Abstract
The NAD(P)H dehydrogenase complex is encoded by 11 ndh genes in plant chloroplast (cp) genomes. However, ndh genes are truncated or deleted in some autotrophic Epidendroideae orchid cp genomes. To determine the evolutionary timing of the gene deletions and the genomic locations of the various ndh genes in orchids, the cp genomes of Vanilla planifolia, Paphiopedilum armeniacum, Paphiopedilum niveum, Cypripedium formosanum, Habenaria longidenticulata, Goodyera fumata and Masdevallia picturata were sequenced; these genomes represent Vanilloideae, Cypripedioideae, Orchidoideae and Epidendroideae subfamilies. Four orchid cp genome sequences were found to contain a complete set of ndh genes. In other genomes, ndh deletions did not correlate to known taxonomic or evolutionary relationships and deletions occurred independently after the orchid family split into different subfamilies. In orchids lacking cp encoded ndh genes, non cp localized ndh sequences were identified. In Erycina pusilla, at least 10 truncated ndh gene fragments were found transferred to the mitochondrial (mt) genome. The phenomenon of orchid ndh transfer to the mt genome existed in ndh-deleted orchids and also in ndh containing species.
Eukaryotic cells arose through the engulfment of bacterial endosymbionts and the subsequent gradual conversion of those bacteria into organelles (mitochondria and chloroplasts)1,2. During this process, there was a massive transfer of genes from the endosymbiont genomes into the nuclear genome of the host cell3. However, plant chloroplasts (cp) possess their own genomes, which contain genes that are involved in photosynthesis, transcription and translation4. The numbers and functions of these cp genes are highly conserved among higher plants. One family of genes that is involved in photosynthesis is the ndh family, of which 11 members encode NADH dehydrogenase subunits. These genes are homologs of those encoding mitochondrial NADH dehydrogenase subunits, which are involved in respiratory electron transport5,6. In angiosperm chloroplasts, these ndh proteins associate with nuclear-encoded subunits to form the NADH dehydrogenase-like complex. This protein complex associates with photosystem I to become a super-complex that mediates cyclic electron transport7, produces ATP to balance the ATP/NADPH ratio, and facilitates chlororespiration when cyclic electron transport pauses overnight8.
Heterotrophic plants, which do not photosynthesize, lack functional ndh genes9. Interestingly, some autotrophic plants, such as pines, Gnetales, Erodium, Melianthus and orchids also lack functional ndh genes in their cp genomes10,11,12,13,14,15,16,17,18. There are five subfamilies in Orchidaceae: Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae and Epidendroideae19,20,21. Chloroplast genomes from four autotrophic Epidendroideae orchid genera, Phalaenopsis, Oncidium, Erycina, and Cymbidium, have been sequenced and are publicly available11,14,16,17. From these orchid cp genomes, only ndhB of Oncidium ‘Gower Ramsey' and ndhE, J, and C of Cymbidium have been predicted to encode functional ndh proteins14,17. All the other ndh gene fragments contain nonsense mutations or are truncated or absent from the plastome11,14,16,17. For example, the cp genome of E. pusilla contains truncated versions of ndhJ, C, D, B, G, and H and lacks sequences for ndhK, F, E, A, and I16. These results indicate that deletion and truncation of ndh gene fragments are common to orchid cp genomes. However, the ndh genes in the cp genome of Apostasia wallichii (Apostasioideae) are transcribed and are predicted to encode functional proteins22. These findings indicate that the orchid common ancestor contained an entire functional set of ndh genes. So far only Epidendroideae cp genome sequences have been published11,14,16,17. Therefore, understanding which orchid subfamilies lack ndh genes in their cp genomes has the potential to considerably increase understanding of photosynthetic evolution in orchids.
Previous studies have demonstrated that plant cpDNA has been transferred to both the mitochondrial (mt)23,24 and nuclear genomes1,25,26,27 Based on PCR results, Chang et al.11 proposed that in Phalaenopsis, the ancestral ndh genes have been transferred to the genome of the nucleus or another organelle. Recently published mitochondrial genomes from different seed plant species contain 1 to 10% cpDNA originating from various regions of the plastome28,29,30. Due to the size and complexity of orchid genomes, it is difficult to obtain these sequences by direct sequencing or genome walking31. To resolve this problem, we previously employed a well-established strategy, using simple PCR to identify BAC clones containing sought-after genes in both E. pusilla and O. ‘Gower Ramsey'16,31. We also used this strategy to identify and sequence BAC libraries that contained cp genomic fragments, which resulted in the sequencing of the complete E. pusilla cp genome16. This targeted method was more cost- and time-effective than whole genome sequencing.
In this report, the evolutionary timings of ndh deletions from orchid cp genomes was investigated. The cp genomes of Vanilla planifolia (Vanilloideae), Paphiopedilum armeniacum, Paphiopedilum niveum, Cypripedium formosanum (Cypripedioideae), Habenaria longidenticulata, Goodyera fumata (Orchidoideae), and Masdevallia picturata (Epidendroideae) were sequenced. In addition, the mitochondrial locations of the ndh genes were investigated in E. pusilla, an Epidendroideae model orchid, by sequencing BAC clones that contained the ndh genes that were missing from the cp genome.
Results
ndh genes among the orchid subfamilies
There are 11 ndh genes in higher plant chloroplast genomes. To understand the differential expression of these genes among the orchid subfamilies, ndh transcripts were identified in 16 species from five Orchidaceae subfamilies (Supplementary Table S1 and S2). Neuwiedia malipoensis (Apostasioideae), Cypripedium singchii (Cypripedioideae), Habenaria delavayi, Goodyera pubescens (Orchidoideae), Masdevallia yuangensis and Cymbidium sinense (Epidendroideae) had all 11 ndh genes. However, Vanilla shenzhenica, V. planifolia, Galeola faberi (Vanilloideae), Drakaea elastica (Orchidoideae), and E. pusilla (Epidendroideae), had only less 5 ndh gene sequences (Supplementary Table S1). The number of ndh gene sequences in each transcriptome did not correlate with known orchid evolutionary relationships (Supplementary Table S1). The fewest ndh transcripts were found in Vanilloideae. Previous studies have reported ndh deletions in Epidendroideae species, but an analysis of the M. yuangensis transcriptome database revealed highly conserved sequences (79.4%) for all of the ndh genes. The same phenomenon also occurred in C. sinense (Supplementary Table S1). These results indicate that the loss of ndh genes from the orchid species occurred independently within the subfamilies.
