Abstract
We review the recent development of novel biochemical and spectroscopic methods to determine the site-specific phosphorylation, expression, mutation, and structural dynamics of phospholamban (PLB), in relation to its function (inhibition of the cardiac calcium pump, SERCA2a), with specific focus on cardiac physiology, pathology, and therapy. In the cardiomyocyte, SERCA2a actively transports Ca2+ into the sarcoplasmic reticulum (SR) during relaxation (diastole) to create the concentration gradient that drives the passive efflux of Ca2+ required for cardiac contraction (systole). Unphosphorylated PLB (U-PLB) inhibits SERCA2a, but phosphorylation at S16 and/or T17 (producing P-PLB) changes the structure of PLB to relieve SERCA2a inhibition. Because insufficient SERCA2a activity is a hallmark of heart failure, SERCA2a activation, by gene therapy (Andino et al. 2008; Fish et al. 2013; Hoshijima et al. 2002; Jessup et al. 2011) or drug therapy (Ferrandi et al. 2013; Huang 2013; Khan et al. 2009; Rocchetti et al. 2008; Zhang et al. 2012), is a widely sought goal for treatment of heart failure. This review describes rational approaches to this goal. Novel biophysical assays, using site-directed labeling and high-resolution spectroscopy, have been developed to resolve the structural states of SERCA2a-PLB complexes in vitro and in living cells. Novel biochemical assays, using synthetic standards and multidimensional immunofluorescence, have been developed to quantitate PLB expression and phosphorylation states in cells and human tissues. The biochemical and biophysical properties of U-PLB, P-PLB, and mutant PLB will ultimately resolve the mechanisms of loss of inhibition and gain of inhibition to guide therapeutic development. These assays will be powerful tools for investigating human tissue samples from the Sydney Heart Bank, for the purpose of analyzing and diagnosing specific disorders.
Keywords: Phospholamban, SERCA2a, Phosphorylation, Subunit model, Loss-of-inhibition mutants
Introduction
The Sydney Heart Bank at the University of Sydney, Australia (Li et al. 2013), is a unique and valuable resource that promises to shed needed light on the genetic and biochemical markers that characterize specific heart diseases in human hearts. This information is needed to enhance methods for diagnosing, understanding, and treating these disorders, which together constitute the greatest health problems facing the aging population worldwide. Phospholamban (PLB) is a 52 amino acid sarcoplasmic reticulum (SR) membrane protein that reversibly inhibits the cardiac isoform of the SR Ca2+ ATPase (SERCA2a) in cardiac myocytes (Bers 2002). Active SERCA2a transports two Ca2+ per hydrolyzed ATP from the myoplasm into the SR during diastole (Zhang et al. 2000) to create a concentration gradient across the SR (Bers 2002). During systole, the passive efflux of Ca2+ from the SR to the myoplasm allows Ca2+ to bind and activate the contractile apparatus. Unphosphorylated PLB (U-PLB) decreases the apparent Ca2+ affinity of SERCA2a, thus reducing diastolic Ca2+ sequestration in the SR (Cantilina et al. 1993) and depleting the Ca2+ gradient required for muscle contraction. β-adrenergic-mediated phosphorylation of PLB at S16 by PKA, or at T17 by CaMKII, partially restores SERCA2a activity (Bers 2002; Karim et al. 2006; Wegener et al. 1989). Recent studies show that PLB remains bound to SERCA2a, even after phosphorylation (Dong and Thomas 2014; James et al. 2012), so SERCA2a activity can be restored only by phosphorylating PLB, ablating PLB, overexpressing SERCA2a, or expressing a loss-of-inhibition mutant of PLB.
A hallmark of heart failure is aberrant Ca2+ handling, typically resulting in elevated diastolic Ca2+ levels (Gwathmey et al. 1991; Mandinov et al. 2000; Wang and Goldhaber 2004), and often correlating with a reduction in PLB phosphorylation (Marks 2013) and/or SERCA2a expression (Bers 2002; Kranias and Hajjar 2012). Therefore, drug or gene therapy has focused on activating SERCA2a. In clinical trials, percutaneous administration of adeno-associated virus type 1 (AAV1) carrying the SERCA2a gene has been shown to decelerate pathological processes in advanced heart failure (Jessup et al. 2011). Thus, SERCA2a activation is a valid therapeutic strategy in heart failure. Since SERCA2a is responsible for 70 % of the Ca2+ removal from the cytoplasm of human cardiomyocytes (Bers 2002), it is likely that SERCA2a activation can compensate for other aberrant Ca2+ handling proteins and can improve the prognosis of heart failure of many etiologies. Increasing PLB phosphorylation and decreasing PLB expression are also valid strategies for SERCA2a activation. For example, AAV9 delivery of an inhibitor of protein phosphatase 1 (PP1), the phosphatase that dephosphorylates PLB at both sites, has been shown to be cardioprotective in mice (Pritchard et al. 2013). Decreasing PLB expression with microRNA has successfully rescued rats from left ventricular heart failure (Tsuji et al. 2009).
New approaches to PLB-based therapy require a more detailed understanding of the biophysical mechanisms of SERCA2a inhibition and relief. Initial models proposed that dissociation of PLB from SERCA2a was necessary to relieve SERCA2a inhibition (Fig. 1a). This model further validated the strategies of decreasing PLB expression and increasing PLB phosphorylation, and motivated the search for a drug that uncouples PLB from SERCA2a (Cornea et al. 2013; Ferrandi et al. 2013; Huang 2013; Khan et al. 2009; Rocchetti et al. 2008). Recent electron paramagnetic resonance (EPR) (James et al. 2012), nuclear magnetic resonance (NMR) (Gustavsson et al. 2013), and fluorescence resonance energy transfer (FRET) (Dong and Thomas 2014) studies have revealed that PLB is essentially a subunit of SERCA2a, remaining tightly bound even after phosphorylation (Fig. 1b). Phosphorylation shifts the PLB equilibrium from a rigid structural state (T-state), which inhibits SERCA2a, to an extended and dynamic (partially unfolded) R-state (Karim et al. 2006), which attenuates SERCA2a inhibition without dissociation of the SERCA2a/PLB complex (Dong and Thomas 2014; James et al. 2012) (Fig. 1b). Since relief of inhibition does not require dissociation, drugs that stabilize the R-state can be effective without dissociating PLB. Also, this subunit model raised the possibility that loss-of-inhibition mutants with R-state properties might bind to SERCA2a with sufficient affinity to displace endogenous PLB and thus relieve SERCA2a inhibition (Gruber et al. 2012; Ha et al. 2007, 2012; Hoshijima et al. 2002; Lockamy et al. 2011).
Fig. 1.
Therapeutic strategies based on two models of SERCA2a/PLB regulation. Fluorescence resonance energy transfer (yellow arrows) from a donor fluorophore (yellow sun, indicating emission) bound to SERCA2a (blue) to an acceptor fluorophore (orange triangle) bound to PLB (red) is indicated by decreased emission (smaller yellow sun). The circled P is phosphate and the purple diamond is a drug. Red PLB is endogenous PLB (WT-PLB) and green PLB is mutant PLB. a Dissociation model: PLB phosphorylation causes PLB to dissociate from SERCA2a, thus relieving inhibition, which should eliminate FRET. Therapeutic strategies: decrease PLB expression (not shown), displace PLB from SERCA2a with drug, or increase PLB phosphorylation. b Subunit model: PLB is essentially a subunit of SERCA2a. Phosphorylation causes PLB to adopt a more extended structure (R-state) without dissociation from SERCA2a, which should increase FRET. Therapeutic strategies include stabilizing PLB R-state with drug, or using gene therapy to deliver a loss-of-inhibition PLB mutant that binds to SERCA2a, thus displacing WT PLB
The evaluation of PLB-based therapies requires site-specific quantitation of changes in the mole fraction of PLB that is phosphorylated (XP) and total PLB expression (T-PLB). The goal of complete PLB ablation has been discounted, because a null PLB phenotype causes lethal, dilated cardiomyopathy in humans (Haghighi et al. 2003). Comparisons of XP and T-PLB values between healthy and failing hearts should lead to more accurate and specific diagnosis of the stage of heart failure. Comparisons of XP and T-PLB values among different heart-failure etiologies are needed to focus diagnosis. XP and T-PLB measurements are needed to normalize drugs and mutants to PLB phosphorylation and expression in biophysical studies and to measure the response to gene therapy.
XP and T-PLB measurements must be paired with SERCA2a activity assays to determine the functional effects of PLB phosphorylation and to quantitate how specific therapeutic strategies alter SERCA2a activity. For example, complete phosphorylation of PLB, at either or both sites, may be functionally equivalent to PLB ablation. The combined measurements of XP, T-PLB, and SERCA2a activity will aid in the understanding of regulatory mechanisms necessary to design therapies that optimize the interplay between exogenous drugs, peptides, antibodies, or nucleotides and endogenous mechanisms of Ca2+ homeostasis.
This review discusses the biochemical methods that measure PLB expression and phosphorylation, the biophysical studies that elucidate the structural dynamics of PLB and the PLB/SERCA2a complex, and the therapeutic strategies that emerge from the biochemical and biophysical properties of PLB.
