Abstract
Purpose
Mutations in the NYX gene are known to cause complete congenital stationary night blindness (CSNB1), which is always accompanied by high myopia. In this study, we aimed to investigate the association between NYX mutations and high myopia with or without CSNB1.
Methods
Four Chinese families having high myopia with or without CSNB1 and 96 normal controls were recruited. We searched for mutations in the NYX gene using Sanger sequencing. Further analyses of the detected variations in the available family members were performed, and the frequencies of the detected variations in 96 normal controls were determined to verify our deduction. The effect of each variation on the nyctalopin protein was predicted using online tools.
Results
Four potential pathogenic variations in the NYX gene were found in four families with high myopia with or without CSNB1. Three of the four variants were novel (c.626G>C; c.121delG; c.335T>C). The previously identified variant, c.529_530delGCinsAT, was found in an isolated highly myopic patient and an affected brother, but the other affected brother did not carry the same variation. Further linkage analyses of this family showed a coinheritance of markers at MYP1. These four mutations were not identified in the 96 normal controls.
Conclusions
Our study expands the mutation spectrum of NYX for cases of high myopia with CSNB1; however, more evidence is needed to elucidate the pathogenic effects of NYX on isolated high myopia.
Introduction
Myopia is one of the most prevalent ocular disorders [1-3] and a major cause of low vision worldwide [4]. Myopia affects 50%–70% of the population in certain urban areas of East Asia [2,5-7], and it is expected to increase in prevalence [1,8,9]. High myopia is defined as a refractive error above −6.0 D, with an axial eyeball length above 26 mm. To date, the true underlying basis of high myopia remains unclear, though development of the disorder has been attributed to both environmental and genetic factors [10,11]. However, although numerous molecular genetic studies have identified over 40 genes as candidate genes for myopia [11-28], there is no single gene that has been consistently found to be a crucially pathogenic factor for myopia worldwide.
The majority of cases of myopia are non-syndromic. However, high myopia is commonly accompanied by other eye disorders, such as Stickler syndrome, Marfan syndrome, Cohen syndrome, Knobloch syndrome, or complete congenital stationary night blindness (CSNB1). Congenital stationary night blindness (CSNB) is a genetically and clinically heterogeneous disorder. Multiple inheritance patterns have been recognized in this disease, including autosomal dominant, autosomal recessive, and X-linked recessive [29]. CSNB has been commonly divided into two types, the Riggs and the Schubert-Bornschein, based on negative electroretinogram (ERG) waveforms. For the Riggs type, rod function is diminished whereas cone function is normal [30]. A shaped, dark-adapted ERG response to a bright flash can be detected in the Schubert-Bornschein type [31]. Furthermore, we classified the Schubert-Bornschein type into two sub-types: complete CSNB (cCSNB or CSNB1) with the complete absence of rod-pathway function and incomplete CSNB (icCSNB or CSNB2) with abnormal rod- and cone-pathway functions [31]. Patients having CSNB1 always demonstrate impaired night vision, strabismus, nystagmus, and high myopia beginning in early childhood. The NYX gene (OMIM 300278), which encodes the protein nyctalopin that belongs to a small leucine-rich repeat (LRR) protein family and is detected in the inner and outer plexiform layers [32], has been identified as a gene responsible for X-linked CSNB1. Previous studies have indicated that the nob gene in mice, which is a classical model for CSNB1 [33,34], and the NYX gene in humans are orthologs [35], which further indicated that NYX is associated with CSNB1. Recently, two studies indicated that the NYX gene was associated with isolated high myopia without CSNB1 [36,37]. These findings suggest that the NYX gene may have a vital effect on isolated myopia. In this study, we recruited four families with high myopia with or without CSNB1 as well as 96 normal controls to investigate the association between NYX mutations and high myopia with or without CSNB1.
Methods
Subjects
We recruited four families having high myopia (refractive error < −6.00 DS) with or without CSNB1 and 96 healthy individuals (+0.5 DS < refractive error < −0.5 DS). All individuals received a comprehensive ophthalmic examination, including vision acuity (Topcon KR-8000, Paramus, Japan), color vision, slit-lamp(SL-1E, Topcon, Japan), axial length (IOL master V5.0, Carl Zeiss Meditec AG, German), power corneal curvature, full-field ERG (ESPION-E2, Diagnosys, Littleton, MA) and fundus examination (CNAN-CR-2, Japan), by the same experienced ophthalmologists. The inclusion criteria for the participants in this study were as follows: 1) myopia occurred before school age and 2) spherical refraction < −6.00 DS. Patients having eye disorders other than nystagmus, strabismus, and night blindness or having systemic diseases were excluded. Written informed consent conforming to the tenets of the Declaration of Helsinki was obtained from each participating individual or his or her guardian before the collection of clinical data and venous blood.
