 Defect occurrence within polymers was successfully predicted in 85% of cases using a PLS regression model based upon molecular descriptors.
Defect occurrence within polymers was successfully predicted in 85% of cases using a PLS regression model based upon molecular descriptors.
Abstract
Polymer microarrays are a key enabling technology for the discovery of novel materials. This technology can be further enhanced by expanding the combinatorial space represented on an array. However, not all materials are compatible with the microarray format and materials must be screened to assess their suitability with the microarray manufacturing methodology prior to their inclusion in a materials discovery investigation. In this study a library of materials expressed on the microarray format are assessed by light microscopy, atomic force microscopy and time-of-flight secondary ion mass spectrometry to identify compositions with defects that cause a polymer spot to exhibit surface properties significantly different from a smooth, round, chemically homogeneous ‘normal’ spot. It was demonstrated that the presence of these defects could be predicted in 85% of cases using a partial least square regression model based upon molecular descriptors of the monomer components of the polymeric materials. This may allow for potentially defective materials to be identified prior to their formation. Analysis of the PLS regression model highlighted some chemical properties that influenced the formation of defects, and in particular suggested that mixing a methacrylate and an acrylate monomer and/or mixing monomers with long and linear or short and bulky pendant groups will prevent the formation of defects. These results are of interest for the formation of polymer microarrays and may also inform the formulation of printed polymer materials generally.
Introduction
The use of combinatorial libraries to uncover materials with unique properties and applications is an area of increasing importance in a number of fields. 1–9 A key aspect of this technology is the creation of micro-scale samples of materials that can be assessed in parallel, for example as a microarray, then scaled up once a ‘hit’ composition has been identified. Polymeric materials have been a focus of recent material development endeavours due to their ubiquitous use and ease of synthesis. High throughput materials discovery using polymer microarrays has identified numerous novel materials with varied applications including substrates or matrices that are permissive for cell attachment and growth, 10–16 modulate biomolecular binding, 7,17,18 support the attachment and expansion of stem cells, 5,19,20 prevent bacterial colonisation, 6,9 sort cell populations, 21 exhibit thermo-responsive properties, 22,23 deliver DNA for cell transfection, 24–26 and modulate platelet activation. 8 The printing of monomer/polymer solutions is not limited to the formation of material arrays but has become an important microfabrication technique, involved in the synthesis of electronic components, microlenses, pharmaceutical formulations, light emitting diode displays and solar cells. 27–32 Polymer microarrays have been formed by either printing pre-synthesised polymers 11,16 or using an in situpolymerisation methodology with acrylate, methacrylate, acrylamide or methacrylamide monomers. 33,34 The in situpolymerisation method enables the simple and rapid generation of combinatorial libraries of polymers by premixing monomers and allows for the inclusion of cross-linking reagents that provide control over physical properties of the resultant materials such as stiffness. 5 This approach has successfully identified polymeric formulations that preserve their biological and physical properties upon scale up. 23 The retention of surface chemical properties on scale up is one of the motivations for developing a full range of high throughput surface characterisation methodologies compatible with the microarray format using techniques including X-ray photoelectron spectroscopy (XPS), surface plasmon resonance (SPR), water contact angle (WCA) measurements, atomic force microscopy (AFM) and time-of-flight secondary ion mass spectroscopy (ToF-SIMS). 7,35–38 This suite of techniques also allows the comprehensive analysis of many chemical and physical properties, enabling the establishment of structure–function relationships. 5,35
A key requirement for the continued development of polymer microarrays in materials discovery is further expansion of the combinatorial space explored. To date, up to 24 monomers have been used for the creation of a library of up to 576 materials. However, in the case of acrylates and methacrylates there are over 100 monomers that are commercially available and an even greater number that could be chemically synthesised and exploited for the formation of a polymer microarray. This would allow for the creation of libraries of thousands of materials, for example 100 monomers could be mixed pairwise at a 2 : 1 ratio to create a library of 10 000 materials. We recently explored the inclusion of a greater number of monomers (95) for polymer microarray formation, 39 however, a number of defects in resultant spots was observed. Defects alter the performance of materials, convoluting the assessment of structure–function relationships and often inhibiting the successful scale up of materials. Furthermore, the appearance of defects in some spots requires additional analysis to determine whether the appearance of a defect alters the performance of a material. In order to construct a library of thousands of materials it is necessary to be able to effectively predict which formulations are suitable and thus remove the time and associated cost of preparation, analysis and identification of defective polymer spots.
In order to understand the limiting factors in monomer solution printing, and therefore predict formulations that can be successfully printed, we have explored a library of materials that have previously been observed to produce defective polymer spots. These have been screened for physical and chemical irregularities using light microscopy and chemical surface imaging using ToF-SIMS. A strategic aim of this work was to guide the future design of material microarrays and novel material formulations.
