Abstract
Pilocytic astrocytoma (PA) is the most common primary brain tumor in children; various signaling pathways have been implicated in its biology. The Notch signaling pathway has been found to play a role in development, stem cell biology, and the pathogenesis of several cancers but its role in PA has not been investigated. We studied alterations in Notch signaling components in tumor tissue from 18 patients with PA and 4 with other low-grade astrocytomas to identify much needed therapeutic targets. We found that Notch pathway members were overexpressed at the mRNA (NOTCH1, NOTCH2, HEY1, HEY2) and protein (HES1) levels in PAs at various anatomical sites compared to non-neoplastic brain samples. These changes were not associated with specific BRAF alterations. Inhibiting the Notch pathway in the pediatric low-grade astrocytoma cell lines Res 186 and Res 259 using either RNA interference or a γ-secretase inhibitor resulted in variable but significant reduction in cell growth and migration. This study suggests a potential role for Notch signaling in pediatric low-grade astrocytoma tumorigenesis and that Notch signaling may be a viable pathway therapeutic target.
Keywords: BRAF, Notch, Pediatric low-grade astrocytoma, Pilocytic astrocytoma
INTRODUCTION
Pediatric low-grade astrocytomas (PLGA) encompass the most frequent primary brain tumors afflicting children. These tumors are characterized by slow growth and relatively circumscribed architecture, which allows for gross total resection and surgical cures in many patients. However, a subset of these tumors progress, particularly when they are located in inaccessible surgical sites, and may then be associated with significant morbidity and even mortality over time. Given the toxicities and relatively inefficacy associated with conventional therapies (i.e. chemoradiation), novel rational targeted therapies based on our increasing knowledge of the biology of these tumors are needed.
Recent applications of high-resolution platforms have uncovered important somatic genetic drivers that are relatively specific to PLGA subtypes. For instance, duplications of the BRAF kinase domain resulting in BRAF-KIAA1549 fusions are present in most pilocytic astrocytomas (PAs), the predominant histologic subtype of PLGA (1–5). These alterations lead to increased downstream signaling pathways that have been found to be active in PLGA, particularly MAPK (2). The mTOR pathway is also often active in these tumors (6, 7). Other PLGA subtypes have different alterations. For example, MYB, MYBL1, and FGFR1 mutations or rearrangements are typical of pediatric diffuse astrocytomas (8, 9).
The Notch signaling pathway plays important roles in development and, when they are deregulated, in disease (10). Notch activity also has critical functional roles in a variety of cancers, including those of hematopoietic origin, skin, breast, ovarian, lung, prostate, and pancreas (11–16). Although Notch receptors and ligands function as bona fide oncoproteins in many cancers, tumor suppressor roles may also predominate in specific subtypes or contexts (14).
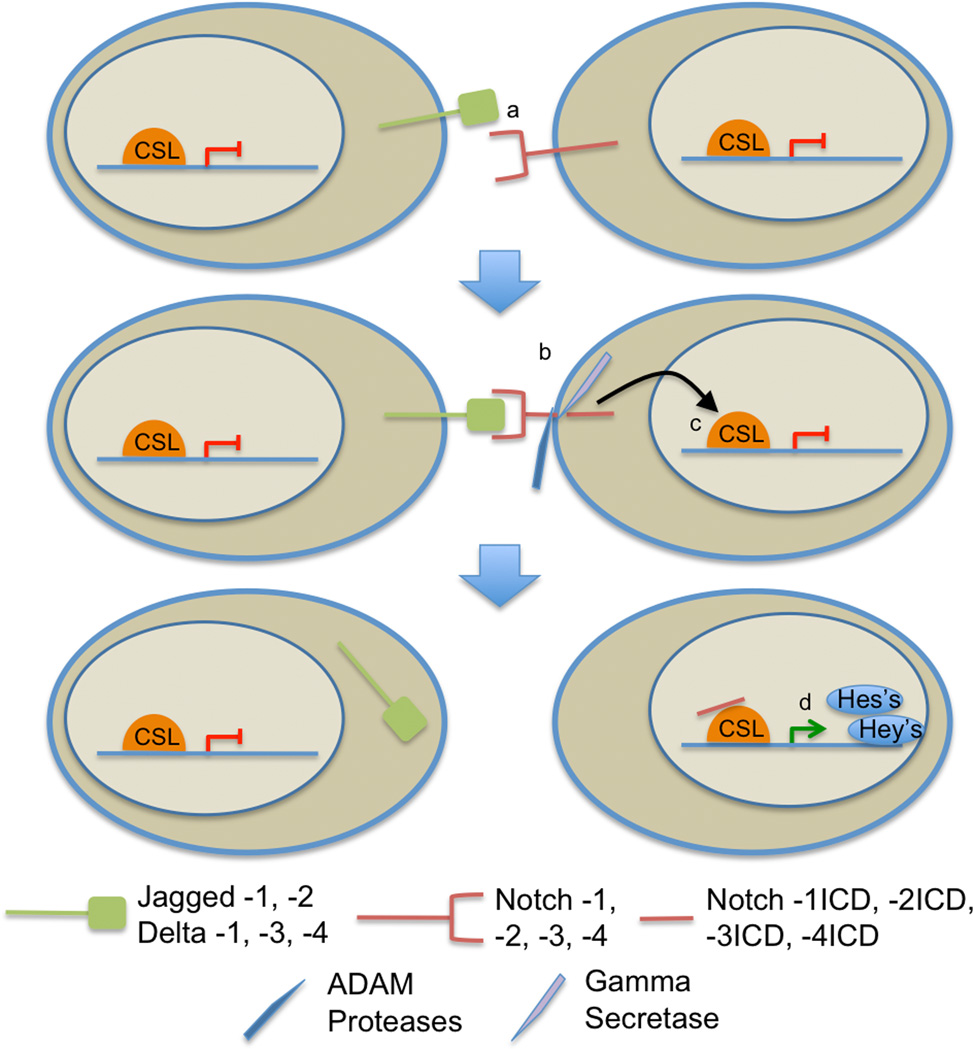
Canonical Notch signaling functions through direct cell-to-cell interactions. The pathway is turned on when a Notch ligand on 1 cell binds to a Notch receptor on an adjacent cell (Fig. 1). Once bound, the Notch receptor goes through 2 proteolytic cleavages. It is first cut extracellularly by an ADAM protease and then within the membrane by γ-secretase. Following the second cleavage, the notch intracellular domain translocates to the nucleus, where it binds to a CBF1/Suppressor of Hairless/LAG-1 family DNA-binding protein. This binding then activates transcription of pathway targets, primarily members of the HES and HEY families, which in turn function as transcriptional repressors. These repressors affect cellular proliferation and differentiation in both embryogenesis and carcinogenesis (17). In addition, crosstalk with other oncogenic canonical pathways has also been described (10), including in the context of astrocytic tumors (18).
