Abstract
Human embryonic stem cells (hESCs) hold great biomedical promise, but experiments comparing them produce heterogeneous results, raising concerns regarding their reliability and utility, although these variations may result from their disparate and anonymous origins. To determine whether primate ESCs have intrinsic biological limitations compared with mouse ESCs, we examined expression profiles and pluripotency of newly established nonhuman primate ESC (nhpESCs). Ten pedigreed nhpESC lines, seven full siblings (fraternal quadruplets and fraternal triplets), and nine half siblings were derived from 41 rhesus embryos; derivation success correlated with embryo quality. Each line has been growing continuously for ~1 year with stable diploid karyo-type (except for one stable trisomy) and expresses in vitro pluripotency markers, and eight have already formed teratomas. Unlike the heterogeneous gene expression profiles found among hESCs, these nhpESCs display remarkably homogeneous profiles (>97%), with full-sibling lines nearly identical (>98.2%). Female nhpESCs express genes distinct from their brother lines; these sensitive analyses are enabled because of the very low background differences. Experimental comparisons among these primate ESCs may prove more reliable than currently available hESCs, since they are akin to inbred mouse strains in which genetic variables are also nearly eliminated. Finally, contrasting the biological similarities among these lines with the heterogeneous hESCs might suggest that additional, more uniform hESC lines are justified. Taken together, pedigreed primate ESCs display homogeneous and reliable expression profiles. These similarities to mouse ESCs suggest that heterogeneities found among hESCs likely result from their disparate origins rather than intrinsic biological limitations with primate embryonic stem cells.
Keywords: Embryonic stem cells, Pluripotency, Differentiation, Gene expression, Primates
Introduction
Human embryonic stem cells (hESCs) may prove invaluable for investigating the molecular mechanisms of development and disease, but results from the available hESC lines are heterogeneous, and comparisons among hESCs produce varying results [1–4]. Transcriptional profile comparisons among different hESC lines, even lines derived and grown within the same laboratory, show concordant rates of 60.2% [1] and 80% [5], quite different from the >99% concordance found after profiling mouse ESCs (mESCs) [6, 7]. Because the available hESC lines have disparate origins both in their genetic and biological backgrounds as well as in the isolating laboratory [4, 8, 9] and are derived from clinically discarded embryos donated by infertile couples, it is difficult to know whether the observed variability is due to these differences or if hESCs have biological variability intrinsically different from mESCs.
ESCs from mice remain unique in their ability to generate chimeric fetuses and offspring after reaggregation with diploid or tetraploid mouse embryos, and mESCs are capable of transmission through the germ line to form either fertile sperm or eggs [10, 11]. Nuclear reprogramming with either abnormal mouse eggs or in vitro has just been reported using mouse cells [12–15]. Embryos from various mammals differ markedly in their biological capacities, with domestic species succeeding in somatic cell nuclear transfer [16] before mice [17], and yet ESC lines from other species, including rats, have yet to be characterized unequivocally [18, 19]. Mice are unrivalled for their experimental utilities, and mouse embryos are also capable of arresting development in utero at the blastocyst stage, a process known as diapause [20]. These properties may also contribute to the remarkable features of mESCs to remain stable in the undifferentiated state while continuing to proliferate. This also raises questions as to whether ESC cells from other mammals, including primates, will share some or all of these properties.
Notwithstanding the invaluable research contributions of mESCs, there are still some limitations for modeling human diseases with mESCs, prompting primate ESC derivations first in monkeys in 1995 (from in vivo embryos flushed after mating) [21] and then in humans in 1998 [22]. Since the original five hESC lines published by Thomson et al. [22], dozens of new hESC lines have been established [5, 23–26] from clinically discarded embryos generated by Assisted Reproductive Technologies (ART) clinics around the world and donated anonymously by infertile patients/couples. The journal Nature reports >275 hESC lines worldwide [27], and the International Society for Stem Cell Research's registry cites well over 100 hESC lines (http://isscr.org/science/sclines.htm). Nonhuman primates afford research opportunities beyond those available from human ART specimens, including the deliberate production of embryos with defined genetics using pedigreed fertile gametes as well as chimera and germ-line transmission testing for pluripotency. The sequencing of the rhesus genome [28] now permits direct genomic comparison among monkeys, apes, and humans. American and Japanese researchers have derived several macaque nonhuman primate ESC (nhpESC) lines [29–31], including a parthenogenetic ESC line [32]. Pluripotency has been demonstrated in nhpESCs using hESC criteria, that is, ability to form tissues representative of all three germ layers in vitro and in teratomas. To address whether primate ESCs are biologically heterogeneous or whether variations among hESCs might be due to their varying origins, we established pedigreed nhpESCs using gametes from fertile rhesus and compared expression profiles and pluripotency within and among these family groups.
