Summary
Advanced basal cell carcinomas (BCCs) frequently acquire resistance to Smoothened (SMO) inhibitors through unknown mechanisms. Here, we identify SMO mutations in 50% (22/44) of resistant BCCs and show that these mutations maintain Hedgehog signaling in the presence of SMO inhibitors. Alterations include four ligand binding pocket mutations defining sites of inhibitor binding and four variants confering constitutive activity and inhibitor resistance, illuminating pivotal residues that ensure receptor autoinhibition. In the presence of a SMO inhibitor, tumor cells containing either class of SMO mutants effectively outcompete cells containing the wild type SMO. Finally, we show that both classes of SMO variants respond to aPKC-ι/λ or GLI2 inhibitors that operate downstream of SMO, setting the stage for the clinical use of GLI antagonists.
Introduction
Uncontrolled activation of the Hedgehog (HH) pathway drives tumor progression in a number of cancers, including basal cell, medulloblastoma, pancreatic, colon, lung, breast, prostate, and blood (Amakye et al., 2013). Normally, HH ligand activates the pathway by binding to and inhibiting the receptor Patched1 (PTCH1), derepressing G-protein–coupled receptor (GPCR) Smoothened (SMO) and activating the GLI transcription factors. In oncogenic contexts, loss of PTCH1 and mutagenic activation of SMO are the most common alterations that induce inappropriate activation of the HH pathway. Basal cell carcinomas (BCCs) represent the most common cancer in the United States with approximately two million new cases per year (Rogers et al., 2010). Advanced BCCs, a small but significant proportion of total BCCs, lead to functional impairment, invasiveness, metastasis, and increased mortality. HH pathway antagonists are under development to combat HH-driven cancers with most therapies directed at inhibiting SMO. Like other heptahelical transmembrane proteins (7-TM), SMO is believed to be autoinhibited in its baseline state through both interactions with a PTCH1-dependent mechanism and through an unidentified ligand binding in its ligand binding pocket (LBP). All current pathway inhibitors targeting SMO bind the LBP and stabilize the autoinhibited state, although the details of these interactions remain unexplored.
As part of the Stanford BCC Consortium, we have enrolled and treated patients for advanced BCCs that led to the approval of the SMO inhibitor vismodegib by the FDA for treatment of advanced/inoperable and metastatic BCCs (Sekulic et al., 2012; Tang et al., 2012). All syndromic BCCs in patients with Basal Cell Nevus Syndrome (Gorlin, caused by inherited PTCH1 loss) respond to vismodegib and have a low rate of acquired resistance (Tang et al., 2012). In contrast, advanced and metastatic BCCs have an overall response rate of 48% (Axelson et al., 2013; Sekulic et al., 2012) with an additional 20% of patients developing resistance during the first year (Chang and Oro, 2012). Vismodegib and other SMO inhibitors have also shown promising results in early clinical trials for medulloblastoma (Gajjar et al., 2013). Despite these successes, many tumors acquire clinical resistance during therapy (Atwood et al., 2012), reinforcing the critical need to understand the basis of inherent resistance at the time of diagnosis and how these tumors evolve resistance during drug treatment. In contrast to visceral tumors, patients with advanced BCCs have low mortality and often develop multiple resistant tumors that are accessible to sequential biopsies (Atwood et al., 2012), providing a unique opportunity to assess spatially and temporally distinct clones during the evolutionary process using genomic tools.
Studies in mice and humans have provided initial insight into mechanisms of resistance to SMO inhibitor therapy. Specific to the HH pathway, germline loss of SUFU, which encodes a GLI inhibitor downstream of PTCH1, was shown to bestow primary resistance to vismodegib in pediatric patients with medulloblastoma (Kool et al., 2014). Additional mechanisms of acquired resistance found in medulloblastoma include amplification of GLI2 (Dijkgraaf et al., 2011), MYCN (Kool et al., 2014), CCND1 (Dijkgraaf et al., 2011), and a missense mutation in SMO (D473H) that confers resistance through disruption of vismodegib binding (Yauch et al., 2009). In BCCs, activation of the GLI kinase atypical Protein Kinase C ι/λ (aPKC-ι/λ) was found to be elevated in vismodegib-resistant tumors and aPKC-ι/λ inhibition in resistant cell lines suppressed growth (Atwood et al., 2013). However, HH-driven medulloblastomas have been shown to evade SMO inhibition by switching their oncogenic signaling pathway and thus losing its addiction to the HH pathway (Buonamici et al., 2010; Kool et al., 2014; Metcalfe et al., 2013). How BCCs evade SMO inhibition remains unknown.
Results
Hedgehog signaling is maintained in vismodegib resistant BCC
As each BCC regardless of patient origin arises from a distinct clone, we interrogated the nature of tumor resistance by sequencing 44 resistant BCCs from 15 patients. “Resistant BCCs” were defined as refractory to vismodegib (91%, 40/44 tumors) or recurrent (9%, 4/44) according to the NCI criteria. “Sensitive BCCs” were defined as BCCs that exhibited a partial or complete response to vismodegib treatment. Histology of resistant tumors was similar to sensitive tumors except for the absence of the superficial subtype (Figure 1A). All biopsies were obtained while patients were undergoing at least three months of continuous vismodegib therapy.
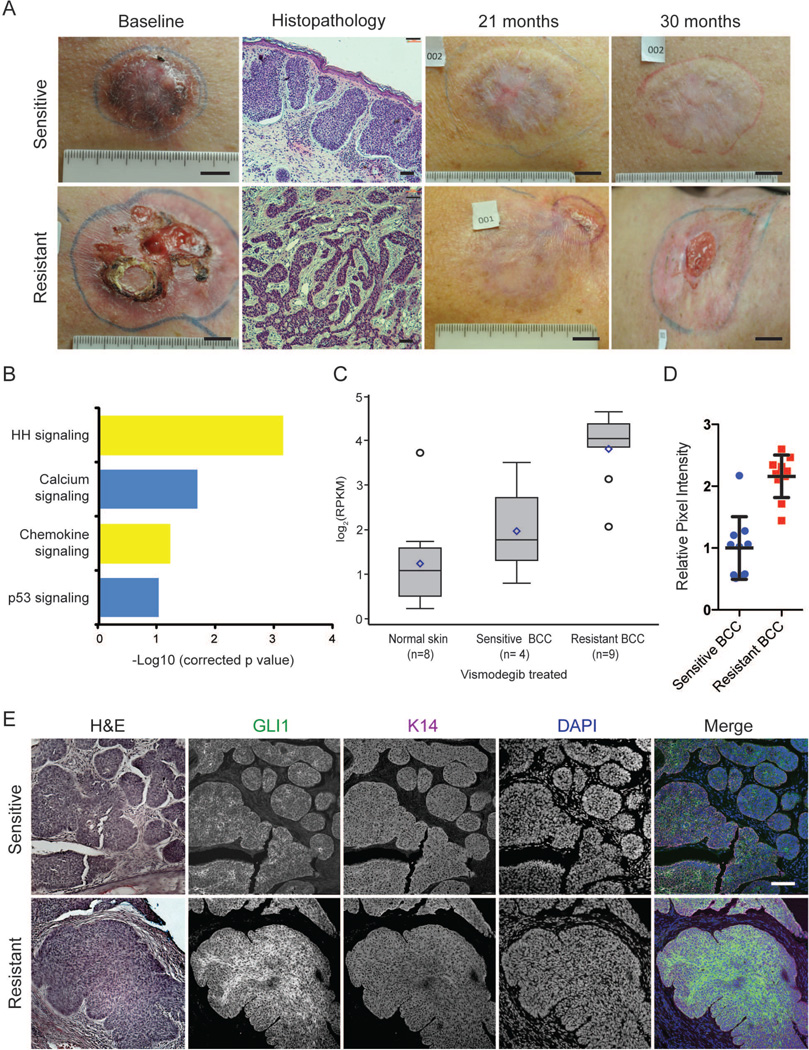
Figure 1. Hedgehog signaling is upregulated in resistant BCCs.