Biochemical analysis
The three factors that determine SERCA2a inhibition by PLB are PLB expression, the mole fraction of each PLB phosphorylation state, and the extent to which each PLB phosphorylation state relieves SERCA2a inhibition. The four phosphorylation states of PLB are U-PLB (unphosphorylated), P16-PLB (phosphorylated at S16), P17-PLB (phosphorylated at T17), and 2P-PLB (phosphorylated at both sites). To quantitate these factors, we prepared purified synthetic standards for all 4 phosphorylation states of human phospholamban, enabling (1) calibration of western blots to account for imperfectly specific antibodies, and (2) control of SERCA2a/PLB stoichiometry and the isolation of each phosphorylation state in SERCA2a activity assays (Ablorh et al. 2014). Human PLB sequences were chosen in anticipation of using them as standards to measure PLB phosphorylation in human tissue samples from the Sydney Heart Bank (Li et al. 2013). SERCA1a was purified from rabbit skeletal muscle, which provides the most reliable source of purified SERCA1a. Rabbit SERCA1a and rabbit SERCA2a genes have 93 % sequence homology (BLAST software), and are quantitatively equivalent in their regulation by PLB (Ji et al. 1999; Zhao et al. 2003). Since the rabbit SERCA2a and human SERCA2a genes have 99 % sequence homology (BLAST software), we are quite confident that our PLB regulation assays are valid for human cardiac muscle.
PLB expression
Northern blots, Southern blots, qPCR, and conventional western blots all measure PLB expression qualitatively. With qPCR, even reproducible standard curves report statistically significant differences in gene numbers between assays (Smith and Osborn 2009). Northern blots and Southern blots are limited by the kinetics of probe hybridization (Reue 1998).
Conventional western blots have failed to measure PLB expression accurately (Calaghan et al. 2008; Grote-Wessels et al. 2008; Guinto et al. 2009; Loyer et al. 2008) because they did not account for the binding of antibodies to both U-PLB and P-PLB, with different affinities (Fig. 2) (Ablorh et al. 2012; Huke and Periasamy 2004; Mayer et al. 2000). If a PLB antibody were to bind U-PLB and P-PLB with the same affinity, it could be used to determine T-PLB expression in samples that contain a mixture of U-PLB and P-PLB. Figure 2 (top) illustrates that PLB antibodies Ab8a3, Ab2D12, and AbA1 bind U-PLB and P-PLB with different affinities. For each antibody, a western blot containing 5, 10, and 15 ng of purified synthetic U-PLB and 5, 10, and 15 ng of purified synthetic P16-PLB was performed, and the sum of the intensities of the group of PLB oligomers (magenta box in Fig. 2) in each lane was measured. These oligomeric bands are indicative of WT-PLB, and appear for both synthetic WT-PLB and in the tissue homogenates discussed below. On each blot, the U-PLB standards display a higher intensity than the P16-PLB standards, because the PLB antibody has a higher affinity for U-PLB than it does for P16-PLB. From these blots, standard curves for U-PLB and P16-PLB for each antibody are plotted (Fig. 2, bottom). Standard slopes for U-PLB and P16-PLB were calculated for each antibody, allowing us to quantitate antibody affinities. In Fig. 2 (bottom), εu is the standard slope for U-PLB for Ab8a3 (green), Ab2D12 (magenta), and AbA1 (blue). εp is the standard slope for P16-PLB for Ab8a3 (green), Ab2D12 (magenta), and AbA1 (blue). Antibody sensitivity can be quantitated by normalizing εu to εp (εu/εp). εu/εp = 7.49 for Ab8a3. Thus, Ab8a3 binds to U-PLB with 7.47 times the affinity that it binds to P16-PLB. By the same calculation, Ab2D12 binds to U-PLB with 4.59 times the affinity that it binds to P16-PLB, and AbA1 binds to U-PLB with 2.11 times the affinity that it binds to P16-PLB (Ablorh et al. 2012). Only an antibody with εu/εp = 1 (i.e., with equal affinity for U-PLB and P16-PLB) could be used to quantitate PLB expression accurately in one measurement, but no such antibody is available.
Fig. 2.
PLB antibody affinities. Top Western blots of purified synthetic U-PLB and P16-PLB (5, 10, and 15 ng) on the same membrane, blotted with one of three PLB antibodies: Ab8a3, Ab2D12, or AbA1. The intensities were measured using a box that encompassed all of the oligomeric states of PLB (magenta box on blot Ab8a3). Bottom Intensities of the 5, 10 and 15 ng of U-PLB (circles) and P16-PLB (squares) versus their concentration and the resulting standard curves from the blot labeled with the same PLB antibody (εu = the slope of the standard curve for U-PLB εp = the slope of the standard curve for P-PLB). This figure was adapted from Ablorh et al. (2012) with permission from Elsevier
Figure 3 uses mixtures of synthetic U-PLB and P16-PLB to illustrate how PLB antibodies with εu/εp ≠ 1 fail to measure PLB expression accurately. In Fig. 3, western blot intensities are measured for 5 mixtures of synthetic U-PLB + P16-PLB. The mole fraction of P16-PLB is different for each sample (0.0, 0.25, 0.50, 0.75, or 1.0), but all samples contain 12 ng of T-PLB (Fig. 3). Mixtures with a higher fraction of phosphorylated PLB have lower intensities on the western blot, suggesting that they have less T-PLB than mixtures with a smaller fraction of P16-PLB, even though all mixtures contain 12 ng of T-PLB. Thus, T-PLB is underestimated in samples with a higher mole fraction of P16-PLB. Figure 3 also shows that the extent of the changes in intensity due to increasing the mole fraction of P16-PLB depends on whether the antibody was Ab8a3 (magenta), Ab2D12 (blue), or AbA1 (green). Thus, data between laboratories is confounded by the use of different antibodies. εu/εp also varies between blots (CV = 26–46 %), when the same antibody is used (Ablorh et al. 2012). Thus, synthetic U-PLB and P-PLB must be used to calibrate each individual blot. In order to determine T-PLB, the mole fraction of phosphorylated PLB (XP) must be known. Thus, PLB expression and phosphorylation must be measured simultaneously, using multiple antibodies.
Fig. 3.
The intensities of mixtures of synthetic U-PLB and synthetic P16-PLB with P16-PLB mole fractions of 0.0, 0.25, 0.50, 0.75, and 1.00 obtained from western blots with either antibody Ab8a3 (magenta square), Ab2D12 (blue circle), or AbA1 (green triangle). Values are means ± SEM. All mixtures contain 12 ng of total PLB. If the antibodies bound U-PLB and P16-PLB with equal affinity, the intensities of the mixtures would be equal (independent of the P16-PLB mole fraction). Instead, the intensities of the mixtures decrease as the mole fraction of P16-PLB increases. This indicates that the PLB antibodies bind U-PLB with greater affinity than they bind P16-PLB, and that none of the 3 PLB antibodies can be used to accurately determine PLB expression in a single blot
Simultaneous measurement of XP and T-PLB
The first study (Ablorh et al. 2012) measured XP as P16-PLB/Total-PLB, but a second study (Ablorh et al. 2014) extended XP measurements to include all four PLB phosphorylation states (Fig. 4). This requires an antibody that binds preferentially to U-PLB (AbU) (either of Ab8a3, Ab2D12, or AbA1), an antibody that binds to P16-PLB (Ab16), an antibody that binds P17-PLB (Ab17), and an antibody that binds to 2P-PLB (Ab2). All PLB antibodies, save Ab2P, bind other phosphorylation states in addition to the state for which they are named with different affinities, as revealed by visibly different intensities of the synthetic PLB standards (U for U-PLB, P16 for P16-PLB, P17 for P17-PLB, and 2P for 2P-PLB) on the blots in Fig. 4a.
Fig. 4.
a Validation of the method: four identical western blots containing 2.5, 5, and 11 ng of synthetic U-PLB (U), P16-PLB (P16), P17-PLB (P17), and 2P-PLB (2P), and mixtures of standards (a–e). blotted with 4 different PLB antibodies (AbU, Ab17, Ab16, and Ab2P). As in Fig. 2, all oligomeric bands were included in the intensity measurements. b Application of the method: mole fraction of each PLB phosphorylation state in pig tissue homogenates with (solid) and without (open) left ventricular hypertrophy (mean ± SEM). The figure was reproduced from Ablorh et al. (2014) with permission from the American Society of Biological Chemistry and Molecular Biology
Our group has developed a method to solve the problem of differential antibody affinities in order to measure the concentrations of each of the 4 phosphorylation states of PLB and then calculate T-PLB in tissues. To validate the method, the concentration of each phosphorylation state was measured in 5 synthetic mixtures where the concentrations were known. For each mixture, 4 identical gels containing 4 standard curves (1 for each phosphorylation state), (lanes 1–12, Fig. 4a) and identical volumes of mixtures a, b, c, d, and e (lanes 13–25, Fig. 4a) were transferred to PVDF membranes. Each membrane was blotted with either AbU, Ab16, Ab17, or Ab2 (Fig. 4a). The nomenclature of the antibodies signifies that they bind preferentially (although not exclusively) to U, P16, P17, or 2P-PLB. The blot in Fig. 4a was used to construct and solve a system of 4 equations with 4 unknowns
| 1 |
where I i is the intensity observed for one of the antibodies (rows in Fig. 4a), j is the PLB phosphorylation state, εij is the slope of the standard curve (from synthetic standards) for phosphorylation state j blotted with antibody i, and cj is the unknown concentration of phosphorylation state j, determined by solving the system of equations (Eq. 1) (Ablorh et al. 2014). These cj values are the concentrations of U-PLB, P16-PLB, P17-PLB, and 2P-PLB in the mixtures. T-PLB was calculated as the sum of the 4 concentrations, and the mole fractions of each phosphorylated state (XP) were then calculated from the ratio of the concentration of each phosphorylation state to T-PLB (Ablorh et al. 2014). This method produces absolute and reproducible numbers, accounting for the imperfect specificities of commercial antibodies. Our method will thus allow comparisons between different experiments, studies, and laboratories.