Results and discussion
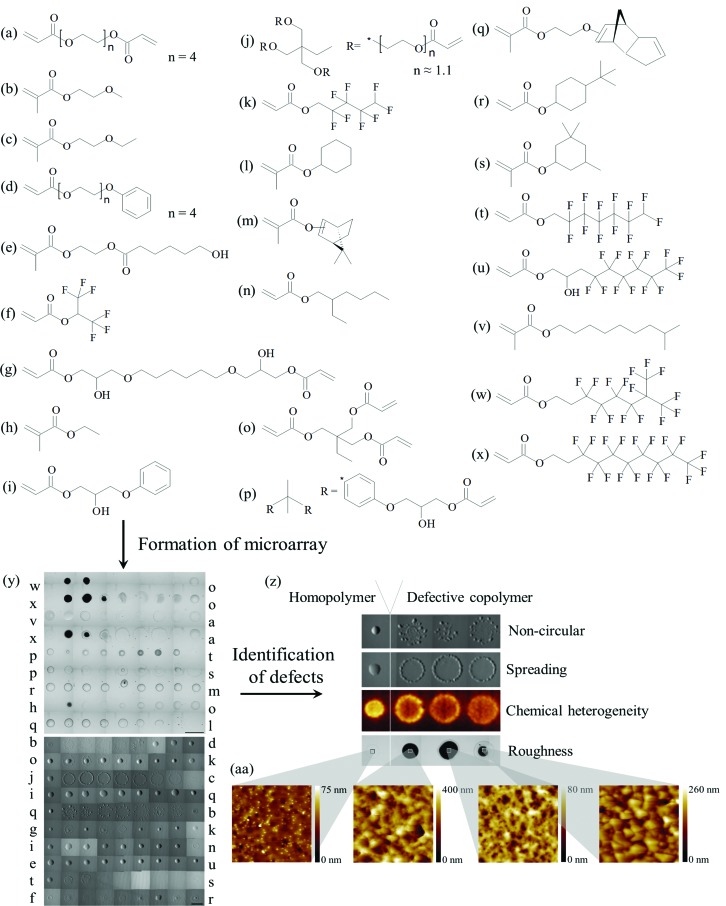
In this study 14 monomer pairs from 24 monomers, shown in Fig. 1a–x, were selected that were previously observed to produce spots with defects that could be observed through a light microscope. In this case, a spot was considered normal if it was visually circular, smooth and continuous. For comparison 4 monomer pairs (monomers p–t, r–m, i–q and e–u) were included that were known to produce defect-free spots. A library of 171 formulations was prepared by producing monomer ratios of 1 : 0, 4 : 1, 3 : 1, 2 : 1, 1 : 1, 1 : 2, 1 : 3, 1 : 4 and 0 : 1 for each of the 19 copolymer pairs. These formulations were printed as a microarray on a poly(2-hydroxyethyl methacrylate) (pHEMA) coated glass slide, which acts as an adhesive matrix for the physical entrapment of printed material. The polymers were distributed over two arrays of 81 and 90 spots each (4 replicate arrays per glass slide) that had different pitches to account for the expected spread of the resultant polymer spots. Light microscopy images of the microarrays are shown in Fig. 1y. Images were taken using transmission mode and therefore opaque spots appear black, whereas to the naked eye these spots appear white. Using the optical image the polymer spots were assessed for circularity and spreading (Fig. 1y) (polymers were considered to have a spreading defect when the diameter of a spot exceeded 700 μm). An increase in surface roughness was visible in the optical image (Fig. 1y) for a number of copolymer series including monomer pairs b–d, j–c, q–b and g–k, and for monomer pairs w–o, x–o and x–a when monomers w or x were at bulk concentrations <100% and >66%. In order to quantitatively assess material roughness all polymers were assessed by AFM, and materials with a root mean square (RMS) roughness greater than 6 nm were considered to have a roughness defect (mean roughness = 6 nm, median roughness = 1.7 nm) (Fig. 1aa). Spots with a high concentration of monomers l, h or s were difficult to observe viaoptical microscopy, thus macro-scale chemical mapping of the array was conducted by ToF-SIMS to assess whether these spots could be detected chemically. All spots could be chemically distinguished from the pHEMA background and, furthermore, micro-scale chemical heterogeneity defects were identified (Fig. 1z). From the SIMS images chemical maps of specific ions were produced, and ions characteristic of specific monomers were identified and grouped. Representative chemical images of characteristic ions are shown in Fig. ESI1.†
Fig. 1. (a–x) Chemical structures of monomers used for formation of the microarray. Monomers were ranked according to their cLogP. (y) The monomers were used to form a microarray. Light microscopy image of a microarray is shown. The letters alongside the image indicate the monomer pairs used in each row. The scale bar is 900 μm. (z) Microarrays were assessed for defects: non-circular (q–bcopolymers shown), spreading (j–ccopolymers shown), chemical heterogeneity (j–ccopolymers shown) and roughness (x–ocopolymers shown). Corresponding homopolymer with no defect are also shown for each defect example presented. Light microscopy images are shown for examples of non-circularity, spreading and roughness, and a ToF-SIMS image of the C3H3 – ion is shown as an example of chemical heterogeneity. (aa) Example AFM images of polymer spots assigned with roughness defect. Images are 5 × 5 μm.
The ion intensity for representative ions can be examined as a function of copolymer composition (Fig. ESI1†). This analysis indicates preferential surface segregation of some monomer components inferred from non-linear intensity versus bulk composition relationships. It is unlikely that these non-linear relationships can be wholly explained by matrix effects 40 as the chemical matrix across a polymer series is similar and the ion intensity trends were observed for multiple characteristic ions. In contrast to monomer pairs such as v–a, where the intensity of characteristic ions for each monomer measured at the surface of the polymer increased linearly with the increase of monomer in the polymer bulk (Fig. ESI1†), large and abrupt changes to the intensity of characteristic ions for monomer pairs x–o, x–a, h–o, q–l, b–d, o–k and q–b