(A) Clinical photographs (scale bar=1 in) and histology (scale bar=100 µm) depicting the time course of a sensitive and a resistant BCC in the same patient during vismodegib therapy. (B) Pathway-driven gene set enrichment analysis (DAVID) in resistant BCCs as compared with sensitive BCCs and normal skin. (C) A box-plot representation comparing the log2 RPKM for GLI1 in resistant BCCs, sensitive BCCs, and normal skin (p=0.0001). Box represents first and third quartiles, with whiskers representing range; center line: median; diamond: mean; circle: outliers (D) Quantification of GLI1 immunofluorescence pixel intensity in Keratin 14 (K14)-positive regions (n=10). Error bars, s.e.m. (E) Representative immunofluorescence staining against GLI1 and K14, as well as DAPI counterstain. Adjacent sections stained with hematoxylin and eosin. Scale bar=100 µm.
Previous work on HH-driven medulloblastomas indicate that medulloblastomas can switch oncogenic pathways to continue tumor growth in the presence of SMO inhibition (Kool et al., 2014), but it is not known whether BCCs behave similarly. Using paired-end high throughput RNA sequencing and pathway analysis with DAVID (Huang da et al., 2009), we identified the HH signaling pathway as the most significantly enriched signaling pathway in resistant BCCs (n=9) as compared with sensitive BCCs (n=4) or normal skin (n=8; p=0.0007)(Figure 1B). Vismodegib sensitive BCCs had slightly elevated GLI1 (a HH target gene) mRNA levels compared to normal skin (Figure 1C). In contrast, resistant BCCs had high GLI1 mRNA levels despite concurrent treatment with vismodegib (p=0.0001). Immunofluorescence for GLI1 revealed elevated levels of GLI1 protein in resistant BCCs compared to sensitive tumors, confirming persistent HH signaling in resistant BCCs at the protein level (Figure 1D, E). These data suggest that resistant BCCs are still addicted to the HH pathway and that genetic alterations that maintain HH pathway output in the presence of vismodegib are the primary mechanisms of resistance.
Exome sequencing identifies recurrent SMO mutations in resistant BCC
As resistant BCCs rely on the HH pathway for continued growth, we wanted to identify the HH specific genetic alterations underlying resistance. We performed whole genome and exome sequencing on 14 resistant BCC tumors along with corresponding matching skin samples with a mean target coverage of 114× (Figure 2A). We identified a mean of 2,364 somatic coding mutations per BCC. While the non-silent SNV rate of 42 per Mb (range: 5–107 per Mb) is somewhat lower than previously reported in BCCs (Jayaraman et al., 2014), it supports the notion that skin cancers carry higher mutation rates than other non-cutaneous tumors. PTCH1 alterations, the most common driver of BCC growth, were detected in 57% (8/14) of samples. Given that TP53 mutations have been reported in BCCs, we investigated whether there was a correlation between TP53 and PTCH1 mutations. We found TP53 mutations in only 4/14 tumors and they had no correlation to the associated PTCH1 mutation (Figure 2B).
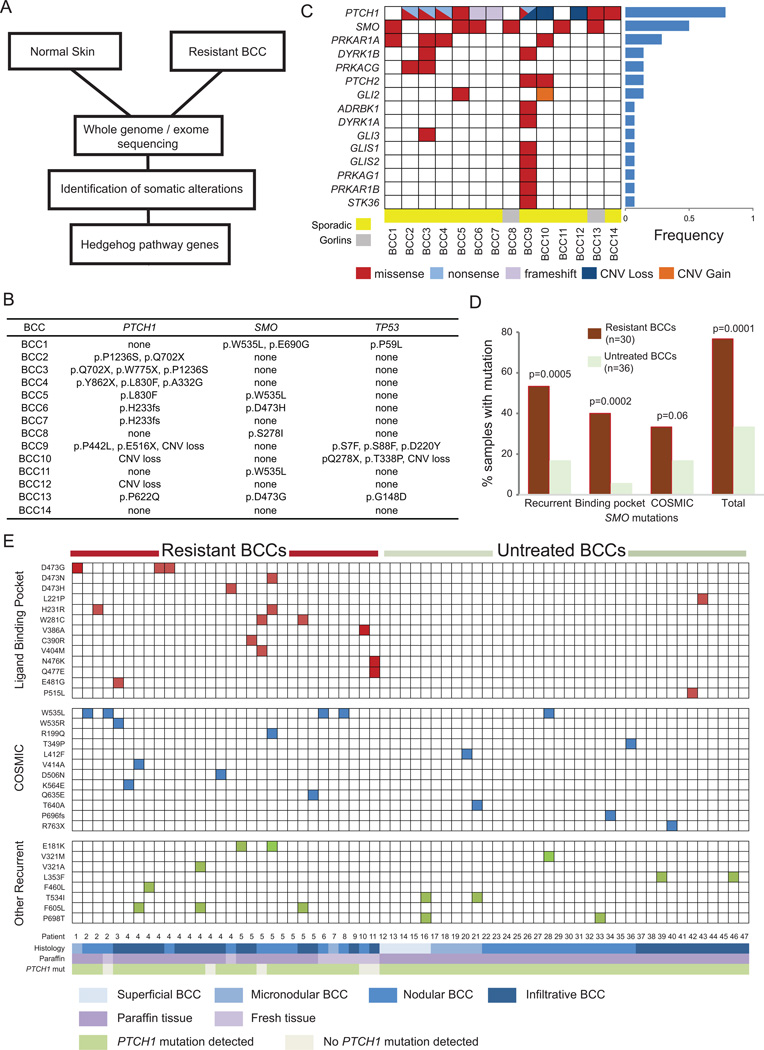
Figure 2. Resistant BCCs harbor recurrent SMO mutations.