Figure 1.
Model of the Notch signaling pathway. (a) The Notch signaling pathway is activated when a Notch ligand and a Notch receptor from adjacent cells bind to one another. (b) Binding of a Notch ligand and receptor initiates sequential cleavage steps, including extracellular ADAM protease followed by intracellular γ-secretase cleavage, of the Notch receptor. (c) An activated form of the receptor (NICD) is released and immediately translocates to the nucleus. (d) The NICD then binds to cofactors, including CSL, to initiate transcription of downstream targets, typically transcriptional repressors from the HES and HEY families. ADAM, a disintegrin and metalloprotease; CSL, CBF1/Suppressor of Hairless/LAG-1 family DNA-binding protein.
Recent studies have also started to uncover a role for Notch signaling in glioma biology. Specifically, Notch signaling appears to play a role in cancer stem cell maintenance in gliomas (19–21), and may also regulate other key glioma cell properties such as migration and invasion (22, 23). Interestingly, Notch3 activation induces gliomas in murine eye and optic nerve (24), a frequent site for PLGA development. Moreover, in a prior gene expression study, NOTCH2 was found to be upregulated in a subset of PAs localized to the hypothalamo-chiasmatic region (25). Therefore, we hypothesized that Notch signaling may play a role in the biology of PLGA and PA in addition to other, more-extensively studied signaling pathways, and may be a therapeutic target.
MATERIALS AND METHODS
Primary Tumor Samples and Cell Lines
Twenty-two primary PLGA and pooled non-neoplastic brain samples were used to quantitate gene expression. Clinicopathologic and BRAF molecular features of these tumors have been published (26); demographic and molecular features are summarized in the Table. A previously characterized tissue microarray containing 61 PAs and 4 non-neoplastic brain controls was also used for immunohistochemical analysis (27). PLGA cell lines Res186 (PA-derived) and Res259 (diffuse astrocytoma-derived) have been previously described (28), and were kindly provided by Dr. Chris Jones (Institute of Cancer Research, Sutton, UK). They were maintained in Dulbecco’s Modified Eagle Medium/F12 Ham’s medium (DMEM/F12), containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin in a humidified 37°C incubator with 5% CO2. This study was performed following institutional review guidelines at Johns Hopkins University.
Table.
Demographic and Molecular Patient Data for Tumor Samples
| Case | Age (y) |
Sex | Pathology | Anatomic Location | BRAF Alteration |
|---|---|---|---|---|---|
| 1 | 15 | M | PA | Cerebellum | BRAF-KIAA1549 |
| 2 | 11 | F | PA | Cerebellum | None1 |
| 3 | 14 | M | PA | Cerebellum | BRAF-KIAA1549 |
| 4 | 4 | F | PA | Posterior fossa | BRAF-KIAA1549 |
| 5 | 20 | M | PA | Thalamus | None1 |
| 6 | 2 | F | Pilomyxoid | Optic pathway | None1 |
| 7 | 21 | M | PA | Optic pathway | None1 |
| 8 | 2 | M | Low grade astrocytoma NOS | Left temporal lobe | None1 |
| 9 | 14 | F | PA | Right parietal lobe | None1 |
| 10 | 19 | F | Low grade astrocytoma, NOS | Spinal cord | BRAF-KIAA1549 |
| 11 | 10 | M | PA | Spinal cord | BRAF-KIAA1549 |
| 12 | 18 | M | PA | 4th ventricle | BRAF-KIAA1549 |
| 13 | 18 | M | PA | Cerebellum | BRAF-KIAA1549 |
| 14 | 21 | M | PA | Cerebellum | BRAF-KIAA1549 |
| 15 | 7 | F | PA | Brainstem | BRAF-KIAA1549 |
| 16 | 6 | M | PA | Posterior fossa | BRAF-KIAA1549 |
| 17 | 10 | F | PA | Cerebellum | BRAF-KIAA1549 |
| 18 | 3 | M | PA | 4th Ventricle | BRAF-KIAA1549 |
| 19 | 11 | M | Low grade astrocytoma, NOS | Cerebellum | BRAF-KIAA1549 |
| 20 | 18 | M | PA | Cerebellum | Not done |
| 21 | 2 | M | PA | Optic pathway | BRAF-KIAA1549 |
| 22 | 12 | M | PA | Cerebellum | BRAF-KIAA1549 |
M, male; F, female NOS, not otherwise specified; PA, pilocytic astrocytoma.
Samples were tested for BRAF (V600E) and the most common BRAF-KIAA1549 fusions only.
Real-time Polymerase Chain Reaction
Primers for each mRNA gene target are listed in Supplemental Table 1. Total mRNA was extracted from the tissue samples and cell lines, converted to cDNA, and normalized to 5 ng/µL with 1 µl of cDNA used per reaction. The amplification protocol was as follows: denaturation at 95°C for 8 minutes, followed by 35 amplification cycles at 95°C for 10 seconds, 58.1°C for 30 seconds, and 65°C for 30 seconds. Melt curve analysis verified the absence of nonspecific PCR products. The relative expression of each gene was compared to β-ACTIN for the primary samples and to HPRT1 for the cell lines.
Immunohistochemistry
Deparaffinized tissue microarray sections were stained with a HES1 antibody. Briefly, antigen retrieval was performed with citrate buffer at pH 6.0 at 95°C to 100°C. Slides were incubated with an anti-HES1 antibody (goat polyclonal, Santa Cruz Biotechnology, Dallas, Texas, 1:100 dilution) for 1 hour, followed by incubation with anti-goat IgG for 1 hour. DAB was used as the chromogen. A neuropathologist (FJR) scored the stained arrays using a 4-tiered scale based on the percentage of tumor cell nuclei showing immunoreactivity as follows: 0 = negative/rare positive cells; 1 = 1–10% positive cells; 2 = 10%– 50% positive cells; and 3 = >50% positive cells. At least 2 intact cores were required for a case to be scored.
Gene Expression Microarray Analysis
Affymetrix HG-U133 Plus 2.0 gene chip mRNA expression data from profiled PA samples and non-neoplastic brain controls were analyzed as previously described (29). In brief, 64 PAs and 20 non-neoplastic brain samples from various anatomical regions were obtained from 3 datasets: a JHH profiled dataset and 2 publically available downloadable files (GSE37307 and GSE5675). Frozen robust multi-array analysis was applied for Raw gene expression data preprocessing and normalization. To correct for multiple comparisons the Benajmini-Hochberg method was used; a false discovery rate <0.001 was considered significant. The Notch pathway components and targets were organized into a heat map using the Pearson’s distance, average clustering method.