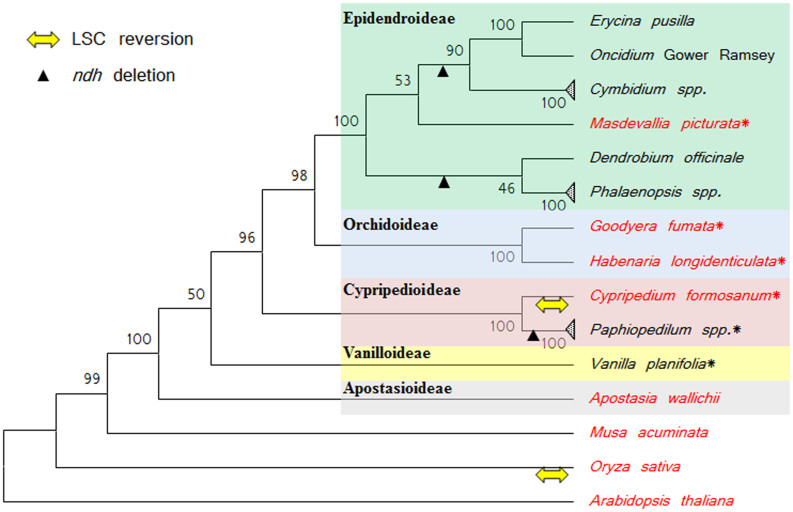
The cp genomes of seven orchids in four subfamilies were determined (Figure 1 and 2, Supplementary Figure S1, Supplementary Tables S3 and S4). The read coverage of the seven assembled chloroplast genomes is shown in Supplementary Table S5. When all 11 ndh DNAs in the cp genome were in frame and encoded the full-length ndh gene, the genomes are classified as ndh-complete type cp genome. If any one of the 11 ndh genes had nonsense mutations, deletions, or was absent, the genomes are classified as ndh-deleted type cp genome. There were both ndh-complete- and ndh-deleted types in Cypripedioideae [Paphiopedilum armeniacum (ndh-deleted), Paphiopedilum niveum (ndh-deleted), and C. formosanum (ndh-complete)] and in Epidendroideae [E. pusilla (ndh-deleted) and M. picturata (ndh-complete)]. Among the only Vanilloideae species investigated, V. planifolia, was an ndh-deleted type, but this result cannot exclude the possibility of ndh-complete species in other Vanilloideae species. The two sequenced cp genomes in Orchidoideae, H. longidenticulata and G. fumata, are both ndh-complete. Based on the transcriptome results, D. elastica, an endangered native Australian orchid, has few ndh transcripts (Supplementary Table S1). This species may be an example of ndh-deleted Orchidoideae.
Figure 1. Phylogenetic analysis of orchids.
The phylogenetic tree was based on the chloroplast rbcL, matK, psaA, psaB, and rpoC2 nucleotide sequences. The species names in red are ndh-complete and in black are ndh-deleted genomes. The asterisk (*) indicates that the cp genomes were sequenced in this report. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.
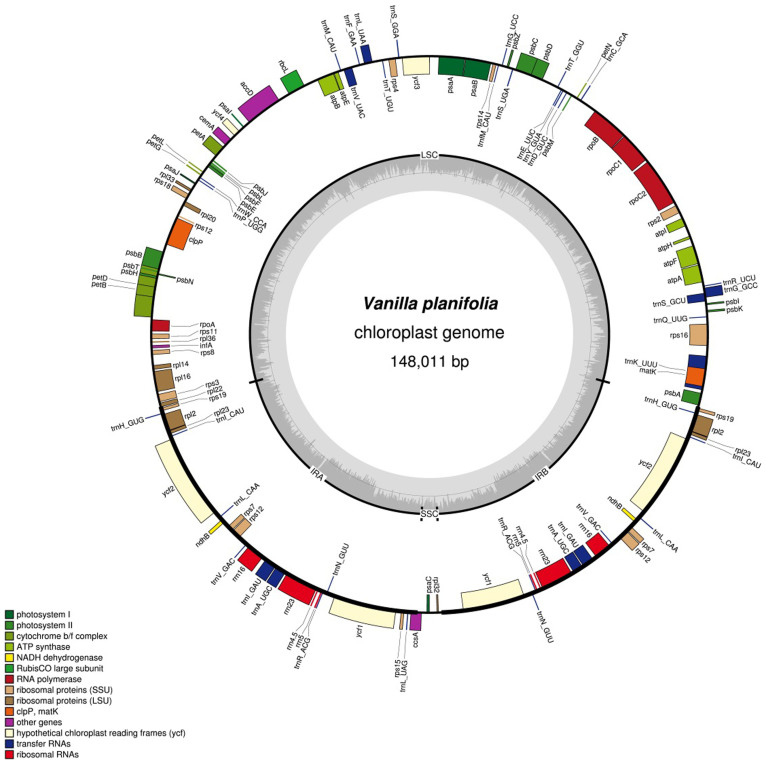
Figure 2. Gene maps of Vanilla planifolia chloroplast genomes.
Genes on the outside of the map are transcribed clockwise whereas genes on the inside of the map are transcribed counterclockwise. Colors indicate genes with different functional groups.
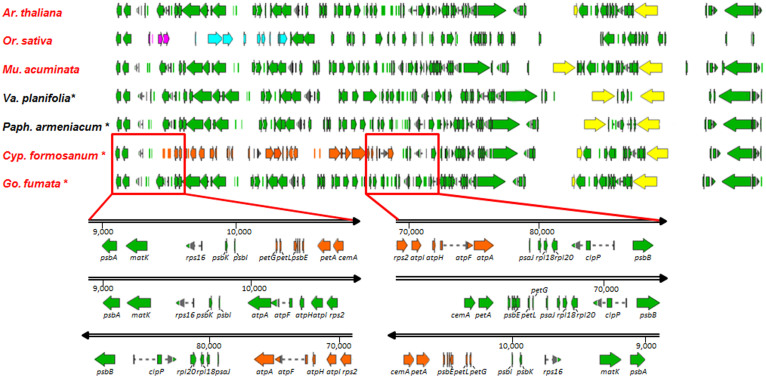
In addition to the ndh gene profiles, other differences were found between these seven orchid cp genomes (Figure 3). Notably, the region from atpA to petG is reversed in the C. formosanum cp genome. These results indicate that the cp genomes of these seven orchids provide useful information on ndh diversity in the family.