(A) Schematic overview of the tumor biopsy and adjacent normal skin collection followed by whole-exome or genome sequencing and analysis. (B) List of SMO, PTCH1, and TP53 mutations identified for each resistant BCC samples subjected to exome sequencing. (C). Spectrum of HH pathway genes with genetic alterations seen in exome sequencing of resistant tumor-normal pairs. The genes are listed on the left hand side and the tumor samples are across the bottom. The fraction of samples with HH pathway mutations is listed in the bar graph to the right. (D) Bar graph showing recurrent, LBP, and COSMIC database SMO mutations in resistant BCCs compared with untreated samples. (E) Schematic showing SMO mutations in resistant BCCs as compared with untreated BCCs. SMO mutations are listed on the left hand side of each row and each column represents a unique sample with patient number and other relevant information listed at the bottom. The mutations are color-coded. Red color: a mutation in an amino acid located in the SMO-LBP. Blue color: a mutation also reported as somatically mutated in cancer in the COSMIC database. Green color: a recurrent mutation in neither the LBP nor the COSMIC database.
We next focused our analysis on genes downstream of PTCH1 that are implicated in HH signaling to assess where along the pathway resistance originates. We identified genetic alterations in 15/29 HH pathway genes including multiple regulatory units of the cAMP/Protein Kinase A signaling pathway and amplification of GLI2, which has previously shown to confer resistance against SMO antagonists in a medulloblastoma allograft model (Dijkgraaf et al., 2011) (Figure 2C). In fact, genetic alterations of the HH pathway downstream of PTCH1 were present in 85% of the resistant BCCs. Of these genes, SMO was the most recurrently mutated gene (42%, 6/14 samples). As one SMO mutation (D473H) was previously identified as a driver of resistance in a medulloblastoma patient (Yauch et al., 2009), we concentrated our efforts on SMO. Interestingly, we detected SMO D473H and D473G in two resistant BCCs originating from one sporadic tumor and one Gorlin’s patient, and W535L in another three resistant BCCs (Figure 2B). Also known as SMO-M2, W535L is a known oncogenic mutation present at low rates in sporadic BCCs and can drive tumor progression in the absence of PTCH1 loss (Xie et al., 1998). The genetic alterations in SMO were significantly more frequent than previously reported in BCCs (Reifenberger et al., 2005) suggesting SMO could be a key driver of BCC resistance.
SMO mutations are enriched in resistant BCCs compared with untreated BCCs In order to interrogate how SMO drives tumor resistance, we sequenced an additional 30 resistant BCCs along with 36 untreated, sporadic BCCs (Figure 2D, E). The coding regions of SMO and PTCH1 were amplified using the Fluidigm Access Array microfluidic device followed by next generation sequencing with a mean coverage of 2365× (+/− 755). This validation set did not have paired germline DNA, thus we cannot exclude the possibility that some of the genetic alterations are germline. However, non-pathogenic mutations present in dbSNP with a minimum allele frequency > 3% were excluded during our analysis. Overall, we detected heterozygous SNV mutations in SMO in 77% (23/30) of the resistant and 33% (12/36) of the untreated BCCs (p=0.0001), suggesting that genetic alterations in SMO may be the predominant mechanism by which tumors evade SMO inhibitor therapy.
Identification of ligand binding pocket mutations that confer vismodegib resistance
The recently reported crystal structure of SMO bound to LY2940680, a SMO inhibitor, revealed that vismodegib putatively binds at the extracellular end of the 7-TM bundle forming extensive contacts with the loops in the LBP (Wang et al., 2013). SMO D473 interacts with a water molecule in the LBP that may have an important role in the conformation of the pocket without directly contacting the inhibitor. We identified mutations at D473 in 17% of our resistant BCCs (5/30). In addition, we detected a significant number of genetic alterations affecting amino acids structurally positioned in the LBP of SMO. LBP mutations were detected in 40% (12/30) of resistant BCCs and 6% (2/36) of untreated BCCs (p=0.0002) (Figure 2D, E).
As D473 mutations are associated with resistance, we first wanted to test whether LBP mutations would confer resistance in BCC by becoming less sensitive to SMO inhibitors such as vismodegib. We expressed human wildtype SMO (SMO WT) or SMO-LBP mutants in Smo−/− mouse embryonic fibroblasts (MEFs) to assess the ability of these mutants to confer drug resistance to vismodegib. Using mRNA levels of the HH target gene Gli1 as a reporter for HH activity, the SMO-LBP mutants did not significantly alter basal HH activity (Figure 3A). However, in contrast to SMO WT, SMO-LBP mutants D473G, H231R, W281C, and Q477E retained high levels of HH activity in the presence of 100 nM vismodegib and SHH-N ligand without altering protein production. Surprisingly, while structural analysis indicates the V386 residue contacts LY2940680 and would be predicted to confer resistance (Wang et al., 2013) (Figure 3B), the V386A variant showed a response similar to SMO WT. This result suggests that vismodegib may bind slightly different residues than LY2940680, with distinct contact points within the SMO-LBP.
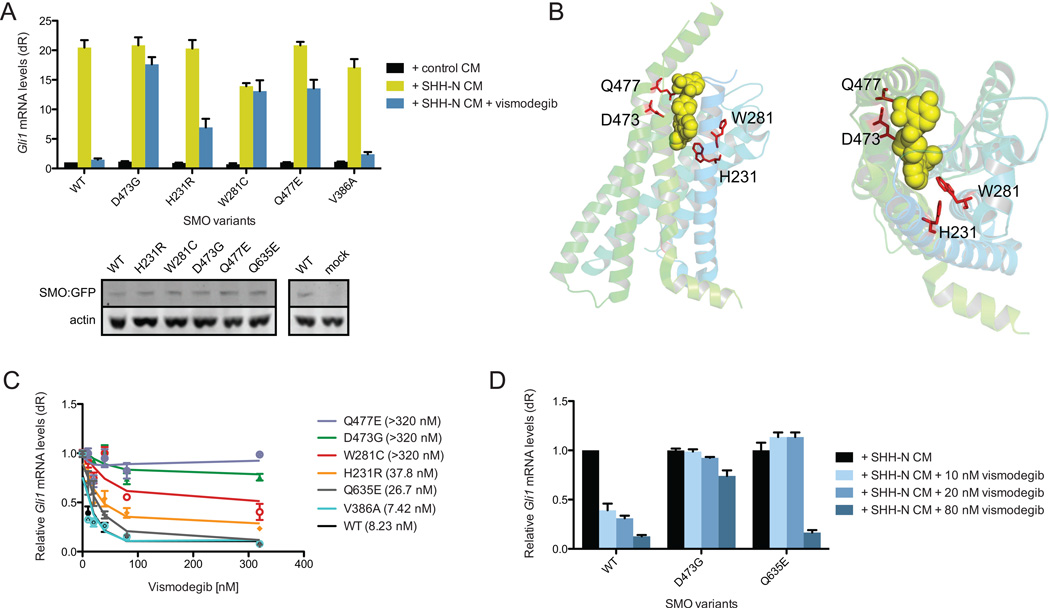
Figure 3. Variation in the responsiveness of SMO ligand binding pocket mutations.