Here, we describe the derivation of ten new nonhuman primate embryonic stem cell lines of known pedigree and demonstrate that they are pluripotent using in vitro and teratoma assays. Derivation success was directly related to embryo quality as measured by rapid preimplantation development where rate of development correlated with likelihood of success in ESC line establishment. The gene expression patterns of these 10 lines are remarkably homogeneous but with still-detectable differences among families and between sexes. Taken together, this suggests that the variability among hESCs is likely the result of their derivation from genetically diverse embryos in disparate research facilities around the world, and that the observed heterogeneity among hESCs is not due to intrinsic biological variability, as compared with mice, but rather is due to the restricted numbers and qualities of the hESC lines widely available.
Materials and Methods
Pedigreed Nonhuman Primate Embryonic Stem Cell Derivations
Embryos were generated from fertile rhesus using intracytoplasmic sperm injection (ICSI) and developed to the blastocyst stage as described [33] following Institutional Animal Care and Use Committee approved protocols. The best embryos were collected from fertilized zygotes observed by Hoffman Modulation Contrast optics for additional in vitro culture between 18–20 hours post-ICSI based on the following criteria: full elicitation of a second polar body; two apposed pronuclei; no cytoplasmic vacuoles, fragmentation, or excessive concentration within the egg center. Additionally, zygotes that failed to fertilize, demonstrated polyploidy (greater than two apposed pronuclei), or were atretic were removed before first division and excluded from this study. Preselected zygotes were transferred to CMRL medium + Buffalo rat liver cell coculture and monitored daily for embryonic development to expanded blastocyst as previously described [34]. Inner cell mass (ICM) cells were isolated from expanded blastocysts using immunosurgery [35]. First, the zonae pellucidae were removed by exposure to acid Tyrode's medium (Specialty Media, Phillipsburg, NJ, http://www.specialtymedia.com) for 30–45 seconds. The ICM was next isolated by exposure first to anti-monkey antiserum (1:3; Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com) for 15–20 minutes at 37°C and then transferred to guinea pig complement (1:3; Biomeda, Foster City, CA, http://biomeda.com) for an additional 15–20 minutes at 37°C. This destroys the outer trophectoderm, leaving the ICM cells intact. Mechanical pipetting with a fine needle (inner diameter 75 μm) was used to dissociate the ICM prior to washing and plating onto mitomycin C-treated mouse embryonic fibroblasts (MEF; Specialty Media) in 80% knockout medium, 20% knockout serum replacer, 1 mM l-glutamine, 0.1 mM nonessential amino acids, 1% penicillin/streptomycin, 12 ng/ml basic fibroblast growth factor, 10 ng/ml Activin A [36], and 10 ng/ml recombinant human leukemia inhibitory factor (hLIF) (all components from Invitrogen [Carlsbad, CA, http://www.invitrogen.com] except hLIF [Chemicon, Temecula, CA, http://www.chemicon.com] and Activin A [Sigma]). After 10–14 days, regions of expanded ICM with characteristic ESC morphology (tightly packed cells with high nuclear/cytoplasm ratio and prominent nucleoli) were dissociated with a fine glass capillary and colony pieces transferred to new MEF for expansion with culture medium changed every 48 hours.