Figure 3. Genome order of the chloroplast-coding regions in 4 orchid genera, Arabidopsis thaliana, Oryza sativa, and Musa acuminata.
The name in red indicates that the cp genome contains complete ndh genes. Black indicates that the ndh genes are deleted. The asterisk (*) indicates the cp genomes were sequenced in this study. P. niveum and P. armeniacum have the same genome structure and order. H. longidenticulata, M. picturata, and G. fumata have the same genome structure and gene order. The arrowheads indicate the direction of the genes. The pink arrows indicate that the rice psbC and psbD are inserted next to psbI but are located behind psbM in the other plant cp genomes. Blue indicates that the sequence region from atpA to psbM is reversed in the rice cp genome. The yellow arrow indicates that there is a full-length ycf1 gene in the IR region. Orange indicates that the atpA-petG region is reversed in C. formosanum. The lower panels are a zoom-in of the box regions. The arrowheads indicate the direction of sequences from 5′ to 3′. The numbers indicate the position in the cp genome.
Identification of non-cp ndh genes in orchids
A comparison of the cp genomic and transcriptomic data showed that only ndhB was found in the V. planifolia cp genome, whereas ndhK, J, and C transcripts were identified in the V. planifolia whole-cell transcriptome. Likewise, in the P. armeniacum cp genome, only the ndhD sequence was encoded in the ndhF-D-E-G-I-A-H region of the cp genome. However, the ndhA and ndhH transcripts were present in the P. armeniacum whole-cell transcriptome, indicating that ndhA and ndhH gene were encoded in either the mt or nuclear genomes in P. armeniacum (Supplementary Table S6). These results, therefore, indicate that non-cp ndh genes exist and may be transcribed. We therefore attempted to identify these genes in mt or nuclear genome sequences.
Many non-cp ndh gene fragments were identified in the orchid whole genomic sequences. In addition to the ndh-deleted species, the ndh-complete species (C. formosanum, H. longidenticulata, G. fumata, and M. picturata) also contained non-cp ndh gene fragments (Supplementary Tables S3 and S4). Because ndh genes in the cp genomes of all plant species are clustered as ndhJ-K-C, ndhF-D-E-G-I-A-H and ndhB, the results below are presented in this order.
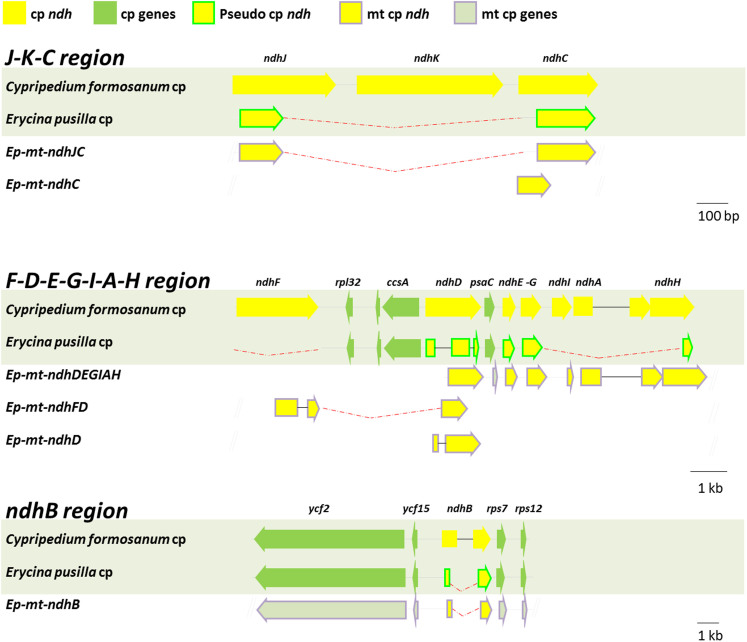
Although flanking DNA sequences can provide information to identify the organelle from which they were derived, some of the sequences were too similar to cp genome sequences or were too short to identify (Supplementary Table S4, Paph_niveum_ndhB1 to Mas_ndhJK). Therefore, it would be important to extend the sequence length in future studies. In recent studies, E. pusilla has been established as a model for orchid genome research16. Using ndh-containing sequences, which were derived from the genomic NGS sequences, specific primers were designed to screen the E. pusilla BAC library16. The BAC clones were identified and sequenced to confirm presence of ndh-like gene fragments and to determine the organelle of origin. Six BAC clones that contained putative ndh gene fragments were identified: Ep-mt-ndhJC, Ep-mt-ndhC, Ep-mt-ndhDEGIAH, Ep-mt-ndhDF, Ep-mt-ndhD and Ep-mt-ndhB (Figure 4). An analysis of the flanking sequences within the BACs revealed that all six of the contigs were derived from the mt genome. According to genome sequencing data, nine ndh genes were transferred to the mt genome in M. picturata, eight in P. niveum and G. fumata, six in P. armeniacum, and four in V. planifolia (Supplementary Table S3, Supplementary Figures S2, S3, and S4). In total, E. pusilla had the greatest number of ndh gene fragments transferred to the mt genome (10 genes in six fragments, Figure 4, Supplementary Table S3). Furthermore, there were eight ndh genes in G. fumata and nine in M. picturata, whose cp genomes contained a complete ndh profile (Supplementary Table S3). These results showed that the number of transferred ndh gene fragments did not correlate with ndh deletions in the cp genome.
Figure 4. Comparison of the Cypripedium formosanum cp, Erycina pusilla cp genomes, and the mt ndh regions.
The arrows indicate coding regions. The arrowheads indicate the direction of the genes.
To understand structural differences between the mt-ndh regions and the original cp region, E. pusilla mt genome sequence was aligned with the corresponding C. formosanum (ndh-complete) and E. pusilla cp genome regions. This alignment showed a massive loss of DNA in the ndh copies that were transferred to the E. pusilla mt genome (Figure 4, red-dotted lines). It is also clear that all of the mt-ndh fragments were truncated or contained large deletions. Interestingly, some of the ndh transfers may have occurred after an ndh deletion within the cp genome (such as Ep-mt-ndhJC, Figure 4). Alternatively, the complete ndhJ-K-C region may have first been transferred to the mt genome and then undergone deletions.