(A) SMO variants expressed in Smo−/− MEFs and treated with SHH-N conditioned media (CM) with or without 100 nM vismodegib. Western blot shows the expression of SMO WT and SMO variants. (B) Side view (left) and top down view (right) of the position of the SMO variants within the SMO crystal structure showing their arrangement relative to an inhibitor (Wang et al., 2013). (C) Response of indicated SMO mutants with different concentrations of vismodegib. IC50 are shown in brackets. (D) HH pathway activity in Smo−/− MEFs expressing the indicated SMO and treated with SHH-N conditioned media (CM) with or without 10, 20, or 80 nM vismodegib. All error bars, s.e.m.
We then quantified the dose-response curve of each mutant to vismodegib. SMO WT and V386A had IC50 concentrations at 8.23 nM and 7.42 nM, respectively (Figure 3C). The rest of the SMO-LBP mutants segregated into two classes: moderate or high drug resistance. The IC50 of H231R (37.8 nM) was 4.5-fold higher compared to SMO WT, whereas D473G, W281C, and Q477E had IC50 concentrations greater than 320 nM (roughly 40-fold more than the IC50), which was the endpoint of our assay. Interestingly, the functional LBP mutants were only observed in resistant BCCs indicating that tumor cells expressing this class of mutants are selected for during therapy (Figure 2E). These experiments demonstrate SMO-LBP mutants present in resistant BCCs that functionally confer resistance to vismodegib while retaining normal regulatory control by PTCH1 and HH ligand.
Because the concentration of vismodegib in our initial screening assay was roughly 12-fold above the IC50 and data from our initial studies demonstrated that even small changes in IC50 appeared to provide a growth advantage, we assessed vismodegib sensitivity of recurrent SMO mutants and SMO mutations in the COSMIC database at low drug concentrations near the IC50 of 10 nM and 20 nM. Using this more sensitive assay, we identified Q635E as a mutant with resistance at low but not high vismodegib concentrations with an IC50 of 26.7 nM (Figure 3C, D). This data suggests that some SMO mutations may confer a partial reduction in vismodegib sensitivity that, in the appropriate context, could contribute to clinical resistance in BCC.
SMO mutations in structural pivot regions of the transmembrane helices confer constitutive activity and drug resistance
We detected a number of SMO mutations outside of the LBP and many were recurrent or detected in other solid tumors, including medulloblastoma, colon carcinoma, and glioma (COSMIC, Sanger Institute) (Forbes et al., 2011). COSMIC and recurrent mutations were found in 47% (14/30) of resistant as compared to 28% (10/36) of untreated BCCs (p=0.05)(Figure 2D, E). In support of this, we found constitutively active (CA) W535 mutants in only one (1/36) of our sporadic but in 5 (5/30) of our resistant tumors. Interestingly, residue W535 maps to transmembrane helix 7 of SMO and structurally aligns near pivot residues involved in activating class A GPCRs through structural conformations (Figure 4A, B) (Wang et al., 2013). 7-TM GPCRs maintain inactivity through multiple autoinhibitory interactions. While previous studies suggest SMO functions like other GPCRs (Ayers and Therond, 2010; Riobo et al., 2006), SMO possesses less than 10% sequence identity at the amino acid level. Moreover, in key domains thought to be pivot regions for activation, SMO lacks key prolines thought to allow transmembrane movement and G protein activation (Wang et al., 2013), bringing into question whether SMO functions similarly to other GPCRs.
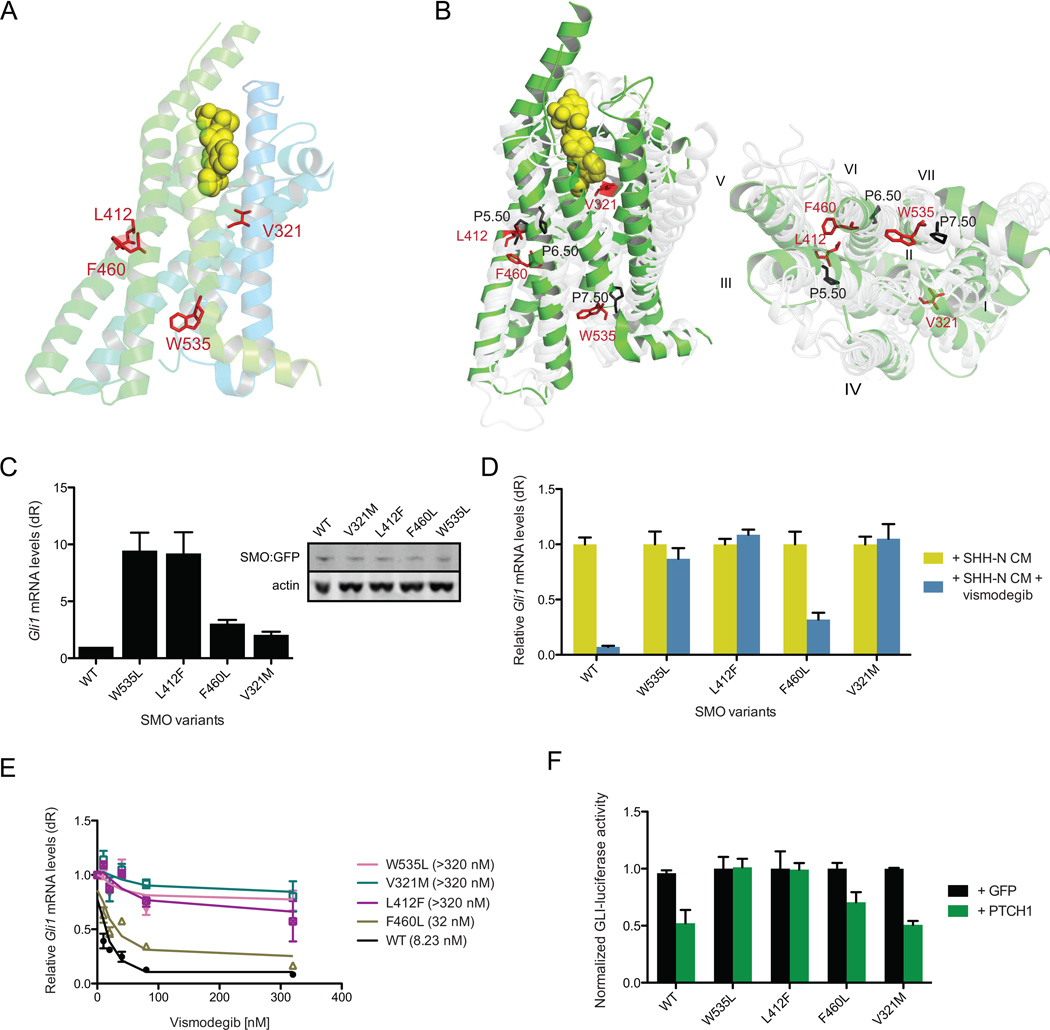
Figure 4. Two distinct mechanisms of SMO-mediated resistance in BCCs.