Cardiac pathology changes XP
The assay was applied to a porcine model of left ventricular hypertrophy, where two- to three-fold increases in P16-PLB (Mills et al. 2006) and P17-PLB (Boknik et al. 2001; Mills et al. 2006) have been reported. The calibration of our blots with synthetic standards reduced underestimation of T-PLB, leading to less dramatic increases in PLB phosphorylation than previous reports (Boknik et al. 2001; Mills et al. 2006). We quantitated XP for all 4 PLB phosphorylation states in hypertrophied and non-hypertrophic porcine left ventricular tissue homogenates (Fig. 4b). The greatest mole fraction was observed for P16-PLB (XP16 = 0.47 ± 0.006), followed by U-PLB (XU = 0.30 ± 0.014), doubly phosphorylated 2P-PLB (X2P = 0.16 ± 0.007), and P17-PLB (XP17 = 0.07 ± 0.007) for the sham controls (Fig. 4b). Hypertrophy caused a significant decrease in the mole fraction of U-PLB (p = 0.005), (Fig. 4b), a calculation made possible by including X2P in our measurements where other groups had not (Ablorh et al. 2014). Quantitation of PLB phosphorylation states in both non-HF and HF hearts from the Sydney Heart Bank (Li et al. 2013) are currently underway. Preliminary data suggest that all four PLB phosphorylation states are also present in human tissue with and without dilated cardiomyopathy.
SERCA2a activity
Synthetic PLB standards also allowed us to evaluate the potency of each phosphorylation state in relieving SERCA2a inhibition. pKCa, (the negative log(10) of the Ca2+ concentration at half-maximal SERCA2a activity) was measured with the NADH-enzyme-linked activity assay (Fig. 5a). In this assay, SERCA2a ATPase activity is coupled to the conversion of PEP to pyruvate by pyruvate kinase. The subsequent dehydrogenation of pyruvate to lactate is coupled to the oxidation of NADH to NAD+, which can be measured by an increase in absorbance at 280 nm (Fig. 5a) (Li et al. 2012; Mueller et al. 2004). The data were fit to the Hill equation (Eq. 2) using Origin software.
| 2 |
Fig. 5.
SERCA2a activity. a The enzyme-linked NADH assay for measuring SERCA2a activity and calculating ∆pKCa. SERCA2a ATPase activity is coupled to phosphorylation of pyruvate by pyruvate kinase. NADH oxidation is coupled to dehydrogenation of pyruvate to form lactate. The oxidation of NADH is measured by absorbance at 280 nm. b ∆pKCa of U-PLB (U), P16-PLB (P16), P17-PLB (P17), and 2P-PLB (2P) with the reference point of pKCa for SERCA2a alone. The figure was reproduced from Ablorh et al. 2014 with permission from the American Society of Biological Chemistry and Molecular Biology
Co-reconstituting synthetic (human sequence) PLB with SERCA2a (rabbit sequence) allowed complete, exclusive, and site-specific phosphorylation of PLB as well as control SERCA2a/PLB stoichiometry. This facilitated the measurement of pKCa shift due to each phosphorylation state from the reference point of SERCA2a alone (Fig. 5b). The most potent phosphorylated (P-PLBs) were at the highest concentration. P16-PLB > > 2P > P17-PLB for both concentration and potency when compared to U-PLB (Figs. 4b, 5b) (Ablorh et al. 2014). Thus, P16-PLB provides coarse adjustments to SERCA2a inhibition, while 2P-PLB and P17-PLB finely tune PLB-mediated SERCA2a regulation. The efficacy of PLB mutants also depends heavily on their pKCa shifts because their potency in relieving SERCA2a inhibition will determine SERCA2a activity (Ablorh et al. 2014).
Structural dynamics of PLB and PLB/SERCA2a
PLB equilibria
PLB phosphorylation shifts the PLB structural equilibrium from the T-state to the R-state. The T-state PLB is L-shaped and rigid, with the cytoplasmic domain helical and membrane-bound. The R-state is more extended and dynamic, with a cytoplasmic domain that is partially unfolded and detached from the membrane (Fig. 6). The T-state conformation inhibits SERCA2a, and the R-state attenuates SERCA2a inhibition. EPR has shown that even P-PLB that is forced into the T-state with a lipid inhibits SERCA2a (Karim et al. 2006). The T-state/R-state equilibrium of U-PLB is 84 % T-state and 16 % R-state (Nesmelov et al. 2007). Conversely, relief of inhibition requires PLB to adopt the R-state. Detaching the cytoplasmic domain of U-PLB from the membrane using positively charged lipids forces PLB into the R-state, and relieves SERCA2a inhibition (Li et al. 2012). Phosphorylation shifts the T/R equilibrium to 65 % T-state and 35 % R-state in reconstituted lipids, as shown by EPR (Karim et al. 2006). PLB undergoes a monomer/pentamer equilibrium. The convergent theory is that the PLB monomer binds SERCA2a, while the pentamer provides storage of excess PLB (Vostrikov et al. 2013). Phosphorylation (Cornea et al. 1997) and phosphomimetic mutation (Hou et al. 2008) relieve SERCA2a inhibition in part by promoting polymerization of PLB (Kimura et al. 1997).
Fig. 6.
PLB equilibria. In the T-state/R-state equilibrium, the T-state dominates in both monomeric and pentameric PLB. In the monomer/pentamer equilibria, the pentamer dominates in both the T-state and R-state equilibrium so that pentameric, T-state PLB is the dominant form
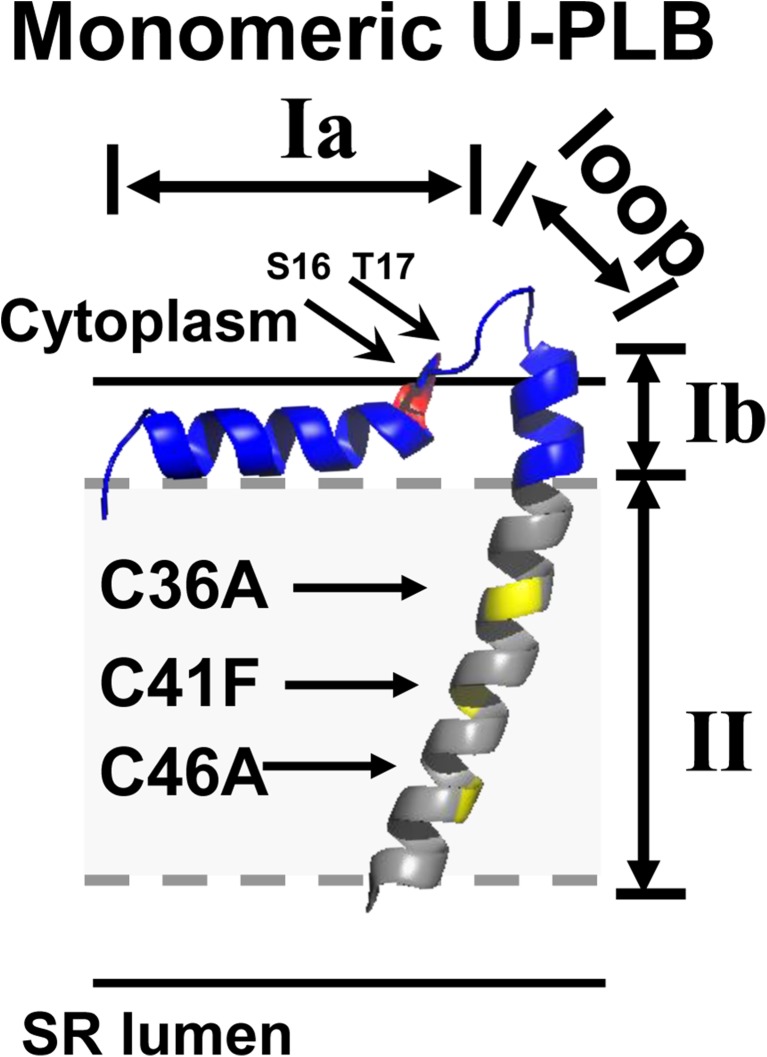
The fully functional PLB monomer, (C36A, C41F, C46A) (Fig. 7) has been employed to study PLB equilibria in the context of SERCA2a inhibition. Each domain of PLB is differentially involved in the T/R transition, the assembly of the PLB pentamer, and the formation of the SERCA2a/PLB complex. PLB has 4 dynamic domains: Ia (1–16), the loop (17–22), Ib (23–30), and II (31–52) (Traaseth et al. 2007) (Fig. 7), and 3 structural domains: the cytoplasmic helix (1–16), the loop (17–22), and the transmembrane helix (23–52) (Zamoon et al. 2003). The transmembrane helix is comprised of domains Ib and II (Fig. 7). The transmembrane helix is necessary and sufficient to bind and inhibit SERCA2a (Metcalfe et al. 2004). In each domain, phosphorylation has a unique effect on the structural dynamics, pentamer assembly, and SERCA2a binding of PLB.