Drug Treatment and Plasmids
For knockdown experiments, lentiviruses, prepared in PLKO.1, encoding small hairpin RNAs (shRNAs) against CBF1 (sequence 1: GCT GGA ATA CAA GTT GAA CAA; sequence 3: GCA CAG ATA AGG CAG AGT ATA) were used, as previously described (30); they were purchased from Thermo Fisher Scientific (Waltham, MA). Infected cells were selected using 5 µg/ml puromycin. For pharmacologic inhibition, cells were treated with the γ-secretase inhibitor (GSI) MRK003, a generous donation from Merck (Whitehouse Station, NJ). During GSI treatment the cells were incubated in DMEM/F12 containing 2% FBS.
Cellular Growth Assays
The cell titer 96 Aqueous One solution cell proliferation assay kit (Promega, Madison, WI) was used to examine cell growth utilizing 3-(4,5--dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS). Viable cells were counted and plated at a density of 1,500 cells per well in triplicate using DMEM/F12 with 2% FBS. Twenty µl of MTS reagent was added to 100 µl medium in 96-well plates and incubated for 1 hour at 37°C in 5% CO2. Cells were counted at day 0, 2, 4 and 6 for each condition: Parental, PLKO control, shScramble control, shCBF1 #1, and CBF1 #3 for molecular inhibition, and DMSO control, and 2 µM, and 5 µM of the GSI inhibitor MRK003 for pharmacologic inhibition. The 490-nm absorbance value (which is proportional to the number of viable cells in culture) was measured using an Epoch Micro-Volume Spectrophotometer Plate Reader (Biotek, Winooski, VT).
Boyden Chamber Invasion Assay
Cellular invasion was assayed by first precoating 6.5-mm-diameter Falcon cell culture inserts (8-µm pore size; Becton Dickinson, Franklin Lakes, NJ), with Matrigel diluted 1:100 with medium in 24-well plates. Cells were then added to serum-free medium in the upper chamber and allowed to invade through the Matrigel-coated pores into 10% FBS-containing medium in the lower chamber for 16 hours. The cells in the upper chamber were then removed, the remaining cells on the bottom-side of the culture insert were fixed in 70% EtOH and counted from 3 random high-powered fields.
Migration Assay
Cells were grown in a 6-well plate until confluence, and then scratched twice with a P1000 pipette tip. The cells were rinsed in PBS 5 times, imaged at 4 separate locations, and grown overnight in DMEM/F12 with 2% FBS. Seventeen hours later, the cells were imaged again at the same 4 locations. ImageJ software (National Institutes of Health, Bethesda, MD) was then used to determine the surface area of the scratch, with the percent migration being determined by: [Total Area (Time 0) – Remaining Area (Time 17)] / Total Area (Time 0).
Western Blot Analysis
TNE buffer, including protease inhibitors diluted to 1:100, was used to lyse the cells. Protein was transferred to nitrocellulose membranes and incubated overnight with anti-CBF1 diluted 1:1000 or anti-β-actin (Cell Signaling Technology, Danvers, MA) at 1:20,000 in 5% non-fat milk. Secondary antibodies (rabbit and/or mouse IgG) (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) were diluted 1:5000 in blocking solution, and the blots were developed using enhanced chemiluminescence reagent. Densitometry analysis was performed using ImageJ software with the gel analysis method described in the ImageJ documentation.
Statistical Analysis
Experiments were performed at least 3 times unless otherwise specified. Statistics were determined using the Student t-test, with p < 0.05 considered significant. Statistical analyses were performed using Prism 6 software (GraphPad, San Diego, CA).
RESULTS
Activation of the Notch Signaling Pathway is Frequent in PLGAs
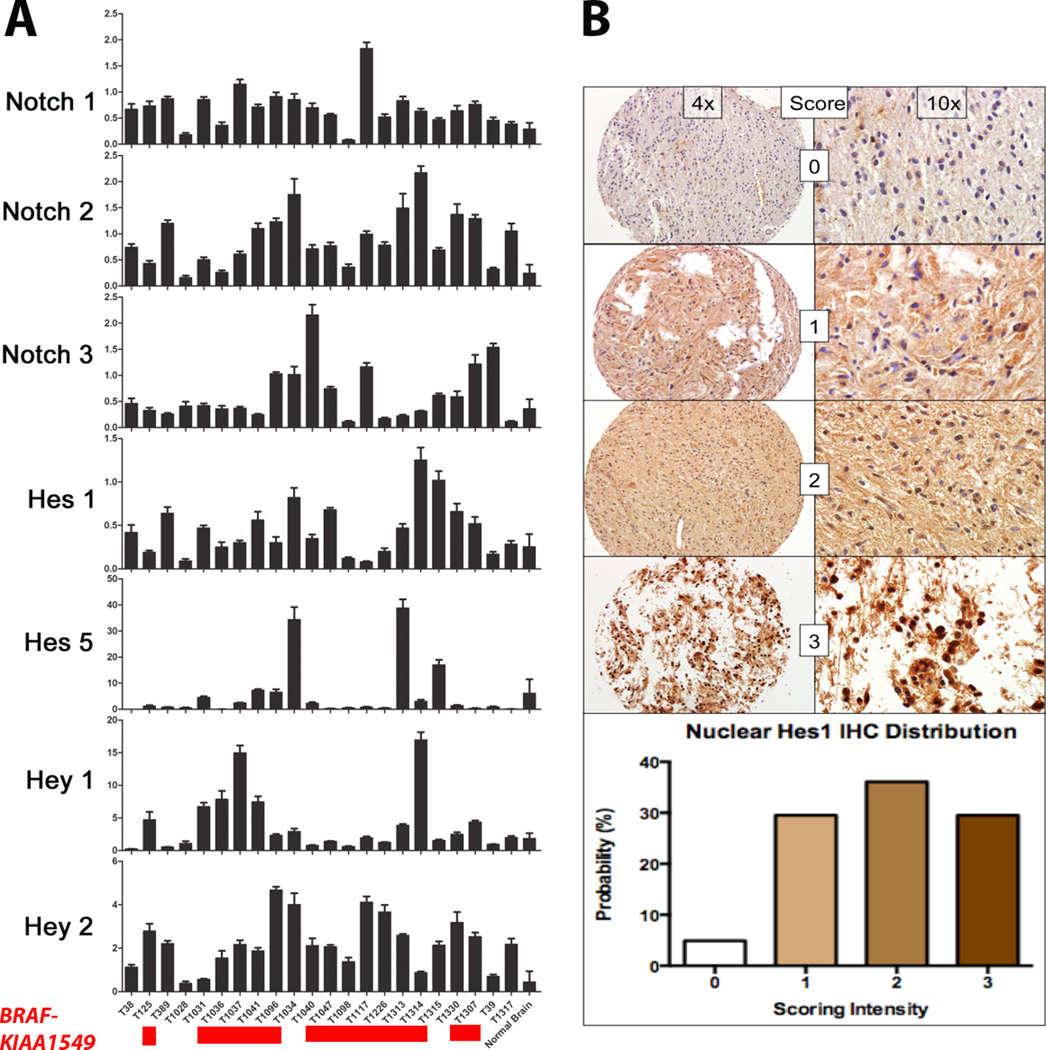
To study the activity of the Notch signaling pathway in PLGA, analysis of the Notch pathway was performed by qRT-PCR in 22 primary PLGAs (predominantly PA) (Table), and compared to pooled non-neoplastic brain samples. BRAF-KIAA1549 fusions were present in the majority of the tumors tested (15 of 21; 71%). The BRAF (V600E) point mutation or a clinical history of neurofibromatosis-1 were absent. Other alterations present in PLGGs lacking BRAF alterations (e.g. FGFR1 mutations, RAF1 fusions) were not tested. Components of the Notch signaling pathway and downstream targets were overexpressed in a majority of the tumors compared to pooled non-neoplastic brain samples, including those with and without BRAF alterations. NOTCH1 mRNA was overexpressed in 91% of the samples tested (20/22), NOTCH2 in 95% (21/22), and NOTCH3 in 55% (12/22). The Notch target HES1 was overexpressed in 68% of the samples tested (15/22), HES5 in 18% (4/22), HEY1 in 50% (11/22), and HEY2 in 95% (21/22) (Fig. 2A).
