Abstract
Discrete Fourier transformations have recently been developed to model the evolution of two-state characters (the Cavender/Farris model). We report here the extension of these transformations to provide invertible relationships between a phylogenetic tree T (with three probability parameters of nucleotide substitution on each edge corresponding to Kimura's 3ST model) and the expected frequencies of the nucleotide patterns in the sequences. We refer to these relationships as spectral analysis. In either model with independent and identically distributed site substitutions, spectral analysis allows a global correction for all multiple substitutions (second- and higher-order interactions), independent of any particular tree. From these corrected data we use a least-squares selection procedure, the closest tree algorithm, to infer an evolutionary tree. Other selection criteria such as parsimony or compatibility analysis could also be used; each of these criteria will be statistically consistent for these models. The closest tree algorithm selects a unique best-fit phylogenetic tree together with independent edge length parameters for each edge. The method is illustrated with an analysis of some primate hemoglobin sequences.
Full text
PDF
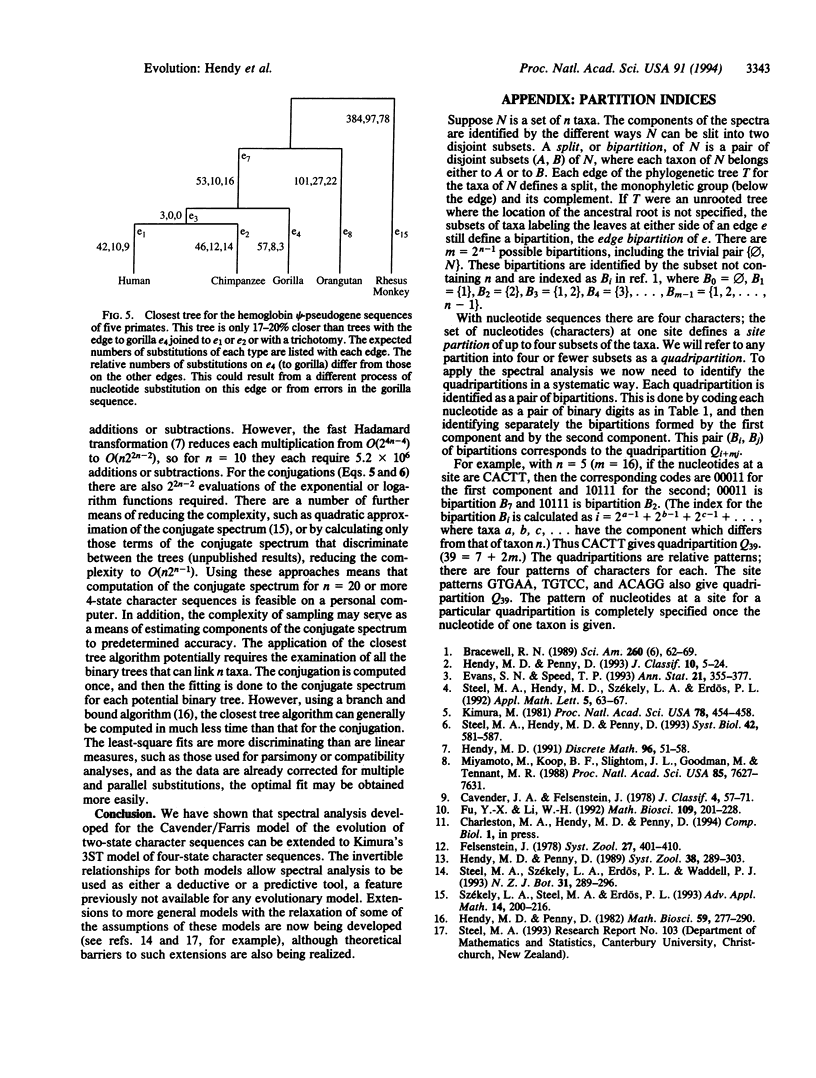
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fu Y. X., Li W. H. Construction of linear invariants in phylogenetic inference. Math Biosci. 1992 May;109(2):201–228. doi: 10.1016/0025-5564(92)90045-x. [DOI] [PubMed] [Google Scholar]
- Henderson I. M., Hendy M. D., Penny D. Influenza viruses, comets and the science of evolutionary trees. J Theor Biol. 1989 Oct 9;140(3):289–303. doi: 10.1016/s0022-5193(89)80087-6. [DOI] [PubMed] [Google Scholar]
- Kimura M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc Natl Acad Sci U S A. 1981 Jan;78(1):454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A. Macromolecular crystals. Sci Am. 1989 Mar;260(3):62–69. doi: 10.1038/scientificamerican0389-62. [DOI] [PubMed] [Google Scholar]
- Miyamoto M. M., Koop B. F., Slightom J. L., Goodman M., Tennant M. R. Molecular systematics of higher primates: genealogical relations and classification. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7627–7631. doi: 10.1073/pnas.85.20.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]