Fig. 7.
AFA-U-PLB shows PLB dynamic domains with identification of phosphorylation sites (S16 in red and T17 in dark red) and sites for monomeric mutations (C36A, C41F, and C46A in yellow). Gray dashes outline the hydrophobic core in the phospholipid bilayer. This structure is adapted from frame 12 of PDB file 2 KB7 (Traaseth et al. 2009). Sequence of human PLB: M E K V Q Y L T R S A I R R A S T I E M P Q Q A R Q K L Q N L F I N F C L I L I C L L L I C I I V M L L
PLB domains
Domain Ia is a rigid, amphipathic α-helix that uses its hydrophobic face to insert itself into the SR membrane (Fig. 7), and its positively charged arginine residues to bind to the negatively charged lipid head-groups on the SR membrane. Domain Ia contains the S16 phosphorylation site (Fig. 7). Phosphorylation at S16 shifts the T-state/R-state equilibrium toward the R-state (Karim et al. 2006) and increases the population of the pentamer (Cornea et al. 1997). The T-state to R-state shift requires changes in the structure and dynamics of PLB, such as unwinding of the cytoplasmic domain Ia helix (Abu-Baker and Lorigan 2006; De Simone et al. 2013). EPR revealed an order-to-disorder (T-to-R) transition within the PLB cytoplasmic domain upon phosphomimetic mutation of PLB (Karim et al. 2006). NMR residual dipolar couplings (RDC) show a reduction of the number of contacts between PLB and the membrane surface upon phosphorylation (De Simone et al. 2013), suggesting that the domain Ia membrane detachment occurs because of the decrease in the net charge of the domain, which perturbs its hydrophobic and electrostatic interactions with the membrane (Abu-Baker and Lorigan 2006). The pentamer has less apparent affinity for SERCA2a than the monomer, so SERCA2a inhibition is reduced (Kelly et al. 2008). Domain Ia loss-of-inhibition mutants exhibit several biophysical properties of P16-PLB.
The loop region (amino acids 17–22) consists of a random coil of hydrophilic amino acids that reside above the membrane surface (Fig. 7). The loop contains the T17 phosphorylation site (Fig. 7). Upon phosphorylation of S16, the loop extends so that P-PLB can adopt the dynamic, extended structure of the R-state. The loop is also responsible for coupling between the cytoplasmic and transmembrane helices (Ha et al. 2012; Li et al. 2005).
Domain Ib (amino acids 23–30) is part of the transmembrane helix. 13C solid-state NMR studies have shown that it is a hydrophobic α-helix that is aligned with domain II (Yu and Lorigan 2014). Upon S16 phosphorylation, domain Ib changes its structure from an α-helix to an uncoiled coil and loses its alignment with domain II (Yu and Lorigan 2014). Alanine scanning of a peptide containing PLB residues 21–30 revealed that domain Ib residues N27 (which is K27 in humans), and N30 were also important for SERCA2a binding (Asahi et al. 2001).
Domain II (amino acids 31–52) (Fig. 7) has one face that contains amino acids that comprise the SERCA2a/PLB transmembrane interface. Mutagenesis studies of a transmembrane peptides co-reconstituted with SERCA2a suggest that L31, L42, and L52 are involved in SERCA2a binding. Molecular modeling suggest that PLB residues P35, I38, I48, and V49 complement a hydrophobic pocket near the N-terminus of SERCA2a and stabilize the SERCA2a/PLB complex (Afara et al. 2006). The opposite face is instrumental in the quaternary structure of PLB. Mutagenesis studies with subsequent SDS-PAGE gel-shift assays have revealed that the other face of PLB (residues L37, I40 L44, I47, and L51) form a leucine/isoleucine zipper (Simmerman et al. 1996) required for self-assembly into pentamers. Mutation of any of the zipper amino acids prevents PLB pentamer assembly (Simmerman et al. 1996). EPR studies have shown that mutations of the cysteine residues in the transmembrane domain destabilize the PLB pentamer (Karim et al. 1998). C41L is tetrameric on SDS-PAGE, and reaction of the remaining cysteines, C36 and C46, causes PLB to become monomeric. The structure, dynamics, and topology of domain II are unaltered upon S16 phosphorylation. Only the T-state was detected by EPR when PLB was TOAC-labeled at position 36 (Gustavsson et al. 2011).
Therapeutic strategies
Therapeutic strategies include (1) decreasing T-PLB levels with micro-RNA or antibodies (Andino et al. 2008; He et al. 1999), (2) increasing PLB phosphorylation by inhibiting PP1, the protein phosphatase that dephosphorylates PLB at both S16 and T17 (Fish et al. 2013; Miyazaki et al. 2012; Oh et al. 2013; Pritchard et al. 2013; Xu et al. 2007; Zhang et al. 2012), (3) uncoupling PLB and SERCA2a with drugs, (4) stabilizing the R state of PLB using drugs, (5) stabilizing the PLB pentamer, and (6) administering PLB loss-of-inhibition mutants using gene therapy (Fig. 1).
Decreasing PLB expression
Antisense RNA (He et al. 1999; Tsuji et al. 2009), siRNA (Fechner et al. 2007) and shRNA (Andino et al. 2008) have been used to decrease PLB expression. Decreasing PLB expression has successfully reversed the effects of heart failure in animal models (Gruber et al. 2012; Ha et al. 2007, 2012; He et al. 1999; Hoshijima et al. 2002; Iwanaga et al. 2004; Lockamy et al. 2011; Schmidt et al. 2001; Trieber et al. 2009). Isolated myocytes from phospholamban knockout mice displayed both accelerated Ca2+ uptake by SERCA2a and improved cardiac performance (Santana et al. 1997). PLB ablation has also rescued rats from ventricular failure induced by Ca2+ overload (Tsuji et al. 2009). In contrast, a human PLB-null phenotype causes lethal, dilated cardiomyopathy (Haghighi et al. 2003). For example, both the L39stop mutation, which translocates PLB to the sarcolemma and nucleus (Haghighi et al. 2003), and the R14del mutation, which translocates PLB to the sarcolemma (Haghighi et al. 2012), abolish PLB regulation of SERCA2a, resulting in lethal cardiomyopathy and premature death (Haghighi et al. 2003, 2012). The detrimental effects of ablating PLB from the SR by various methods suggest that cardiac Ca2+ homeostasis in the human heart requires control of SERCA2a activity by PLB phosphorylation.
Increasing PLB phosphorylation
Protein phosphatase 1 (PP1) dephosphorylates PLB at both S16 and T17 (Steenaart et al. 1992), so PP1 inhibitors have been used to increase PLB phosphorylation. Gene therapy vectors that inhibit PP1 have successfully improved cardiac performance in animals (Fish et al. 2013; Miyazaki et al. 2012; Oh et al. 2013; Pritchard et al. 2013). AAV9 delivery of PP1 inhibitor was cardioprotective in mice (Pritchard et al. 2013). Intracoronary delivery of AAV9.I-1c to pigs after left anterior descending artery occlusion-induced heart failure stayed the progression of heart failure and reduced the size of the scar created by the myocardial infarction (Fish et al. 2013). AAV9 delivery of shRNA against PP1β subunit improved both left ventricular diastolic function and ventricular remodeling in mice with heart failure induced by B-type natriuretic peptide (BNP) (Miyazaki et al. 2012). Drugs and peptides can also increase PLB phosphorylation by inhibiting PP1. The small molecule, astragaloside IV, increases PLB phosphorylation through PKA activation (Xu et al. 2007; Zhang et al. 2012). Decoy peptides for PP1 have also been used to keep PLB phosphorylated. The decoy peptides include the 9 amino-acid PLB sequence 14–22 (which contains the phosphorylation sites S16 and T17), with and without phosphomimetic S16E or T17E mutations (Oh et al. 2013).
Targeting SERCA2a and/or PLB with small molecules
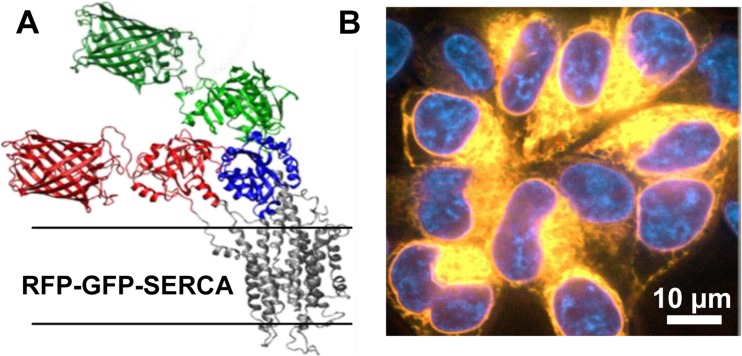
Istaroxime (Ferrandi et al. 2013; Huang 2013; Khan et al. 2009; Rocchetti et al. 2008), which increases diastolic function in humans (Khan et al. 2009), has been shown to partially decouple SERCA2a and phospholamban. Istaroxime altered Vmax and pKCa of SERCA2a only in the presence of PLB, so its regulation of SERCA2a must result from its interaction with PLB (Ferrandi et al. 2013). FRET measurements between SERCA2a and PLB in membranes reconstituted from purified proteins labeled with dyes were used to screen a small-molecule library, with the goal of discovering uncouplers of SERCA2a-PLB. The screen was successful—several useful compounds were discovered and are now under development in preclinical trials (Cornea et al. 2013). However, all of those compounds were direct SERCA2a activators, rather than SERCA2a-PLB uncouplers. Therefore, a subsequent screen was developed, in which FRET is measured within SERCA2a, using a 2-color fluorescent fusion construct of the enzyme (Gruber et al. 2014) (Fig. 8). This assay was further enhanced by employing a novel fluorescence lifetime plate-reader. The results were quite promising for future discovery of direct SERCA2a activators.