Mutation screening
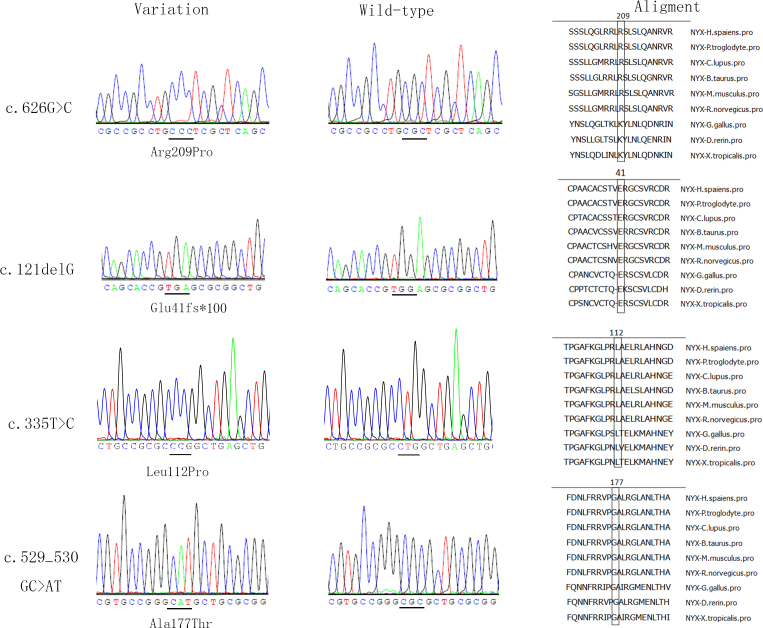
Genomic DNA was extracted from venous blood leukocytes by the phenol/chloroform method. Variations in NYX were detected in all of the recruited families using Sanger sequencing. Further analyses of the detected variations in the available family members and the analysis of the 96 normal controls were then performed to verify our deduction. PCR was used to amplify the coding sequence and the adjacent intronic sequence (NCBI: NC_000023.11, NM_022567). We designed primers using Primer3 (Table 1). Touchdown PCR was performed as follows: 1) 95 °C for 5 min for denaturation; 2) 35 cycles of amplification at 95 °C for 30 s, at 64–58 °C for 30 s (starting from 64 °C and decreasing by 0.5 °C per cycle for 14 cycles and then remaining at 58 °C for 21 cycles), and at 72 °C for 40 s; and 3) 72 °C for 10 min for the final extension. A cycle sequencing kit (ABI BigDye Terminator cycle sequencing kit v3.1, Applied Biosystems) and an ABI3100 Genetic Analyzer (Applied Biosystems) were used for sequencing and electrophoresis, respectively. The final sequences were compared with NYX consensus sequences from the NCBI database using the DNASTAR software (Madison, WI). The descriptions of the variations followed the nomenclature recommendations (HGVS). Polymorphism Phenotyping (Polyphen-2), the Sorting Intolerant From Tolerant Program (SIFT), Condel, and Provean were used to predict the functional consequences of the mutations on the encoded nyctalopin protein (Table 2). The MegAlign program of the DNASTAR package was used to analyze the degree of evolutionary conservation at the amino acid positions altered by the identified mutations (Figure 1).
Table 1. Primers used for the amplification and sequencing of the NYX gene.
| Exon | Direction | Primer sequence (5′-3′) | Size of amplified fragment (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| E1 |
F |
TGGGGAGCTTCTGATTTTCTGTTG |
443 |
58 |
| R |
ATCCCCACCACCTGCTGTTTTCTT |
|||
| E2A |
F |
GCGGGTGTCTTAGGTGGATA |
472 |
58 |
| R |
GCGTGATGAAGGACAGGTTG |
|||
| E2B |
F |
GACCTTTGGCTGACGGTTG |
756 |
58 |
| R |
TTGTCGTTGAGCAGCAGATG |
|||
| E2C |
F |
CTTCGACAACCTGTTCCGC |
559 |
58 |
| R |
CTCCATCCAGTCCCTCAGC |
|||
| E2D |
F |
CTCTACCTGGACCGCAACA |
689 |
58 |
| R | TTTCACCTCTGCCCTCCATT |
F: forward sequence; R: reverse sequence. The primer sequences, sizes of PCR products, and the annealing temperatures used for the amplification are listed. Five pairs of primers were used to amplify and sequence the entire NYX coding sequence.