were observed, compared to relatively small changes in the polymer bulk. For example, the ion intensity measured for the monomer pair x–a of ion C2H3O+, characteristic of monomer a, increased from 40.7% (normalised to value of ion in the homopolymer of monomer a) to 93.0% when the bulk concentration of monomer a was increased from 0 to 33% (Fig. ESI1†). Further increases in the bulk content of monomer a resulted in no further increase in the intensity of ion C2H3O+. Concomitantly, the intensity of the ion F2 –, characteristic of monomer x, decreased from 100.0% to 34.1% when the bulk concentration of monomer x was decreased from 100 to 80% (Fig. ESI1†). Further decreases in monomer x bulk concentration only resulted in small decreases in the intensity of the F2 – ion. This demonstrates that for some monomer pairs, variance in the monomer composition will not result in a concomitant distribution of surface chemistries. For monomer pair h–o, this is likely caused by the high volatility of monomer h, which likely evaporates between printing and fixation. However, for the other monomer pairs the relatively low volatility of the monomers means that the disparity between the surface analysis and bulk compositions can only be explained by the preferential surface segregation of one of monomer components to the surface. Thus, the surface chemistry of polymers cannot be inferred from their bulk composition and must be ascertained from surface chemical analysis as previously noted for polymer microarrays. 37
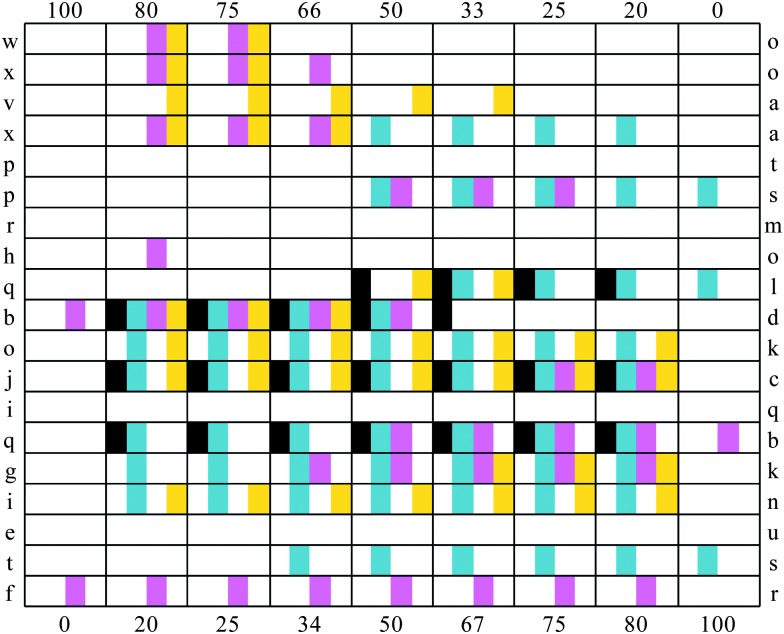
In total, four different defect types were classified (1 from AFM, 2 from light microscopy and 1 from ToF-SIMS): increased roughness, uncontrolled spreading, non-circularity and micro-scale chemical heterogeneity. The defects observed for each polymer are tabulated in Fig. 2. In some cases the occurrence of a defect was related to the presence of a particular monomer. This was evident when the homopolymer of a particular monomer exhibited a particular defect, for example homopolymers of monomer s and l produced spread spots. However, in most cases the occurrence of defects was related to the mixing of a specific pair of monomers.
Fig. 2. Table of the defects observed for each copolymer series. The two monomer components for each material are indicated in the far left and right columns. The % content of the monomer indicated on the left is indicated in the top row, and the % content of the monomer indicated in the right column is indicated in the bottom row. The observation of a particular defect is indicated: non-circular ( ), spread (
), spread ( ), rough (
), rough ( ) and chemical heterogeneity (
) and chemical heterogeneity ( ). White areas indicate that for this polymer a defect was not observed.
). White areas indicate that for this polymer a defect was not observed.
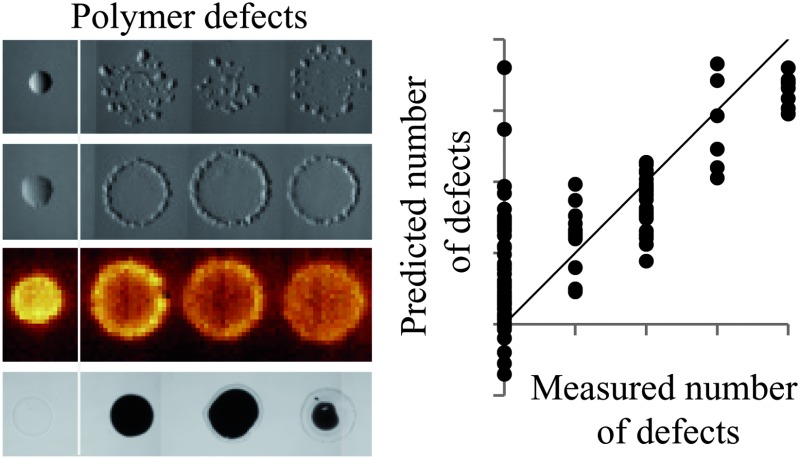
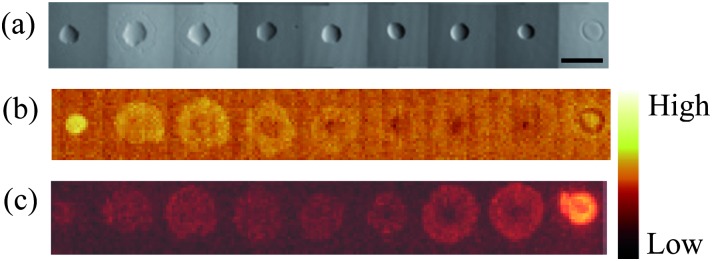
Non-circular spots were observed for monomer pairs q–l, b–d, j–c and q–b. This defect was closely associated with spreading, where all but one of the materials identified as being non-circular also spread. In a number of cases copolymers spread significantly more than the homopolymers of a monomer pair. Examples of this were observed for monomer pairs x–a, o–k, j–c, q–b and i–n. In the case of monomer pair i–n, the light microscopy image of the copolymer series suggested that all polymers were defect free, being smooth and round with an average diameter of approximately 400 μm (Fig. 3a). However, ToF-SIMS maps of ions characteristic of monomers i and n reveal that the diameter of the polymer chemistry was approximately 900 μm (Fig. 3b and c). Thus, the extent of spreading was not observed in the light microscopy image but only in the ToF-SIMS chemical maps. This was also observed for monomer pairs q–l and t–s.
Fig. 3. Analysis of the copolymer series of monomers i and n. From left to right the content (%) of monomer i is 100, 80, 75, 66, 50, 33, 25, 20 and 0. (a) Light microscopy image of the copolymer series. The scale bar is 900 μm. (b and c) ToF-SIMS images of the copolymer series. The ions mapped were (b) C4H– characteristic of monomer i and (c) C5H11O3 – characteristic of monomer n. An intensity scale for the ToF-SIMS images is shown on the right.