Fig. 8.
2-color SERCA2a used for small-molecule screening. a Computational model of GFP and RFP modeled on the crystal structure of SERCA2a1a (PDB 1IWO). b Confocal imaging of live HEK cells expressing 2-color SERCA2a. Yellow co-localization of RFP and GFP fluorescence, Blue DAPI nuclear stain. Thus, 2-color SERCA2a is localized to intracellular ER membranes. This figure is reproduced from Gruber et al. (2014) with permissions from Sage Publishing
PLB mutants
In order for a PLB mutant to be useful for therapeutic activation of SERCA2a (via gene therapy), it must bind tightly enough to SERCA2a to displace endogenous PLB, while having low inhibitory potency (Fig. 1b).
Loss-of-inhibition mutants that mimic R-state PLB
Decreasing the inhibitory potency of PLB, producing loss-of-inhibition mutants, is accomplished by simulating the structural rearrangement of PLB from its T-state to its R-state (Fig. 1b). Since S16 phosphorylation is capable of mounting the maximum β-adrenergic response (Chu et al. 2000), as explained quantitatively by our recent measurements (Figs. 4, 5), EPR, FRET, and NMR have focused on using the biophysical changes that accompany P16-PLB phosphorylation as a template for R-state and monomeric PLB mutants (Brittsan et al. 2003; Ha et al. 2007, 2012; Hoshijima et al. 2002; Li et al. 2005; Marks 2013; Metcalfe et al. 2005). Loss-of-inhibition mutants that mimic R-state PLB include: S16E, R9C, K3E/R14E, P21A, P21G, M20G, Q22G, and M20G + P21G. The S16E mutation mimics the negative charge of the phosphate at position S16. By a separate mechanism, it causes pentamerization, as verified by increased intramolecular FRET between protomers in S16E-PLB in comparison to that of U-PLB (Hou et al. 2008). The K3E/R14E mutation may cause loss of inhibition because of the increase in negative charge of the cytoplasmic domain (from +2 to −2) (MacLennan et al. 1997). K3E/R14E also interacts with endogenous PLB monomers to form heteropentamers that neither bind nor inhibit SERCA2a (He et al. 1999; Ziolo et al. 2005). R9C also decreases the positive charge of the cytoplasmic domain.
Glycine mutations along the loop (P21G, M20G, Q22G, and M20G + P21G) increase the flexibility of the loop to shift the T-state-R-state equilibrium to the R-state, even in the absence of phosphorylation (Ha et al. 2007, 2012). The order of loss of inhibition is: M20G > P21G > Q22G > > M20G-P21G (Ha et al. 2012). Thus, M20G and P21G mutations are additive for loss of inhibition. Mutating three glycines (M20G + P21G + Q22G) is superinhibitory, perhaps due to uncoupling of the cytoplasmic and transmembrane helices (Ha et al. 2012).
The loop mutants display further loss of inhibition upon phosphorylation. In fact, phosphorylated M20G-P21G provides more relief of inhibition than P16-PLB (Ha et al. 2012). The preservation of the S16 phosphorylation site in these loop mutants suggests that they will remain under β-adrenergic control in vivo. For that reason, loop mutations may prove safer than phosphomimetic mutations (such as S16E) in long term heart failure treatment.
Most domain Ib mutations that have been studied rigorously, such as L27A and N30A, are gain-of-inhibition (Trieber et al. 2005). However, glycine mutations along domain Ib may be able to mimic the phosphorylation-induced unwinding of its α-helix and misalignment with domain II (Yu and Lorigan 2014).
The structural dynamics of domain II is virtually unaffected by PLB phosphorylation (Yu and Lorigan 2014), so the domain II residues have not been mutated for the purpose of mimicking R-state PLB.
Competition between WT-PLB and PLB-mutants
For activation of SERCA2a by gene therapy, an ideal PLB mutant (PLBM) should have low inhibitory potency but high affinity for SERCA2a (Fig. 1b). Competitive binding of PLB mutants (PLBM) has been measured by intermolecular FRET (Gruber et al. 2012; Lockamy et al. 2011) (Fig. 9). SERCA2a and PLB were labeled in live cells with fluorescent fusion proteins (donor-CFP and acceptor-YFP, respectively), and the PLB mutant was unlabeled. The competitiveness of the mutant was evaluated from the decrease in FRET efficiency between CFP-SERCA2a and YFP-PLB in the presence of the mutant. Unlabeled S16E, P21G, and I40A all showed a slightly greater decrease in FRET efficiency than WT-PLB, whereas L31A showed an increase in FRET efficiency in comparison to that of WT-PLB (Fig. 9) (Gruber et al. 2012). These results corroborate FRET measurements performed with purified reconstituted proteins (Lockamy et al. 2011) and crosslinking experiments performed with PLB-tethered SERCA2a in microsomes harvested from sf21 cells (Chen 2014). Thus, several promising mutants have already been identified for preclinical gene therapy trials.
Fig. 9.
FRET competition in live cells (Gruber et al. 2012). a FRET is detected from CFP-SERCA2a to YFP-PLB, expressed stably in HEK cells (left), but transient transfection with unlabeled PLB mutant (PLB M, green) displaces YFP-PLB, eliminating FRET and relieving SERCA2a inhibition. b FRET efficiency E (error bars ±SEM). AID (activation-induced deaminase, a cytoplasmic protein) is a transfection control to rule out non-specific transfection artifacts. Decreased FRET (E) between SERCA2a and YFP-PLB in the presence of the unlabeled mutants (PLBM) indicates that unlabeled PLBM constructs compete effectively with YFP-PLB for binding to CFP-SERCA2a. Adapted from Gruber et al. (2012) with permission from Elsevier
L37A and I40A mutations destabilize the PLB pentamer (Cornea et al. 1997; Karim et al. 1998). Fluorescence recovery after photobleaching (FRAP) in AAV-293 cells has shown that I40A (6.8 ± 2.0) has a faster recovery than WT (9.7 ± 0.7), indicating that I40A is less pentameric (Kelly et al. 2008). Pentameric mutant, S16E (Kimura et al. 1997; Simmerman et al. 1996; Toyofuku et al. 1994), displaced WT with the same efficiency of I40A (Gruber et al. 2012), which suggests that the mechanism of S16E competition with WT-PLB is stronger in molecular interactions with SERCA2a (than I40A), as opposed to depolymerization.
Resolution of mutant mechanisms
Both the mechanism of loss of inhibition and competition are essential to predicting the therapeutic value of PLB mutants. L31A, for example, which abolishes SERCA2a inhibition in vitro (Chen et al. 2005), does not do so by simulating the structural rearrangement of PLB accomplished by endogenous phosphorylation. Instead, it binds to SERCA2a with lower affinity (Kd = 9.1) than WT-PLB (Kd = 4.0). Since L31A alone abolishes SERCA2a activity by dissociating from SERCA2a, it cannot compete well with WT-PLB (Fig. 9), and it is not therapeutically promising.
Mutations that decrease Kd may result in gain of inhibition, which is the case with both L37A and I40A. However, when gain-of-inhibition and loss-of-inhibition mutations combine, the resultant, chimeric mutant both relieves SERCA2a inhibition and competes with endogenous PLB. For example, the double mutant N34A/I40A combined N34A, a domain II loss-of-inhibition mutation on the SERCA2a-binding face of domain II, with a monomeric I40A, a gain-of-inhibition mutation that depolymerizes PLB to increase SERCA2a binding by 229 %. The double mutant was both loss-of-inhibition (ΔK Ca = −0.03) and competitive (Asahi et al. 1999). Many gain-of-inhibition mutations are in the transmembrane domain, comprised of domain Ib and domain II. N34A and I40A worked synergistically to decrease inhibition and increase SERCA2a binding, respectively. This suggests that the mechanism of loss-of-inhibition at residue N34 was a structural rearrangement, instead of a decrease in binding (Asahi et al. 1999).
Previous FRET competition experiments did not provide any information about the contribution of structural rearrangement to gain- or loss-of-inhibition. Conversely, EPR experiments that measure structural rearrangements do not provide information about binding.
Resolution of SERCA2a binding and structural rearrangement of PLB in the SERCA2a/PLB complex has been accomplished recently by time-resolved fluorescence resonance energy transfer (TR-FRET) (Dong and Thomas 2014) (Fig. 10). This assay provides the means for investigating mechanisms of action of current mutants and guides the development of chimeric mutants that maximize both competition and inhibitory relief.