Table 2. Variations in the NYX gene detected in 4 families with high myopia with or without CSNB.
| Position | Nucleotide change | Amino acid change | State | Computational prediction |
|||
|---|---|---|---|---|---|---|---|
| Polyphen | Condel | Provean | SIFT | ||||
| chrX-41333332 |
c.626G>C |
p.Arg209Pro, |
hemi |
PrD |
N |
D |
D |
| chrX-41332827 |
c.121delG |
pGlu41Sfs*100, |
hemi |
||||
| chrX-41333041 |
c.335T>C |
p.Leu112Pro, |
hemi |
PrD |
D |
D |
D |
| chrX-41333235_236 | 529_530delGCinsAT | p.Ala177Thr, | hemi | PrD | D | N | D |
Abbreviations: Hemi: hemizygote; PrD: probably damaging; D: deleterious; N: neutral.
Figure 1.
Four NYX variants detected in four highly myopic families with or without CSNB1. This figure shows the sequences of the four probands alongside the normal control sequence, as well as the amino acid sequence alignment of these four variants.
Genotyping and linkage analysis
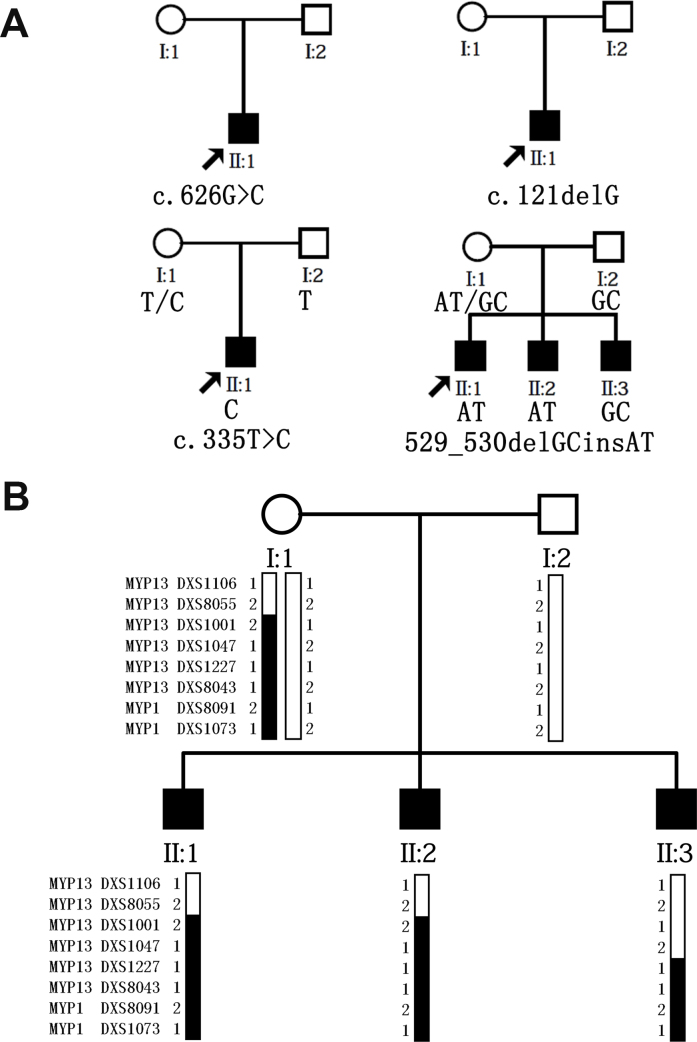
5′-Fluorescently labeled microsatellite markers were used to genotype the family in which the mutations did not co-segregate with high myopia with or without CSNB1. To clarify the myopia loci of this family, polymorphic microsatellite markers covering MYP1 and MYP13 were used in the linkage analysis. Eight markers spaced at intervals of approximately 10 cm (Applied Biosystems, Foster City, CA) were used for the genotyping, and an X-chromosome linkage scan was performed as previously described [38]. Haplotypes were generated using the Cyrillic 2.1 program and were confirmed by inspection (Figure 2).
Figure 2.
Four pedigree charts and a haplotype diagram of the recruited families. A: Pedigree chart of the four families and the co-segregation analysis of the families with the c.529–530delGCinsAT and c.335T>C mutations. B: Family structure and haplotype diagram of the family with the c.529–530delGCinsAT mutation. Solid squares represent the affected family members, and blackened bars indicate disease alleles.
Results
Four families met the criteria of high myopia with or without CSNB1. We detected four hemizygotic variations in the four families, three of which were novel: c.626G>C p. (Arg209Pro), c.121delG p. (Glu41fs*100), and c.335T>C p. (Leu112Pro). One mutation had previously been reported: c.529_530delGCinsAT (p.Ala177Thr) [38]. These detected variations were not found in the 96 normal controls. Further analyses were performed for the families with the c.335T>C and to c.529_530delGCinsAT mutations. However, the proband and an affected male exhibited the previously reported mutation to c.529_530delGCinsAT, whereas another high myopic brother did not carry this mutation. A genotyping and linkage analysis was performed to clarify the pathogenic loci for high myopia in this family (Figure 2). Sequence variations were based on NM_022567.2 for the coding sequence and NP_072089.1 for the amino acid sequence. All detected variants were predicted to be functionally detrimental and occurred in highly conserved regions (Figure 1).
