Abstract
Epigenetic gene silencing is seen in several repeat-expansion diseases. In fragile X syndrome, the most common genetic form of mental retardation, a CGG trinucleotide–repeat expansion adjacent to the fragile X mental retardation 1 (FMR1) gene promoter results in its epigenetic silencing. Here, we show that FMR1 silencing is mediated by the FMR1 mRNA. The FMR1 mRNA contains the transcribed CGG-repeat tract as part of the 5′ untranslated region, which hybridizes to the complementary CGG-repeat portion of the FMR1 gene to form an RNA·DNA duplex. Disrupting the interaction of the mRNA with the CGG-repeat portion of the FMR1 gene prevents promoter silencing. Thus, our data link trinucleotide-repeat expansion to a form of RNA-directed gene silencing mediated by direct interactions of the trinucleotide-repeat RNA and DNA.
Fragile X syndrome (FXS) results from the absence of the fragile X mental retardation protein (FMRP), which is encoded by the fragile X mental retardation 1 (FMR1) gene on the X chromosome (1). Impaired FMRP expression is caused by an inherited CGG trinucleotide–repeat expansion adjacent to the FMR1 promoter (2). FMR1 alleles that contain >200 CGG repeats undergo epigenetic silencing of the FMR1 promoter at ~11 weeks of gestation (2–4). It remains unclear how the CGG-repeat expansion leads to gene silencing.
The mechanism of gene silencing in FXS has been particularly difficult to dissect. Transgenes containing expanded CGG-repeat FMR1 alleles are not silenced in cell lines or mice (5). Furthermore, overexpression of expanded CGG-repeat sequences is complicated by sequence instability in plasmids (6). Recently, human embryonic stem cells (hESCs) harboring an FMR1 allele with >200 CGG repeats (FXS hESCs) were shown to undergo FMR1 gene silencing upon differentiation (7). The switch from active FMR1 gene expression to FMR1 silencing in FXS hESCs resembles the switch that occurs in FXS embryos (4), which provides an in vitro system to study FMR1 silencing.
To monitor FMR1 silencing, we used two FXS lines, WCMC-37 and SI-214 (8, 9) (referred to throughout the text as FXS-1 and FXS-2, respectively); each contains >400 CGG repeats (fig. S1). We monitored FMR1 silencing after inducing neuronal differentiation over 60 days (fig. S2). In neurons derived from FXS hESCs, FMRP and FMR1 mRNA were readily detected until ~48 days, at which point the levels dropped and were absent by day 51 (fig. S3 to S5). The FMR1 promoters in the undifferentiated FXS hESC lines contain high levels of histone H3 dimethylated on lysine 4 (H3K4me2, associated with gene expression) and low levels of histone H3 dimethylated on lysine 9 (H3K9me2, associated with gene repression) (fig. S6). However, in differentiated FXS cells, the FMR1 promoter switched to the repressive H3K9me2 mark, with a concomitant reduction in the levels of H3K4me2 (fig. S6). Taken together, these data indicate that the loss of FMRP and FMR1 mRNA in FXS hESC lines correlates with the epigenetic silencing of the FMR1 promoter.