Relationships between cp-ndh deletions and ndh genes that were transferred to the mt genome
An evolutionary snapshot of ndh gene transfer between the cp and mt genomes was taken by comparing the published mt genomes and the available cp genomes. All of the Tracheophyta (vascular plant) mt genomes contained partial cp genome fragments (Figure 5, species names in black and green). Twenty-two published Tracheophyta mt genomes contained some portion of ndh DNA. The length of the mt-ndh genic regions reflected the taxonomic relationships (Supplementary Table S3). For Oryza, the presence of ndh sequences in the mt genome correlated at the genus level (Supplementary Table S3). Based on a phylogenetic analysis of their cp genomes, Bambusa oldhamii and Oryza sativa were more closely related to each other than the other Poaceae species. They have a similar ndh transfer pattern, and the presence of ndhJ, ndhK and ndhC in both their mt genomes. However, the appearance of mt genome ndh sequences at the species level was different in the Zea genus (Figure 5)32; transferred ndh fragments were found within Zea, except Zea luxurians. A similar pattern was seen in mt-ndhD in P. armeniacum and P. niveum (Supplementary Figure S3). These results indicate that the mt genomes among members of the same family or genera may show different ndh content. This phenomenon is likely due to mtDNA rearrangements28. One possible alternative is that the mt genomes of P. armeniacum and P. niveum are not complete. As more mt genomes are sequenced and completed, the taxonomic relationship between the ndh regions and the cp genome sequences can be further studied.
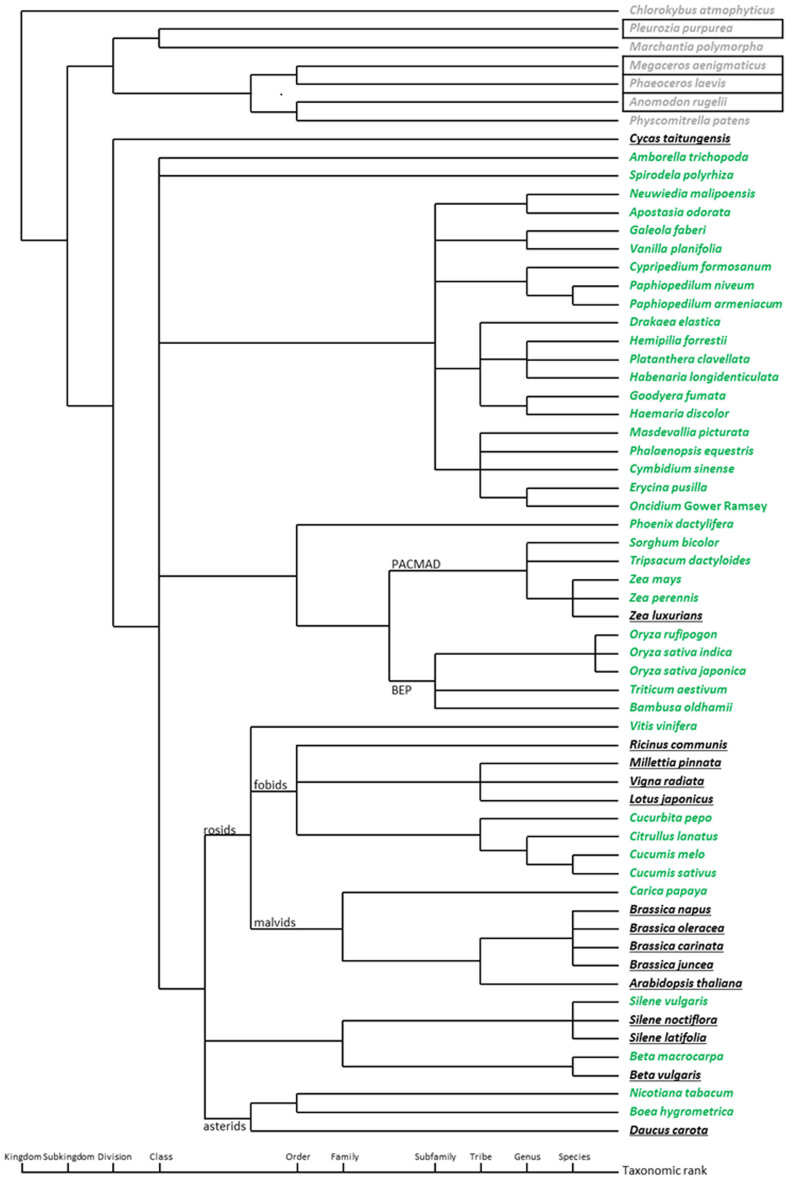
Figure 5. Transfer of chloroplast DNA, including ndh genes, to mt genomes within Kingdom Plantae.
The published mitochondrial and chloroplast genomes were downloaded from NCBI. The species names in the boxes indicate that the cp genome is unavailable. The cp and mt genomes were compared using BLASTN. The names in gray indicate that no cp DNA sequences were found in the mt genome. The names that are underlined in black indicate that cp DNA was found in the mt genome but that the ndh genes were not. The names in green indicate the presence of ndh genes in the mt DNA.
Of all of these mt ndh genes, only mt-ndhD in Vitis vinifera, mt-ndhJ in Phoenix dactylifera, B. oldhamii, and Oryza and mt-ndhB in Triticum aestivum and Zea are full length (Supplementary Table S3)32. Although some mt-ndh gene fragments are in frame and their transcripts have been identified in these species, they cannot complement ndh function in ndh-deleted orchids.
A large portion of the cp genome is found in the mt genome
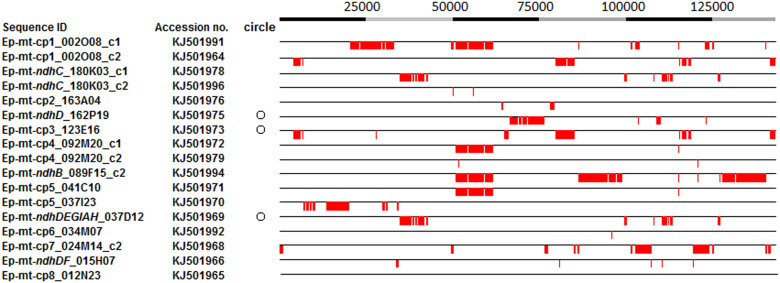
The mt genomes of numerous species also contain other cp sequences in addition to the ndh gene fragments. Some cp-like contigs were identified among the E. pusilla genomic sequences and found to be similar to other plant mt genome sequences through BLASTN. Using BAC library screening, these cp-like mt DNA clones were identified in E. pusilla (Figure 6, Supplementary Table S4). Based on the sequences of these BAC clones, we estimate that more than 130 kb of cp genomic DNA (more that 76% of the cp genome) was transferred into the mt genome of E. pusilla (Figure 6). The largest cp insertion into the mt genome was 12 kb (accession no. KJ501994, containing the cp IR region).