Fig. 10.
Two mechanisms to relieve SERCA2a inhibition by PLB in cardiac SR. Phosphorylation shifts the PLB cytoplasmic domain toward the dynamically disordered R state. Micromolar Ca2+ induces a structural change in the SERCA2a transmembrane domain. The figure was reproduced from Dong and Thomas (2014) with permission from Elsevier
From bench to bedside
PLB knockdown (but not knockout), increasing PLB phosphorylation, and gene therapy with loss-of-inhibition mutants are promising therapeutic approaches to increase SERCA2a activity. The restoration of SERCA2a activity needs to be partial to avoid the lethal effects of complete loss of PLB regulation of SERCA2a (Haghighi et al. 2003); i.e., complete loss of β-adrenergic SERCA2a regulation.
The S16E mutant shows clinical promise. rAAV-mediated transcoronary administration of S16E to hamsters (Hoshijima et al. 2002) and percutaneous administration of S16E to sheep with heart failure (Kaye et al. 2007), both resulted in the restoration of cardiac function (Hou et al. 2008; Kaye et al. 2007). Adenovirus-mediated gene therapy with the K3E/R14E mutant has restored the force–frequency response to rabbits with heart failure induced by aortic constriction (Ziolo et al. 2005).
The R9C mutation has shown that loss of inhibition in vitro does not always equate with therapeutic value in vivo. Though it is loss-of-inhibition in vitro, R9C is an autosomal dominant missense mutation that causes enlarged hearts, and subsequent death at an average of 25 years (Schmitt et al. 2003). R9C-PLB traps PKA and prevents it from phosphorylating endogenous PLB. As a result, SERCA2a escapes β-adrenergic control, causing lethal dilated cardiomyopathy (Schmitt et al. 2003). R9L and R9H mutations have also been observed to trap PKA and cause heart failure and premature death (Ceholski et al. 2012), but their structure, dynamics and topology have not been well studied (Medeiros et al. 2011). Since PKA preferentially binds R-state PLB (Masterson et al. 2011), other R-state mutants may ligate PKA. Measuring the endogenous PLB phosphorylation after delivery of PLB mutants in cells could predict this type of problem. Similarly, L39stop-PLB translocation cautions against mutating the SR-localization sequence at the C-terminus of PLB. Measuring PLB expression in cardiac SR could predict this problem.
Mutants that cannot be phosphorylated may not maintain SERCA2a activity within the narrow window required for homeostasis during both rest and cardiac challenge, because they escape β-adrenergic control. Similarly, monomeric mutants cannot be attenuated by self-association, and they will promote pentamer assembly of endogenous PLB by the law of mass action. Translation from animals to humans may also present unforeseen problems as with PLB ablation, which is compatible with life in mice (Schmidt et al. 2001) but lethal in humans (Hou et al. 2008; Kelly et al. 2008). Thus, next-generation structurally designed PLB mutants, designed to increase SERCA2a affinity but also decrease inhibitory potency, while maintaining PKA-mediated phosphorylation, hold great promise (Gruber et al. 2014; Lockamy et al. 2011). Direct small-molecule activation of SERCA2a is also an attractive approach, since it is likely to preserve β-adrenergic control, and variation of dosage is more feasible than for gene therapy (Gruber et al. 2014).
Conclusion
In vivo, PLB and its phosphorylated derivatives must maintain SERCA2a activity within a narrow window, because complete inhibition and complete disinhibition of SERCA2a both result in lethal dilated cardiomyopathy. The goal of PLB-related therapies is a partial restoration of SERCA2a activity. This can be accomplished by reducing PLB expression, increasing PLB phosphorylation, and uncoupling PLB and SERCA2a, all of which are viable strategies for the long-term activation of SERCA2a. Further study of the biochemical and biophysical properties of U-PLB and P-PLB, and the mechanism of action of current drugs and mutants, will advance the field by guiding the design of new small-molecules, RNAs, and vectors delivering competitive PLB mutants to restore SERCA2a function, and ultimately reverse the progression of heart failure.
Acknowledgments
This project used the facilities of the Biophysical Spectroscopy Center and the Peptide Synthesis Facility, University of Minnesota. We thank Simon J. Gruber for expert advice and Octavian Cornea for preparing the manuscript. Many others contributed to the studies reviewed here, especially Christine B. Karim, Razvan L. Cornea, Jesse E. McCaffrey, Zachary M. James, and Xiaoqiong Dong.
Compliance with ethical standards
ᅟ
Funding
This work was supported by National Institutes of Health grants to D.D.T. (AG26160, AG042996) and N.D.A. (T32 HL069764).
Conflict of interest
Naa-Adjeley Dromoh Ablorh declares that she has no conflict of interest. David Dale Thomas declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Special Issue: Biophysics of Human Heart Failure
References
- Ablorh NA, Miller T, Nitu F, Gruber SJ, Karim C, Thomas DD. Accurate quantitation of phospholamban phosphorylation. Immunoblot Anal Biochem. 2012;425:68–75. doi: 10.1016/j.ab.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablorh NA, Dong X, James ZM, Xiong Q, Zhang J, Thomas DD, Karim CB. Synthetic Phosphopeptides Enable Quantitation of the Content and Function of Phospholamban's four Phosphorylation States. Cardiac Muscle J Biol Chem. 2014 doi: 10.1074/jbc.M114.556621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Baker S, Lorigan GA. Phospholamban and its phosphorylated form interact differently with lipid bilayers: A 31P, 2H, and 13C solid-state NMR spectroscopic study. Biochemistry. 2006;45:13312–13322. doi: 10.1021/bi0614028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afara MR, Trieber CA, Glaves JP, Young HS. Rational design of peptide inhibitors of the sarcoplasmic reticulum calcium pump. Biochemistry. 2006;45:8617–8627. doi: 10.1021/bi0523761. [DOI] [PubMed] [Google Scholar]
- Andino LM, Takeda M, Kasahara H, Jakymiw A, Byrne BJ, Lewin AS. AAV-mediated knockdown of phospholamban leads to improved contractility and calcium handling in cardiomyocytes. J Gen Med. 2008;10:132–142. doi: 10.1002/jgm.1131. [DOI] [PubMed] [Google Scholar]
- Asahi M, Kimura Y, Kurzydlowski K, Tada M, MacLennan DH. Transmembrane helix M6 in sarco(endo)plasmic reticulum Ca(2+)-ATPase forms a functional interaction site with phospholamban. Evidence for physical interactions at other sites. J Biol Chem. 1999;274:32855–32862. doi: 10.1074/jbc.274.46.32855. [DOI] [PubMed] [Google Scholar]
- Asahi M, Green NM, Kurzydlowski K, Tada M, MacLennan DH. Phospholamban domain IB forms an interaction site with the loop between transmembrane helices M6 and M7 of sarco(endo)plasmic reticulum Ca2+ ATPases. Proc Natl Acad Sci U S A. 2001;98:10061–10066. doi: 10.1073/pnas.181348298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- Boknik P, et al. Enhanced protein phosphorylation in hypertensive hypertrophy. Cardiovasc Res. 2001;51:717–728. doi: 10.1016/S0008-6363(01)00346-7. [DOI] [PubMed] [Google Scholar]
- Brittsan AG, et al. Chronic SR Ca2 + -ATPase inhibition causes adaptive changes in cellular Ca2+ transport. Circ Res. 2003;92:769–776. doi: 10.1161/01.RES.0000066661.49920.59. [DOI] [PubMed] [Google Scholar]
- Calaghan S, Kozera L, White E. Compartmentalisation of cAMP-dependent signalling by caveolae in the adult cardiac myocyte. J Mol Cell Cardiol. 2008;45:88–92. doi: 10.1016/j.yjmcc.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Cantilina T, Sagara Y, Inesi G, Jones LR. Comparative studies of cardiac and skeletal sarcoplasmic reticulum ATPases. Effect of a phospholamban antibody on enzyme activation by Ca2+ J Biol Chem. 1993;268:17018–17025. [PubMed] [Google Scholar]
- Ceholski DK, Trieber CA, Holmes CF, Young HS. Lethal, hereditary mutants of phospholamban elude phosphorylation by protein kinase A. J Biol Chem. 2012;287:26596–26605. doi: 10.1074/jbc.M112.382713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Competitive displacement of wild-type phospholamban from the Ca-free cardiac calcium pump by phospholamban mutants with different binding affinities. J Mol Cell Cardiol. 2014;76C:130–137. doi: 10.1016/j.yjmcc.2014.08.020. [DOI] [PubMed] [Google Scholar]
- Chen Z, Stokes DL, Jones LR. Role of leucine 31 of phospholamban in structural and functional interactions with the Ca2+ pump of cardiac sarcoplasmic reticulum. J Biol Chem. 2005;280:10530–10539. doi: 10.1074/jbc.M414007200. [DOI] [PubMed] [Google Scholar]
- Chu G, Lester JW, Young KB, Luo W, Zhai J, Kranias EG. A single site (Ser16) phosphorylation in phospholamban is sufficient in mediating its maximal cardiac responses to beta -agonists. J Biol Chem. 2000;275:38938–38943. doi: 10.1074/jbc.M004079200. [DOI] [PubMed] [Google Scholar]