Figure 2.
The Notch pathway is upregulated in pediatric low-grade astrocytomas (PLGA) at the mRNA and protein levels. (A) Components of the Notch signaling pathway are upregulated at the mRNA level in the majority of the primary PLGAs tested vs. pooled non-neoplastic brain samples. Samples carrying the BRAF:KIAA1549 fusion are highlighted in red below the tumor labels. (B) HES1 protein levels are increased in pilocytic astrocytomas (n = 61) vs. non-neoplastic brain samples by immunohistochemistry. The scoring and percentage of samples at each level of expression are listed. The tissue for the score of 0 is from a non-neoplastic brain sample.
To study activation of the Notch signaling pathway in PAs at the protein level, the Notch target HES1 was evaluated by immunohistochemistry in 61 PAs on tissue microarrays. Ninety-five percent (58/61) of the PA samples exhibited some degree of nuclear HES1 staining, with 66% (40/61) showing moderate to strong expression (Fig. 2B). There was no significant association between HES1 immunohistochemistry and anatomic site, the presence or absence of BRAF alterations, BRAF alteration type, or outcome (p > 0.05).
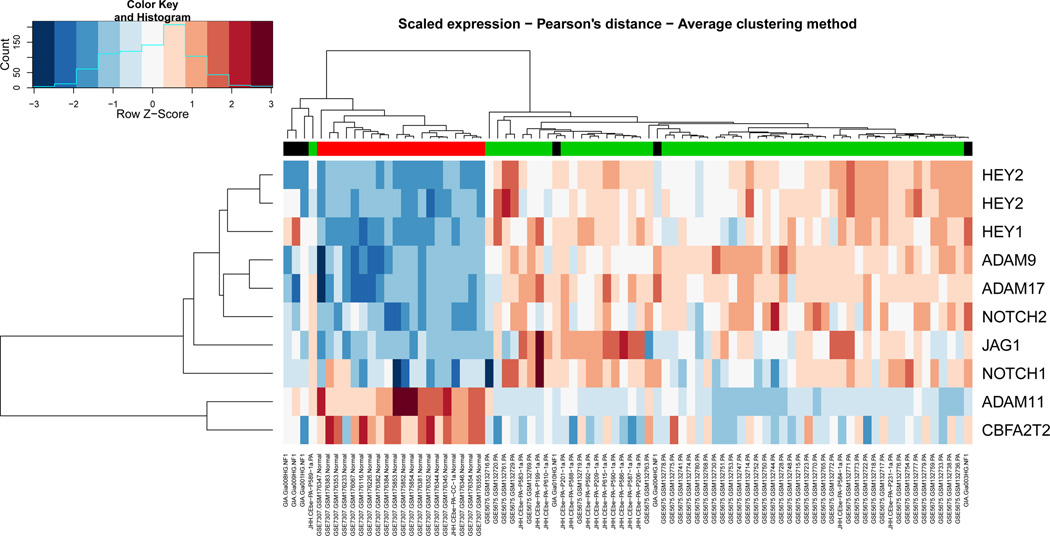
To confirm that the Notch pathway was active in PAs, the most common subtype of PLGA, we studied global gene expression in a larger cohort of PAs profiled at Johns Hopkins combined with others obtained from publically available datasets (Affymetrix HG-U133 Plus 2.0). Using a false discovery rate < 0.001, and excluding gene expression changes overlapping between the 3 different dataset batches (to adjust for batch effect), a total of 2,682 genes were found to be differentially expressed between PAs and non-neoplastic brain tissues. This list included gene members of the Notch signaling pathway (Fig. 3; Supplemental Table 2). Of interest, 2 separate probes showed that the known Notch target HEY2 was overexpressed >100-fold in tumor samples compared to non-neoplastic controls. The related family-member HEY1 was overexpressed ~60-fold. Other genes encoding for components of the canonical Notch signaling pathway proper that were also overexpressed included the Notch ligand JAGGED1 55-fold), and receptors NOTCH1 and NOTCH2 (12– and 6-fold, respectively). Conversely, genes encoding for other Notch ligands (e.g. Jagged2, Delta 1, 3 and 4) were not found to be differentially expressed.
Figure 3.
Components of the Notch signaling pathway are overexpressed in pilocytic astrocytomas (PA) vs. non-neoplastic brain. Heat map constructed using Affymetrix array mRNA datasets highlights components of the Notch signaling pathway that are differentially expressed in PA vs. control non-neoplastic brain samples (PA = 64, non-neoplastic brain = 20).