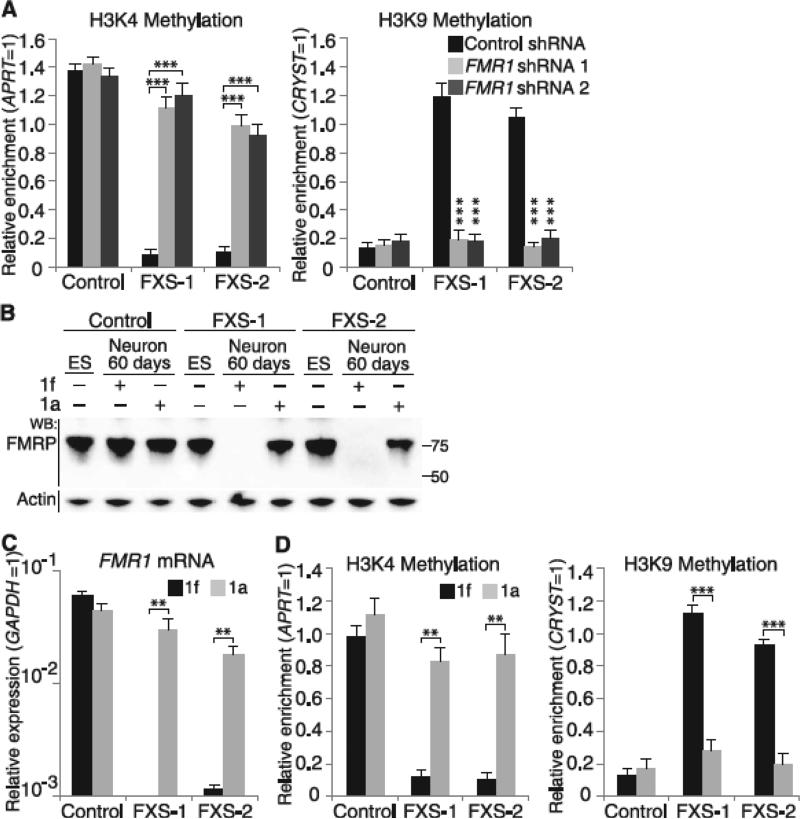
The expanded CGG repeat influences DNA structure, which may be recognized by pathways that induce epigenetic silencing (10). However, the expanded CGG repeat is also transcribed as the 5′ untranslated region (UTR) of the FMR1 mRNA, which makes it possible that the mRNA could induce promoter silencing. We therefore tested whether the FMR1 transcript is required for silencing. Knockdown of FMR1 mRNA in FXS hESCs prevented differentiation-induced FMR1 silencing, as measured by the retention of transcriptionally active histone marks at the FMR1 promoter (Fig. 1A). The effect of the FMR1-specific short hairpin RNAs (shRNAs) was not due to knockdown of a previously described antisense FMR1 transcript that partially overlaps with the FMR1 mRNA (11) (fig. S7B). These data indicate that the FMR1 transcript is required for FMR1 silencing.
Fig. 1.
The FMR1 transcript and its CGG-repeat tract are required for FMR1 silencing. (A) FMR1 mRNA is required for FMR1 silencing in differentiating FXS hESCs. shRNA-expressing lentivirus was applied at day 1, and histone marks at FMR1 promoters were measured at day 60. FXS hESCs expressing control shRNA showed high levels of transcriptionally repressive marks (H3K9me2) and low levels of transcriptionally active marks (H3K4me2). FMR1-specific shRNA prevented the appearance of repressive marks and maintained the expression of transcriptionally active marks (n = 4 per condition). ES, hESCs; WB, Western blot; APRT, adenine phosphoribosyltransferase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CRYST, crystallin. (B to D) The CGG-repeat RNA-binding small molecule 1a blocks FMR1 silencing. Differentiating FXS hESCs treated with 10 μM 1a did not lose FMRP (B) or FMR1 mRNA (C) (n = 3 per condition) and retained active FMR1 promoters (D) (n = 3 per condition). Data are means ± SEM. Statistical analysis was performed using Student's t test (two-tailed distribution, **P < 0.01, ***P < 0.001). When comparing different cell lines, we considered the samples as two samples with unequal variance. When comparing different conditions on the same cell line, we considered the samples as two samples with equal variance.
CGG repeats in mRNA form a hairpin structure (10) (fig. S7C), which may be unfolded and linearized in order to mediate FMR1 silencing. To test the role of the mRNA CGG repeat in FMR1 gene silencing, we used 1a, a small molecule that selectively binds the repeating G-G internal loops in the RNA hairpin and inhibits its thermal melting (12) (fig. S7D). Application of 1a (10 μM) throughout the differentiation prevented differentiation-induced FMR1 silencing in FXS hESCs (Fig. 1, B to D, and fig. S8). Application of the structurally related control compound 1f (10 μM), which does not bind CGG repeats (12), did not affect FMR1 silencing. The effect of 1a was not due to impaired differentiation, as the 1a-treated cells expressed the neuronal marker β-III tubulin (fig. S9). Together, these data suggest that linearization of the CGG-repeat hairpin in the FMR1 transcript is required for silencing.