- Cornea RL, Jones LR, Autry JM, Thomas DD. Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biochemistry. 1997;36:2960–2967. doi: 10.1021/bi961955q. [DOI] [PubMed] [Google Scholar]
- Cornea RL, et al. High-throughput FRET assay yields allosteric SERCA2a activators. J Biomol Screen. 2013;18:97–107. doi: 10.1177/1087057112456878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone A, Gustavsson M, Montalvao RW, Shi L, Veglia G, Vendruscolo M. Structures of the excited states of phospholamban and shifts in their populations upon phosphorylation. Biochemistry. 2013;52:6684–6694. doi: 10.1021/bi400517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Thomas DD. Time-resolved FRET reveals the structural mechanism of SERCA2a-PLB regulation. Biochem Biophys Res Commun. 2014;449:196–201. doi: 10.1016/j.bbrc.2014.04.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechner H, et al. Highly efficient and specific modulation of cardiac calcium homeostasis by adenovector-derived short hairpin RNA targeting phospholamban. Gene Ther. 2007;14:211–218. doi: 10.1038/sj.gt.3302872. [DOI] [PubMed] [Google Scholar]
- Ferrandi M, et al. Istaroxime stimulates SERCA2a and accelerates calcium cycling in heart failure by relieving phospholamban inhibition. Br J Pharmacol. 2013;169:1849–1861. doi: 10.1111/bph.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish KM, et al. AAV9.I-1c delivered via direct coronary infusion in a porcine model of heart failure improves contractility and mitigates adverse remodeling. Circ Heart Fail. 2013;6:310–317. doi: 10.1161/CIRCHEARTFAILURE.112.971325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote-Wessels S, et al. Inhibition of protein phosphatase 1 by inhibitor-2 exacerbates progression of cardiac failure in a model with pressure overload. Cardiovasc Res. 2008;79:464–471. doi: 10.1093/cvr/cvn113. [DOI] [PubMed] [Google Scholar]
- Gruber SJ, Haydon S, Thomas DD. Phospholamban mutants compete with wild type for SERCA2a binding in living cells. Biochem Biophys Res Commun. 2012;420:236–240. doi: 10.1016/j.bbrc.2012.02.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber SJ, et al. Discovery of enzyme modulators via high-throughput time-resolved FRET in living cells. J Biomol Screen. 2014;19:215–222. doi: 10.1177/1087057113510740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinto PJ, Haim TE, Dowell-Martino CC, Sibinga N, Tardiff JC. Temporal and mutation-specific alterations in Ca2+ homeostasis differentially determine the progression of cTnT-related cardiomyopathies in murine models. Am J Physiol Heart Circ Physiol. 2009;297:H614–626. doi: 10.1152/ajpheart.01143.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M, Traaseth NJ, Karim CB, Lockamy EL, Thomas DD, Veglia G. Lipid-mediated folding/unfolding of phospholamban as a regulatory mechanism for the sarcoplasmic reticulum Ca2 + -ATPase. J Mol Biol. 2011;408:755–765. doi: 10.1016/j.jmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M, Verardi R, Mullen DG, Mote KR, Traaseth NJ, Gopinath T, Veglia G. Allosteric regulation of SERCA2a by phosphorylation-mediated conformational shift of phospholamban. Proc Natl Acad Sci U S A. 2013;110:17338–17343. doi: 10.1073/pnas.1303006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwathmey JK, et al. Diastolic dysfunction in hypertrophic cardiomyopathy. Effect on active force generation during systole. J Clin Invest. 1991;87:1023–1031. doi: 10.1172/JCI115061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KN, et al. Controlling the inhibition of the sarcoplasmic Ca2 + -ATPase by tuning phospholamban structural dynamics. J Biol Chem. 2007;282:37205–37214. doi: 10.1074/jbc.M704056200. [DOI] [PubMed] [Google Scholar]
- Ha KN, Gustavsson M, Veglia G. Tuning the structural coupling between the transmembrane and cytoplasmic domains of phospholamban to control sarcoplasmic reticulum Ca(2+)-ATPase (SERCA2a) function. J Muscle Res Cell Motil. 2012;33:485–492. doi: 10.1007/s10974-012-9319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi K, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111:869–876. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi K, et al. The human phospholamban Arg14-deletion mutant localizes to plasma membrane and interacts with the Na/K-ATPase. J Mol Cell Cardiol. 2012;52:773–782. doi: 10.1016/j.yjmcc.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, et al. Effects of mutant and antisense RNA of phospholamban on SR Ca(2+)-ATPase activity and cardiac myocyte contractility. Circulation. 1999;100:974–980. doi: 10.1161/01.CIR.100.9.974. [DOI] [PubMed] [Google Scholar]
- Hoshijima M, et al. Chronic suppression of heart-failure progression by a pseudophosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery. Nat Med. 2002;8:864–871. doi: 10.1038/nm739. [DOI] [PubMed] [Google Scholar]
- Hou Z, Kelly EM, Robia SL. Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex. J Biol Chem. 2008;283:28996–29003. doi: 10.1074/jbc.M804782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL. SERCA2a stimulation by istaroxime: a novel mechanism of action with translational implications. Br J Pharmacol. 2013;170:486–488. doi: 10.1111/bph.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huke S, Periasamy M. Phosphorylation-status of phospholamban and calsequestrin modifies their affinity towards commonly used antibodies. J Mol Cell Cardiol. 2004;37:795–799. doi: 10.1016/j.yjmcc.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Iwanaga Y, et al. Chronic phospholamban inhibition prevents progressive cardiac dysfunction and pathological remodeling after infarction in rats. J Clin Invest. 2004;113:727–736. doi: 10.1172/JCI18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James ZM, McCaffrey JE, Torgersen KD, Karim CB, Thomas DD. Protein-protein interactions in calcium transport regulation probed by saturation transfer electron paramagnetic resonance. Biophys J. 2012;103:1370–1378. doi: 10.1016/j.bpj.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup M, et al. Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2 + -ATPase in patients with advanced heart failure. Circulation. 2011;124:304–313. doi: 10.1161/CIRCULATIONAHA.111.022889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Loukianov E, Loukianova T, Jones LR, Periasamy M. SERCA2a1a can functionally substitute for SERCA2a in the heart. Am J Physiol. 1999;276:H89–97. doi: 10.1152/ajpheart.1999.276.1.H89. [DOI] [PubMed] [Google Scholar]
- Karim CB, Stamm JD, Karim J, Jones LR, Thomas DD. Cysteine reactivity and oligomeric structures of phospholamban and its mutants. Biochemistry. 1998;37:12074–12081. doi: 10.1021/bi980642n. [DOI] [PubMed] [Google Scholar]
- Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA2a. J Mol Biol. 2006;358:1032–1040. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Kaye DM, et al. Percutaneous cardiac recirculation-mediated gene transfer of an inhibitory phospholamban peptide reverses advanced heart failure in large animals. J Am Coll Cardiol. 2007;50:253–260. doi: 10.1016/j.jacc.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Kelly EM, Hou Z, Bossuyt J, Bers DM, Robia SL. Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells. J Biol Chem. 2008;283:12202–12211. doi: 10.1074/jbc.M707590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, et al. Istaroxime, a first in class new chemical entity exhibiting SERCA2a-2 activation and Na-K-ATPase inhibition: a new promising treatment for acute heart failure syndromes. Heart Fail Rev. 2009;14:277–287. doi: 10.1007/s10741-009-9136-z. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Kurzydlowski K, Tada M, MacLennan DH. Phospholamban inhibitory function is activated by depolymerization. J Biol Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- Kranias EG, Hajjar RJ. Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res. 2012;110:1646–1660. doi: 10.1161/CIRCRESAHA.111.259754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Boschek CB, Xiong Y, Sacksteder CA, Squier TC, Bigelow DJ. Essential role for Pro21 in phospholamban for optimal inhibition of the Ca-ATPase. Biochemistry. 2005;44:16181–16191. doi: 10.1021/bi051075o. [DOI] [PubMed] [Google Scholar]
- Li J, James ZM, Dong X, Karim CB, Thomas DD. Structural and functional dynamics of an integral membrane protein complex modulated by lipid headgroup charge. J Mol Biol. 2012;418:379–389. doi: 10.1016/j.jmb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, et al. Heart research advances using database search engines. Human Protein Atlas and the Sydney Heart Bank. Heart Lung Circ. 2013;22:819–826. doi: 10.1016/j.hlc.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Lockamy EL, Cornea RL, Karim CB, Thomas DD. Functional and physical competition between phospholamban and its mutants provides insight into the molecular mechanism of gene therapy for heart failure. Biochem Biophys Res Commun. 2011;408:388–392. doi: 10.1016/j.bbrc.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyer X, et al. Cardiomyocyte overexpression of neuronal nitric oxide synthase delays transition toward heart failure in response to pressure overload by preserving calcium cycling. Circulation. 2008;117:3187–3198. doi: 10.1161/CIRCULATIONAHA.107.741702. [DOI] [PubMed] [Google Scholar]