Core Binding Factor Inhibition Reduces the Growth and Migration of Res186 and Res259
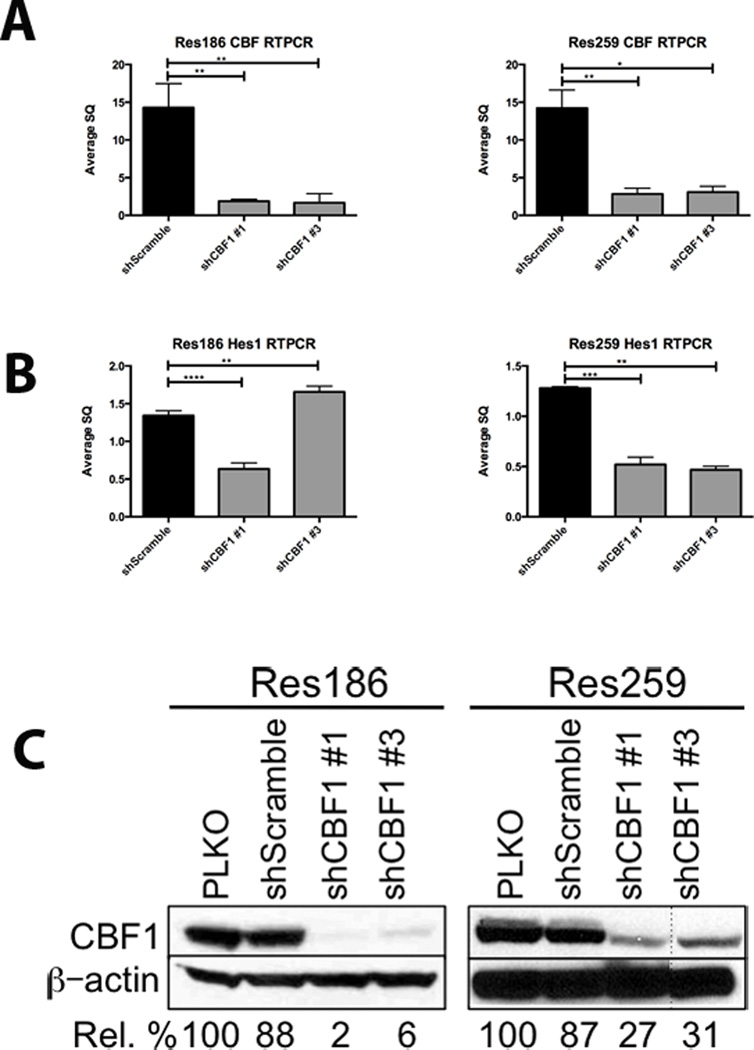
To evaluate the role of Notch signaling in PLGA, lentiviruses containing short hairpins directed against CBF1, a cofactor necessary for Notch receptor binding and activation, were used to infect the PLGA-derived cell lines Res186 and Res259. We chose to inhibit core binding factor (CBF) because it is a co-binding factor for the transcriptional activation of all 4 Notch receptors, regardless of which Notch receptor is activated (i.e. global Notch signaling pathway inhibition). This approach has previously been used to inhibit canonical Notch signaling in neural stem cells (31). CBF1 mRNA was reduced an average of 10-fold in Res186 and 5-fold in Res259 using 2 separate hairpins, compared to a scrambled short hairpin control (Fig. 4A). CBF1 knockdown resulted in a concomitant, though less pronounced, decrease in HES1 mRNA (Fig. 4B). Protein expression of CBF1 was also significantly reduced (Fig. 4C).
Figure 4.
Notch pathway inhibition through CBF1 knockdown in Res186 and Res259 cell lines. (A) RT-PCR demonstrates a significant decrease in CBF1 mRNA in cell lines Res186 and Res259 using 2 separate short-hairpins. (B) RT-PCR for HES1 mRNA demonstrates a concomitant decrease. (C) Western blot after 7 days of selection shows that CBF1 is also inhibited at the protein level. Representative images of each experiment are shown; each was performed in triplicate after 7 days of selection in 5 µg/ml puromycin. Densitometry analyses representing percent CBF1 knockdown are listed below the images (*p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001). PLKO, control.
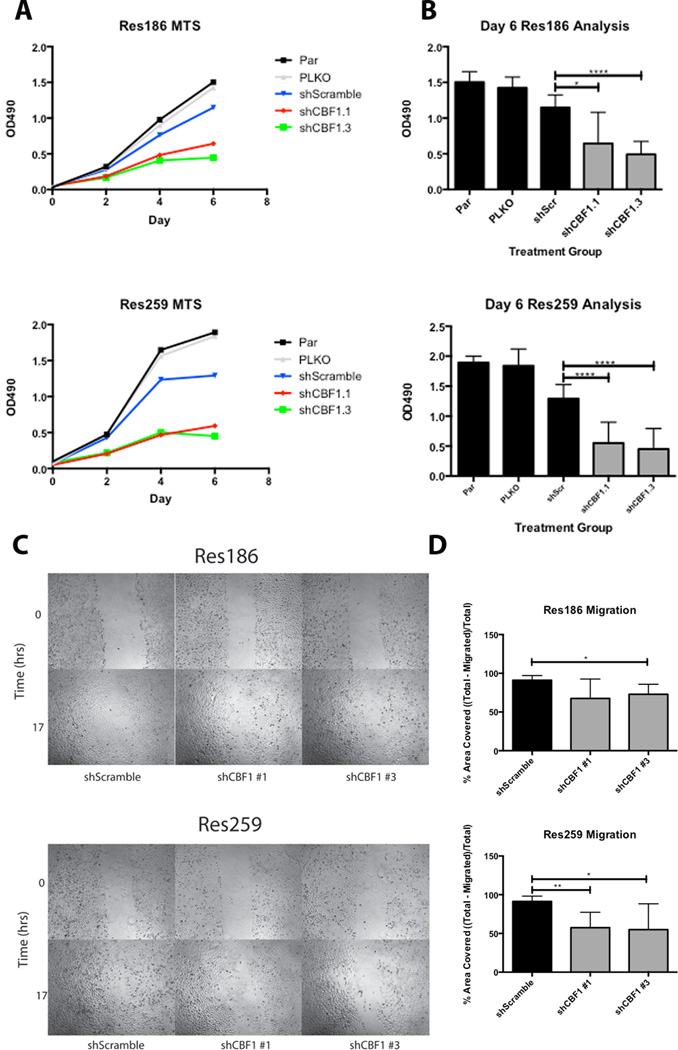
To study the effect of Notch inhibition on cellular properties, the MTS assay for cell growth was performed. shCBF1 #1 and shCBF1 #3 inhibition of Res186 and Res259 resulted in a discernable decrease in cellular growth after just 2 days (Fig. 5A). By 6 days, Res186 was inhibited by 44% and 63% using shCBF1 #1 and shCBF1 #3, respectively, compared to the scramble short-hairpin control; Res259 was inhibited by 58% and 64%, respectively, under the same conditions (Fig. 5B).
