched and papillate curved | 41.5 ± 1.5 × 5.4 ± 0.2 | 45.5 ± 2.5 × 5.8 ± 0.2 |
aMean values of 30 random conidia ± standard deviation
In the present study, 27 strains isolated from different crops and soil in Malaysia (Table 1) belonging to the F. cf. solani (USM FSSC-K600T) were examined and phylogenetically analyzed on the basis of ITS regions and partial TEF-1α sequences. Strains of F. cf. solani (USM FSSC-K600T) exhibited morphological variability in culture. Cultures grew fast, the growth rate (mm⁄day) on PDA at 25 °C in intermittent light ranged from 7.8 to 8.6 mm⁄day. The hyphae initially hyaline and mycelium became yellowish white; green to bluish-gray, and purple in reverse after 1–2 weeks (Fig. 1 (1)). At 25 °C, aerial conidiophores formed abundantly on CLA, unbranched or branched, up to 200 μm long, 3.5–7.0 μm at base. Phialides were more or less erect, subcylindrical, or cylindrical arising from conidiophores. Sporodochial conidiophores formed on distinctive collarete at the tip. The macroconidia arising from sporodochia are falcate, dorsiventral, with 3–5-septate and mostly 5-septate, with pointed, tapered, and curved apical cell and well-developed foot cell. The macroconidia are typically falcate, widest in the middle of their length (Fig. 2a–c). The microconidia are oval, reniform, elongated oval to sometimes obovoid with a truncate base, mostly 0-septate (Fig. 2d, e). The size of conidia measures as follows: 0-septate = (6–) 8–12 (−13.5) × (3–) 3.4–5.3 μm (−5.6); 1-septate = (9–) 14–16 (−26) × (4–) 4–4.6 μm (−5.8); 3-septate = (35–) 42–48 (−52) × (4.8–) 5.4–5.9 μm (−6.5); 4-septate (40–) 48–54 (−58) × (5.0–) 5.5–6.0 μm (−6.5); and 5-septate (44–) 51–57 (−63) × (5.0–) 5.5–6.0 μm (−6.5) (Table 2). Chlamydospores formed relatively abundant in mycelium, mostly globose, subglobose, intercalary or terminal and rough walled, 5–15 μm in diameter, and may occasionally be found within the macroconidia. Chlamydospores formed singly, and in clusters, or in chains (Fig. 2f).
Fig. 1.
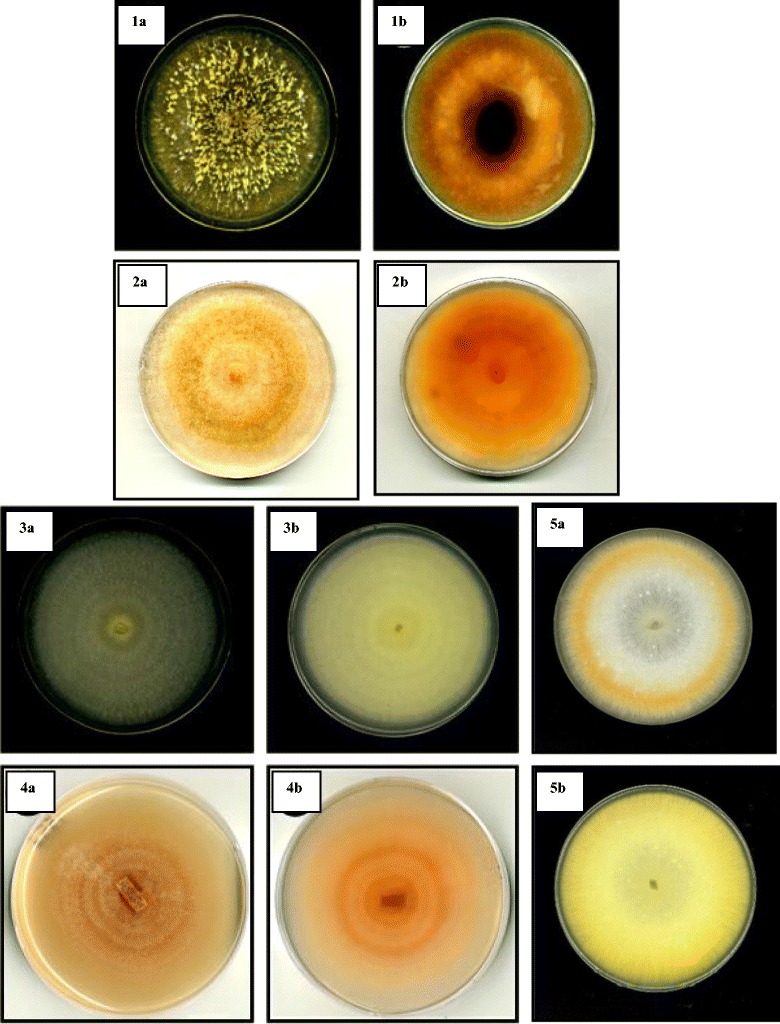
Colonies of Malaysian members of the FSSC grown on PDA incubated under 12 h alternating light (black/white) at 25 ± 2 °C for 2 weeks. a Top view, b reverse view: 1, Fusarium cf. solani (USM FSSC-K600T); 2, Fusarium keratoplasticum (USM FSSC-P2108S); 3, FSSC 5 (USM FSSC-C3496Gr); 4, Fusarium cf. ensiforme (USM FSSC-C4651Tb); 5, F. falciforme (USM FSSC-S2216Ru)
Fig. 2.