Heterogeneous surface chemistry was observed for monomer pairs w–o, x–o, v–a, o–k, j–c, g–k and i–n. In the case of the monomer pair q–b, the copolymers had a blotchy appearance in the light microscopy images (Fig. 1y) that suggested multiple polymer spots instead of only one. However, the ToF-SIMS chemical maps of q-bcopolymers revealed a continuous, homogeneous chemistry. Together with the observation of chemical heterogeneity, this result further demonstrates the need to assess the polymer quality not only by light microscopy but also using a chemical-mapping technique. 39,41
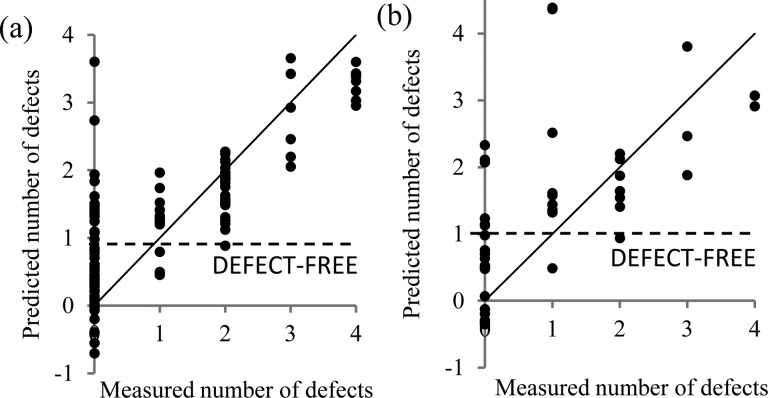
In order to understand the cause of the spot defects observed and to enable their prediction, each spot was assigned a score based upon how many of the four defects it exhibited (Fig. 2). A list of molecular descriptors was calculated for each monomer structure as a quantitative assessment of the chemical properties of the materials that could be acquired prior to polymer synthesis. For each polymer spot, a molecular descriptor value was calculated using a linear combination based upon the molar content of each monomer to represent a homogeneous molecular mixture. Additionally, for each monomer pair the difference in the value of each molecular descriptor between the monomer constituents was calculated as a measure of their difference. A total of 244 molecular descriptors were organised for each polymer as the multivariate explanatory dataset with the defect score as the univariate dataset. Partial least square (PLS) regression 42 was used to explore correlations between molecular descriptor values for each polymer with the number of spot defects 43 and to assess whether the presence of a defect in a polymer could be predicted from the molecular descriptors. A random group of 42 polymers were selected as a ‘test’ set and the remaining 129 polymers were used as a training set to build the PLS model. PLS regression has previously been useful for correlating multivariate ToF-SIMSspectra with univariate datasets such as water contact angle, the probability of stem cell colony formation, or bacterial attachment. 5,9,42 To avoid overfitting, 44 after building an initial model those descriptors with a regression coefficient below 10% of the largest regression coefficient calculated for the model were excluded and the model was calculated again with a total of 166 descriptors. The predicted number of defects for the ‘training’ and ‘test’ sets from this model are shown in Fig. 4. In the training set 85% of the materials were correctly predicted as either having or not having a defect (prediction ≥1 considered as defect present whilst a prediction <1 considered as no defect present). An identical success rate at predicting defects was also achieved in the test set, suggesting that the model is not overfitted. Thus, the model successfully predicted whether a spot formulation was likely to be defective, and demonstrated a relationship between the formation of defects in printed materials and the material properties described by the molecular descriptors of homo- and co-polymers. This offers a significant advantage to materials design, whereupon defects could be predicted without the experimental requirements of identifying defects through light microscopy, ToF-SIMS and AFM.
Fig. 4. PLS regression model prediction of the number of defects of polymer spots, calculated for the (a) training set and (b) test set. A line is drawn indicating the defect-free cut-off, below which all samples were predicted to be defect-free. The y = x line is drawn as a guide.
In the PLS model, each descriptor value was assigned a positive or negative regression coefficient depending on whether the descriptor correlated positively or negatively with the number of spot defects. The magnitude of a regression coefficient informs how strongly a particular descriptor influences the model and, thus, its correlation with the occurrence of a defect. The key molecular descriptors and their corresponding regression coefficient for the PLS model are listed in Table 1. The top 20 molecular descriptors with the most positive or negative regression coefficients are listed in Table ESI1.† Definitions of the key molecular descriptors are listed in Table 2. Differences in carbon chain length (khs.ssCH2, khs.sCH3), increased number and differences in C–F bonds (BCUTw.1l), increased number of methacrylate groups (C3sp2) and increased size of monomers or decreased number of C O or C C bonds correlated positively with the occurrence of defects whilst differences in the number of methacrylate groups and differences in the anisotropy of monomers (topoShape) correlated negatively with the occurrence of defects. This suggests that mixing a methacrylate and an acrylate monomer and/or mixing monomers with long and linear or short and bulky pendant groups are not associated with the formation of defects. This general observation may help inform the design of future systems, both monomers for formation of polymer microarrays and for other multi-component printed systems such as polymer/drug systems, hybrid polymeric resists or other polymer blends. Although the defects described occurred in micro-scale samples, it is likely that the general observations can also be applied to scaled-up applications where novel polymeric formulations are desired.
Table 1. The molecular descriptors that gave the largest positive or negative regression coefficients for PLS models describing the number of defects. Descriptors prefixed with ‘Δ’ relate to the difference in the molecular descriptor of the monomer components, whereas non-prefixed descriptors relate to the average value of the molecular descriptors of the monomer components.
| Regression coefficient | Molecular descriptor |
| 0.56 | Δkhs.ssCH2 |
| 0.52 | BCUTw.1l |
| 0.50 | Δkhs.sCH3 |
| 0.45 | C3sp2 |
| 0.45 | HybRatio |
| 0.44 | ΔBCUTw.1l |
| –1.11 | ΔC3sp2 |
| –0.97 | ΔtopoShape |
Table 2. Definitions of the key molecular descriptors listed in Table 1 and 3 .