Figure 5.
CBF1 inhibition reduces cellular growth and migration in Res186 and Res259 cell lines. (A) Slower cellular growth of Res186 and Res259 was seen as early as 2 days into an MTS assay in the CBF1 inhibited cells vs. the PLKO and Scramble controls. (B) The difference in growth curves became significant after 6 days. Results are the compilation of 3 separate experiments. (C) Slower cellular migration at 17 hours following the generation of a scratch across a confluent plate of cells. (D) The column graph is a compilation of 6 separate scratches represented in panel C. (*p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001).
Next, we studied the effect of Notch pathway inhibition on other cellular properties, including invasion and migration. In a scratch test assay, both Res186 and Res259 cell migration were inhibited compared to the shScramble control (Fig. 5C). Combining 6 separate scratch assays, the migration of Res186 was inhibited by 23% and 18% in cells expressing shCBF1 #1 and shCBF1 #3, respectively, although the results were only statistically significant using shCBF1 #3. Migration of Res259 was inhibited by 34% and 36% under the same conditions (Fig. 5D). Conversely, we did not observe a consistent effect on invasion using a Boyden chamber assay (data not shown).
Pharmacological Inhibition of Notch Signaling Using a GSI
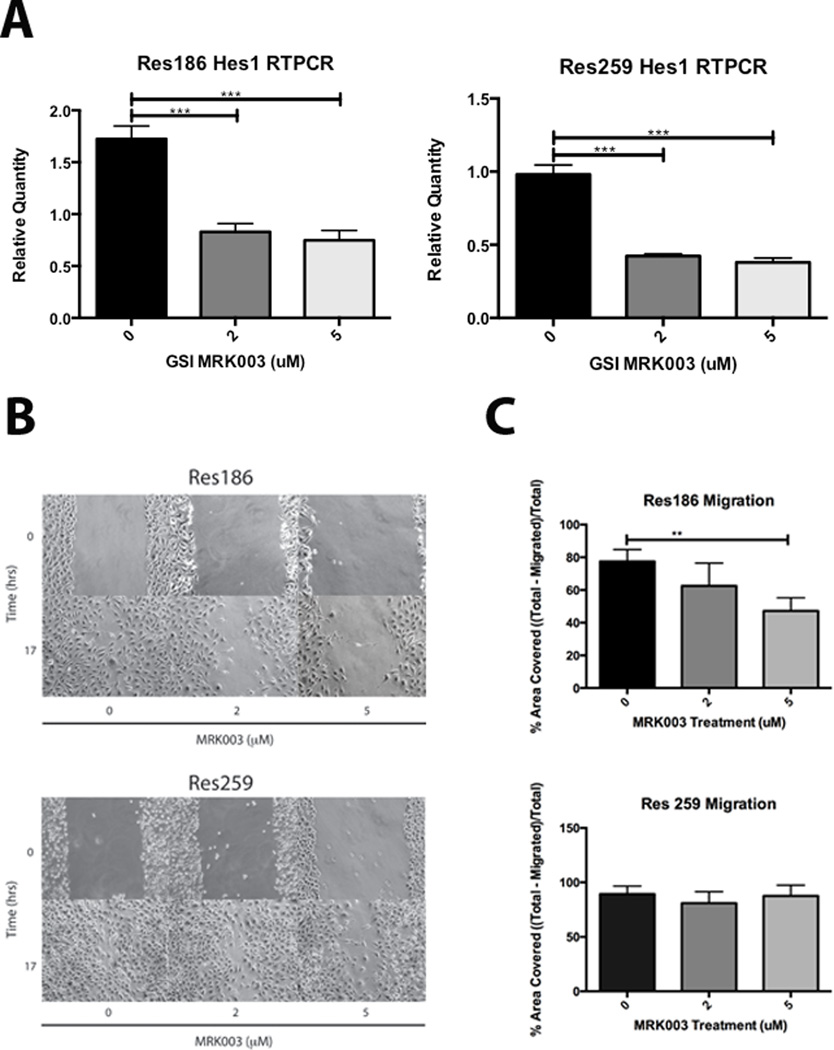
We sought to complement our genetic inhibition of Notch with a more clinically relevant pharmacological approach. Treatment of Res186 with the GSI MRK003 reduced HES1 mRNA expression by 52% at 2 µM and 57% at 5 µM; in Res259, HES1 expression is inhibited by 58% at 2 µM and 61% at 5 µM (Fig. 6A), consistent with molecular Notch pathway inhibition. GSI inhibition trended toward decreased growth on MTS, although it was not statistically significant (Supplementary Fig. 1). Migration of Res186 cells was reduced by 15% and 30% in the presence of 2 and 5 µM GSI, respectively (Fig. 6B, C). However, GSI inhibition had no discernable effect on the migration of Res259 cells.
Figure 6.
Pharmacologic inhibition of the Notch signaling pathway using the γ-secretase inhibitor (GSI) MRK003. (A) HES1 in cell lines Res186 and Res259 is inhibited at the mRNA level after 48-hour treatment with GSI MRK003. (B, C) Scratch test assays demonstrate that Res186 migration is significantly inhibited after 17 hours at 5 µM GSI MRK003, although Res259 trends slightly slower, it is not statistically significant. All experiments were performed in triplicate; cells were treated with GSI MRK003 in 2% fetal bovine serum (*p < 0.05, **p < 0.005, ***p < 0.0005).
DISCUSSION
In this study, we extend prior observations on the oncogenic role of the Notch signaling pathway to an important group of pediatric neoplasms, i.e. PLGA. We sought to explore the role of Notch signaling in PLGA tumorigenesis and to determine whether Notch inhibition could potentially function as a therapeutic target.
The functional significance of Notch signaling in gliomas has been previously studied (32). Notch signaling has been found to play a key role during brain development. In addition, Notch signaling also plays a role in the adult brain neural stem cell niche. Specifically, Notch seems to regulate adult neural stem cell maintenance (33), and prevent astrocytic differentiation in the adult subventricular zone through downstream effectors (e.g. EphB2) (34). Dysregulation of the Notch pathway occurs through various means, and in certain contexts can specifically facilitate gliomagenesis and lead to tumor initiation (24). Furthermore, Notch pharmacologic blockade specifically leads to decreased glial tumorigenicity, decreased stem cell marker expression, and induction of glial differentiation (19).