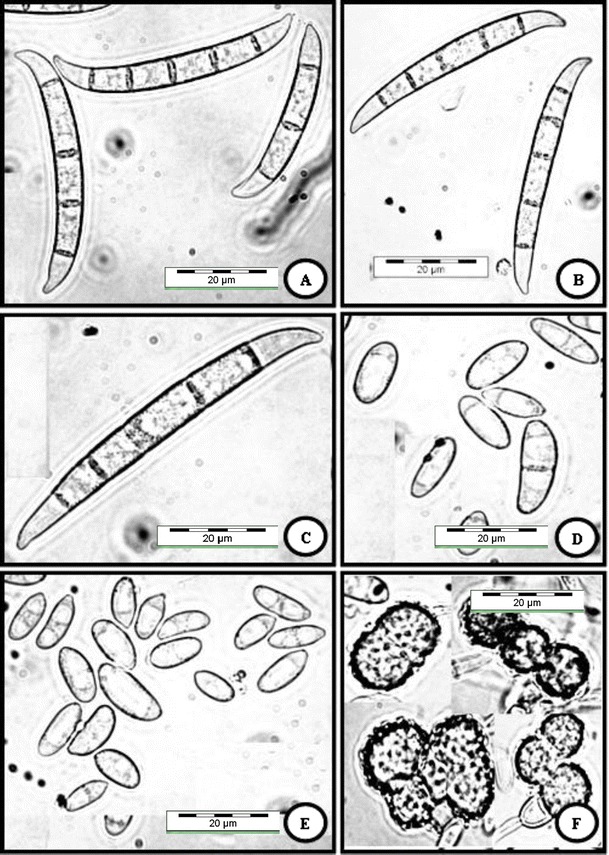
Fusarium cf. solani (USM FSSC-K600T) grown on CLA, 2 weeks, 25 °C, cool white fluorescent light 12 h/darkness 12 h. a–c Multiseptate macroconidia produced from sporodochia. d, e, Oval-shaped and reniform conidia formed on conidiophores in hyphae. f Terminal and intercalary chlamydospores. Bar = 20 μm for all pictures
The growth rate (mm⁄day) on PDA at 25 °C in intermittent light ranged from 5.5 to 6.0 mm⁄day. The hyphae initially were hyaline and sparsely formed. Mycelium tend to becomes cream to orange in reverse after 1–2 weeks (Fig. 1 (4)). At 25 °C, abundant, erect, unbranched, or once-branched conidiophores are formed on the surface of the agar. Cream slimy sporodochia is produced on the surface of the agar. Phialides are more or less erect, somewhat swollen in the middle or cylindrical, arising from conidiophores, mainly (28–) 48–85 mm (−115) long and (2.6–) 3.1–4.4 μm (−5.3) at the base. Phialides produced in sporodochia are cylindrical, (13–) 16–21 mm (−26) long, (3.0–) 3.7–5.0 mm (−5.2) in diameter. The macroconidia arising from sporodochia are long, slightly curved with 3–8-septate mostly 5–7-septate, with papillate and somewhat curved apical cell and well-developed foot cell (Fig. 3a, b). The microconidia are oval and elongated oval, mostly 0-septate (Fig. 3c, d). The size of conidia measures as follows: 0-septate = (5–) 6–11 (−15) × (2.8–) 3.3–4.3 μm (−5.0); 5-septate = (50–) 56–63 (−75) × (5.3–) 6–6.5 μm (−6.8); 6–7-septate = (59–) 64–75 (−86) × (5.0–) 6–6.8 μm (−7.2); and 8-septate = 70–80 × 5.7–6.8 μm. Chlamydospores are smooth walled (Fig. 3e).
Fig. 3.