Establishment, Characterizations, and Confirmations of Pedigreed nhpESC Lines
Pluripotency Markers Detected by Immunocytochemistry
nhpESCs were assayed for characteristic pluripotency markers Oct-4, Nanog, stage-specific embryonic antigen (SSEA)-4, TRA 1-81, and TRA 1-60 as well as the negative hESC-nhpESC marker SSEA-1 (Oct-4, TRA 1-81, and TRA 1-60 from [Santa Cruz Bio-technology Inc., Santa Cruz, CA, http://www.scbt.com]; Nanog from [R&D Systems Inc., Minneapolis, http://www.rndsystems.com]; SSEA-4 and SSEA-1 from [Developmental Studies Hybrid-oma Bank, Iowa City, IA, http://www.uiowa.edu/~dshbwww]). Immunocytochemistry was performed on undifferentiated colonies after fixation by 2% paraformaldehyde in phosphate-buffered saline (PBS) for 40 minutes. After washes in PBS + 1% Triton X-100 (PBS-Tx; Sigma) for 15 minutes, nonspecific binding of the primary, excluding Nanog, and secondary antibodies was blocked by a 30-minute incubation in PBS containing 5% goat serum and 0.3% bovine serum albumin. Nanog staining was carried out without the blocking step. Primary antibodies were applied for 40 minutes at 37°C in a humidified chamber, washed extensively in PBS-Tx, and then incubated in appropriate fluorescently labeled secondary antibodies (40 minutes). DNA was detected with 5 μM TOTO-3 (Molecular Probes, Eugene, OR, http://probes.invitrogen.com) added at room temperature 20 minutes prior to mounting coverslips in Vectashield antifade medium (Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com) to retard photobleaching. Slides were examined using laser scanning confocal microscopy [37].
Pluripotency Markers Detected by Reverse Transcription-Poly merase Chain Reaction
Pluripotent nhpESCs were collected by scraping and then pelleted by centrifugation at 200g for 5 minutes. RNA was isolated using TRIzol (Invitrogen). Briefly, 100 μl of TRIzol was added per 100–1,000 cells and vortexed to lyse cells to homogeneity; 200 μl of chloroform (Sigma) was added followed by mixing and centrifugation for 5 minutes at 25,000g. The RNA containing supernatant was removed and the RNA pelleted using 600 μl of 100% isopropanol added to the supernatant and incubated at −20°C for at least 4 hours. The sample was centrifuged twice at 13,000g for 30 minutes at 4°C, with an ethanol wash in between, followed by air-drying. The RNA pellet was reconstituted in nuclease-free water and treated with 1 μl of DNase I for 30 minutes at 37°C. cDNA was prepared using the ImProm-II Reverse Transcription System (Progen, Heidelberg, Germany, http://www.progen.de) according to manufacturer's directions. Primers used were Oct-4, forward cgaccatctgccgctttgag and reverse ccccctgtcccccattccta, Nanog forward ctgtgatttgtgggcctgaa and reverse tgtttgcctttgggactggt, Rex1 forward gcgtacgcaaattaaagtccaga and reverse cagcatcctaaacagctcgcagaat, and Sox-2 forward cccccggcggcaatagca and reverse tcggcgccggggagatacat.
Pluripotency Demonstrated in Teratomas
Please see supplemental online Methods.
Independent External Genetic Testing for Pedigreed Confirmations
Pedigree of each cell line described was confirmed independently by DNA genotyping by Dr. Cecilia Penedo, Veterinary Genetics Laboratory, University of California, Davis (supplemental online Table 3).
Cytogenetic Analysis to Assay Normal Karyotype
Independent cytogenetic investigations were performed by the clinical cytogenetics facility of the University of Pittsburgh Cancer Institute under the supervision of Dr. Susanne Gollin and also within our lab [35].
Gene Expression Profiling
RNA Extraction
Total RNA was extracted using the TRIzol protocol (100 microliters per 104–105 cells) [38] followed by the Qiagen RNeasy Micro Kit (Hilden, Germany, http://www.qiagen.com) according to the manufacturer's recommendations to “clean up” the RNA isolated using TRIzol. RNA quality and quantity were determined using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE, http://www.nanodrop.com) and Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, http://www.agilent.com).