The missense mutation c.626G>C p. (Arg209Pro), which was predicted to be damaging by Polyphen-2, Provean, and SIFT, was detected in a seven-year-old boy. At the age of five, he exhibited spherical equivalents of −9.0 D (OD) and −10.0 D (OS; Table 3). His guardians said it was difficult for him to walk in a dim environment. Typical changes of high myopia and a “tigroid” appearance of the posterior retina and optic nerve head crescent were detected in the fundus of the boy (Figure 3). During an examination by an experienced ophthalmologist, horizontal, continuous, oblique, pendular, and dysconjugate eye movements typical of nystagmus were also observed. Furthermore, an absent dark-adapted, rod-mediated b-wave response, a deficient electronegative configuration of the combined rod response, absent scotopic oscillatory potentials, and an abnormal response of the cones were detected in the boy. These signs indicated that he had CSNB1 with high myopia (Figure 4).
Table 3. Clinical information of eight subjects with the mutations in our study.
| Mutation | Gender | Age (years) at |
Spherical refraction (diopters) |
Axial length (mm) |
BCVA |
ERG responses |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| exam | onset | OD | OS | OD | OS | OD | OS | rod | cone | ||
| c.626G>C |
M |
5 |
EC |
−9.00 |
−10.00 |
25.28 |
24.93 |
0.3 |
0.2 |
absent |
diminished |
| c.121delG |
M |
31 |
EC |
−16.00 |
−9.50 |
29.87 |
27.66 |
0.2 |
0.4 |
absent |
diminished |
| c.335T>C |
M |
11 |
EC |
−10.00 |
−11.00 |
N/A |
N/A |
0.5 |
0.5 |
N/A |
N/A |
| c.529–530GC>AT |
M |
15 |
EC |
−21.00 |
−20.00 |
30.71 |
30.38 |
0.5 |
0.6 |
normal |
SD |
| c.529–530GC>AT I1 |
M |
37 |
EC |
−5.50 |
−5.25 |
24.39 |
24.37 |
1.0 |
1.0 |
normal |
normal |
| c.529–530GC>AT I2 |
F |
39 |
None |
−0.25 |
−0.12 |
23.88 |
24.01 |
1.0 |
1.0 |
normal |
normal |
| c.529–530GC>AT II2 |
M |
13 |
EC |
−7.00 |
−4.50 |
26.12 |
25.18 |
0.6 |
1.0 |
normal |
SD |
| c.529–530GC>AT II3 | M | 9 | EC | −11.37 | −7.50 | 27.93 | 26.95 | 0.2 | 0.6 | normal | SD |
Abbreviations: BCVA: best corrected visual acuity; ERG: electrophysiology; EC: early childhood; M: male; F: female; OD: right eye; OS: left eye; SD: slightly diminished; N/A: not available.
Figure 3.
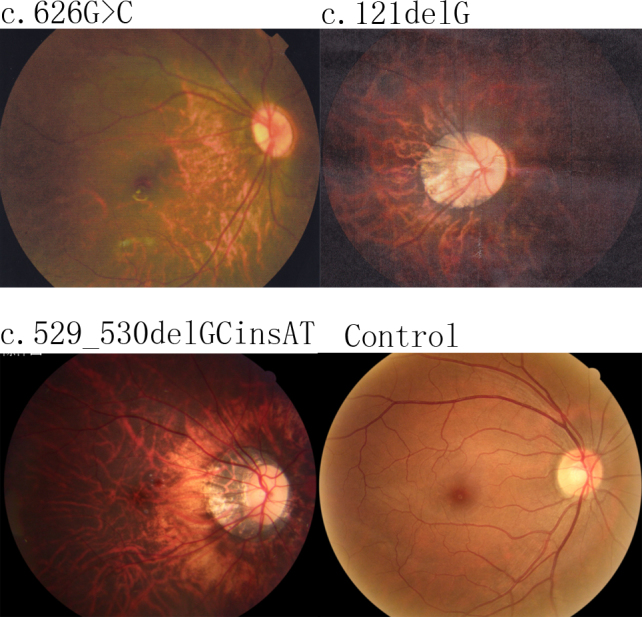
Fundus photography results for the right eyes of the probands carrying the c.626G>C, c.121delG, and c.529–530delGCinsAT variants and of a normal control. Typical changes that occur due to high myopia, including the “tigroid” appearance of the posterior retina, are shown; the optic nerve head crescent is shown in all of the fundus photographs of the probands.