| Molecular descriptor | Definition | Chemical significance for monomers in study |
| BCUTp.1h | Eigenvalue based descriptor that describes a chemical diversity by atomic weight, partial charge and polarisability. High polarisability gives low value | Increases with low molecular weight and cyclic or aromatic structures. Decreases with number of C–F groups |
| BCUTw.1l | Eigenvalue based descriptor that describes a chemical diversity by atomic weight, partial charge and polarisability. High atomic weight gives high value | Increases with number of C–F bonds and atomic weight |
| C1sp3 | Counts the number of connected sp3 hybridised carbon | Increases with the number of ether linkages or branching |
| C3sp2 | Counts the number of connected sp2 hybridised carbon | Counts number of methacrylate groups |
| FMF | The descriptor is the ratio of heavy atoms in the framework to the total number of heavy atoms in the molecule. By definition, acyclic molecules which have no frameworks, will have a value of 0 | Measures the extent of cyclic structures within a monomer |
| HybRatio | Characterizes molecular complexity in terms of carbon hybridization states | Score increases with a decreased number of sp2 hybridised carbon centres (C O or C C groups) and larger monomers |
| khs.dO | Counts the occurrences of double-bonded oxygen | Counts the number of (meth)acrylate groups |
| khs.sCH3 | Counts the occurrences of CH3 groups (with one single bond) | Counts the number of CH3 groups |
| khs.ssCH2 | Counts the occurrences of CH2 groups (with two single bonds) | Counts the number of CH2 groups |
| LipinskiFailures | This Class contains a method that returns the number failures of the Lipinski's Rule Of Five (has more than 5 H-bond donors, more than 10 H-bond acceptors, a molecular weight above 500 and a LogP over 5) | Larger values for large, hydrophobic monomers |
| topoShape | A measure of the anisotropy in a molecule | Larger values for straight long molecules |
| VC.4 | Evaluates the Kier and Hall chi clusters of order 4 | Larger values for monomers with many side groups (excluding H) |
The influence of a molecular descriptor on a specific defect was convoluted within the initial PLS regression model that predicted the number of defects. Thus, in order to identify which molecular descriptors correlated with the formation of specific defects PLS models were produced for each of the four defects (Fig. ESI2†). For each of these models each polymer was assigned a 1 (defect observed) or 0 (defect not observed) for each defect separately. For the training sets the presence of defects was correctly predicted for between 90.7% and 97.7% of samples, whilst correct predictions were slightly lower for the test sets (between 81.0% and 92.9% of samples) (Fig. ESI2†). The regression coefficients produced for each model were assessed to identify which molecular descriptors most strongly correlated with a particular defect (Table 3 and ESI2†). PLS models were also produced to predict quantitatively the defects of roughness and spot diameter, however, a poor correlation was observed between the measured and predicted values (R 2 < 0.42) (Fig. ESI3†).
Table 3. The molecular descriptors that gave the largest positive or negative regression coefficients for PLS models formed describing each defect. Descriptors prefixed with ‘Δ’ relate to the difference in the molecular descriptor of the monomer components, whereas non-prefixed descriptors relate to the average value of the molecular descriptors of the monomer components.
| Non-circular |
Spreading |
Rough |
Chemical heterogeneity |
||||
| Regression coefficient | Molecular descriptor | Regression coefficient | Molecular descriptor | Regression coefficient | Molecular descriptor | Regression coefficient | Molecular descriptor |
| 0.20 | C3sp2 | 0.26 | Δkhs.ssCH2 | 0.38 | ΔFMF | 0.16 | HybRatio |
| 0.12 | Δkhs.ssCH2 | 0.23 | HybRatio | 0.15 | ΔC1sp3 | ||
| 0.10 | ΔBCUTw.1l | 0.23 | khs.sCH3 | 0.14 | ΔBCUTp.1h | ||
| 0.21 | Δkhs.sCH3 | 0.11 | Δkhs.dO | ||||
| 0.20 | VC.4 | ||||||
| –0.26 | ΔC3sp2 | –0.43 | ΔC3sp2 | –0.27 | ΔtopoShape | –0.21 | ΔFMF |
| –0.24 | ΔtopoShape | –0.24 | LipinskiFailures | –0.25 | ΔC3sp2 | ||
| –0.21 | ΔtopoShape | ||||||
For materials observed to be non-circular, molecular descriptors describing the number of methacrylate groups and the difference in the number of CH2 groups (khs.ssCH2) correlated positively with the formation of the defect. This is supported by the disproportionate number of methacrylate monomers (b, c, q and l) associated with non-circular spots (compared with two acrylate monomers d and j). The difference in CH2 groups for the monomer pairs studied is indicative of mixing monomers with different lengths of ethylene glycol chains for monomer pairs b–d and j–c. Differences in length and number of C–F bonds (BCUTw.1l) also correlated with the occurrence of non-circular spots, as observed for monomer pairs b–d and q–b.
For a liquid on a surface the liquid–solid (γ SL), liquid–gas (γ) and solid–gas (γ S0 dry substrate) surface tensions are related to a spreading coefficient (S) according to eqn (1). 45
| S = γ S0 – (γ SL + γ) | 1 |
Spreading will occur when S > 0, thus, when γ S0 > γ SL + γ. 46 γ S0 is assumed to be constant for all monomer systems as all printing was conducted onto the same substrate at equivalent environmental conditions. Therefore, molecular descriptors associated with spreading are also associated with a low γ SL and/or γ. Increased spreading was correlated with differences in the number of CH2 groups (khs.ssCH2). For the monomer pairs where spreading was observed, a difference in CH2 groups is mainly associated with differences in the lengths of ethylene glycol groups, seen for monomer pairs b–d and j–c, and mixingfluorocarbons with non-fluorinated monomers as is seen for monomer pairs g–k and x–a. Monomer pairs with both a fluorinated and non-fluorinated member that did not spread included a viscous non-fluorinated di- or tri acrylate monomer such as monomer o or p. As ethylene glycol moieties are hydrophilic and fluorocarbon groups are hydrophobic, monomer mixtures with differences in these moieties will result in a net amphiphilic monomer bulk. This combination may assist with lowering γ, hence resulting in spreading. The abundance of CH3 groups (khs.sCH3) and differences in the number of CH3 groups also correlated with spreading, and is mainly associated with monomer s, which is one of two monomers which spread as a homopolymer. However, no spreading was associated with monomers m and r that also contain a number of CH3 groups. A decreased number of sp2 hybridised carbon and increased number of atoms (HybRatio) also correlated with spreading. As most of the monomers used in this study are mono(meth)acrylates, this descriptor is most strongly influenced by the number of atoms in the molecule. Thus, this descriptor suggests spreading is associated with larger monomers that are mono(meth)acrylates, which is observed for monomers d and s and monomer pair x–a. Increased complexity in molecular shape (VC.4) also correlated with spreading, observed for monomer g, j, o and monomer pair x–a.