Preparation of Labeled cRNA
One microgram of total RNA was used to start the manual target preparation using the CodeLink Expression Bioarray System (Amersham Biosciences, Piscataway, NJ, http://www.amersham.com). Briefly, double-stranded cDNA synthesis was performed with a T7 oligo(dT) primer followed by purification. This cDNA was used as a template for in vitro transcription with biotin labeled nucleotides. Fifteen micrograms of the labeled cRNA were hybridized to Affymetrix rhesus macaque genome 49 format arrays (GeneChip Rhesus Macaque Genome Array, catalog number 900656; Affymetrix, Santa Clara, CA, http://www.affymetrix.com), followed by washing and staining with streptavidin phycoerythrin as recommended by the manufacturer. Arrays were scanned on an Affymetrix GeneChip 3000 Scanner. The arrays contain 52,303 probe sets that represent ~30,000 human orthologs and expressed sequence tags (ESTs). Affymetrix GCOS software was used for the scanning of the probe arrays, and the probe intensity analysis and normalization were performed using RMAExpress [39].
Microarray Data Analysis
The gene expression analysis protocol has been described previously [40]. Statistical analysis was performed using the ScoreGene gene expression package (http://www.cs.huji.ac.il/labs/compbio/scoregenes), and data visualization was performed using Genomica (http://genomica.weizmann.ac.il) [41], Spotfire Decision Site 8.0 (Spotfire Inc., Göteborg, Sweden, http://spotfire.tibco.com), and TreeView (http://jtreeview.sourceforge.net). The RMA output for every gene was divided by the geometric mean of all the values for the same gene and was log base 2 transformed. In the analysis, we only included transcripts with locus link numbers. To determine the differentially expressed genes, we used t test or the nonparametric threshold number of misclassifications [42]. In the clustering, we included only genes that had a p value <.01 in both scoring methods and a ratio more than twofold in any of the pairwise comparisons. False discovery rate (FDR) analysis was carried out as described previously [43].
Results
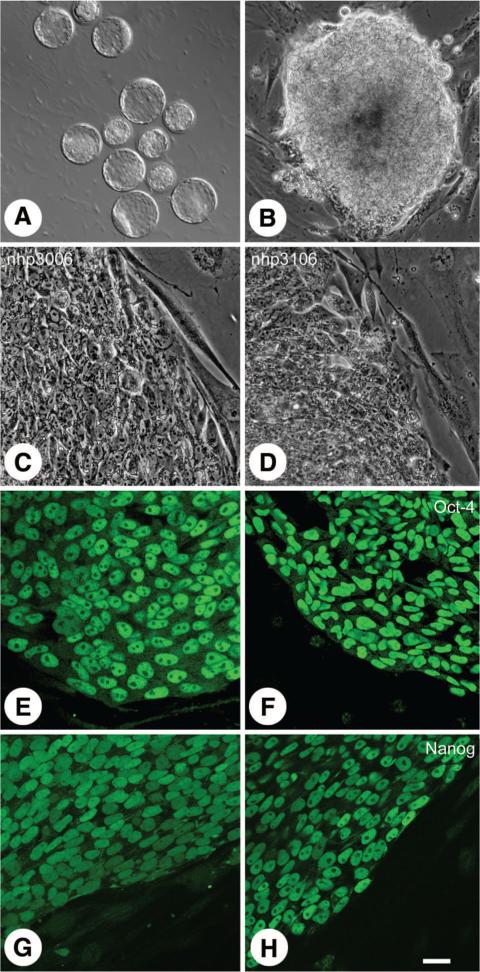
We have isolated 10 new embryonic pedigreed stem cell lines from old world Rhesus monkeys (Macaca mulatta, five male and five female). Monkey embryos were generated using intracytoplasmic sperm injection into isolated monkey oocytes and the resulting embryos cultured to the expanded blastocyst stage (Fig. 1A). Forty-one expanded blastocysts were used for these derivations. Inner cell masses isolated by immunosurgery were plated onto inactivated mouse embryonic fibroblasts. Success of isolation did not appear to be related to the size of the isolated ICM; however, within 1 week of plating, ICMs could be identified as promising by rapid expansion in culture (Fig. 1B). Ten to fourteen days after plating, putative embryonic stem cells with characteristically high nuclear/cytoplasm ratio and prominent nucleoli were passaged onto fresh feeders for propagation (Fig. 1C, 1D).