(A) Position of the SMO variants within the SMO crystal structure showing their arrangement relative to an inhibitor (Wang et al., 2013) in TM3 (V321), TM5 (F460), TM6 (L412F), and TM7 (W535L). (B) Side view (left) and top down view (right) of the overlay of pivot regions of B2 adrenergic receptor (grey) with those of SMO (green). Black numbers represent prolines in the B2 adrenergic receptor structure around which the lower receptor pivots. (C) Baseline HH pathway activity in Smo−/− MEFs under serum-starvation conditions expressing SMO WT or indicated SMO-CA variants. Western blot of expression of SMO WT compared to SMO variants. (D) HH pathway activity in Smo−/− MEFs expressing the indicated SMO treated with SHH-N CM with or without 100 nM vismodegib. (E) Response of indicated SMO with different concentrations of vismodegib. IC50 are shown in brackets. (F) Coexpression of SMO-CA variants and PTCH1 or GFP in a GLI-luciferase reporter assay. All error bars, s.e.m.
Intriguingly, functional studies of the B2 adrenergic receptor indicate the existence of key activating residues in pivot regions of transmembrane helices 3, 5, and 6 (Katritch et al., 2013). In the SMO crystal structure, these regions correspond to residues 320–340, 410–415, and 455–465, respectively. Several SMO mutations (V321M, L412F, and F460L) mapped to the pivot regions and led us to hypothesize that these residues may play critical roles in enabling conformational changes between active and inactive states (Figure 4A, B). Confirming our hypothesis, when we expressed these SMO mutants into Smo−/− MEFs, we observed constitutive HH activation in the absence of HH ligand without an increase in protein production (Figure 4C). These mutants were also partially or completely unresponsive to vismodegib suggesting these residues play an important role in the transmission of the inhibitory signal (Figure 4D). These mutants also separated into two classes of drug sensitivity with F460L moderately responding to vismodegib at an IC50 of 32 nM, whereas W535L, V321M, and L412F had IC50 concentrations greater than 320 nM, which was the endpoint of our assay (Figure 4E). In addition, these mutants displayed a range of PTCH1 inhibition states where high amounts of constitutive activity in GLI-luciferase assays correspond to strong resistance to PTCH1 inhibition, suggesting SMO-CA mutant activity is at least partly based on their ability to prevent PTCH1 catalytic signal (Figure 4F). Moreover, examination of the distribution of these CA and vismodegib resistant mutants revealed that they were found in both untreated and SMO inhibitor resistant tumors and were not paired with PTCH1 CNV loss or frame-shift mutations, suggesting this class of mutants drive initial BCC tumorigenesis and confer inherent resistance at the time of treatment (Figure 2B). These results identify SMO-CA mutants, in addition to W535L, that impart dual roles in tumorigenesis and acquired resistance, pointing to a class of mutations that would cause inherent resistance to SMO inhibition.
SMO mutations confer both intrinsic and acquired resistance to vismodegib
In order to better understand tumor evolution, we identified twelve resistant BCCs in which we had obtained paired pre-treatment biopsies and interrogated these samples for the presence of SMO mutations. Eight of the post-treatment samples had functionally-proven resistant SMO mutations (Figure 5A). Four of the resistant BCCs harbored either D473G or Q477E LBP mutations that were undetectable in the matched pre-treatment tumors. In fact, we were unable to detect any functionally validated LBP mutations in untreated BCCs (Figure 2E), suggesting that tumor cells expressing this class of mutant are selected for during therapy. Interestingly, one patient developed two spatially distinct resistant clones during treatment with vismodegib. Both clones arose from a single sporadic BCC that harbored the original PTCH1 H233fs driver mutation. One clone acquired a D473H LBP mutation whereas the second clone had no detectable SMO mutation, illustrating the heterogeneity of tumor evolution and acquired drug resistance (Figure 5B). In contrast, three pretreatment BCCs harbored subclones of the W535L allele that was then enriched in the post-treatment resistant BCCs (Figure 5A). Additionally, another resistant BCC acquired a S533N clone that is a putative SMO-CA variant and has been shown to cause medulloblastoma in mice (Dey et al., 2012). As SMO-CA mutants are present in both untreated and resistant tumors in the larger BCC cohort (Figure 2E), this suggests that SMO-CA mutants may confer intrinsic resistance prior to treatment and may represent a significant population in untreated samples. The apparent low allele fraction of the SMO-CA mutants prior to treatment may point to robust heterogeneity of tumor clones within these large advanced tumors that constantly compete and evolve with only the drug-resistant clones enriching upon drug treatment. Taken together, our results show 50% (22/44) of resistant BCCs harbor SMO mutations that are functionally shown to confer vismodegib resistance either through disruption of ligand responsiveness or release of autoinhibition (Figure 2E, 5C).
Figure 5. SMO mutations drive tumor evolution and drug resistance.
(A) Bar graph showing the allele fraction (red) of the indicated SMO-LBP or CA mutants in pre-treated (Pre) or treated and resistant (Post) BCCs. (B) A schema showing sequencing of two resistant clones arising from the same sporadic BCC under vismodegib selection. (C) Frequencies of BCC having functional SMO mutations shown to either impart constitutive activity or confer resistance to vismodegib. (D, E) Representative fluorescent images (D, scale bar 100 µm) and quantitation (E) of competition assay with stable ASZ001 BCC cell lines coexpressing SMO WT and mCherry or SMO variants and GFP with or without vismodegib. (F) HH pathway activity in Smo−/− MEFs expressing the indicated SMO variant and treated with SHH-N CM with or without 32 nM vismodegib, 20 µM PSI, or 8 µM ATO. All error bars, s.e.m.