Utilizing Notch signaling as a therapeutic target has been shown to be efficacious in a variety of research settings. Our group and others have shown that clinically relevant levels of GSI administration were able to significantly reduce tumor burden and prolong the survival of mice with glioblastoma xenografts (19, 35). Similar results using clinically relevant GSI levels were also found in breast cancer (36). Others have studied Notch blockade as a strategy in pediatric brain tumors. For example, a Phase I clinical trial using the γ-secretase inhibitor MK-0752 in patients with pediatric brain tumors was tolerated, although it failed to show a significant response (37). Promising results were found using the same drug on a larger cohort of adult patients with high-grade gliomas, suggesting that GSIs have the potential to be effective against gliomas (38).
In the current study, we have shown that the Notch signaling pathway is active in most PLGA/PA tumors studied, and that it may affect cell characteristics such as growth and migration. The availability of PLGA models for functional experiments is fairly limited due in part to the phenomenon of oncogene-induced senescence that is likely secondary to BRAF activation in these tumors (27, 39). Xenografts models of PLGG with clinically relevant attributes are particularly scant. Currently, understanding how Notch signaling is specifically activated in these tumors is unclear. In T-cell acute lymphoblastic leukemia, activating NOTCH1 mutations are frequent (40) but similar alterations have not been identified in PLGA. Two relevant signaling pathways activated in PLGA, MAPK and mTOR could potentially affect Notch signaling. For example, MAPK activity has been shown to regulate Notch signaling in papillary thyroid carcinoma (41). Most of the tumors directly tested in our study had a BRAF-KIAA1549 fusion (~71%), which is typical of PAs. Alternative alterations that also affect MAPK signaling have been described in PA (e.g. FGFR1 mutations, RAF1 fusions, and BRAF deletions or insertions) (9, 42). Additionally, some non-glial tumors with FGFR1 rearrangements have demonstrated increased Notch constitutive activity (43). These are relatively rare in PLGA, but future studies with larger cohort numbers will elucidate whether alterations in Notch expression may vary according to specific genetic aberrations leading to increased MAPK activity.
We used 2 currently available PLGA cell lines (28), i.e. Res186 (PA-derived) and Res259 (diffuse astrocytoma-derived), to study the effects of Notch signaling inhibition using shRNA and pharmacologic GSI inhibition. Both approaches effectively downregulated the expression of downstream Notch targets in these cells and shRNA-mediated inhibition of CBF1 also significantly affected cell growth. Interestingly, cell migration (but not invasion) was affected by both approaches. Although most PLGAs are circumscribed and display little tumor spread, a subset of these tumors may extend into the subarachnoid space and disseminate through the CSF, causing significant morbidity for which efficacious therapies are lacking. Inhibiting migration may, therefore, represent a complementary approach in the subset of PLGA that disseminate in this manner. Conversely, a prior study from our group using these cell lines showed that mTOR inhibition affects proliferation (6), a phenomenon studied by Kaul et al in neural stem cells carrying BRAF fusions (7). Of interest, Notch activity may also affect the mTOR pathway in other tumors (44). One caveat regarding pharmacologic inhibition using GSIs is that these compounds also affect other cellular dynamics, and the effects of GSI inhibition may not be completely ascribed to Notch blockade. However, GSIs have been found to inhibit Notch in preclinical studies and early phase clinical trials of patients with solid tumors (19, 45, 46). It is of interest that the anti-tumor effects were more pronounced by CBF1 knockdown, which should more specifically inhibit Notch, although effects on other cell pathways cannot be completely excluded.
Similar to the earlier mTOR study utilizing these cell lines (6), Res186 seems to be more responsive to drug dosing than Res259. Currently, it is unclear whether this reflects a difference in drug sensitivity related to tumor type (PA vs. diffuse astrocytoma) or whether it is an intrinsic property of these cell lines. As mentioned above, it is difficult to develop PLGA lines. One caveat of the current study is that the PLGA lines used in these experiments contain genetic alterations that are relatively rare in PLGA (e.g. homozygous PTEN deletion in Res186, PDGFRA gain in Res259) (28), which could explain some of the negative pharmacologic results. We used a drug dosage that has been found to be effective in prior glioma studies and that has been shown to inhibit Notch signaling and multiple Notch targets (19, 45). Future studies, and greater availability of PLGA models, should provide answers to these questions, and clarify the feasibility of Notch inhibition as a viable target.
In summary, this study demonstrates a high level of Notch activity in the majority of PLGAs studied; targeting this pathway could be feasible, alone or in combination, for these tumors in the future. As newer drugs with increased specificity and decreased toxicity become available, as well as more faithful in vivo models, additional studies of Notch inhibition in PLGA may be warranted.
Supplementary Material
Acknowledgments
This work was supported in part by the Knights Templar Eye Foundation and the Childhood Brain Tumor Foundation (FJR), the National Institute of Health grant RO1NS055089 (CGE), Lauren’s First and Goal and the Pilocytic/Pilomyxoid Fund (CGE, FJR).