- MacLennan DH, Toyofuku T, Kimura Y. Sites of regulatory interaction between calcium ATPases and phospholamban. Basic Res Cardiol. 1997;92(Suppl 1):11–15. doi: 10.1007/BF00794063. [DOI] [PubMed] [Google Scholar]
- Mandinov L, Eberli FR, Seiler C, Hess OM. Diastolic heart failure. Cardiovasc Res. 2000;45:813–825. doi: 10.1016/S0008-6363(99)00399-5. [DOI] [PubMed] [Google Scholar]
- Marks AR. Calcium cycling proteins and heart failure: Mechanisms and therapeutics. J Clin Invest. 2013;123:46–52. doi: 10.1172/JCI62834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson LR, Yu T, Shi L, Wang Y, Gustavsson M, Mueller MM, Veglia G. cAMP-dependent protein kinase A selects the excited state of the membrane substrate phospholamban. J Mol Biol. 2011;412:155–164. doi: 10.1016/j.jmb.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer EJ, Huckle W, Johnson RG, Jr, McKenna E. Characterization and quantitation of phospholamban and its phosphorylation state using antibodies. Biochem Biophys Res Commun. 2000;267:40–48. doi: 10.1006/bbrc.1999.1920. [DOI] [PubMed] [Google Scholar]
- Medeiros A, et al. Mutations in the human phospholamban gene in patients with heart failure. Am Heart J. 2011;162(1088–1095):e1081. doi: 10.1016/j.ahj.2011.07.028. [DOI] [PubMed] [Google Scholar]
- Metcalfe EE, Zamoon J, Thomas DD, Veglia G. (1)H/(15)N heteronuclear NMR spectroscopy shows four dynamic domains for phospholamban reconstituted in dodecylphosphocholine micelles. Biophys J. 2004;87:1205–1214. doi: 10.1529/biophysj.103.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe EE, Traaseth NJ, Veglia G. Serine 16 phosphorylation induces an order-to-disorder transition in monomeric phospholamban. Biochemistry. 2005;44:4386–4396. doi: 10.1021/bi047571e. [DOI] [PubMed] [Google Scholar]
- Mills GD, Kubo H, Harris DM, Berretta RM, Piacentino V, 3rd, Houser SR. Phosphorylation of phospholamban at threonine-17 reduces cardiac adrenergic contractile responsiveness in chronic pressure overload-induced hypertrophy. Am J Physiol Heart Circ Physiol. 2006;291:H61–70. doi: 10.1152/ajpheart.01353.2005. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, et al. Heart failure-inducible gene therapy targeting protein phosphatase 1 prevents progressive left ventricular remodeling. PLoS ONE. 2012;7:e35875. doi: 10.1371/journal.pone.0035875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Karim CB, Negrashov IV, Kutchai H, Thomas DD. Direct detection of phospholamban and sarcoplasmic reticulum Ca-ATPase interaction in membranes using fluorescence resonance energy transfer. Biochemistry. 2004;43:8754–8765. doi: 10.1021/bi049732k. [DOI] [PubMed] [Google Scholar]
- Nesmelov YE, Karim CB, Song L, Fajer PG, Thomas DD. Rotational dynamics of phospholamban determined by multifrequency electron paramagnetic resonance. Biophys J. 2007;93:2805–2812. doi: 10.1529/biophysj.107.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JG, et al. Decoy peptides targeted to protein phosphatase 1 inhibit dephosphorylation of phospholamban in cardiomyocytes. J Mol Cell Cardiol. 2013;56:63–71. doi: 10.1016/j.yjmcc.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Pritchard TJ, et al. Active inhibitor-1 maintains protein hyper-phosphorylation in aging hearts and halts remodeling in failing hearts. PLoS ONE. 2013;8:e80717. doi: 10.1371/journal.pone.0080717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue K. mRNA quantitation techniques: Considerations for experimental design and application. J Nutr. 1998;128:2038–2044. doi: 10.1093/jn/128.11.2038. [DOI] [PubMed] [Google Scholar]
- Rocchetti M, et al. Modulation of sarcoplasmic reticulum function by PST2744 [istaroxime; (E, Z)-3-((2-aminoethoxy)imino) androstane-6,17-dione hydrochloride)] in a pressure-overload heart failure model. J Pharmacol Exp Ther. 2008;326:957–965. doi: 10.1124/jpet.108.138701. [DOI] [PubMed] [Google Scholar]
- Santana LF, Kranias EG, Lederer WJ. Calcium sparks and excitation-contraction coupling in phospholamban-deficient mouse ventricular myocytes. J Physiol. 1997;503(Pt 1):21–29. doi: 10.1111/j.1469-7793.1997.021bi.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Edes I, Kranias EG. Phospholamban: A promising therapeutic target in heart failure. Cardiovasc Drugs Ther. 2001;15:387–396. doi: 10.1023/A:1013381204658. [DOI] [PubMed] [Google Scholar]
- Schmitt JP, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- Simmerman HK, Kobayashi YM, Autry JM, Jones LR. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol. 2009;67:6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- Steenaart NA, Ganim JR, Di Salvo J, Kranias EG. The phospholamban phosphatase associated with cardiac sarcoplasmic reticulum is a type 1 enzyme. Arch Biochem Biophys. 1992;293:17–24. doi: 10.1016/0003-9861(92)90359-5. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Kurzydlowski K, Tada M, MacLennan DH. Amino acids Glu2 to Ile18 in the cytoplasmic domain of phospholamban are essential for functional association with the Ca(2+)-ATPase of sarcoplasmic reticulum. J Biol Chem. 1994;269:3088–3094. [PubMed] [Google Scholar]
- Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G. Spectroscopic validation of the pentameric structure of phospholamban. Proc Natl Acad Sci U S A. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci U S A. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieber CA, Douglas JL, Afara M, Young HS. The effects of mutation on the regulatory properties of phospholamban in co-reconstituted membranes. Biochemistry. 2005;44:3289–3297. doi: 10.1021/bi047878d. [DOI] [PubMed] [Google Scholar]
- Trieber CA, Afara M, Young HS. Effects of phospholamban transmembrane mutants on the calcium affinity, maximal activity, and cooperativity of the sarcoplasmic reticulum calcium pump. Biochemistry. 2009;48:9287–9296. doi: 10.1021/bi900852m. [DOI] [PubMed] [Google Scholar]
- Tsuji T, et al. Rescue of Ca2+ overload-induced left ventricular dysfunction by targeted ablation of phospholamban. Am J Physiol Heart Circ Physiol. 2009;296:H310–317. doi: 10.1152/ajpheart.00975.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov VV, Mote KR, Verardi R, Veglia G. Structural dynamics and topology of phosphorylated phospholamban homopentamer reveal its role in the regulation of calcium transport. Structure. 2013;21:2119–2130. doi: 10.1016/j.str.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Goldhaber JI. Return of calcium: Manipulating intracellular calcium to prevent cardiac pathologies. Proc Natl Acad Sci U S A. 2004;101:5697–5698. doi: 10.1073/pnas.0401518101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener AD, Simmerman HK, Lindemann JP, Jones LR. Phospholamban phosphorylation in intact ventricles. Phosphorylation of serine 16 and threonine 17 in response to beta-adrenergic stimulation. J Biol Chem. 1989;264:11468–11474. [PubMed] [Google Scholar]
- Xu XL, Ji H, Gu SY, Shao Q, Huang QJ, Cheng YP. Modification of alterations in cardiac function and sarcoplasmic reticulum by astragaloside IV in myocardial injury in vivo. Eur J Pharmacol. 2007;568:203–212. doi: 10.1016/j.ejphar.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Yu X, Lorigan GA. Secondary structure, backbone dynamics, and structural topology of phospholamban and its phosphorylated and Arg9Cys-mutated forms in phospholipid bilayers utilizing 13C and 15 N solid-state NMR spectroscopy. J Phys Chem B. 2014;118:2124–2133. doi: 10.1021/jp500316s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoon J, Mascioni A, Thomas DD, Veglia G. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys J. 2003;85:2589–2598. doi: 10.1016/S0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lewis D, Strock C, Inesi G, Nakasako M, Nomura H, Toyoshima C. Detailed characterization of the cooperative mechanism of Ca(2+) binding and catalytic activation in the Ca(2+) transport (SERCA2a) ATPase. Biochemistry. 2000;39:8758–8767. doi: 10.1021/bi000185m. [DOI] [PubMed] [Google Scholar]
- Zhang DW, et al. Astragaloside IV alleviates hypoxia/reoxygenation-induced neonatal rat cardiomyocyte injury via the protein kinase a pathway. Pharmacology. 2012;90:95–101. doi: 10.1159/000339476. [DOI] [PubMed] [Google Scholar]
- Zhao W, et al. Combined phospholamban ablation and SERCA2a1a overexpression result in a new hyperdynamic cardiac state. Cardiovasc Res. 2003;57:71–81. doi: 10.1016/S0008-6363(02)00609-0. [DOI] [PubMed] [Google Scholar]
- Ziolo MT, Martin JL, Bossuyt J, Bers DM, Pogwizd SM. Adenoviral gene transfer of mutant phospholamban rescues contractile dysfunction in failing rabbit myocytes with relatively preserved SERCA2a function. Circ Res. 2005;96:815–817. doi: 10.1161/01.RES.0000163981.97262.3b. [DOI] [PubMed] [Google Scholar]