Increased roughness of spots strongly correlated with differences in monomer components for the molecular descriptor FMF, which for this set of monomers is mainly associated with the cyclic nature of the monomers. All monomer pairs in this study that showed a significant change (>0.15) in the descriptor FMF also exhibited roughness that could be observed optically. This included monomer pairs p–s, b–d, i–q, q–b, t–s and f–r. This result strongly suggests that mixing a cyclic monomer with a non-cyclic monomer can increase the macroscale roughness of a polymer formed.
Micro-scale chemical heterogeneity correlated with differences in the number of (meth)acrylate groups (khs.dO), increased number of sp2 hybridised carbon and increased number of atoms (HybRatio), differences in the number of ether linkages or branching (C1sp3), and differences in the number of C–F bonds, molecular weight and linearity (BCUTp.1h). Differences in the number of (meth)acrylate groups is associated with mixing mono(meth)acrylates with the triacrylate monomers j or o, where chemical heterogeneities were observed in each of the copolymers that included these monomers, except for the monomer pair h–o. The occurrence of chemical heterogeneities with monomers of large molecular size was observed for monomer pairs v–a, x–a and i–n, and monomers w and x. Differences in the molecular descriptor BCUTp.1h are observed for monomer pairs w–o, x–o, x–a and g–k, all of which contain only one monomer with C–F bonds. Thus, it is likely that the correlation of this molecular descriptor with chemical heterogeneities is largely due to differences in the number of C–F bonds, which will strongly influence the monomers' hydrophobicity. The occurrence of chemical heterogeneities with monomer pairs with differences in ether linkages or branching was observed for monomer pairs v–a, x–a, b–d, j–c, g–k and i–n. In all these cases, it is the disparity in ether linkages between the two monomers that results in the increased difference in the molecular descriptor C1sp3. For these monomers, the presence of ether linkages is associated with ethylene glycol functionalities, which is strongly linked with increased hydrophilicity. Phase separation of monomers prior to polymerisation is one mechanism by which chemical heterogeneities could arise. It is, therefore, unsurprising that a number of molecular descriptors associated with a molecule's hydrophilicity/hydrophobicity were associated with the occurrence of chemical heterogeneities. However, this analysis suggests that other monomer properties such as the poor mixing of mono-(meth)acrylates with di- or tri-(meth)acrylates can also induce this defect. The use of multifunctional monomers as cross-linking agents to modulate material elasticity has been previously reported. 47 Furthermore, the rate of polymerisation and conversion vary significantly for monoacrylate and multiacrylates monomers. 48 This may tend towards the formation of block copolymers, as opposed to ideal or alternate copolymers. Thus, the chemical heterogeneities may arise through the polymerisation kinetics in addition to phase separation, although further analysis of the resultant polymers is required to verify this.
Similar to the PLS model that predicted the number of total defects, a difference in the number of methacrylate groups and differences in the anisotropy of the monomer components correlated negatively for all defects, identifying these descriptors as a strong indicator of monomer pairs which will produce defect-free polymers.
In summary, a method has been developed to assess and predict various defects that occur within printed monomer/polymer formulations. To assess the spots successfully both optical assessment and the use of ToF-SIMS chemical mapping were necessary. For 85% of the materials studied, a PLS regression model was able to successfully predict the occurrence of defects from molecular descriptors that can be calculated prior to material formation. This should offer a significant time benefit, with the possibility of predicting defects without ToF-SIMS, AFM or light microscopy analysis. Insight into the cause of defects was provided by assessing the regression coefficients calculated for each molecular descriptor within the PLS regression model, which highlighted that a difference in the number of methacrylate groups and differences in the anisotropy of the monomers components correlated negatively with the occurrence of all defects. It is expected that these results will be able to inform the future judicious choice of monomer components for formation of novel polymeric materials.
Experimental
Substrate production
Epoxy coated glass slides (Genetix) were dip-coated with a 4% (w/v) pHEMA solution in 95% ethanol to create a low fouling substrate that also enables the attachment of printed spots through physical entrapment. 33 Slides were rapidly dipped into the pHEMA solution, and withdrawn within 1 s, blotted, inverted and then maintained in a near horizontal position for 10 min before being placed into a slide rack. Slides were kept at ambient conditions for 3 days before use.
Polymer microarray formation
A selection of monomers were used as purchased from Sigma. Polymer microarrays were formed using a XYZ3200 pin printing workstation (Biodot) as described previously. 33,34 Printing conditions were O2 <1300 ppm, 25 °C, 40% humidity. Slotted metal pins (946MP6B, Arrayit) with a diameter of 220 μm were used to transfer approximately 2.4 nL of polymerisation solution onto 10 substrates before slides were irradiated with a long wave UV source for 10 s. Polymerisation solution was composed of 75% (v/v) monomer in dimethylformamide with 1% (w/v) photoinitiator 2,2-dimethoxy-2-phenylacetophenone. Once formed arrays were dried at <50 mTorr for 7 days.
Light microscopy
Light microscopy images were acquired using an Olympus IX51 light microscope and a Smart Imaging System (IMSTAR S.A.) with a 4× objective lens. Image mosaics were reconstructed and spot diameters were measured using Pathfinder™ software.
AFM
High throughput AFM measurements were taken using a Nanoscope 3000 A instrument in tapping mode as previously described. 35 Silicon tips with a resonant frequency of approximately 300 kHz and a force constant of 40 N m–1 were used (Tap300Al, Budget Sensors). 5 μm regions of the polymer were taken and the RMS roughness was measured across this region. For each polymer, 3 replicate polymers were measured and the roughness across the 3 measurements averaged.