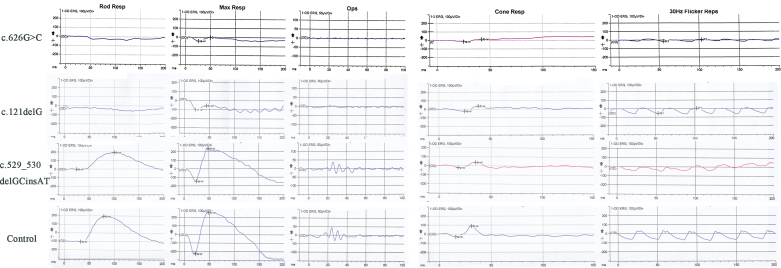
Figure 4.
ERG results for the c.626G>C, c.121delG, and to c.529–530delGCinsAT probands and for a normal control. The subjects with the c.626G>C and c.121delG mutations show a complete absence of rod pathway function and a diminished cone waveform. The subject with the c.529–530delGCinsAT mutation shows normal rod pathway function and a slightly diminished cone waveform.
The novel mutation c.121delG p. (p.Glu41fs*100) is a frameshift change that was found in a 31-year-old female having high myopia, with spherical equivalents of −16.00 D (OD) and −9.50 D (OS). Her corrected visual acuity was only 0.2 (OD) and 0.4 (OS), and she had worn glasses since early childhood. Not only did she have difficulty walking in dim environments, but she also displayed obvious symptoms of nystagmus, i.e., horizontal, oblique, pendular, jerky, and dysconjugate eye movements. From the ERG, we could detect an extinguished, dark-adapted b-wave response, a diminished electronegative mixed rod-cone response, decreased scotopic oscillatory potentials, and a mildly reduced amplitude of the cone response, which was typical of CSNB1 (Figure 4). Taken together, the novel mutations c.626G>C p. (Arg209Pro) and c.121delG p. (p.Glu41fs*100) were detected in two highly myopic patients with CSNB1.
The c.335T>C p. (Leu112Pro) variation is a missense mutation found in a 14-year-old boy who carried spherical equivalents of –9.00 D (OD) and −10.00 D (OS) when he was eleven. A funduscopic observation by an experienced ophthalmologist revealed myopia fundus changes typical of high myopia. However, given the incomplete clinical data, we could not determine whether he had CSNB1. Through a telephone interview, we learned that the unaided vision of his parents was normal and that they did not have night blindness. This NYX mutation co-segregated with high myopia in our analysis (Figure 2), and c.335T>C p. (Leu112Pro) was predicted to be damaging by SIFT, Condel, Provean, and Polyphen-2.
As well, to c.529_530delGCinsAT p. (Ala177Thr) was previously reported to affect the development of isolated high myopia [38]. This mutation was detected in a 15-year-old boy from an X-linked, recessive, high myopia family who exhibited spherical equivalents of −21.0 D (OD) and −20.0 D (OS). A “tigroid” appearance of the posterior retina and optic nerve head crescent were detected in the fundus of the boy (Figure 3). We did not observe symptoms of nystagmus or strabismus or other signs of CSNB1. The boy’s ERG showed a normal rod response and a slightly diminished cone waveform. It must be noted that the dark-adapted b-wave response, electronegative mixed rod-cone response, and scotopic oscillatory potentials were within normal ranges. The ERG of this boy was completely different from a patient with CSNB1. It was easy for him to play games and walk alone in dim light, and his family indicated no signs of night blindness. After careful examination of his family members, we found that two of the boy’s brothers exhibited the same symptoms: high myopia, normal rod function, and no signs of nyctalopia (Table 3). In summary, we found a proband with the to c.529–530delGCinsAT variant in brothers who were diagnosed with isolated high myopia. This mutation was predicted by Polyphen-2, Condel, and SIFT to be damaging. However, after the co-segregation analysis of this family, we found that a brother of the proband had isolated high myopia but did not carry this mutation. As the loci of X-linked myopia had been mapped to Xq28 and Xq23–27.2, called MYP1 and MYP13, respectively, we performed a linkage analysis of MYP1 and MYP13 for this family (Figure 2). It was conserved between DXS1227 and DXS1073 in the haplotype analysis, which was present in the males with high myopia and the unaffected female carrier. That is, the high myopia in this family might be mapped to MYP1.
As high myopia is the most common ocular disorder in human beings and the etiology is heterogeneous, the pathogenesis of this disorder may differ among the three affected siblings. Therefore, the high myopia of another brother of the proband, who does not have the c.529_530delGCinsAT to mutation, might be caused by other reasons. However, we could not neglect the possible role of MYP1. Taken together, a further analysis is needed to illuminate the cause of high myopia in this family, and the role of NYX in this case of isolated high myopia is ambiguous.