Dicer processes CGG-repeat RNAs into small RNAs in vitro (13) and may contribute to FMR1 silencing (14). However, knockdown of Dicer, Ago1, or Ago2 did not prevent differentiation-induced FMR1 silencing (fig. S10). Thus, FMR1 silencing does not require a Dicer-directed pathway.
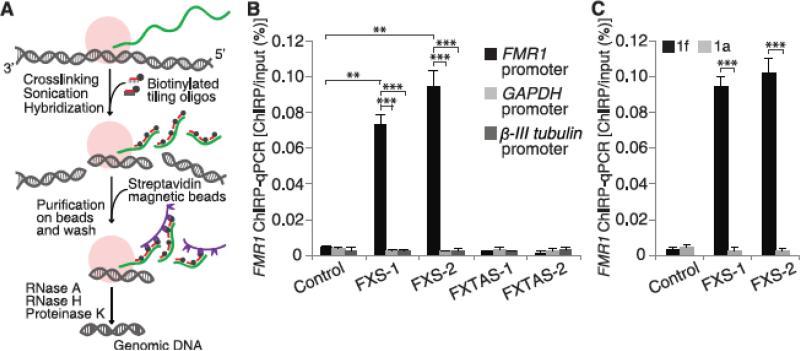
We next asked if the FMR1 transcript directs silencing by binding, directly or indirectly, to the promoter. To test this, we measured FMR1 mRNA binding to the FMR1 gene using the chromatin isolation by RNA purification (ChIRP) technique (Fig. 2A) (15, 16). After cross-linking endogenous RNA to its binding partners, we pulled down FMR1 mRNA using biotinylated oligonucleotides that hybridize along the length of the transcript. The amount of FMR1 promoter pulled down was quantified by quantitative polymerase chain reaction (qPCR) with promoter-specific primers. In control hESC-derived neurons, there was minimal FMR1 transcript bound to the FMR1 promoter at any time point during differentiation (Fig. 2B and fig. S11). Similarly, in FXS hESC-derived neurons at 12, 24, 36, and 60 days, minimal FMR1 transcript was bound to the promoter (fig. S11). However, at 45 days in FXS hESC-derived neurons, FMR1 mRNA was readily detectable on the FMR1 promoter (Fig. 2B). Linearization of the CGG hairpin is required for binding of the FXS FMR1 mRNA to the FXS FMR1 promoter, as treatment of both FXS hESC lines with 1a, but not 1f, reduced binding of the FMR1 RNA to the promoter during differentiation (Fig. 2C).
Fig. 2.
FMR1 mRNA interacts with the FMR1 promoter in a CGG repeat–dependent manner. (A) Schematic representation of ChIRP technique [adapted from (15)]. RNA (green) and protein complexes (pink) are cross-linked to the DNA (gray) in cells by glutaraldehyde. The cell lysate is sonicated to shear DNA to ~500 bp. Streptavidin beads (purple) are used to pull down biotinylated oligonucleotides hybridized to RNA. Bound DNA sequences are detected by qPCR. (B) FMR1 mRNA interacts with the FMR1 promoter. FMR1 mRNA bound to the FMR1 gene was measured by ChIRP at day 45 of differentiation (see fig. S11 for other time points). FMR1 mRNA was readily detectable on the FMR1 promoter in FXS neurons but not control neurons (n = 3 per condition). FMR1 mRNA does not bind to FMR1 promoters in FMR1 premutation lines, FXTAS-1 and FXTAS-2, that contain 70 and 73 CGG repeats, respectively. GAPDH and β-III tubulin promoters were used as controls. (C) The CGG-repeat portion of the transcript is required for the binding of FMR1 mRNA to the FMR1 gene. FMR1 binding to the FMR1 gene was markedly reduced in FXS hESC-derived neurons cultured in the presence of 1a. The control compound 1f did not block the FMR1 transcript–FMR1 gene interaction. Data are means ± SEM; Student's t test (two-tailed distribution, **P < 0.01, ***P < 0.001). Different conditions on the same cell line were considered as two samples with equal variance; different cell lines were considered as two samples with unequal variance.