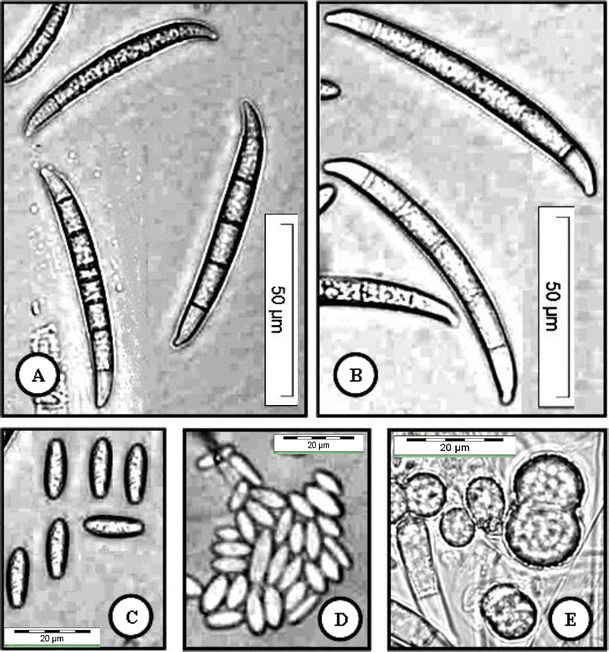
Fusarium cf. ensiforme (USM FSSC-C4651Tb) grown on CLA, 2 weeks, 25 °C, cool white fluorescent light 12 h/darkness 12 h. a, b Macroconidia (5–7-septate) produced from sporodochia. c, d Oval and elongated oval microconidia. e Terminal and intercalary chlamydospores. a, b Bar = 50 μm; c, d Bar = 20 μm
Cultures grew fast; the growth rate (mm⁄day) on PDA at 25 °C in intermittent light ranged from 8 to 9 mm⁄day (Fig. 1 (2)). The hyphae initially were hyaline. Mycelium becomes yellowish white and yellowish in reverse after 1–2 weeks. At 25 °C, aerial conidiophores formed abundantly on CLA, unbranched or branched, up to 100 μm long, 3.5–6.5 μm at base. Phialides are subcylindrical or cylindrical arising from conidiophores. Sporodochial conidiophores are formed on distinctive collarete at the tip. The macroconidia arising from sporodochia are small in length, falcate, dorsiventral, with 3-septate but rarely 4-septate with blunt apical cell and notched basal cell (Fig. 4a, b). The macroconidia are typically falcate of their length. The microconidia are oval, elongated oval to sometimes clavate with a truncate base mostly 0–1-septate (Fig. 4c, d). The size of conidia measures as follows: 0-septate = (3.6–) 6–9.15 (−11.5) × (2.8–) 3.7–5.9 μm (−6.3); 1-septate = (10–) 12–15 (−23) × (4–) 4.6–5 μm (−6.0); 3-septate = (27–) 30–38 (−44) × (4–) 4.8–5.8 μm (−6.4); and 4-septate = 36–40 × 5.7–6.2 μm. Chlamydospores are formed relatively abundant in the mycelium, mostly globose, subglobose, intercalary or terminal, and smooth to rough walled, 8–15 μm in diameter. Chlamydospores are formed singly, and in cluster, or in chains (Fig. 4e–g).
Fig. 4.
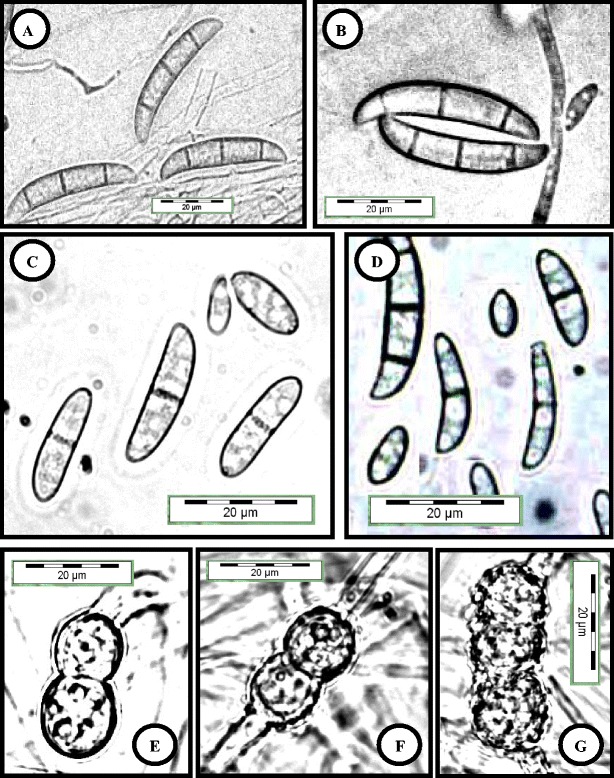
Fusarium keratoplasticum (USM FSSC-P2108S) grown on CLA, 2 weeks, 25 °C, cool white fluorescent light 12 h/darkness 12 h. a, b Three septate macroconidia produced from sporodochia. c, d Oval-shaped and curved cylindrical conidia were formed on conidiophores in hyphae. e–g Terminal and intercalary chlamydospores. Bar = 20 μm for all pictures
Cultures grew fast, and the growth rate (mm⁄day) on PDA at 25 °C in intermittent light ranged from 8 to 9 mm⁄day (Fig. 1 (5)). The hyphae initially were hyaline. Mycelium becomes yellowish white and yellowish in reverse after 1–2 weeks. At 25 °C, aerial conidiophores formed abundantly on CLA, unbranched or branched, up to 200 μm long, 3.7–7.0 μm at base. Phialides are subcylindrical or cylindrical arising from conidiophores. Sporodochial conidiophores were formed on distinctive collarete at the tip. The macroconidia are arising from sporodochia falcate, dorsiventral, with 3–4-septate, with pointed apical cell and barely notched basal cell. The macroconidia are slightly curved of their length (Fig. 5a–c). The microconidia were oval, elongated oval to sometimes obovoid with a truncate base mostly 0–1-septate (Fig. 5d, e). The size of conidia measures as follows: 0-septate = (5.6–) 6–12.15 (−13) × (2.8–) 3.7–4.8 μm (−5.3); 1-septate = (10–) 12–15 (−26) × (4–) 4.2–5 μm (−5.4); 3-septate = (26–) 31–42 (−44) × (4.4–) 4.5–5 μm (−5.5); and 4-septate = 39–44 × 4.6–5.2 μm. Chlamydospores are formed relatively abundant in the mycelium, mostly globose, subglobose, intercalary or terminal, and rough walled, 8–15 μm in diameter. Chlamydospores are formed singly, and in cluster, or in chains (Fig. 5f).