SMO mutations impart a growth advantage in the presence of vismodegib
Our data suggests that tumor clones that can maintain high HH activation in the presence of SMO antagonist gain a selective growth advantage and become overrepresented within the tumor. To quantify the selective advantage of identified SMO mutations, we designed a red:green competition assay where ASZ001 BCC cells expressing SMO WT are marked with mCherry and those expressing SMO-LBP or SMO-CA mutants are marked by GFP to determine which alleles confer a growth advantage and outcompete the other in the presence of vismodegib. Interestingly, in the absence of selection SMO-D473G, W535L, L412F, and W281C grew at approximately the same rate as wild type SMO containing cells, presumably due to the high pathway activation already present due to PTCH1 loss. However, in the presence of vismodegib, tumor cells expressing any of the variants gained a significant growth advantage and outcompeted SMO WT-containing tumor cells, indicating that these SMO mutations can selectively grow during SMO antagonist therapy to cause drug resistance (Figure 5D, E).
HH antagonists downstream of SMO are effective in the presence of SMO variants
A subset of SMO variants have slightly elevated IC50 concentrations suggesting that higher SMO inhibitor concentrations may be therapeutically beneficial. However, many other variants do not significantly respond to drug even at high inhibitor concentrations. This led us to explore whether previously identified GLI antagonists that act downstream of SMO may be effective in suppressing the HH pathway in the presence of SMO inhibitor resistant variants. We expressed SMO-LBP and SMO-CA variants into Smo−/− MEFs and observed a loss of Gli1 mRNA in the presence of SHH-N ligand and inhibitor concentrations 4-fold greater than their respective IC50. As expected, all variants had partial or complete resistance to vismodegib (Figure 5F). However, the aPKC-ι/λ/GLI inhibitor PSI (Atwood et al., 2013) and the GLI2 antagonist ATO (Kim et al., 2013) were both effective at suppressing HH pathway activation in the presence of any SMO variant, suggesting that GLI antagonists may be useful against SMO inhibitor resistant tumors.
Discussion
The nature of acquired resistance in advanced BCCs has been largely unexplored despite skin tumors representing an easily accessible model system to study tumor evolution. We have made the surprising discovery that despite the ability of other cancers to feed on oncogenic signals originating from multiple pathways, BCCs exclusively rely on the HH pathway for growth. This unique property of BCCs allowed us to use SMO inhibitor-resistant tumors as a robust system to uncover how tumors evolve to bypass SMO inhibition and maintain high levels of HH activity. Our results indicate 50% of resistant BCCs operate under two distinct modes of resistance: disruption of ligand responsiveness and release of autoinhibition. In addition, HH antagonists downstream of SMO are effective at suppressing HH activation and may present viable therapies to treat resistant tumors.
Despite the high mutational load in BCCs that make it one of the most mutated human cancers (Jayaraman et al., 2014), a finding we confirm in our study, the inherently low rate of resistance to SMO antagonists are surprising. A likely reason may be the limited repertoire of variants that could confer pathway maintenance in the presence of vismodegib. While HH-dependent medulloblastomas use multiple signaling pathways for growth and differentiation (Metcalfe et al., 2013), we find that BCCs have an absolute dependence on the HH pathway. This limitation appears to reduce the chance that another mutation outside the HH pathway would cause drug resistance and may be the reason why we observe a high proportion of SMO mutations in resistant BCCs. Another reason may be that only one copy of SMO is required to transduce the active signal. Low selective pressure and slow growth potential of BCCs may naturally suppress any need to bypass SMO as the active signal transducer of the HH pathway.
Our data provides strong structural support that a conserved autoinhibitory mechanism exists in SMO despite less than 10% sequence identity at the amino acid level (Wang et al., 2013). This structural conservation allowed us to predict which SMO variants would confer resistance. We identified five SMO mutations that mostly decorate the LBP and conferred resistance to vismodegib in our assays. The functionally relevant mutations were not found in untreated BCCs presumably as they did not confer additional HH activity in the presence of ligand and thus would have no selective pressure, suggesting that LBP mutations are acquired after drug treatment. Moreover, as CA mutants would confer a growth advantage to BCCs in addition to resistance to therapy, it is not surprising that we find these inherently resistant variants in both untreated and resistant tumors. Consistent with this idea, we have found that non-advanced BCCs from Gorlin patients that contain PTCH1 mutations lack SMO mutations and respond to vismodegib (Sarin et al., in preparation).
In addition, SMO contains 7-TM alpha helices that act in concert to transduce activity with helices three, five, six, and seven having pivotal roles in activation of the receptor. W535L is a previously described CA mutant found on helix seven (Xie et al., 1998) and is believed to interact with helices five and six to prevent activation. We have found CA mutants on helix three (V321M), helix five (L412F), and helix six (F460L) that complement W535L. The proximity within the SMO structure of L412 and F460 suggest they interact to reinforce autoinhibition through helix five and six interaction. As V321 lies at the interface between the LBP and the autoinhibitory loops, we postulate that this residue may help LBP inhibition with helices five and six. Our study defines key interface residues for SMO activation that may be hot spots for resistance alleles in other HH-dependent cancers.
As a subset of functionally validated SMO variants is present in untreated BCCs, our data presents an opportunity for genetic prescreening to determine the optimal personal therapy to evade drug resistance. All functionally validated SMO variants operated on a spectrum of vismodegib sensitivity that point to two important criteria for treatment options. Tumors harboring mutations that partially suppress vismodigib sensitivity may be treatable with higher concentrations of drug to overcome their elevated IC50. We find that HH target gene expression can be effectively suppressed at higher drug concentrations but not at low concentrations. Tumors harboring mutations that show near complete vismodegib resistance may be better candidates for drugs that inhibit HH activity outside the SMO-LBP. In fact, we find that HH antagonists such as PSI, which targets the GLI kinase aPKC-ι/λ (Atwood et al., 2013), or ATO, which targets GLI2 (Kim et al., 2013), are quite effective at suppressing HH activation associated with any SMO variant. Other inhibitors that target at the level of GLI, such as the BET bromodomain family of chromatin modifiers or S6K1 that have been shown to function in medulloblastoma or esophageal adenocarcinoma respectively, may also be useful in counteracting resistance (Tang et al., 2014; Wang et al., 2012). Knowledge of the genetic alterations present in resistant BCCs improves our understanding of SMO structure and function, enables personalized therapy based on pre-existing mutations, and informs the development and application of future treatments.