Figure 6. BLASTN comparison of the Erycina pusilla mt DNA and the E. pusilla cp genome.
The numbers at the top are the positions of the E. pusilla cp genome. The red boxes indicate the aligned sequences. The numbers in the first column are the clone IDs in the E. pusilla BAC library. The circles indicate that the clones are circular chromosomes and have been confirmed by PCR. c1: contig 1; c2: contig 2.
There were 60 cp-like protein-coding genes in the E. pusilla mt genome BAC clones, with notable exceptions being the matK, atpA, atpF, ycf3, rps4, psaI, petA, and psbJ genes (Supplementary Table S8). According to the transcriptome results detailed above, the ndh DNA regions in the orchid mt genome do not encode functional proteins. Interestingly, nine cp-like mt genes (psbI, rps2, petN, atpE, psaJ, psbH, rpl36, rpl22, and psaC, Supplementary Table S8) contained intact gene sequences with start and stop codons but may not have required regulatory sequences.
To determine which of the cp-like mt gene fragments could be transcribed, BLASTN was used to search the E. pusilla transcriptome. There were 28 cp-like mt gene transcripts (Supplementary Table S8), including three intact genes: psaJ, rpl36, and rpl22. The mt-psbI sequence shared 100% identity with the cp genome sequence; therefore, we could not conclude the derivation of the transcript. The cp-like mt transcripts could be divided into two types: those containing only cp-like gene fragments and those containing mt genes (KJ501986 and KJ501987). These results indicate that these gene fragments can be transcribed but translation requires further investigation.
Discussion
According to our results, a whole-cell transcriptome database can provide useful information for investigation of ndh genes in orchid genomes. However, transcriptome data should be interpreted with caution for several reasons. First, it is difficult to determine the source genome (i.e., nuclear or organellar) from transcriptome data. Second, the transcripts identified may not be full-length, may contain untranslated portions, or may not correlate with the cp genomic sequences. Third, transcriptome profiles differed between species within a subfamily. For example, in the Cypripedioideae, all 11 ndh genes were identified in Cypripedium but only 7 were identified in Paphiopedilum. Fourth, source materials (roots, different stages of leaves, etc.) also influence a transcription profile. Although some of the transcriptome data did not show full-length ndh genes, this does not necessarily mean that these genes were lost or truncated from these species. Because of these vagaries, results deduced from transcriptomes need to be confirmed by targeted sequencing of genomic DNA.
Comparison of the transcriptomes and cp genomes yielded some information; however, cp genome sequences from additional orchid species across the five subfamilies will be required to better understand the structures of ndh genes among orchid subfamilies. Our data also indicate that species within the same subfamily may have different ndh gene profiles.
According to the orchid cp genome data, cp ndh-deleted and ndh-complete species exist in the Orchidaceae family. Martin and Sabater hypothesized that the function of ndh genes are related to terrestrial adaptations of photosynthesis33. However, no significant differences were observed between the cp ndh-deleted and ndh-complete orchids in our study in terms of biogeography or growth conditions (including light and water requirements). Leitch et al.34 reported that orchid genome size is related to taxonomy. However, terrestrial orchids have larger genome sizes than epiphytic orchids and no significant differences were observed in our study between the ndh-deleted and ndh-complete orchids in terms of genome size34.
Contemporary cyanobacteria encode several thousand genes, but only 20 to 200 of these genes have been retained in modern plastid genomes1. A massive number of chloroplast-originating genes, including those for photosynthesis, were transferred to the nucleus after the endosymbiosis of the cyanobacterial ancestor1,35. According to our results, cp ndh gene fragments exist in the mt genome of cp ndh-deleted orchids that are not translated to functional proteins. One possible reason for this phenomenon could be that these cp ndh genes were transferred to the nucleus. This phenomenon has been observed in other cases of cp gene deletion. The cp rpl22 genes have been lost in some rosid species36; however, the nuclear rpl22 gene of these rosid species remains functional. This gene can be transcribed and translated into a protein that is targeted to the chloroplast, where it functions in protein synthesis36. Three distinct pathways for loss of cp genes have been studies so far: a) transfer to the nucleus (infA, rpl22, rpl32, rpoA), b) substitution of a nuclear encoded mitochondrial targeted gene (rps16) and c) substitution of a nuclear gene for a plastid gene (accD, rpl23)36. Therefore, we have investigated these different possibilities and transfer of ndh genes to the mitochondrial genome.
Another possibility is that there are no ndh genes in some species of orchid. No complete and functional ndh transcripts have been identified in the ndh-deleted orchid transcriptomes21,37,38. Although ndh proteins play an important role in cyclic electron transport in tobacco and M. polymorpha, there are no significant growth differences between the wild type and ndh-deleted transformants6,39,40,41. This is because there is an alternative PSI cyclic electron transport pathway: the proton gradient regulation 5 (PGR5)/PGR5-like photosynthetic phenotype 1 (PGRL1)-dependent antimycin A-sensitive pathway7,42,43. This pathway is partly redundant with the NDH complex-dependent pathway6. Therefore, the ndh-deleted transformants may be able to use this pathway for cyclic electron transport.
Not all of the ndh genes were found in the transcriptome, likely because these genes are expressed at low levels and/or only during specific developmental stages or under specific growth conditions. Genome sequences may be more useful because they represent complete genetic complements. We performed NGS sequencing of the E. pusilla genome and obtained 15 Gb of sequence data, which corresponds to about 10 times coverage of the genome. No nuclear ndh gene could be identified in the assembled NGS data. Nonetheless, because we have not completed the assembly of a high-quality draft genome, it is difficult to conclude whether any functional ndh genes exist in the nuclear genome. A concrete conclusion can only be made when the whole genome sequences are available for E. pusilla.