Figure 1.
Derivation and characterization of pedigreed nonhuman primate embryonic stem cells (nhpESC). Rhesus blastocysts (A) developed in vitro from embryos fertilized by intracytoplasmic sperm injection and were cultured until fully expanded. After immunosurgery to remove the outer trophectoderm cells, the isolated inner cell masses were cultured and grown in vitro (B). Both female (nhp20006: [C, E, G]) and male (nhp3106: [D, F, H]) lines were established equally well. nhpESCs had characteristic colony morphology including well-defined borders, high nuclear to cytoplasmic ratio, and prominent nucleoli (C, D). All lines displayed pluripotency markers, such as Oct-4 (E, F) and Nanog (G, H) examined by immunocytochemistry and laser scanning confocal microscopy. Green = Oct-4 (E, F) or Nanog (G, H). Bar = 20 μm (C–H), 28 μm (B), and 130 μm (A).
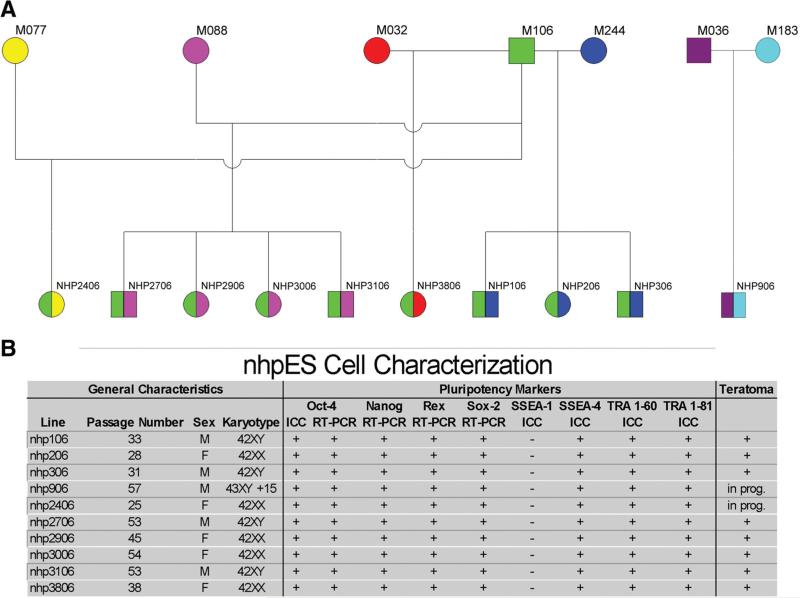
Unlike the anonymous origins of the federally approved hESCs, these nhpESC lines were generated with known pedigrees (Fig. 2A). The 10 isolated lines include two families of full siblings (quadruplet lines nhp2706, -2906, -3006, and -3106 as well as triplet lines nhp106, -206, and -306), and nine of the lines are half siblings of one another sharing the same sire (all but 906). The pedigree was independently confirmed with blinded DNA fingerprinting analysis of each line along with the presumptive gamete donors and unrelated samples at the University of California, Davis (courtesy of Dr. Cecilia Penedo; supplemental online Table 3). Additional confirmation of pedigree was evident as a result of the separate transcriptomics microarray analysis that generated an identical tree of familial relatedness using independent, blinded data and different algorithms (Fig. 3).
Figure 2.