The length of the CGG-repeat tract is the major determinant for FMR1 gene silencing (17). Normal alleles (less than 55 repeats) and “premutation” alleles (55 to 200 repeats) do not lead to FMR1 silencing (17). We therefore asked if the FMR1 transcript binds to the FMR1 promoter in normal and premutation lines. In normal (~30 repeats) and premutation (70 and 73 repeats) hESC lines (fig. S1), the FMR1 transcripts were not bound to the promoter (Fig. 2B). Thus, the lack of FMR1 silencing in normal and premutation hESC lines may reflect the absence of FMR1 mRNA binding to the promoter.
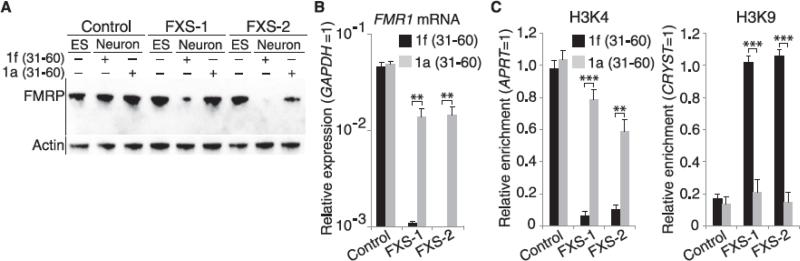
To more precisely define the temporal sensitivity of FMR1 silencing to 1a, we selectively applied 1a during days 1 to 30 (fig. S12) or 31 to 60 of differentiation (Fig. 3). Only application of 1a during days 31 to 60 blocked silencing (Fig. 3). Similarly, application of FMR1-specific shRNA during days 31 to 60 also prevented differentiation-induced silencing (fig. S13). These data suggest that FMR1 mRNA does not trigger gene silencing during the first 30 days but is required in the second 30-day period when it binds to the FMR1 gene and leads to gene silencing.
Fig. 3.
Temporal requirement for FMR1 mRNA binding to the FMR1 promoter. (A) The small molecule 1a blocks the drop in FMRP levels during days 31 to 60 of differentiation. To determine when FMR1 mRNA is required for FMR1 silencing, we applied 1a to FXS hESCs during days 31 to 60 of differentiation. 1a maintained FMRP expression in FXS neurons. (B and C) 1a prevents FMR1 silencing. In FXS neurons, application of 1a during days 31 to 60 of differentiation was sufficient to maintain FMR1 mRNA levels [(B), quantitative reverse transcription polymerase chain reaction (qRT-PCR), n = 4 per condition], high levels of H3K4me2 and low levels of H3K9me2 histone modifications [(C), chromatin immunoprecipitation, n = 4 per condition]. Control hESCs were unaffected by 1a. Application of 1a (10 μM) during days 1 to 30 of differentiation failed to prevent FMR1 silencing (see fig. S12). Data are means ± SEM; Student's t test (two-tailed distribution, **P < 0.01, ***P < 0.001). Different conditions on the same cell line were considered as two samples with equal variance.
We next asked whether FMR1 gene silencing can occur after the 48- to 51-day time point of the differentiation protocol. In these experiments, we inhibited FMR1 silencing by culturing differentiating FXS hESCs in the presence of 1a for 60 days. Withdrawal of 1a for 10 days triggered FMR1 silencing (fig. S14), which suggests that sustained 1a treatment is required to maintain FMR1 in the transcriptionally active state. The small molecule 1a appears to function to prevent silencing, rather than reverse silencing, as application of 1a to cells with an already silenced FMR1 promoter did not reverse silencing (fig. S15).