According to our result, most of the genes transferred to the mt genome are highly similar to those within the cp genome, indicating that these DNA sequences were transferred to the mt genome more recently than the sequences that contained more insertions/deletions and mutations. The nine proteins (psbI, rps2, petN, atpE, psaJ, psbH, rpl36, rpl22, and psaC, Supplementary Table S8) that are encoded by these genes are smaller than are the other cp genome-encoded proteins (29 to 236 a.a., average = 88 a.a.); therefore, their genetic integrity may be easier to maintain. In most Tracheophyta, small cp genome fragments have been identified in mt genomes. In rice, there are 16 cp genome segments in the mt genome with sizes ranging from 32 bp to 6.8 kb44. In maize, many short cp segments (17 to 187 bp) have been transferred to the mt genome24. In palm, there are more than 100 fragments of chloroplast origin ranging in size from 50 bp to 6 kb45. This large amount of synteny is one of the reasons that the ndh-like pseudogenes could be identified. The transfer of cp DNA to mt DNA is widespread in seed plants45,46,47,48,49,50,51,52,53. Based on currently published mt genomes, the Amborella mt genome has the highest amount of cp DNA of all of the ndh genes (138 kb)48. Two Cucurbitaceae mt genomes contain long cp genome transfers: C. pepo (>113 kb, five ndh genes)49 and C. sativus (71 kb, four ndh genes)46. In the palm P. dactylifera mt genome, there are 74 kb of cp genomic DNA, which encompass six ndh pseudogenes45.
More than 130 kb of cp genomic DNA has been transferred into the mt genome in E. pusilla. A previous study indicated that larger mt genomes contain more cp DNA46. We propose that the size and structure of the E. pusilla mt genome may be large and complicated. A complete sequence of the mt genome of E. pusilla will increase our understanding of this phenomenon. In the near future, NGS sequencing of the BAC library will continue to facilitate investigations of the genomic complexity in E. pusilla.
Most cp-like mt genes cannot translate functional proteins. In the Amborella mt genome, which contains 197 foreign protein-coding genes, only 50 (25%) are full-length and contain open reading frames48. These 50 genes are predominantly short, suggesting that many remain intact by chance. The function of cp-like mt DNA sequences include tRNA-encoding genes50,51, the promoter of NADH dehydrogenase subunit 9 (nad9)52 and new chimeric mitochondrial genes that were generated by recombination53. It is still unknown why large cp DNA fragments (>1 kb) are transferred to mt DNA or why this phenomenon is frequent in orchids. Although some of the transferred tRNA genes are functional in mitochondria50,51, fates of protein coding genes require further investigations.
Previous studies have indicated that autotrophic Epidendroideae orchid cp genomes no longer encode all of the ndh genes. Using current orchid transcriptome databases and cp genomic sequences, we demonstrated that the complete ndh gene fragment set still exists in some orchid family members. During evolution, several orchids experienced ndh gene deletions, but these deletions are not correlated with orchid taxonomy. Based on the sequences and gene structures of the ndh genes and gene fragments that remain in the cp genomes, we conclude that these deletion events were independent. Because cyclic electron transport pathway is redundant with the NDH complex-dependent pathway, there is no evolutionary pressure to maintain functional copies of ndh genes in other genomes in sharp contrast to essential genes required for chloroplast protein synthesis that were transferred from chloroplast to the nuclear genome. In all of the sequenced orchid genomes, the ndh gene fragments were transferred to the mitochondrial genomes. These transfers were not directly linked to the ndh deletions in the cp genome. The ndh genes that were transferred to the mt genome have been mutated and truncated and do not complement the ndh gene function. Furthermore, the E. pusilla orchid mt genome contains the largest number of cp genome sequences among the published genomes. This result suggests that in E. pusilla, cp-to-mt gene fragment transfers are more frequent. Together, these findings indicate that the mt genomes of orchids (or at the very least, of E. pusilla) are unique and warrant further investigations.
Methods
Identification of 16 orchid ndh transcripts from public orchid transcriptome databases
To understand the ndh genes in the current public orchid transcriptome database, a well-studied monocot species, banana (Musa acuminata), was used to blast target sequences. Eleven ndh amino acid sequences were used as templates for TBLASTN searches (Accession no. HF677508)54. These amino acid sequences were used to identify the putative ndh gene transcripts from public transcriptome databases21,37,38. The cDNA sequences that were identified in this study were targeted from BLAST searches. The sequence ID is listed in Supplementary Table S2.
Plant and BAC DNA preparation
V. planifolia, H. longidenticulata, G. fumata, P. armeniacum, P. niveum, C. formosanum, M. picturata, and E. pusilla leaves were used as the material for DNA extraction via the cetyltrimethyl ammonium bromide method55. The BAC plasmids for Illumina sequencing were isolated using the NucleoBond BAC 100 Kit (NucleoSpin Blood, Macherey-Nagel, Düren, Germany).
High-throughput sequencing
The above orchid genomic and BAC DNA fragments were sequenced by an Illumina high-throughput sequencing platform (Illumina, San Diego, CA, USA). The paired-end sequencing libraries were constructed by a Nextera XT DNA Sample Preparation Kit according to the instruction manual (Illumina). Briefly, 10 μg of purified DNA was fragmented using the Amplicon Augment Mix (Illumina). Fragmentation reactions were achieved by incubation at 55°C for 5 min, followed by neutralization in buffer at room temperature for 5 min. The neutralized DNA fragments were amplified via a limited-cycle PCR program involving 12 cycles of denaturation at 95°C for 10 s, primer annealing at 55°C for 30 s, and extension at 72°C for 30 s, during which the index primers that were required for cluster formation were also added. The amplified DNA was purified using AMPure XP beads (Beckman Coulter, Indianapolis, IN, USA). The fragment sizes and concentrations of the libraries were determined by a 2100 Bioanalyzer with a High Sensitivity DNA Assay Kit (Agilent Technologies, Santa Clara, CA, USA) and quantitative PCR (Applied Biosystems, Carlsbad, CA, USA). Subsequently, the libraries with insert sizes of 500 to 600 bp were denatured with NaOH and sequenced at read lengths of 250 bases from both ends using a MiSeq Personal Sequencer (Illumina).
Because the mean insert size of each library was approximately 500 to 600 bp, the ends of many reads (250 bp*2) overlapped due to the variability in the insert size. Therefore, the cleaned reads were classified into two groups: overlapping paired-end reads and non-overlapping paired-end reads. The overlapping paired-end reads were further merged into long reads (400 to 500 bp) by FLASH (version 1.2.7) before assembly56. The remaining paired-end reads were used to link the contigs into scaffolds. Newbler (version 2.9) was used to assemble the long reads along with the non-overlapping paired-end reads into contigs and scaffolds. Detailed information on the high-throughput sequencing is provided in Supplementary Table S5. The assembly sequences from the genomic DNA of seven orchids are defined as whole genome sequences of seven orchids for the further analysis.