Familial pedigree and characterizations of 10 nhpES cell lines. Familial relationships among each line are shown in (A). Males (squares) and female (circles) were produced in equal number. Nine of the ten lines are half-siblings (same sire), demonstrated by sharing the same left-side color. Two groups of full-sibling lines were generated (both sides having the same color); nhp2706, -2906, -3006, and -3106 comprise one quadruplet family, and nhp106, -206, and -306 comprise another triplet set. Both families include male and female siblings. (B): All lines were positive by immunocytochemistry for the pluripotency markers Oct-4, stage-specific embryonic antigen (SSEA)-4, TRA 1-60, and TRA 1-81 and negative for the differentiation marker SSEA-1. Additionally, they were positive by RT-PCR for Oct-4, Nanog, Rex-1, and Sox-2. All lines have maintained a stable karyotype. Nine of the lines have a normal diploid chromosome number [42]; nhp906 has a stable trisomy of chromosome 15, the only aneuploidy observed in these lines. Eight of the ten lines have been confirmed by teratoma analysis, and the other two are underway. Abbreviations: F, female; ICC, immunocytochemistry; M, male; nhpES, nonhuman primate embryonic stem; prog., progress; RT-PCR, reverse transcription-polymerase chain reaction.
Figure 3.
Gene expression clustering of nonhuman primate embryonic stem cell lines. Nine lines were analyzed for gene expression using Affymetrix monkey gene expression chips. Each analysis was performed in triplicate except for nhp206, which was duplicated. Cluster analysis of the average value across the replicates showed striking similarity across all lines with similarity >95% among all lines. However, within that small variation not only did the full siblings cluster together but the lines also clustered according to their sex (females: circles; males: squares).
All lines are positive for the characteristic pluripotency markers Oct-4 (Fig. 1E, 1F), Nanog (Fig. 1G, 1H), SSEA-4, TRA 1-60, and TRA 1-81 assessed by immunocytochemistry (Fig. 2) and for Oct-4, Nanog, Rex-1, and Sox-2 by reverse transcriptase polymerase chain reaction (Fig. 2B). All lines maintain a diploid complement of chromosomes including 20 autosomes and 2 sex chromosomes except for nhp906, which has a stable trisomy of chromosome 15 (Fig. 2B). In addition to labeling with consensus pluripotency markers, eight of the lines have produced teratomas when injected into immunocompromised mice. Studies on the remaining two lines are underway.
Growth characteristics of the isolated lines are variable but seemingly independent of sex, family relatedness, or passage number. Each line has a cell cycle time of approximately 20 hours, and individual cultures are passaged weekly. Cell lines isolated most recently (2406, 2706, 2906, 3006, 3106, and 3806) grow the most robustly regardless of passage number, suggesting perhaps that greater experience deriving ESC lines not only improves derivation efficiency but also quality of the newest lines.
The overall derivation success rate of these nhpESCs was 24.4% of the total number of blastocysts (Table 1) and 34.3% of those ICMs plated successfully (10 of 35). Importantly, the success rate was strongly dependent on the age of the blastocyst when immunosurgery was performed. We chose only fully expanded blastocysts for derivations. Embryos reached full expansion between days 9 and 11 of culture, counting the day of ICSI as day 1; 46.7% of the blastocysts that reached full expansion on day 9 successfully established nhpESC lines, whereas those that reached full expansion on day 10 resulted in stable nhpESC lines only 10.5% of the time, although no difference was observed in the plating efficiency between these two groups. These results were especially apparent when embryos were compared within the same clutch. Supplemental online Table 1 cites results comparing nhpESC lines derived from the same ovulation event within a single experiment where nhpESCs were established from day 9 blastocysts but in none of these cases from blastocysts at day 10. We also noted that Indian origin rhesus macaques (captive bred in the U.S. to other Indian rhesus) were more successful in nhpESC establishment compared with rhesus of Chinese origin (first generation imports from China).
Table 1.
Prime embryos produce embryonic stem cell lines more successfully than developmentally delayed embryos
| Day | No. of blastocysts for ESC derivations | No. of inner cell mass outgrowths (% blastocysts) | No. of lines still growing (% blastocysts) |
|---|---|---|---|
| 9 | 15 | 13 (86.7) | 7 (46.7) |
| 10 | 19 | 17 (89.5) | 2 (10.5) |
| 11 | 7 | 5 (71.4) | 1 (14.3) |
| Total | 41 | 35 (85.4) | 10 (24.4) |
Seven of the ten nonhuman primate ESC (nhpESC) lines established were derived from the fastest developing fifteen embryos (46.7%), whereas the other three nhpESC lines were established from twenty-six delayed embryos (11.5% success rate per embryo).