Discussion
In this study, two novel NYX mutations (c.626G>C and c.121delG) were detected in two highly myopic families with CSNB1. Another novel mutation (c.335T>C) was found in a boy with high myopia who may or may not have CSNB1, and a fourth previously reported mutation (c.529_530delGCinsAT) [37] was detected in a patient with isolated high myopia. All four mutations were predicted to be detrimental by online algorithms, and the affected amino acids were conserved across nine species. A further analysis was conducted of the available members of these four families. The c.335T>C mutation co-segregated with high myopia with or without CSNB1, but an affected brother did not carry the same mutation as the proband; instead, this brother had a c.529_530delGCinsAT to mutation in the NYX gene. Overall, three mutations were found in highly myopic families with or without CSNB1. However, we did not obtain enough evidence to conclude that NYX plays an independent role in isolated high myopia.
The NYX gene is located on chromosome Xp11.4 (OMIM 300278) and encodes the 481-amino acid nyctalopin protein, which contains 11 consecutive LRRs [39]. Currently, 59 mutations detected in NYX have been associated with CSNB1, and most of these mutations are located in the LRRs. The c.626G>C, c.121delG, c.335T>C, and to c.529_530delGCinsAT mutations are located in the regions encoding the sixth, first, third, and fifth LRRs of nyctalopin, respectively. Furthermore, these four affected amino acids are conserved in several species. Although the exact function of nyctalopin is unknown, these four mutations in the LRRs may play important roles in CSNB1. Recently, nyctalopin has been reported to interact with TRPM1 and GRM6, where it plays a significant role in mediating protein–protein interactions, e.g., in retinal processing to transmit the biochemical signal from photoreceptors to bipolar cells [40-42]. CSNB1 patients with NYX mutations have functional deficits in synaptic transmission that can be detected using full-field ERG. The clinical diagnosis of CSNB1 is typically based on an ERG with a reduced rod waveform. The patients carrying the c.626G>C and c.121delG mutations presented ERGs consistent with the typical ERG associated with CSNB1. As for the slightly diminished cone waveform, several studies showed that the cone-mediated ON pathway is associated with myopia [36,37]. The clinical data of the participants carrying the c.626G>C and c.121delG variants were in accord with those of highly myopic patients with CSNB1. Furthermore, although c.626G>C was a missense mutation and c.121delG was a truncation mutation, we did not find much difference in the severities of their phenotypes. Our data therefore provide additional data indicating the complexity of the relationship between NYX mutations, high myopia, and CSNB1, and it can be useful as we continue to develop our understanding of the structure–function relationships of nyctalopin. Two recent studies have reported that the c.144C>G, c.572_573delGCinsAA, and c.529_530delGCinsAT mutations had vital effects on isolated high myopia [36,37]. The small indel mutation to c.529–530delGCinsAT was also found in one of our families. The patients in this family were diagnosed with isolated high myopia; however, we found that the proband and one affected brother had the c.529_530delGCinsAT mutation, while another high myopic brother did not carry the same mutation. As previous studies identified two loci (MYP1 and MYP13) associated with X-linked high myopia [19,28,43,44], we performed a genotyping and linkage study on these two loci and found that the high myopia of this family may map to MYP1. There was no doubt that the etiology of high myopia in this study was heterogeneous, resulting in different family members in one high myopic family developing the disease due to different causes. In this family, the affected brother carrying the different mutation might also carry another mutation in another gene that causes high myopia. Generally speaking, we could not confirm that the high myopia in this family was associated with NYX or MYP1, and the deduction that NYX is a candidate gene for isolated high myopia requires more evidence to be confirmed.
Taken together, we expanded the mutation spectrum of NYX for high-myopic patients with CSNB1, but more studies are needed to elucidate the association between isolated high myopia and the NYX gene. Our data therefore provide additional evidence concerning the complex relationship among NYX mutations, high myopia, and CSNB1, and it will be useful as we continue to develop our understanding of the structure–function relationships of nyctalopin.
Acknowledgments
We thank the family members for actively participating in our study. We also thank Qingjiong Zhang, department of Ophthalmic Genetics & Molecular Biology, Zhongshan Ophthalmic Center, Sun Yat-sen University, for his helpful guidance. The study was supported by Hubei Province's Outstanding Medical Academic Leader program; grant 81,360,154 from the National Nature Science Foundation of China; and grant 2013KF04 from the open project of the State Key Laboratory of Ophthalmology.