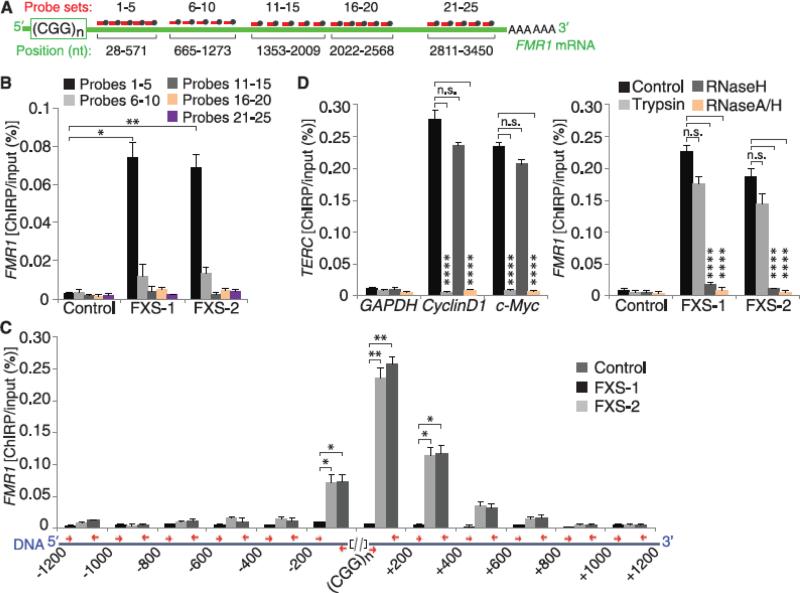
We next mapped the part of the FMR1 transcript that binds to the FMR1 gene. To do this, we performed ChIRP using biotinylated primers that hybridize to different regions of the FMR1 transcript (Fig. 4A). Primers that hybridize adjacent to the CGG repeat pulled down the FMR1 promoter, whereas primers directed elsewhere along the FMR1 transcript resulted in markedly reduced pull down (Fig. 4B). Together with the finding that 1a blocks the binding of FMR1 mRNA to the FMR1 gene, these data suggest that the CGG-repeat region of the FMR1 transcript interacts with the FMR1 gene.
Fig. 4.
The CGG-repeat portion of the FMR1 mRNA hybridizes to the complementary region of the FMR1 DNA. (A) Schematic of the hybridization sites of the different biotinylated-oligonucleotide sets used to pull down sheared RNA in ChIRP experiments (see methods for further details on probe design). (B) The 5′ UTR portion of the FMR1 transcript binds to the FMR1 gene. ChIRP was performed using the probe sets shown in (A). Only probes that bind the CGG repeat–proximal portion of the FMR1 transcript pulled down the FMR1 promoter in FXS neurons (n = 4 per condition). (C) The FMR1 transcript binds to the CGG-repeat portion of the FMR1 gene. To determine where the FMR1 mRNA binds on the FMR1 gene, we measured the ChIRP signal along a 1200-bp region both upstream and downstream of the genomic CGG repeat. The positions of the primers used to amplify portions of the FMR1 gene (blue) are indicated (red arrows) (referred to as bp relative to the 5′ or 3′ end of the CGG repeat). The ChIRP signal was highly enriched adjacent to the genomic CGG repeat (n = 3 per condition) (see also fig. S16). (D) The FMR1 mRNA binds to the FMR1 DNA in a protein-independent and RNase H–sensitive manner. The binding of the control noncoding-RNA TERC to its target promoters (CyclinD1 and c-Myc) was abolished after trypsin treatment (n = 3), whereas the binding of the FMR1 transcript to the FMR1 gene was unaffected by trypsin in FXS hESC-derived neurons (n = 3 per condition). In contrast, RNase H treatment only blocked the FMR1 ChIRP signal (n = 3 per condition). RNase A–RNase H treatment, which digests all RNA, is used as a control to demonstrate the RNA-dependence of the ChIRP signal. Data are means ± SEM; Student's t test (two-tailed distribution, *P < 0.05, **P < 0.01, ****P < 0.0001). Different conditions on the same cell line were considered as two samples with equal variance; different cell lines were considered as two samples with unequal variance.