Fig. 5.
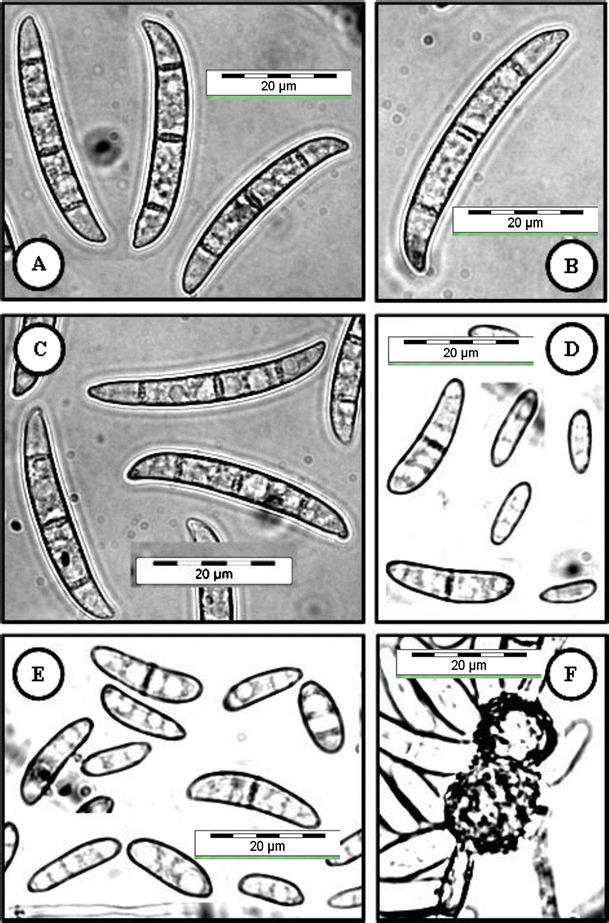
Fusarium falciforme (USM FSSC-S2216Ru) grown on CLA, 2 weeks, 25 °C, cool white fluorescent light 12 h/darkness 12 h. a–c Multiseptate macroconidia produced from sporodochia. d, e Oval-shaped and reniform conidia were formed on conidiophores in hyphae. f An intercalary chlamydospores. Bar = 20 μm for all pictures
The growth rate (mm⁄day) on PDA at 25 °C in intermittent light ranged from 6.0 to 7.5 mm⁄day. Aerial mycelium is sparsely formed, arranged in concentric rings, and richly conidial. The mycelium becomes white and cream in reverse after 1–2 weeks (Fig. 1 (3)). At 25 °C, abundant conidiophores, erect or verticillate in the upper part, with two to three phialides arising at each node. On CLA, conidiophores formed abundantly and sometimes on coiled hyphae in the aerial mycelium. Cream to bluish slimy sporodochia is produced adjacent to leaf agar. Phialides more or less erect, somewhat swollen in the middle or cylindrical, arising from conidiophores, mainly 22–150 × 4–7 μm at base. The macroconidia arising from sporodochia are medium in length, somewhat straight especially in the ventral side with 3–5-septate, with papillate and somewhat curved apical cell and barely notched basal cell (Fig. 6a, b). The microconidia are oval, elongated oval, clavate and reniform, and mostly 0–1-septate (Fig. 6c, d). The size of conidia measures as follows: 0-septate = (3.4–) 6–9.15 (−12.5) × (2.8–) 3.7–5.3 μm (−5.6); 1-septate = (10–) 14–19 (−27) × (4–) 4.7–5.3 μm (−6.0); 3-septate = (30–) 35–42 (−44) × (5–) 5.4–5.9 μm (−6.5); 4-septate = (38–) 40–44(−48) × (5.0–) 5.5–6.0 μm (−6.5); and 5-septate = 43–50 × 5.7–6.5 μm. Chlamydospores are formed singly, and in cluster, or in chains (Fig. 6e, f).
Fig. 6.