Complete chloroplast genomes of seven orchids
The cp DNA contigs were identified using the E. pusilla cp genome sequence to screen the whole genome sequences of seven orchids by BLASTN19. Those contigs with a high coverage were used as the skeleton of the cp genomes. The gaps between the contigs were filled by sequences that were derived from PCR. The cp genome was annotated using the Dual Organellar GenoMe Annotator (DOGMA)57. For genes with a low sequence identity, manual annotation was performed after identifying the positions of the start and stop codons as well as the translated amino acid sequence using the chloroplast/bacterial genetic code. The annotated genome sequences were submitted to NCBI. The accession numbers for the assembled chloroplast genomes are listed in Supplementary Table S4.
Identification of orchid non-cp ndh genes
Because no whole nuclear genome sequence of an orchid has been published, regions containing ndh-coding sequences in the genomes of the nucleus and other organelles (non-cp ndh genes) have not been identified. To identify the non-cp ndh genes, 11 banana (M. acuminata) ndh amino acid sequences were used as templates for TBLASTN searches of the orchid genomic sequences (Accession no. HF677508)54. The identified contigs were analyzed by BLASTN searches with the sequences of their corresponding cp genomes to confirm that these sequences were not cp genomic sequences. The mt DNA sequences were determined by comparisons of the regions flanking the ndh gene fragments with the mt genome sequences in the NCBI database using BLASTN. In addition, the raw read count was used to distinguish the source of the DNA using the correlation between the number of high-throughput sequencing reads per sequence and the source (multiple copies of organelles vs. single genome, Supplementary Table S5) of the DNA when total DNA was used to exclude the cp genome sequence58. The remaining candidate contigs were confirmed by NCBI BLASTN to ensure the sequences were mt DNA.
The non-cp ndh sequences which derived from genomic sequencing in E. pusilla were used to design the specific primers. These primers were used to screen the BAC library. Using this strategy, there was no need to isolate the nuclei or mitochondria or to purify the genomic DNA without cp genome contamination. The sequences were annotated by BLASTX using the Arabidopsis protein database. The figures of the 446 annotated and aligned DNA positions were drawn using PowerPoint (Microsoft, 14 447 Redmond, WA). The flow chart is shown in Supplementary Figure S5.
Phylogenetic analysis
To investigate the relationships between the orchids in this report, the combined chloroplast rbcL, matK, psaA, and psaB nucleotide sequences were used for the phylogenetic analysis. The consensus sequences were aligned using ClustalW2 (http://www.clustal.org/) and concatenated. Subsequently, the maximum-likelihood phylogeny was estimated using a web version of PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/) using the multiple sequence alignments59. The General Time Reversible model was used as a substitution model to build the phylogenetic tree. The bootstrap values were obtained from 1000 bootstrap replicates and are presented as percentages. Finally, the phylogenetic tree was plotted by TreeVector (http://supfam.cs.bris.ac.uk/TreeVector/).
Identification of the ndh genes and cp-like DNA from the published plant mt genomes
To identify the ndh genes in the published plant mt genomes, the 43 complete mt genomes and the cp genomes of these species or related species in the same genus were downloaded from the NCBI database. The species and accession nos. are listed in Supplementary Table S9. The cp and mt genomes were compared using BLASTN to identify the cp DNA in the mt genomes.
Identification of the BAC clones that contain the ndh genes and cp-like DNA in their mitochondrial DNA
The specific primers were designed to identify the BAC clones carrying the ndh genic regions and cp-like DNA (Supplementary Table S10). The BAC clones containing the ndh regions and cp-like DNA of interest were obtained by PCR screening from the super pool, plate, row and spot31. These primers were also used to screen the O. ‘Gower Ramsey' BAC library to identify ndh clones.
Author Contributions
M.C.S., M.T.C., H.D. and C.S.L. designed the research. J.J.W.C., Y.T.H., S.Y.L., M.C.S. and M.T.C. performed next generation sequencing. J.J.W.C., Y.T.H., S.Y.L., M.C.S., M.T.C., C.F.L. and C.Y.C. performed bioinformatics analysis. H.D., W.J.C., C.T.H., D.C.L., F.H.W. and M.L.C. performed chloroplast genome annotation and uploaded to NCBI. W.J.C., C.T.H., D.C.L. and F.H.W. performed BAC library screening and sequencing. G.K.S.W. and M.K.D. established the 1000 plants transcriptome database. M.C.S., W.J.C., H.D., M.K.D., V.A.A. and C.S.L. wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Jer-Ming Hu (Department of Ecology and Evolutionary Biology, National Taiwan University) for his helpful comments. We also thank Der-Ming Yeh, Li-Ya Chao, Hsiao-Feng Lo (Highland Experimental Farm, National Taiwan University), Jin-Jun Yue (Research Institute of Subtropical Forestry, Chinese Academy of Forestry, China), and Yung-I Lee (National Museum of Natural Science, Taiwan) for providing the orchid materials. We thank Anita K. Snyder and Miranda Loney for correcting the English. We also thank the High Throughput Sequencing Core hosted in the Biodiversity Research Center at Academia Sinica and Mei-Yeh Lu for performing the NGS experiments. The 1000 Plants (1 KP) initiative, led by GKSW, is funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates Technology Futures (AITF) Innovates Centres of Research Excellence (iCORE), Musea Ventures, and BGI-Shenzhen. This work was supported by the Academia Sinica, Taiwan.