We next mapped the part of the FMR1 gene that is bound to the FMR1 mRNA. In our previous ChIRP experiments, we measured the binding of the FMR1 mRNA to a portion of the FMR1 promoter that lies 92 to 196 base pairs (bp) upstream of the CGG-repeat sequence. To more precisely define the binding site, we measured the ChIRP signal both upstream and downstream of the genomic CGG-repeat sequence in differentiating FXS hESCs. Because of the high G/C content of the repeat sequence, binding to this region cannot be tested. We found that the ChIRP signal was detectable on both sides of the ~1200-bp genomic CGG repeat and is markedly reduced at sites away from the repeat (Fig. 4C). This pattern of binding is consistent with the genomic CGG-repeat sequence being the binding site for the FMR1 mRNA (see fig. S16).
Some noncoding RNAs have been shown to interact with promoters indirectly through a protein intermediate (18). To determine if FMR1 mRNA binds to its promoter through a protein intermediate, we performed ChIRP experiments, except we treated the cross-linked lysate with trypsin to digest any protein intermediates. For the control TERC noncoding RNA (15), the binding to its target promoters was abolished after trypsin treatment (Fig. 4D). However, FMR1 mRNA binding to the FMR1 gene was not affected by trypsin (Fig. 4D).
We next asked if FMR1 mRNA binds to the FMR1 gene by forming an RNA•DNA heteroduplex. To test this, we used ribonuclease H (RNase H), which selectively degrades RNA hybridized to DNA. Treatment of the cross-linked DNA fragments with RNase H selectively prevented the pull down of the FMR1 promoter but did not affect the TERC pull downs (Fig. 4D). These data indicate that FMR1 mRNA binds the FMR1 gene through a direct RNA·DNA duplex and does not require a protein intermediate.
The FMR1 mRNA CGG repeat may bind to the complementary CCG portion of the DNA that becomes accessible while it is being transcribed. Indeed, transcription through G-rich sequences causes stalling in vitro and in vivo (19, 20). Conceivably, the nascent FMR1 CGG-repeat RNA interacts with the template strand of the unwound DNA to form a RNA•DNA duplex that is highly stabilized by its G/C content. This would require that the CGG-repeat sequence in the RNA achieves a sufficient length to reach back and interact with the DNA and may contribute to the requirement for >200 CGG repeats for silencing. In addition, the length of the resulting RNA•DNA duplex may need to be of sufficient length to activate downstream pathways that induce FMR1 silencing.
The initial step in FMR1 silencing is the binding of the FMR1 mRNA to the genomic repeat. The inability of the FMR1 transcript to bind to the DNA before day 45 may relate to DNA accessibility during transcription. The expression of diverse DNA helicases, which are known to regulate DNA accessibility (21, 22), is reduced during hESC differentiation (23). Conceivably, helicase activity may contribute to the temporal interaction of FMR1 mRNA and DNA. However, the exact mechanisms underlying the temporal nature of FMR1 silencing remain unknown.
The formation of the FMR1 RNA•DNA duplex coincides with the initiation of epigenetic silencing in the FMR1 gene. Because this causes a drop in FMR1 mRNA expression, subsequent maintenance of FMR1 silencing is unlikely to be FMR1 mRNA–dependent. It remains to be determined which mechanisms maintain epigenetic silencing of FMR1 throughout the patient's lifetime.
FXS hESCs allow the characterization of the endogenous FMR1 transcript transcribed from the endogenous promoter. This is important because RNA-directed gene silencing frequently occurs in cis with the nascent transcript affecting a promoter within the gene locus (24). Indeed, only FMR1 and not other CGG repeats in the genome are silenced in FXS (25). Thus, rather than overexpressing CGG-repeat RNAs, which is complicated by plasmid instability, pharmacologic targeting of the endogenous CGG repeat provides insight into its role in promoter silencing.