ToF-SIMS
Measurements were conducted using a ToF-SIMS (IONTOF GmbH) instrument operated using a 25 kV Bi3 + primary ion source exhibiting a pulsed target current of ∼1 pA. Samples were scanned at a pixel density of 100 pixels per mm, with 8 shots per pixel over a given area. An ion dose of 2.45 × 1011 ions per cm2 was applied to each sample area ensuring static conditions were maintained throughout. Both positive and negative secondary ion spectra were collected (mass resolution of >7000), over an acquisition period of fifteen scans (the data from which were added together). Owing to the non-conductive nature of the samples, charge compensation, in the form of a low energy (20 eV) electron floodgun, was applied.
Generation of molecular descriptors
Molecular descriptors for all 24 monomers were calculated from SMILES input codes using the cdk toolkit, 49,50 as previously implemented in “R” 51 via the package “rcdk”. 52
PLS regression analysis of ToF-SIMS data relative to the number of polymer defects
Multivariate PLS regression analysis of the molecular descriptors relative to the number of polymer defects, spot diameter or roughness was performed as previously described. 42 Each material in the array was catalogued according to its specific observed defects (excessive spreading, increased roughness, no circularity or chemical heterogeneities). In each case, the number of defects for each polymer (0–4) was determined to produce a univariate dataset. A set of 122 molecular descriptors were calculated for each of the monomers used for polymer microarray synthesis. For each copolymer the average molecular descriptor and difference in molecular descriptor between the two monomer counterparts was calculated, creating a multivariate set of data with 244 components. A PLS model for each defect was created using the Eigenvector PLS Toolbox 3.5 for Matlab. Datasets were mean-centred prior to processing. The ‘leave one out’ method was used for cross-validation. 42 The number of latent variables used in the model was determined as the minimum in the root mean square error of cross validation (RMSECV) curve, shown in Fig. ESI4.† After initially calculating the model, descriptors with a regression coefficient less than 10% of the maximum regression coefficient calculated in the model were excluded and the model was calculated again, this time with a total of 166 descriptors.
Acknowledgments
Wellcome Trust is kindly acknowledged for funding this work (grant number 085245). Morgan Alexander acknowledges the Royal Society for the provision of a Wolfson Merit Award.
Footnotes
References
- Webster D. C. Macromol. Chem. Phys. 2008;209:237–246. [Google Scholar]
- Xiang X. D., Sun X. D., Briceno G., Lou Y. L., Wang K. A., Chang H. Y., Wallacefreedman W. G., Chen S. W., Schultz P. G. Science. 1995;268:1738–1740. doi: 10.1126/science.268.5218.1738. [DOI] [PubMed] [Google Scholar]
- Brocchini S., James K., Tangpasuthadol V., Kohn J. J. Biomed. Mater. Res. 1998;42:66–75. doi: 10.1002/(sici)1097-4636(199810)42:1<66::aid-jbm9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Hook A. L., Anderson D. G., Langer R., Williams P., Davies M. C., Alexander M. R. Biomaterials. 2010;31:187–198. doi: 10.1016/j.biomaterials.2009.09.037. [DOI] [PubMed] [Google Scholar]
- Mei Y., Saha K., Bogatyrev S. R., Yang J., Hook A. L., Kalcioglu Z. I., Cho S. W., Mitalipova M., Pyzocha N., Rojas F., Van Vliet K. J., Davies M. C., Alexander M. R., Langer R., Jaenisch R., Anderson D. G. Nat. Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernagallo S., Wu M., Gallagher M. P., Bradley M. J. Mater. Chem. 2011;21:96–101. [Google Scholar]
- Hook A. L., Thissen H., Voelcker N. H. Langmuir. 2009;25:9173–9181. doi: 10.1021/la900735n. [DOI] [PubMed] [Google Scholar]
- Hansen A., McMillan L., Morrison A., Petrik J., Bradley M. Biomaterials. 2011;32:7034–7041. doi: 10.1016/j.biomaterials.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Hook A. L., Chang C., Yang J., Luckett J., Cockrayne A., Atkinson S., Mei Y., Bayston R., Irvine D. J., Langer R., Anderson D. G., Williams P., Davies M. C., Alexander M. R. Nat. Biotechnol. 2012;30:868–875. doi: 10.1038/nbt.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkuri M. D., Driever C., Johnson G., McFarland G., Thissen H., Voelcker N. H. Biomacromolecules. 2009;10:1163–1172. doi: 10.1021/bm801417s. [DOI] [PubMed] [Google Scholar]
- Anderson D. G., Putnam D., Lavik E. B., Mahmood T. A., Langer R. Biomaterials. 2005;26:4892–4897. doi: 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Flaim C. J., Chien S., Bhatia S. N. Nat. Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- Hay D. C., Pernagallo S., Diaz-Mochon J. J., Medine C. N., Greenhough S., Hannoun Z., Schrader J., Black J. R., Fletcher J., Dalgetty D., Thompson A. I., Newsome P. N., Forbes S. J., Ross J. A., Bradley M., Iredale J. P. Stem Cell Res. 2011;6:92–102. doi: 10.1016/j.scr.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Khan F., Tare R. S., Kanczler J. M., Oreffo R. O. C., Bradley M. Biomaterials. 2010;31:2216–2228. doi: 10.1016/j.biomaterials.2009.11.101. [DOI] [PubMed] [Google Scholar]
- Khan F., Tare R. S., Oreffo R. O. C., Bradley M. Angew. Chem., Int. Ed. 2009;48:978–982. doi: 10.1002/anie.200804096. [DOI] [PubMed] [Google Scholar]
- Tourniaire G., Collins J., Campbell S., Mizomoto H., Ogawa S., Thaburet J. F., Bradley M. Chem. Commun. 2006:2118–2120. doi: 10.1039/b602009g. [DOI] [PubMed] [Google Scholar]