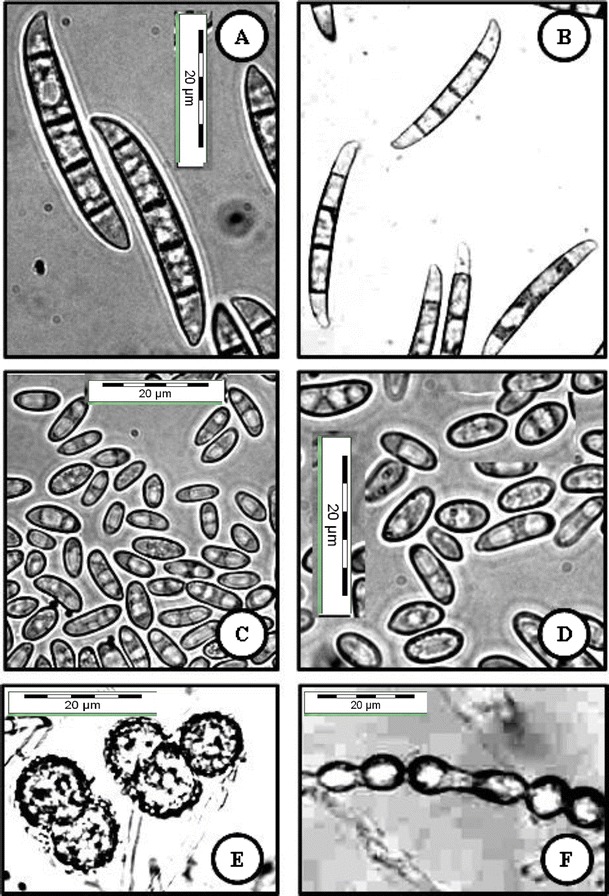
FSSC 5 (USM FSSC-C3496Gr) grown on CLA, 2 weeks, 25 °C, cool white fluorescent light 12 h/darkness 12 h. a, b Macroconidia produced from sporodochia. c, d Oval-shaped and reniform conidia formed on conidiophores in hyphae. e, f Terminal and intercalary chlamydospores. Bar = 20 μm for all pictures
Phylogenetic Analysis
The tree generated from the combined dataset of ITS regions and TEF-α supported previously inferred clade 3. This also highlighted the fact that members of clade 3 represent a monophyletic lineage (Fig. 7). MP of 91 taxa (including outgroup) of the Fusarium in FSSC inferred from combined ITS and tef1 sequences also revealed diverse phylogenetic affinities among members of FSSC clade 3. All strains isolated from soils and plants represented five different lineages in the clade 3 (Fig. 7). Phylogenetic tree demonstrated that 27 strains isolated from different sources in Malaysia (Table 1) were placed in F. cf. solani (USM FSSC-K600T) group with a strong bootstrap support (95 %). All 27 strains are closely related to, but phylogenetically distinct from typical F. falciforme and doubtlessly represent a potentially novel phylogenetic species.
Fig. 7.
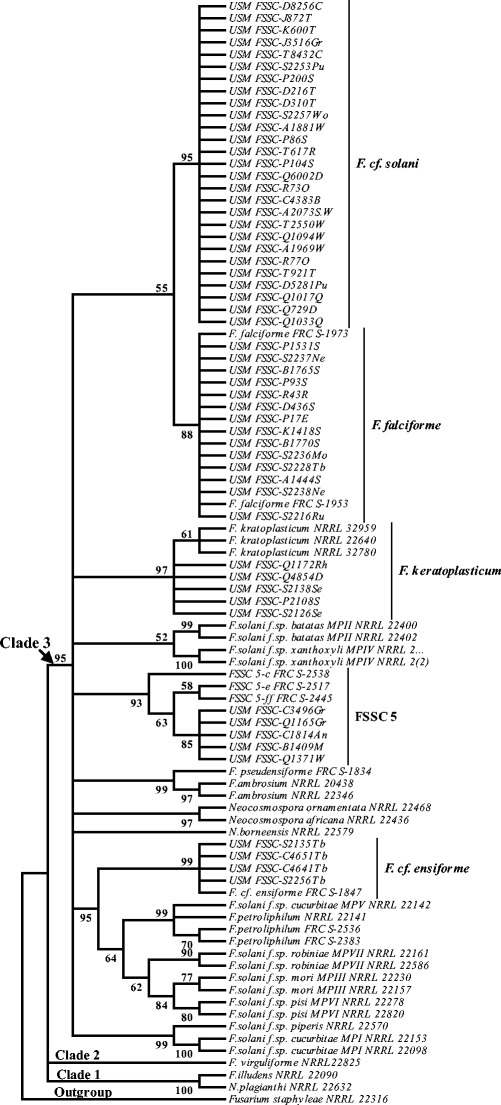
A maximum parsimony phylogeny for 91 taxa of the FSSC inferred from combined ITS and tef1 sequences. Bootstrap tests were performed with 1,000 replications. Fusarium staphyleae (NRRL22316) obtained from GenBank was treated as the outgroup
The tree showed a sister relationship between F. keratoplasticum (NRRL 32959, NRRL 22640, and NRRL 32780) and five new strains included in Table 1, and based on morphological features, all strains were identified as F. keratoplasticum. The tree showed a well-supported relationship (88 % MP bootstrap) between F. falciforme (FRC S-1973 and FRC S-1953) obtained from GenBank and all 14 new strains included in Table 1, which were identified as F. falciforme based on morphological features. Also, the tree showed a monophyly between undescribed species FSSC 5 and five strains isolated from different plants (Table 1). All these five strains represented a putative new lineage within clade 3 with strong phylogenetic affinity (93 % MP bootstrap). The phylogenetic tree also represented strains FSSC-C4651Tb, FSSC-C4641Tb, FSSC-S2135Tb, and FSSC-S2256Tb isolated from a tree bark placed in separated lineage within F. ensiforme clade with a strong bootstrap support (99 % MP) with Fusarium sp. (FRC S-1847). Based on morphological characters, all these strains were identified as F. cf. ensiforme.