Our data demonstrate that an mRNA can mediate promoter silencing and links trinucleotide repeat expansion to a novel form of RNA-directed promoter silencing. Epigenetic changes are seen in diverse repeat-expansion diseases (26, 27). The prevalence of repeat expansion–associated epigenetic changes raises the possibility that aspects of the mRNA-directed gene silencing pathway described here may contribute to gene expression alterations in other repeat-expansion diseases as well.
Supplementary Material
Acknowledgments
We thank members of the Jaffrey lab for helpful comments and suggestions, E. Ferretti for assistance with chromatin immunoprecipitation, Q. Zhan for help with hESC cultures, S.L. Nolin and C. Dobkin for PCR on premutation lines. hESC lines are available from N.Z. and Z.R. (Weill Cornell Medical College hESC lines, nizanin@med.cornell.edu, zrosenw@med.cornell.edu) and S.R.J. (SI-214 hESC line, srj2003@med.cornell.edu), subject to approval by the Embryonic Stem Cell Research Oversight Committee (ESCRO) of Weill Cornell Medical College. M.D.D. previously consulted for SMaRT (Small Molecule Approach to Targeting RNA) Therapeutics. A patent (U.S. Patent and Trademark Office application no. 61/694,977) was filed on 30 August 2012 by the Scripps Research Institute governing composition and use of 1f and 1a. This work was supported by the Tri-Institutional Stem Cell Initiative (Tri-I SCI) grant 2008-019 (S.R.J., N.Z., and Z.R.), New York Stem Cell Foundation-Druckenmiller Fellowship (D.C.), Life Sciences Research Foundation Fellowship and Tri-I SCI postdoctoral fellowship (M.S.C.), and a FRAXA postdoctoral fellowship (W.-Y.Y.). Portions of this project not involving non-NIH registry stem cells were supported by NIH R01 MH80420 (S.R.J.) and NIH R01 GM079235 (M.D.D.).
References and Notes
- 1.Verkerk AJ, et al. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 2.Oberlé I, et al. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 3.Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Am. J. Hum. Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willemsen R, Bontekoe CJ, Severijnen LA, Oostra BA. Hum. Genet. 2002;110:601–605. doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- 5.Brouwer JR, et al. Exp. Cell Res. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandberg G, Schalling M. Nucleic Acids Res. 1997;25:2883–2887. doi: 10.1093/nar/25.14.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eiges R, et al. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Gerhardt J, et al. Mol. Cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verlinsky Y, et al. Reprod. Biomed. Online. 2005;10:105–110. doi: 10.1016/s1472-6483(10)60810-3. [DOI] [PubMed] [Google Scholar]
- 10.Usdin K, Woodford KJ. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladd PD, et al. Hum. Mol. Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 12.Disney MD, et al. ACS Chem. Biol. 2012;7:1711–1718. doi: 10.1021/cb300135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handa V, Saha T, Usdin K. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin P, Alisch RS, Warren ST. Nat. Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 15.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon MD, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y, Lakkis L, Devys D, Warren ST. Am. J. Hum. Genet. 1995;56:106–113. [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JT. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 19.Belotserkovskii BP, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabczyk E, Fishman MC. J. Biol. Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
- 21.Bochman ML, Paeschke K, Zakian VA. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackintosh SG, Raney KD. Nucleic Acids Res. 2006;34:4154–4159. doi: 10.1093/nar/gkl501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu JQ, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guil S, Esteller M. Nat. Struct. Mol. Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 25.Alisch RS, et al. BMC Med. Genet. 2013;14:18. doi: 10.1186/1471-2350-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans-Galea MV, Hannan AJ, Carrodus N, Delatycki MB, Saffery R. Trends Mol. Med. 2013;19:655–663. doi: 10.1016/j.molmed.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Xi Z, et al. Am. J. Hum. Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.