Experimental Procedures
Case samples
After Stanford Human Subjects panel approval, written informed consent was obtained from patients 18 years or older with advanced BCCs for tumor sequencing (protocol #18325). BCCs were defined as resistant or sensitive to vismodegib therapy using the following criteria: 1) “Resistant BCC”: continuous treatment with vismodegib at therapeutic doses of 150 mg per day with stable disease or progressive disease as defined by the Response Evaluation Criteria in Solid Tumors (RECIST v. 1.1); 2) “Sensitive BCC”: partial or complete response to vismodegib therapy at doses of 150 mg per day as defined by RECIST.
RNA sequencing
RNA-sequencing was performed on 9 resistant BCCs, 4 sensitive BCCs and 8 normal skin biopsies. 2 ug of total RNA was extracted from tissue samples stored in RNA later using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA integrity was confirmed with the Agilent 2001 bioanalyzer. cDNA were prepared using the Ovation RNA-Seq System V2 (NuGen) per the manufacturer’s protocol. cDNA libraries were sheared by sonication (Covaris model S1) and purified using the Qiagen Minelute Kit. End repair was performed with T4 DNA Polymerase, T4 Polynucleotide Kinase, and Klenow DNA polymerase (New England Biolabs) at 20°C for 30 min and purified using the Qiagen Minelute Kit. dA-tailing was performed with Klenow fragment 3’ to 5’ exonuclease (New England Biolabs) at 37°C for 30 min and purified using the Qiagen Minelute Kit. Adapter ligation was performed with Illumina adapters and T4 DNA Ligase (New England Biolabs), purified with Qiagen Minelute Kit, and 150–400 basepair fragments were gel purified on a 3% GTG low melting point agarose gel. RNA-Seq libraries were PCR amplified 18 cycles with Phusion DNA polymerase (New England Biolabs), purified with Qiagen Minelute Kit, and size-selected on a 3% GTG low melting point agarose gel. RNA-Seq libraries were analyzed with the Agilent 2001 bioanalyzer and were sequenced paired-end at 100bp using an Illumina HiSeq 2500.
RNA-Seq reads were aligned to the human reference genome sequence (hg19) with TopHat. We obtained an average of 250 milliion reads per sample and 88% alignment to the human genome. Uniquely genomic and split-mapped reads were used to quantify the expression levels for GLI1. The National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) databases were used as reference annotations to calculate values of reads per kilobase of transcript per million mapped reads for known transcripts (RPKM) (Mortazavi et al., 2008). RPKM values were then log2-transformed and box plot analysis was used to visualize differential expression of GLI1 among the normal skin, sensitive BCC, and resistant BCC tissue samples. The general linear model (GLM) was used to assess statistical differences among the groups.
Whole-exome sequencing and analysis
Fresh tissue samples of 14 resistant BCCs and adjacent normal skin were obtained and stored in RNALater at −20°C (Ambion). DNA was isolated using the DNeasy Blood & Tissue kit according to manufacturer’s protocols (Qiagen). Capture libraries were constructed from 2 µg of DNA from BCC and normal skin using the Agilent SureSelect XT Human All Exon V4 kit according to manufacturer’s specifications. Enriched exome libraries were multiplexed and sequenced on the Illumina HiSeq 2500 platform to generate 100 basepair paired-end reads. Sequencing reads were aligned to the human reference genome sequence (hg19) using Burrows-Wheeler Aligner (BWA). SAM to BAM conversion and marking of PCR duplicates were performed using Picard tools (version 1.86), followed by local realignment around indels and base quality score recalibration using the Genome Analysis Toolkit (GATK) (v2.3.9). We obtained mean target coverage of 114× over the coding regions. Somatic SNVs and indels were called using both Samtools mpileup and GATK. Variants were annotated for standard quality metrics, for presence in dbSNP138, for presence in the National Heart, Lung, and Blood Institute exome sequencing project (ESP6500, Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/) and COSMIC database v.64. Variants were filtered if they did not result in a predicted change to the protein coding sequence. Genetic alterations were selected if they occurred in genes listed in the human HH signaling pathway in the KEGG database. To determine copy number variation for PTCH1 and TP53, read counts were calculated for each exon and scaled to 10 Million. Log2 (tumor/normal) was calculated and the average log2 (tumor/normal) was calculated for each tumor. A copy number gain or loss was called when the average log2 (tumor/normal) for a given gene was less than or greater than 0.35.
Targeted resequencing of SMO and PTCH1 in FFPE samples
5–8 10 µm sections were obtained from the FFPE tumor block and DNA was isolated using the Qiagen DNeasy Blood and Tissue kit according to manufacturer’s protocol (Qiagen). The exonic regions of PTCH1 and SMO were amplified using the Access Array platform (Fluidigm). The samples were amplified in a multiplex format with genomic DNA (100 ng) according to the manufacturer's recommendation (Ambry Genetics). Subsequently, the multiplexed library pools were subjected to deep sequencing using the Illumina MiSeq platform. After demultiplexing and FASTQ file generation for the raw data, 150 basepair reads were aligned to the human reference genome sequence (hg19) using the BWA aligner. Samtools mpileup was used to call variants. Only bases meeting the minimum base quality score of 20 from reads meeting the minimum mapping quality score of 20 were considered. A minimum allele frequency of 5% at a position with a read depth > 100 was required to make calls. Identified variants were annotated using SeattleSeq138 to exclude non-pathogenic variants reported in dbSNP138 and to identify variants that had nonsynonymous consequences or affected splice sites.
Hedgehog signaling assays
Variants were inserted into the full-length human SMO gene by standard mutagenesis and cloned into peGFP-C1 (Clontech). SMO variants were nucleofected (Amaxa) into Smo-null mouse embryonic fibroblasts and plated at confluence in DMEM + 10% FBS. To test for HH response, serum was removed after 48 hrs and SHH-N conditioned media was added. Cells were treated with or without varying concentrations of vismodegib (LC Labs), PSI (Atwood et al., 2013), or ATO (Sigma) for 48 hr and RNA was harvested using the RNeasy Minikit (Qiagen). Quantitative RT-PCR was performed using the Brilliant II SYBR Green QRT-PCR Master Mix Kit (Agilent Technologies) on a Mx3000P qPCR System (Agilent Technologies). Fold change in mRNA expression of the HH target gene Gli1 was measured using ΔΔCt analysis with Gapdh as an internal control gene. For luciferase assays, vectors expressing SMO variants and either PTCH1 or GFP were transfected (Fugene6, Promega) into CH310T1/2 cells with pGL3B 6xGliCS (Atwood et al., 2013) and serum-starved for 48 hrs. Cells were lysed and luciferase expression was determined using the Dual-Luciferase Assay System (Promega) and a TD-20/20 Luminometer. For competition assays, lentiviral pCDHGFP or pCDHmCherry (SBI) vector expressing SMO WT or SMO variants were used to generate stable lines in ASZ001 BCC cells using puromycin selection. Cells coexpressing SMO WT and GFP was mixed with cells coexpressing a SMO variant and mCherry and grown in the presence or absence of vismodegib for the specified amount of time. Live fluorescent images (Zeiss Axio Observer with 10× objective) of four random fields per competition assay per biological replicate were taken at each time point and the number of green and red cells counted to generate green-to-cherry ratios.