- Mei Y., Gerecht S., Taylor M., Urquhart A., Bogatyrev S. R., Cho S. W., Davies M. C., Alexander M. R., Langer R. S., Anderson D. G. Adv. Mater. 2009;21:2781–2786. doi: 10.1002/adma.200803184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourniaire G., Diaz-Mochon J. J., Bradley M. Comb. Chem. High Throughput Screening. 2009;12:690–696. doi: 10.2174/138620709788923692. [DOI] [PubMed] [Google Scholar]
- Yang J., Mei Y., Hook A. L., Taylor M., Urquhart A. J., Bogatyrev S. R., Langer R., Anderson D. G., Davies M. C., Alexander M. R. Biomaterials. 2010;31:8827–8838. doi: 10.1016/j.biomaterials.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epa V. C., Yang J., Mei Y., Hook A. L., Langer R., Anderson D. G., Davies M. C., Alexander M. R., Winkler D. A. J. Mater. Chem. 2012;22:20902–20906. doi: 10.1039/C2JM34782B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare R. S., Khan F., Tourniaire G., Morgan S. M., Bradley M., Oreffo R. O. C. Biomaterials. 2009;30:1045–1055. doi: 10.1016/j.biomaterials.2008.10.038. [DOI] [PubMed] [Google Scholar]
- Zhang R., Liberski A., Sanchez-Martin R., Bradley M. Biomaterials. 2009;30:6193–6201. doi: 10.1016/j.biomaterials.2009.07.055. [DOI] [PubMed] [Google Scholar]
- Hook A. L., Scurr D. J., Anderson D. G., Langer R., Williams P., Davies M., Alexander M. R. Surf. Interface Anal. 2013;45:181–184. doi: 10.1002/sia.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook A. L., Thissen H., Voelcker N. H. Biomacromolecules. 2009;10:573–579. doi: 10.1021/bm801217n. [DOI] [PubMed] [Google Scholar]
- Unciti-Broceta A., Diaz-Mochon J. J., Mizomoto H., Bradley M. J. Comb. Chem. 2008;10:179–184. doi: 10.1021/cc7001556. [DOI] [PubMed] [Google Scholar]
- Hook A. L., Creasey R., Thissen H., Hayes J. P., Voelcker N. H. Biofabrication. 2009;1:045003. doi: 10.1088/1758-5082/1/4/045003. [DOI] [PubMed] [Google Scholar]
- Aernouts T., Aleksandrov T., Girotto C., Genoe J., Poortmans J. Appl. Phys. Lett. 2008;92:033306. [Google Scholar]
- Basirico L., Cosseddu P., Scida A., Fraboni B., Malliaras G. G., Bonfiglio A. Org. Electron. 2012;13:244–248. [Google Scholar]
- Hebner T. R., Wu C. C., Marcy D., Lu M. H., Sturm J. C. Appl. Phys. Lett. 1998;72:519–521. [Google Scholar]
- Kim J. Y., Pfeiffer K., Voigt A., Gruetzner G., Brugger J. J. Mater. Chem. 2012;22:3053–3058. [Google Scholar]
- Paul K. E., Wong W. S., Ready S. E., Street R. A. Appl. Phys. Lett. 2003;83:2070–2072. [Google Scholar]
- Scoutaris N., Alexander M. R., Gellert P. R., Roberts C. J. J. Controlled Release. 2011;156:179–185. doi: 10.1016/j.jconrel.2011.07.033. [DOI] [PubMed] [Google Scholar]
- Anderson D. G., Levenberg S., Langer R. Nat. Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- Hook A. L., Chien Yi C., Yang J., Scurr D., Langer R., Anderson D. G., Atkinson S., Williams P., Davies M. C., Alexander M. R. J. Visualized Exp. 2012 doi: 10.3791/3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook A. L., Yang J., Chen X., Roberts C. J., Mei Y., Anderson D. G., Langer R., Alexander M. R., Davies M. C. Soft Matter. 2011;7:7194–7197. doi: 10.1039/C1SM06063E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M., Urquhart A. J., Zelzer M., Davies M. C., Alexander M. R. Langmuir. 2007;23:6875–6878. doi: 10.1021/la070100j. [DOI] [PubMed] [Google Scholar]
- Urquhart A. J., Anderson D. G., Taylor M., Alexander M. R., Langer R., Davies M. C. Adv. Mater. 2007;19:2486–2491. [Google Scholar]
- Tweedie C. A., Anderson D. G., Langer R., Van Vliet K. J. Adv. Mater. 2005;17:2599–2604. [Google Scholar]
- Celiz A. D., Hook A. L., Scurr D. J., Anderson D. G., Langer R., Davies M. C., Alexander M. R. Surf. Interface Anal. 2013;45:202–205. doi: 10.1002/sia.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Eynde X., Bertrand P., Penelle J. Macromolecules. 2000;33:5624–5633. [Google Scholar]
- Scurr D. J., Hook A. L., Burley J., Williams P. M., Anderson D. G., Langer R., Alexander M. R., Davies M. C. Surf. Interface Anal. 2013;45:466–470. doi: 10.1002/sia.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M., Urquhart A. J., Anderson D. G., Langer R., Davies M. C., Alexander M. R. Surf. Interface Anal. 2009;41:127–135. doi: 10.1002/sia.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoutaris N., Hook A. L., Gellert P. R., Roberts C. J., Alexander M. R., Scurr D. J. J. Mater. Sci.: Mater. Med. 2012;23:385–391. doi: 10.1007/s10856-011-4474-5. [DOI] [PubMed] [Google Scholar]
- Le T., Epa V. C., Burden F. R., Winkler D. A. Chem. Rev. 2012;112:2889–2919. doi: 10.1021/cr200066h. [DOI] [PubMed] [Google Scholar]
- de Gennes P. G. Rev. Mod. Phys. 1985;57:827–863. [Google Scholar]
- Bonn D., Eggers J., Indekeu J., Meunier J., Rolley E. Rev. Mod. Phys. 2009;81:739–805. [Google Scholar]
- Pedron S., Peinado C., Bosch P., Benton J. A., Anseth K. S. J. Biomed. Mater. Res., Part A. 2011;96:196–203. doi: 10.1002/jbm.a.32974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C., Moussa K. Macromolecules. 1989;22:4455–4462. [Google Scholar]
- Steinbeck C., Han Y. Q., Kuhn S., Horlacher O., Luttmann E., Willighagen E. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck C., Hoppe C., Kuhn S., Floris M., Guha R., Willighagen E. L. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- R-Development-Core-Team, R Foundation for Statistical Computing, Vienna, 2012, vol. 2011. [Google Scholar]
- Guha R. J. Stat. Software. 2007;18 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.