References
- 1.He M, Zheng Y, Xiang F. Prevalence of myopia in urban and rural children in mainland China. Optom Vis Sci. 2009;86:40–4. doi: 10.1097/OPX.0b013e3181940719. [DOI] [PubMed] [Google Scholar]
- 2.Sawada A, Tomidokoro A, Araie M, Iwase A, Yamamoto T, Tajimi Study Group Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology. 2008;115:363–73. doi: 10.1016/j.ophtha.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 3.Wong TY, Foster PJ, Hee J, Ng TP, Tielsch JM, Chew SJ, Johnson GJ, Seah SK. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Invest Ophthalmol Vis Sci. 2000;41:2486–94. [PubMed] [Google Scholar]
- 4.Sliva R. Myopia maculopathy: a review. Opthalmologica. 2012;228:197–213. doi: 10.1159/000339893. [DOI] [PubMed] [Google Scholar]
- 5.Fan DS, Lam DS, Lam RF, Lau JT, Chong KS, Cheung EY, Lai RY, Chew SJ. Prevalence, incidence, and progression of myopia of school children in Hong Kong. Invest Ophthalmol Vis Sci. 2004;45:1071–5. doi: 10.1167/iovs.03-1151. [DOI] [PubMed] [Google Scholar]
- 6.He M, Zeng J, Liu Y, Xu J, Pokharel GP, Ellwein LB. Refractive error and visual impairment in urban children in southern China. Invest Ophthalmol Vis Sci. 2004;45:793–9. doi: 10.1167/iovs.03-1051. [DOI] [PubMed] [Google Scholar]
- 7.Matsumura H, Hirai H. Prevalence of myopia and refractive changes in students from 3 to 17 years of age. Surv Ophthalmol. 1999;44:S109–15. doi: 10.1016/s0039-6257(99)00094-6. [DOI] [PubMed] [Google Scholar]
- 8.Bloom RI, Friedman IB, Chuck RS. Increasing rates of myopia: the long view. Curr Opin Ophthalmol. 2010;21:247–8. doi: 10.1097/ICU.0b013e328339f1dd. [DOI] [PubMed] [Google Scholar]
- 9.Bar Dayan Y, Levin A, Morad Y, Grotto I, Ben-David R, Goldberg A, Onn E, Avni I, Levi Y, Benyamini OG. The changing prevalence of myopia in young adults: a 13-year series of population-based prevalence surveys. Invest Ophthalmol Vis Sci. 2005;46:2760–5. doi: 10.1167/iovs.04-0260. [DOI] [PubMed] [Google Scholar]
- 10.Ip JM, Huynh SC, Robaei D, Rose KA, Morgan IG, Smith W, Kifley A, Mitchell P. Ethnic differences in the impact of parental myopia: findings from a population-based study of 12-year-old Australian children. Invest Ophthalmol Vis Sci. 2007;48:2520–8. doi: 10.1167/iovs.06-0716. [DOI] [PubMed] [Google Scholar]
- 11.Lam CY, Tam PO, Fan DS, Fan BJ, Wang DY, Lee CW, Pang CP, Lam DS. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 12.Hawthorne FA, Young TL. Genetic contributions to myopic refractive error: insights from human studies and supporting evidence from animal models. Exp Eye Res. 2013;114:141–9. doi: 10.1016/j.exer.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–6. [PubMed] [Google Scholar]
- 14.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oettingm WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–24. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naiglin L, Gazagne C, Dallongeville F, Thalamas C, Idder A, Rascol O, Malecaze F, Calvas P. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002;39:118–24. doi: 10.1136/jmg.39.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–6. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–60. [PubMed] [Google Scholar]
- 18.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–7. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23-q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007;13:229–36. [PMC free article] [PubMed] [Google Scholar]
- 21.Nishizaki R, Ota M, Inoko H, Meguro A, Shiota T, Okada E, Mok J, Oka A, Ohno S, Mizuki N. New susceptibility locus for high myopia is linked to the uromodulin-like 1 (UMODL1) gene region on chromosome 21q22.3. Eye (Lond) 2009;23:222–9. doi: 10.1038/eye.2008.152. [DOI] [PubMed] [Google Scholar]
- 22.Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes T, Ciner E, Bailey-Wilson JE. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet. 2004;75:448–59. doi: 10.1086/423789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wojciechowski R, Moy C, Ciner E, Ibay G, Reider L, Bailey-Wilson JE, Stambolian D. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–99. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008;32:454–63. doi: 10.1002/gepi.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nürnberg G, Jacobi FK, Broghammer M, Becker C, Blin N, Nurnberg P, Stephani U, Pusch CM. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008;21:429–38. [PubMed] [Google Scholar]