Statistical analysis
P values for the comparison of the log2 RPKM for GLI1 among the normal skin, resistant, and sensitive BCCs was calculated using the generalized linear model (GLM) test. P values for the percent of samples with SMO mutations in resistant BCCs as compared with the untreated BCCs was calculated using Z test for 2 population portions. P values were calculated using two-tailed comparisons.
Immunofluorescence staining
Immunofluorescence staining carried out using antibodies against GLI1 (1:100; R&D, AF3455) and Keratin 14 (1:500; Abcam, ab7800). Secondary antibodies were from Invitrogen. GLI1 staining quantified using pixel intensity measurements in Image J. Pixel intensity was measured in K14-positive regions in 10 fields for both sensitive and resistant BCCs. Adjacent tumor sections stained with hematoxylin and eosin.
Significance.
Advanced BCCs acquire resistance to SMO inhibitors through two distinct mechanisms that explain the majority of resistance in BCCs and structurally elucidate SMO-mediated Hedgehog signaling. These genetic alterations suggest that SMO functions similarly to other class A GPCRs despite less than 10% sequence identity. Furthermore, this work offers strategies tumors use to evade drug resistance prior to treatment and helps inform the development of second line therapies.
Acknowledgements
We wish to thank members of the Oro lab for guidance and F. deSauvage for sharing data prior to publication. The work was funded by the V Foundation Translational Award, NIAMS (5ARO54780, 2AR046786), NIH Pathway to Independence Award 1K99CA176847 (SXA), the Damon Runyon Clinical Investigator Award (JT), the American Skin Association Clinical Scholar Award (AC), the Stanford Cancer Institute Grant, and the Dermatology Foundation Career Development Award (KS). AO and AC are investigators in Genentech, Novartis, and Eli Lilly clinical trials. JT is a consultant to Genentech.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE58377.
Author Contributions
S.X.A., K.Y.S., A.E.O., and J.Y.T. designed experiments and wrote the manuscript. S.X.A. and K.Y.S. performed the majority of the experiments. S.X.A., K.Y.S., and J.R.L. performed and analyzed sequencing. K.Y.S., A.L.S.C., J.Y.T., M.R., M.S.A., and J.K. collected and annotated clinical samples. K.Y.S. and A.E.O. worked on structure modeling. R.J.W. performed immunofluorescence staining and aided in competition experiments. G.K. and M.R. aided in tumor DNA isolation. C.Y. aided in functional experiments.
References
- Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- Atwood SX, Chang AL, Oro AE. Hedgehog pathway inhibition and the race against tumor evolution. J Cell Biol. 2012;199:193–197. doi: 10.1083/jcb.201207140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature. 2013;494:484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelson M, Liu K, Jiang X, He K, Wang J, Zhao H, Kufrin D, Palmby T, Dong Z, Russell AM, et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin Cancer Res. 2013;19:2289–2293. doi: 10.1158/1078-0432.CCR-12-1956. [DOI] [PubMed] [Google Scholar]
- Ayers KL, Therond PP. Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends Cell Biol. 2010;20:287–298. doi: 10.1016/j.tcb.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AL, Oro AE. Initial assessment of tumor regrowth after vismodegib in advanced Basal cell carcinoma. Arch Dermatol. 2012;148:1324–1325. doi: 10.1001/archdermatol.2012.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey J, Ditzler S, Knoblaugh SE, Hatton BA, Schelter JM, Cleary MA, Mecham B, Rorke-Adams LB, Olson JM. A distinct Smoothened mutation causes severe cerebellar developmental defects and medulloblastoma in a novel transgenic mouse model. Molecular and cellular biology. 2012;32:4104–4115. doi: 10.1128/MCB.00862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkgraaf GJ, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, Burdick D, Goldsmith R, Robarge K, Sutherlin D, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71:435–444. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajjar A, Stewart CF, Ellison DW, Kaste S, Kun LE, Packer RJ, Goldman S, Chintagumpala M, Wallace D, Takebe N, et al. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: a pediatric brain tumor consortium study. Clin Cancer Res. 2013;19:6305–6312. doi: 10.1158/1078-0432.CCR-13-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009;Chapter 13(Unit 13):11. doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- Jayaraman SS, Rayhan DJ, Hazany S, Kolodney MS. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Invest Dermatol. 2014;134:213–220. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, Chen B, King EM, Borodovsky A, Riggins GJ, et al. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell. 2013;23:23–34. doi: 10.1016/j.ccr.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Alicke B, Crow A, Lamoureux M, Dijkgraaf GJ, Peale F, Gould SE, de Sauvage FJ. PTEN loss mitigates the response of medulloblastoma to Hedgehog pathway inhibition. Cancer Res. 2013;73:7034–7042. doi: 10.1158/0008-5472.CAN-13-1222. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Reifenberger J, Wolter M, Knobbe CB, Kohler B, Schonicke A, Scharwachter C, Kumar K, Blaschke B, Ruzicka T, Reifenberger G. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol. 2005;152:43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Saucy B, Dilizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A. 2006;103:12607–12612. doi: 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HW, Weinstock MA, Harris AR, Hinckley MR, Feldman SR, Fleischer AB, Coldiron BM. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch Dermatol. 2010;146:283–287. doi: 10.1001/archdermatol.2010.19. [DOI] [PubMed] [Google Scholar]
- Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JY, Mackay-Wiggan JM, Aszterbaum M, Yauch RL, Lindgren J, Chang K, Coppola C, Chanana AM, Marji J, Bickers DR, Epstein EH., Jr Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med. 2012;366:2180–2188. doi: 10.1056/NEJMoa1113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, Bergthold G, Masoud S, Nguyen B, Vue N, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–740. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding Q, Yen C-J, Xia W, Izzo JG, Lang J-Y, Li C-W, Hsu JL, Miller SA, Wang X, et al. The Crosstalk of mTOR/S6K1 and Hedgehog Pathways. Cancer Cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]