Discussion
Due to high morphological variation among members of FSSC in this study and in order to help in defining the species, both molecular and morphological data were taken into consideration. O’Donnell [15] and Aoki et al. [18, 19] found that DNA sequences of ITS regions can clearly present the evolutionary relationships among this species complex and sequence of TEF-1α gene always offered a finer resolution and separated strains of most Fusarium complex species at species rank [30]. Therefore, in this study for accurate identification of FSSC, a molecular systematic study using sequence divergence of ITS regions and TEF-1α gene was used.
As shown by the molecular analysis of combined sequence data of ITS regions and TEF-1α (Fig. 7), members of F. cf. solani (USM FSSC-K600T) formed a distinct group. The type of macroconidia in F. cf. solani (USM FSSC-K600T) was specific among members of clade 3. The macroconidia arising from sporodochia was typically falcate and mostly 5-septate with papillate, tapered, and curved apical cell and well-developed foot cell. F. cf. solani (USM FSSC-K600T) was found to be the most dominant species from different substrates in Malaysia. The dominance of this species in all sampling sites from different sources was indicative of the revolutionary status of this imperfect fungus in Malaysia. However, molecular phylogeny showed a sister relationship between F. falciforme (USM FSSC-S2216Ru) and F. cf. solani (USM FSSC-K600T), but the macroconidia of F. cf. solani were larger than those of F. falciforme and their aerial conidiophores were branched. Fusarium cf. ensiforme (FSSC-C4641Tb) had a close morphological resemblance to Fusarium sp. (JF433047) that had been described by Nalim et al. [17]. Molecular phylogeny, based on ITS regions and TEF-1α, confirmed that all of these strains formed a monophyletic group with the typical strain of Fusarium sp. (JF433047) obtained from GenBank. Based on clear morphological differences and phylogenetic analysis of combined sequence data of ITS regions and TEF-1α (Fig. 7), this group represented a potentially novel phylogenetic species within F. ensiforme clade.
F. keratoplasticum (USM FSSC-P2108S) is characterized by production of dense hyphae and short 3-septate macroconidia (Tables 2 and 3). Morphological data showed a close morphological resemblance between strains USM FSSC-Q1172Rh, USM FSSC-Q4854D, USM FSSC-S2138Se, USM FSSC-P2108S, and USM FSSC-S2126Se included in Tables 2 and 3 with F. keratoplasticum [21]. This result was confirmed by the phylogenetic analysis based on sequence divergent of combined sequence data of ITS regions and TEF-1α. F. keratoplasticum (USM FSSC-P2108S) had a close morphological resemblance to F. falciforme (USM FSSC-S2216Ru), but the macroconidia in F. falciforme was larger than those of F. keratoplasticum. This differentiation was confirmed by combined sequence data of ITS regions and TEF-1α (Fig. 7). In this study, F. falciforme and F. keratoplasticum were reported for the first time for Malaysian mycoflora. The phylogenetic tree presented the monophyly of undescribed species FSSC 5 with strains C3496Gr, Q1165Gr, C1814An, B1409M, and Q1371W (93 % MP bootstrap) (Fig. 7).
Table 3.
Comparison of growth rates and range in sporodochial conidia size of individual strains of FSSC used in this study and common species in the FSSC [17, 21]
| Characters | FSSC 5 | F. cf. solani | F. keratoplasticum | F. keratoplasticum | F. falciforme | F. falciforme | F. cf. ensiforme | F. cf. ensiforme | |
|---|---|---|---|---|---|---|---|---|---|
| ᅟ | USM FSSC-C3496Gr | USM FSSC-K600T | FRC S-2391 | USM FSSC-P2108S | USM FSSC-S2216Ru | USM FSSC-C4651Tb | FRC S-1847 | ||
| Colony diametera | Mean = 3 cm | Mean = 3.5 cm | Mean = 3 cm | Mean = 3 cm | Mean = 3 cm | Mean = 3 cm | Mean = 1.18 cm | Mean = 1.18 cm | |
| Pigmentation on PDA | White to cream | White to yellow or green | – | White to yellow | White to yellow | White to yellow | Dark reddish brown | Dark reddish brown | |
| Aerial mycelium | Present | Present | Present | Present | – | Present | Present | Present | |
| Sporodochia | Shape | Oval, elongated oval, clavate, and reniform | Oval, reniform, elongated oval, and obovoid with a truncate base | Oval, elongated oval and obovoid with a truncate base | Oval, elongated oval and obovoid with a truncate base | Elongated oval and obovoid with a truncate base | Elongated oval and obovoid with a truncate base | Ellipsoidal | Ellipsoidal |
| Microconidia | Shape | Straight to slightly curved | Typically curved | Falcate and dorsiventral | Falcate and dorsiventral | Falcate and dorsiventral | Falcate and dorsiventral | Slightly curved | Slightly curved |