References
- Martin W. et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. P. Natl. Acad. Sci. USA 99, 12246–12251 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald J. M. Origin of eukaryotic cells: 40 years on. Symbiosis 54, 69–86 (2011). [Google Scholar]
- Fuentes I., Karcher D. & Bock R. Experimental reconstruction of the functional transfer of intron-containing plastid genes to the nucleus. Curr. Biol. 22, 763–771 (2012). [DOI] [PubMed] [Google Scholar]
- Zurawski G. & Clegg M. T. Evolution of higher-plant chloroplast DNA-encoded genes: implications for structure-function and phylogenetic studies. Ann. Rev. Plant Physiol. 38, 391–418 (1987). [Google Scholar]
- Matsubayashi T. et al. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol. Gen. Genet. 210, 385–393 (1987). [DOI] [PubMed] [Google Scholar]
- Ueda M. et al. Composition and physiological function of the chloroplast NADH dehydrogenase-like complex in Marchantia polymorpha. Plant J. 72, 683–693 (2012). [DOI] [PubMed] [Google Scholar]
- Munekage Y. et al. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582 (2004). [DOI] [PubMed] [Google Scholar]
- Peltier G. & Cournac L. Chlororespiration. Annu. Rev. Plant Biol. 53, 523–550 (2002). [DOI] [PubMed] [Google Scholar]
- Barrett C. F. et al. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 31, 3095–3112 (2014). [DOI] [PubMed] [Google Scholar]
- Wakasugi T. et al. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. P. Natl. Acad. Sci. 91, 9794–9798 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C. et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23, 279–291 (2006). [DOI] [PubMed] [Google Scholar]
- McCoy S. R., Kuehl J. V., Boore J. L. & Raubeson L. A. The complete plastid genome sequence of Welwitschia mirabilis: an unusually compact plastome with accelerated divergence rates. BMC Evol. Biol. 8, 130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann T. W. A., Kuzmina M. & Stefanović S. Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr. Genet. 55, 323–337 (2009). [DOI] [PubMed] [Google Scholar]
- Wu F. H. et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 10, 68 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier J., Guisinger M. M. & Jansen R. K. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol. Biol. 76, 263–272 (2011). [DOI] [PubMed] [Google Scholar]
- Pan I. C. et al. Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical oncidium breeding. PloS one 7, e34738 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. B., Tang M., Li H. T., Zhang Z. R. & Li D. Z. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 13, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M. L., Blazier J. C., Govindu M. & Jansen R. K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659 (2013). [DOI] [PubMed] [Google Scholar]
- Górniak M., Paun O. & Chase M. W. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: Congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 56, 784–795 (2010). [DOI] [PubMed] [Google Scholar]
- Kocyan A., Qiu Y. L., Endress P. & Conti E. A phylogenetic analysis of Apostasioideae (Orchidaceae) based on ITS, trnL-F and matK sequences. Plant Syst. Evol 247, 203–213 (2004). [Google Scholar]
- Tsai W. C. et al. OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 54, E7 (2013). [DOI] [PubMed] [Google Scholar]
- Givnish T. J. et al. Assembling the tree of the monocotyledons: plastome sequence phylogeny and evolution of Poales. Ann. MO Bot. Gard. 97, 584–616 (2010). [Google Scholar]
- Stern D. B. & Palmer J. D. Tripartite mitochondrial genome of spinach: physical structure, mitochondrial gene mapping, and locations of transposed chloroplast DNA sequences. Nucleic Acids Res. 14, 5651–5666 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D. S., Nielsen B. L. & Daniell H. A 7.5-kbp region of the maize (T cytoplasm) mitochondrial genome contains a chloroplast like trnI (CAT) pseudo gene and many short segments homologous to chloroplast and other known genes. Curr. Genet. 32, 125–131 (1997). [DOI] [PubMed] [Google Scholar]
- Huang C. Y., Ayliffe M. A. & Timmis J. N. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422, 72–76 (2003). [DOI] [PubMed] [Google Scholar]
- Bock R. & Timmis J. N. Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays 30, 556–566 (2008). [DOI] [PubMed] [Google Scholar]
- Lloyd A. H. & Timmis J. N. The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol. Biol. Evol. 28, 2019–2028 (2011). [DOI] [PubMed] [Google Scholar]
- Wang D., Rousseau-Gueutin M. & Timmis J. N. Plastid sequences contribute to some plant mitochondrial genes. Mol. Biol. Evol. 29, 1707–1711 (2012). [DOI] [PubMed] [Google Scholar]
- Wang D. et al. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol. Biol. Evol. 24, 2040–2048 (2007). [DOI] [PubMed] [Google Scholar]
- Smith D. R. Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol. Evol. 3, 743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. T. et al. Integration of molecular biology tools for identifying promoters and genes abundantly expressed in flowers of Oncidium Gower Ramsey. BMC Plant Biol. 11, 60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton S. W. et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiology 136, 3486–3503 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín M. & Sabater B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 48, 636–645 (2010). [DOI] [PubMed] [Google Scholar]
- Leitch I. et al. Genome size diversity in orchids: consequences and evolution. Ann. Bot. 104, 469–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J. N., Ayliffe M. A., Huang C. Y. & Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- Jansen R. K., Saski C., Lee S.-B., Hansen A. K. & Daniell H. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 28, 835–847 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. T. J. et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PloS one 7, e50226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C. L. et al. Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol. 54, E11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows P. A., Sazanov L. A., Svab Z., Maliga P. & Nixon P. J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17, 868–876 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofer W., Koop H. U., Wanner G. & Steinmüller K. Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD (P) H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol. Gen. Genet. 258, 166–173 (1998). [DOI] [PubMed] [Google Scholar]
- Shikanai T. et al. Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. P. Natl. Acad. Sci. 95, 9705–9709 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y. et al. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110, 361–371 (2002). [DOI] [PubMed] [Google Scholar]
- DalCorso G. et al. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132, 273–285 (2008). [DOI] [PubMed] [Google Scholar]
- Nakazono M. & Hirai A. Identification of the entire set of transferred chloroplast DNA sequences in the mitochondrial genome of rice. Mol. Gen. Genet. 236, 341–346 (1993). [DOI] [PubMed] [Google Scholar]
- Fang Y. J. et al. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PloS one 7, e37164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson A. J., Rice D. W., Dickinson S., Barry K. & Palmer J. D. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 23, 2499–2513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Moreno L. et al. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics 12, 424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D. W. et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342, 1468–1473 (2013). [DOI] [PubMed] [Google Scholar]
- Alverson A. J. et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 27, 1436–1448 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronico P., Gallerani R. & Ceci L. Compilation and classification of higher plant mitochondrial tRNA genes. Nucleic Acids Res. 24, 2199–2203 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld J., Unseld M. & Brennicke A. The mitochondrial genome of Arabidopsis is composed of both native and immigrant information. Trends Plant Sci. 4, 495–502 (1999). [DOI] [PubMed] [Google Scholar]
- Nakazono M., Nishiwaki S., Tsutsumi N. & Hirai A. A chloroplast-derived sequence is utilized as a source of promoter sequences for the gene for subunit 9 of NADH dehydrogenase (nad9) in rice mitochondria. Mol. Gen. Genet. 252, 371–378 (1996). [DOI] [PubMed] [Google Scholar]
- Hao W. & Palmer J. D. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. P. Natl. Acad. Sci. 106, 16728–16733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Baurens F. C., Cardi C., Aury J. M. & D'Hont A. The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PloS one 8, e67350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. G. & Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T. & Salzberg S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K. & Boore J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhang X., Hu S. & Yu J. An efficient procedure for plant organellar genome assembly, based on whole genome data from the 454 GS FLX sequencing platform. Plant methods 7, 38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information