REFERENCES
- 1.Bar EE, Lin A, Tihan T, et al. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol. 2008;67:878–887. doi: 10.1097/NEN.0b013e3181845622. [DOI] [PubMed] [Google Scholar]
- 2.Forshew T, Tatevossian RG, Lawson AR, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218:172–181. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 3.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfister S, Janzarik WG, Remke M, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sievert AJ, Jackson EM, Gai X, et al. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19:449–458. doi: 10.1111/j.1750-3639.2008.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutt-Cabezas M, Karajannis MA, Zagzag D, et al. Activation of mTORC1/mTORC2 signaling in pediatric low-grade glioma and pilocytic astrocytoma reveals mTOR as a therapeutic target. Neuro-oncology. 2013;15:1604–1614. doi: 10.1093/neuonc/not132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaul A, Chen YH, Emnett RJ, et al. Pediatric glioma-associated KIAA1549:BRAF expression regulates neuroglial cell growth in a cell type-specific and mTOR-dependent manner. Genes Dev. 2012;26:2561–2566. doi: 10.1101/gad.200907.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramkissoon LA, Horowitz PM, Craig JM, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci USA. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Wu G, Miller CP, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan K, Brown AM. Is there a role for Notch signalling in human breast cancer? Breast Cancer Res. 2003;5:69–75. doi: 10.1186/bcr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang TP, Gazdar AF, Virmani AK, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Nat Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 13.Hopfer O, Zwahlen D, Fey MF, et al. The Notch pathway in ovarian carcinomas and adenomas. Br J Cancer. 2005;93:709–718. doi: 10.1038/sj.bjc.6602719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobry C, Oh P, Mansour MR, et al. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123:2451–2459. doi: 10.1182/blood-2013-08-355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto Y, Maitra A, Ghosh B, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 16.Santagata S, Demichelis F, Riva A, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 17.Kageyama R, Masamizu Y, Niwa Y. Oscillator mechanism of Notch pathway in the segmentation clock. Devel Dynam. 2007;236:1403–1409. doi: 10.1002/dvdy.21114. [DOI] [PubMed] [Google Scholar]
- 18.Xu P, Qiu M, Zhang Z, et al. The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo. J Neuro-oncology. 2010;97:41–51. doi: 10.1007/s11060-009-0007-1. [DOI] [PubMed] [Google Scholar]
- 19.Chu Q, Orr BA, Semenkow S, et al. Prolonged inhibition of glioblastoma xenograft initiation and clonogenic growth following in vivo Notch blockade. Clinical Cancer Res. 2013;19:3224–3233. doi: 10.1158/1078-0432.CCR-12-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulasov IV, Nandi S, Dey M, et al. Inhibition of Sonic hedgehog and Notch pathways enhances sensitivity of CD133(+) glioma stem cells to temozolomide therapy. Molec Med. 2011;17:103–112. doi: 10.2119/molmed.2010.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Wang C, Meng Q, et al. siRNA targeting Notch-1 decreases glioma stem cell proliferation and tumor growth. Molec Biol Rep. 2012;39:2497–2503. doi: 10.1007/s11033-011-1001-1. [DOI] [PubMed] [Google Scholar]
- 22.Sivasankaran B, Degen M, Ghaffari A, et al. Tenascin-C is a novel RBPJkappa-induced target gene for Notch signaling in gliomas. Cancer Res. 2009;69:458–465. doi: 10.1158/0008-5472.CAN-08-2610. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Chen T, Zhang J, et al. Notch1 promotes glioma cell migration and invasion by stimulating beta-catenin and NF-kappaB signaling via AKT activation. Cancer Science. 2012;103:181–190. doi: 10.1111/j.1349-7006.2011.02154.x. [DOI] [PubMed] [Google Scholar]
- 24.Pierfelice TJ, Schreck KC, Dang L, et al. Notch3 activation promotes invasive glioma formation in a tissue site-specific manner. Cancer Res. 2011;71:1115–1125. doi: 10.1158/0008-5472.CAN-10-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchoghandjian A, Fernandez C, Colin C, et al. Pilocytic astrocytoma of the optic pathway: a tumour deriving from radial glia cells with a specific gene signature. Brain. 2009;132:1523–1535. doi: 10.1093/brain/awp048. [DOI] [PubMed] [Google Scholar]
- 26.Lin A, Rodriguez FJ, Karajannis MA, et al. BRAF alterations in primary glial and glioneuronal neoplasms of the central nervous system with identification of 2 novel KIAA1549:BRAF fusion variants. J Neuropathol Exp Neurol. 2012;71:66–72. doi: 10.1097/NEN.0b013e31823f2cb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raabe EH, Lim KS, Kim JM, et al. BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clinical Cancer Res. 2011;17:3590–3599. doi: 10.1158/1078-0432.CCR-10-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bax DA, Little SE, Gaspar N, et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PloS one. 2009;4:e5209. doi: 10.1371/journal.pone.0005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho CY, Bar E, Giannini C, et al. MicroRNA profiling in pediatric pilocytic astrocytoma reveals biologically relevant targets, including PBX3, NFIB, and METAP2. Neuro-oncology. 2013;15:69–82. doi: 10.1093/neuonc/nos269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dull T, Zufferey R, Kelly M, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizutani K, Yoon K, Dang L, et al. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 32.Stockhausen MT, Kristoffersen K, Poulsen HS. Notch signaling and brain tumors. Adv Exp Med Biol. 2012;727:289–304. doi: 10.1007/978-1-4614-0899-4_22. [DOI] [PubMed] [Google Scholar]
- 33.Faigle R, Song H. Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochimica et biophysica acta. 2013;1830:2435–2448. doi: 10.1016/j.bbagen.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura T, Goritz C, Catchpole T, et al. EphB signaling controls lineage plasticity of adult neural stem cell niche cells. Cell Stem Cell. 2010;7:730–743. doi: 10.1016/j.stem.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert CA, Daou MC, Moser RP, et al. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 2010;70:6870–6879. doi: 10.1158/0008-5472.CAN-10-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schott AF, Landis MD, Dontu G, et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin Cancer Res. 2013;19:1512–1524. doi: 10.1158/1078-0432.CCR-11-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouladi M, Stewart CF, Olson J, et al. Phase I trial of MK-0752 in children with refractory CNS malignancies: a pediatric brain tumor consortium study. J Clin Oncol. 2011;29:3529–3534. doi: 10.1200/JCO.2011.35.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krop I, Demuth T, Guthrie T, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30:2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 39.Jacob K, Quang-Khuong DA, Jones DT, et al. Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas. Clinical Cancer Res. 2011;17:4650–4660. doi: 10.1158/1078-0432.CCR-11-0127. [DOI] [PubMed] [Google Scholar]
- 40.Lee SY, Kumano K, Masuda S, et al. Mutations of the Notch1 gene in T-cell acute lymphoblastic leukemia: analysis in adults and children. Leukemia. 2005;19:1841–1843. doi: 10.1038/sj.leu.2403896. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita AS, Geraldo MV, Fuziwara CS, et al. Notch pathway is activated by MAPK signaling and influences papillary thyroid cancer proliferation. Trans Oncol. 2013;6:197–205. doi: 10.1593/tlo.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT, Hutter B, Jager N, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren M, Cowell JK. Constitutive Notch pathway activation in murine ZMYM2-FGFR1-induced T-cell lymphomas associated with atypical myeloproliferative disease. Blood. 2011;117:6837–6847. doi: 10.1182/blood-2010-07-295725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuhashi Y, Itoh M, Nara N, et al. NOTCH knockdown affects the proliferation and mTOR signaling of leukemia cells. Anticancer research. 2013;33:4293–4298. [PubMed] [Google Scholar]
- 45.Asnaghi L, Ebrahimi KB, Schreck KC, et al. Notch signaling promotes growth and invasion in uveal melanoma. Clin Cancer Res. 2012;18:654–665. doi: 10.1158/1078-0432.CCR-11-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Messersmith WA, Shapiro GI, Cleary JM, et al. A Phase I, Dose-finding Study in Patients With Advanced Solid Malignancies of the Oral Gamma-Secretase Inhibitor PF-03084014. Clin Cancer Res. 2014 Sep 17; doi: 10.1158/1078-0432.CCR-14-0607. pii: clincanres.0607.2014. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.