- 27.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Confirmation of linkage to ocular refraction on chromosome 22q and identification of a novel linkage region on 1q. Arch Ophthalmol. 2007;125:80–5. doi: 10.1001/archopht.125.1.80. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Li S, Xiao X, Jia X, Guo X. Confirmation of a genetic locus for X-linked recessive high myopia outside MYP1. J Hum Genet. 2007;52:469–72. doi: 10.1007/s10038-007-0130-9. [DOI] [PubMed] [Google Scholar]
- 29.Xiao X, Jia X, Guo X, Li S, Yang Z, Zhang Q. CSNB1 in Chinese families associated with novel mutations in NYX. J Hum Genet. 2006;51:634–40. doi: 10.1007/s10038-006-0406-5. [DOI] [PubMed] [Google Scholar]
- 30.Riggs LA. Electroretinography. Vision Res. 1986;26:1443–59. doi: 10.1016/0042-6989(86)90167-7. [DOI] [PubMed] [Google Scholar]
- 31.Schubert G, Bornschein H. Analysis of the human electroretinogram. Ophthalmologica. 1952;123:396–413. doi: 10.1159/000301211. [DOI] [PubMed] [Google Scholar]
- 32.Morgans CW, Ren G, Akileswaran L. Localization of nyctalopin in the mammalian retina. Eur J Neurosci. 2006;23:1163–71. doi: 10.1111/j.1460-9568.2006.04647.x. [DOI] [PubMed] [Google Scholar]
- 33.Pardue MT, McCall MA, Lavail MM, Gregg RG, McCall MA, Peachey NS. A naturally occurring mouse model of X-linked congenital stationary night blindness. Invest Ophthalmol Vis Sci. 1998;39:2443–9. [PubMed] [Google Scholar]
- 34.Gregg RG, Mukhopadhyay S, Candille SI, Ball SL, Pardue MT, McCall MA, Peachey NS. Identification of the gene and the mutation responsible for the mouse nob phenotype. Invest Ophthalmol Vis Sci. 2003;44:378–84. doi: 10.1167/iovs.02-0501. [DOI] [PubMed] [Google Scholar]
- 35.Candille SI, Pardue MT, McCall MA, Peachey NS, Gregg RG. Localization of the mouse nob (no b-wave) gene to the centromeric region of the X chromosome. Invest Ophthalmol Vis Sci. 1999;40:2748–51. [PubMed] [Google Scholar]
- 36.Zhang Q, Xiao X, Li S, Jia X, Yang Z, Huang S, Caruso RC, Guan T, Sergeev X, Hejtmancik JF. Mutations in NYX of indiviuals with high myopia, but without night blindness. Mol Vis. 2007;13:330–6. [PMC free article] [PubMed] [Google Scholar]
- 37.Yip SP, Li CC, Yiu WC, Hung WH, Lam WW, Lai PW, Fung WY, Chu PH, Jiang B, Chan HH, Yap MK. A novel missense mutation in the NYX gene associated with high myopia. Ophthalmic Physiol Opt. 2013;33:346–53. doi: 10.1111/opo.12036. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–60. [PubMed] [Google Scholar]
- 39.Bech-Hansen NT, Naylor MJ, Maybaum TA, Sparkes RL, Koop B, Birch DG, Bergen AA, Prinsen CF, Polomeno RC, Gal A, Drack AV, Musarella MA, Jacobson SG, Young RS, Weleber RG. Mutations in NYX, encoding the leucine rich proteoglycan nyctalopin, cause X -linked complete congenital stationary night blindness. Nat Genet. 2000;26:319–23. doi: 10.1038/81619. [DOI] [PubMed] [Google Scholar]
- 40.Gregg RG, Kamermans M, Klooster J, Lukasiewicz PD, Peachey NS, Vessey KA, McCall MA. Nyctalopin expression in retinal bipolar cells restores visual function in a mouse model of complete X-linkedcongenital stationary night blindness. J Neurophysiol. 2007;98:3023–33. doi: 10.1152/jn.00608.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leroy BP, Budde BS, Wittmer M, De Baere E, Berger W, Zeitz C. A common NYX mutation in Flemish patients with X linked CSNB. Br J Ophthalmol. 2009;93:692–6. doi: 10.1136/bjo.2008.143727. [DOI] [PubMed] [Google Scholar]
- 42.Pearring JN, Bojang P, Jr, Shen Y, Koike C, Furukawa T, Nawy S, Gregg RG. A role for nyctalopin, a small leucine-rich repeat protein, in localizing the TRP melastatin 1 channel to retinaldepolarizing bipolar cell dendrites. J Neurosci. 2011;31:10060–6. doi: 10.1523/JNEUROSCI.1014-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young TL, Deeb SS, Ronan SM, Dewan AT, Alvear AB, Scavello GS, Paluru PC, Brott MS, Hayashi T, Holleschau AM, Benegas N, Schwartz M, Atwood LD, Oetting WS, Rosenberg T, Motulsky AG, King RA. X-linked high myopia associated with cone dysfunction. Arch Ophthalmol. 2004;122:897–908. doi: 10.1001/archopht.122.6.897. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease: linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–6. [PubMed] [Google Scholar]