| Septa | 3–5 | 3–5 (mostly 5-septate) | 3–4 | 3–4 (mostly 3-septate) | 3–4 (mostly 4-septate) | 3–4 (mostly 4-septate) | 2–7 | 2–8 | |
| μmb | Length | (30–) 35–44 (–48) | (35–) 42–54 (–58) | 41.15 | (27–) 30–42 (–44) | 44.3 ± 2.6 | (28–) 31–45 (–47) | (52–) 56–66 (–71) | (52–) 56–66 (–71) |
| μmc | Width | (5–) 5.4–5.9 (–6.5) | (4.8–) 5.4–5.9 (–6.5) | 6.05 | (4–) 4.8–5.8 (–6.4) | 5.9–6.1 | 5.5–6.1 | (5.5–) 6.0–6.7 (–7.0) | (5.5–) 6.0–6.7 (–7.0) |
| Length | 43–50 | (44–) 51–57 (–63) | – | – | – | – | (50–) 64–73 (–75) | (62–) 66–78 (–89) | |
| width | 5.7–6.5 | (5.0–) 5.5–6.0 (–6.5) | – | – | – | – | (5.3–) 6–6.5(–6.8) | (5.0–) 5.7–7.0 (–7.5) | |
| μmd | Length | – | – | – | – | – | – | (59–) 64–75(–86) | (62–) 66–78 (–89) |
| Width | – | – | – | – | – | – | (5.0–) 6–6.8 (–7.2) | (5.0–) 5.7–7.0 (–7.5) | |
| Chlamydospores | Present | Present | Present | Present | Present | Present | Present | Present | |
FRC Fusarium Research Center at Pennsylvania State University, Pennsylvania, USM Fusarium culture collection of school of Biological Science, Universiti Sains Malaysia
aColonies on PDA at 25 °C in intermittent light after 4 days
bMean length of macroconidia (3–4-septate)
cMean length of macroconidia (5-septate)
dMean length of macroconidia (6–7-septate)
According to Short et al. [21, 22], members of F. falciforme, F. keratoplasticum, and FSSC 5 usually were associated with human infectious diseases, while these strains were associated with plants, and this would be a very interesting finding and would provide phylogenetic context to all of the strains. Combined sequence data of ITS regions and TEF-1α offered higher resolutions and high levels of polymorphism within many species of fungi [3, 15, 30–32]. Phylogenetic analysis of combined sequences of ITS regions and TEF-1α in this study has shown to be a useful marker for identification and characterisation of taxa in FSSC. Since members of the FSSC have caused various types of diseases on plants and notable infections of humans, therefore accurate identification of pathogenic members of FSSC is very important in order to develop proper management practices in phytopathological and medical communities that work on this group of Fusarium species.
Acknowledgments
Khosrow Chehri acknowledges the Universiti Sains Malaysia, Penang, Malaysia for providing necessary facilities to carry out this research (RU Research grants 1001/PBIOLOGI/811182 and FRGS 203/PBIOLOGI/6711311).
References
- 1.Booth C. The genus Fusarium. Kew, Surrey, United Kingdom: Commonwealth Mycological Institute; 1971. [Google Scholar]
- 2.Zhang N, O’Donnell K, Sutton DA, Nalim FA, Summerbell RC, Padhye AA, Geiser DM. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol. 2006;44:2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, Zhang N, Geiser DM. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol. 2008;46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendel Z, Protasov A, Sharon M, Zveibil A, Ben Yehuda S, O’Donnell K, Rabaglia R, Wysoki M, Freeman S. An Asian ambrosia beetle Euwallacea fornicatus and its novel symbiotic fungus Fusarium sp. pose a serious threat to the Israeli avocado industry. Phytoparasitica. 2012;40:235–238. doi: 10.1007/s12600-012-0223-7. [DOI] [Google Scholar]
- 5.Wollenweber HW, Reinking OA. Die Fusarien, ihre Beschreibung. Berlin, Germany: Schadwirkung und Bekampfung. Verlag Paul Parey; 1935. [Google Scholar]
- 6.Snyder WC, Hansen HN. The species concept in Fusarium with reference to section Martiella. Am J Bot. 1941;28:738–742. doi: 10.2307/2436658. [DOI] [Google Scholar]
- 7.Burgess LW (1981) General ecology of the fusaria. In: PE Nelson, TA Toussoun, RJ Cook (eds) Fusarium: diseases, biology, and taxonomy. Pennsylvania State University Press, University Park, PA. pp. 225–235
- 8.Leslie JF, Summerell BA. The Fusarium laboratory manual. UK: Blackwell Publish Ltd. p; 2006. p. 388. [Google Scholar]
- 9.Sakurai Y, Matuo T. On the form name and race of Hypomyces solani (Rke. et Berth.) Snyd. et Hans. which is pathogenic to the mulberry trees. Ann. Phytopathol. Soc. Jpn. 1959;24:219–223. doi: 10.3186/jjphytopath.24.219. [DOI] [Google Scholar]
- 10.Sakurai Y, Matuo T. Studies on the intraspecific groups in Fusarium solani (1). On mating populations and morphological groups in the species. Res Rep Fac Textile Sericulture, Shinshu Univ. 1960;10:21–32. [Google Scholar]
- 11.Sakurai Y, Matuo T. Studies on the intraspecific groups in Fusarium solani (2). On the differentiation in utilization of carbon sources among the intraspecific groups. Ann Phytopathol Soc Jpn. 1961;26:118–124. doi: 10.3186/jjphytopath.26.118. [DOI] [Google Scholar]
- 12.Matuo T, Snyder WC. Use of morphology and mating populations in the identification of Formae speciales in Fusarium solani. Phytopathology. 1973;63:562–565. doi: 10.1094/Phyto-63-562. [DOI] [Google Scholar]
- 13.O’Donnell K, Gray LE. Phylogenetic relationships of the soybean sudden death syndrome pathogen Fusarium solani f. sp. phaseoli from rDNA sequence data and PCR primers for its identification. Mol Plant-Microbe Interact. 1995;5:709–714. doi: 10.1094/MPMI-8-0709. [DOI] [PubMed] [Google Scholar]
- 14.Guadet J, Julien J, Lafay JF, Brygoo Y. Phylogeny of some Fusarium species as determined by large subunit rRNA sequences comparison. Mol Biol Evol. 1989;6:27–242. doi: 10.1093/oxfordjournals.molbev.a040548. [DOI] [PubMed] [Google Scholar]
- 15.O’Donnell K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia. 2000;92:919–938. doi: 10.2307/3761588. [DOI] [Google Scholar]
- 16.O’Donnell K, Kistler HC, Cigelnike E, Ploetz RC. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nalim FA, Samuels GJ, Wijesundera RL, Geiser DM. New species from the Fusarium solani species complex derived from perithecia and soil in the Old World tropics. Mycologia. 2011;103:1302–1330. doi: 10.3852/10-307. [DOI] [PubMed] [Google Scholar]
- 18.Aoki T, O’Donnell K, Homma Y, Lattanzi A. Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex-F. virguliforme in North America and F. tucumaniae in South America. Mycologia. 2003;954:660–684. doi: 10.2307/3761942. [DOI] [PubMed] [Google Scholar]
- 19.Aoki T, O’Donnell K, Scandiani MM. Sudden death syndrome of soybean in South America is caused by four species of Fusarium: Fusarium brasiliense sp. nov., F. cuneirostrum sp. nov., F. tucumaniae, and F. virguliforme. Mycoscience. 2005;46:162–183. doi: 10.1007/S10267-005-0235-Y. [DOI] [Google Scholar]
- 20.Aoki T, Scandiani MM, O’Donnell K. Phenotypic, molecular phylogenetic, and pathogenetic characterization of Fusarium crassistipitatum sp. nov., a novel soybean sudden death syndrome pathogen from Argentina and Brazil. Mycoscience. 2011;53:167–186. doi: 10.1007/S10267-011-0150-3. [DOI] [Google Scholar]
- 21.Short DPG, O’Donnell K, Thrane U, Nielsen KF, Zhang N, Juba JH, Geiser DM. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet Biol. 2013;53:59–70. doi: 10.1016/j.fgb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Short DPG, O’Donnell K, Geiser DM. Clonality, recombination, and hybridization in the plumbing-inhabiting human pathogen Fusarium keratoplasticum inferred from multilocus sequence typing. BMC Evol Biol. 2014;14:91. doi: 10.1186/1471-2148-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasson MT, O’Donnell K, Rooney AP, Sink S, Ploetz RC, Ploetz JN, et al. An inordinate fondness for Fusarium: phylogenetic diversity of fusaria cultivated by ambrosia beetles in the genus Euwallacea on avocado and other plant hosts. Fungal Genet Biol. 2013;56:147–157. doi: 10.1016/j.fgb.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Guarro J. Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment. Eur J Clin Microbiol Infect Dis. 2013;32:1491–1500. doi: 10.1007/s10096-013-1924-7. [DOI] [PubMed] [Google Scholar]
- 25.Fisher NL, Burgess LW, Toussoun TA, Nelson PE. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology. 1982;72:151–153. doi: 10.1094/Phyto-72-151. [DOI] [Google Scholar]
- 26.Summerbell R, Schroers H-J. Analysis of phylogenetic relationship of Cylindrocarpon lichenicola and Acremonium falciforme to the Fusarium solani species complex and a review of similarities in the spectrum of opportunistic infections caused by these fungi. J Clin Microbiol. 2002;40:2866–2875. doi: 10.1128/JCM.40.8.2866-2875.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lui D, Coloe S, Baird R, Pedersen J. Rapid mini-preparation of fungal DNA for PCR. J Clin Microbiol. 2000;38:471–477. doi: 10.1128/jcm.38.1.471-471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 29.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 30.Geiser DM, Jimeénez-Gasco MDM, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 2004;110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 31.Overton BE, Stewart EL, Geiser DM. Taxonomy and phylogenetic relationships of nine species of Hypocrea with anamorphs assignable to Trichoderma section Hypocreanum. Stud Mycol. 2006;56:39–65. doi: 10.3114/sim.2006.56.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zalar P, Gostinčar C, de Hoog GS, Uršič V, Sudhadham M, Gunde-Cimerman N. Redefinition of Aureobasidium pullulans and its varieties. Stud Mycol. 2008;61:21–38. doi: 10.3114/sim.2008.61.02. [DOI] [PMC free article] [PubMed] [Google Scholar]