To evaluate the similarity of these new nhpESCs, we compared the gene expression of the 10 lines using Affymetrix rhesus macaque genome 49 format arrays. Samples were collected over multiple passages ranging from passage 5 through passage 12. These arrays contain 52,303 probe sets. We analyzed ~31,000 annotations that represent all genes with a known gene ID or human orthologs and including ESTs (all raw data can be found at Geo National Center for Biotechnology Information accession number GSE7534). Each analysis was performed in triplicate except nhp206 (only two samples) and nhp2906 (single sample). To avoid the risk of introducing arti-factual variability into the analysis, we chose to minimize variability among samples by preparing all samples using the same reagents at the same time. This precluded repeating nhp206 and nhp2906 in separate experiments. Since nhp2906 was represented only by a single sample, it was excluded from the average cluster analysis. However, all samples are included and compared in supplemental online Figure 1. We performed hierarchical clustering of the average gene expression using optimal leaf order analysis [44] and found that all of these nhpESC lines were extremely similar to each other in their gene expression (Fig. 3, black indicates no difference). As an indication of the consistency of this analysis, similar relationships were also observed when individual samples were clustered (supplemental online Fig. 1).
To further quantitate the similarity, we calculated the correlation values among individual samples. The lowest value was 94.4% between samples 1a and 31c and was as high as 99% between many of the samples (supplemental online Table 3), indicating very little variability in the individual samples and confirming the similarity among these 10 ESC lines. Even among these highly similar lines, it was observed that full siblings had expression patterns more similar to each other than to their half siblings, indicating a familial influence on expression (Fig. 3).
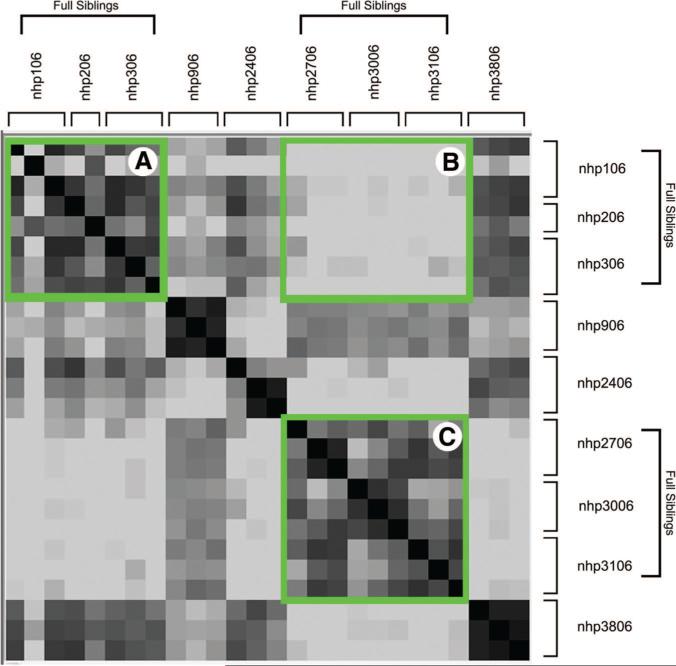
This relationship can be seen more clearly in the heat map depicted in Figure 4. This figure was generated by using Genomica software and calculating the log ratio of the correlation among samples (supplemental online Table 2). As expected, samples from the same line are closely related (darker colors). Moreover, the similarity is observed not only among individual samples of the same line, but also among lines that are full siblings. Box A and Box C represent the comparisons of samples within a family; note the outlined dark area and the surrounding lighter areas indicating similarity within the family. Box B highlights the dissimilarity when comparing samples between the two families; note the light color within this box, clearly distinguishing full siblings from half siblings.
Figure 4.
Heat map analysis of gene expression in nine nonhuman primate embryonic stem cell lines. Familial relatedness is demonstrated when the individual replicates are compared in a heat map analysis. In this analysis, similarity is represented by darker colors (identity is black), and dissimilar comparisons are light. Samples from full siblings compared within the family produce relatively dark areas (Boxes A and C), and comparisons among the families produce a light area in the heat map (Box B).
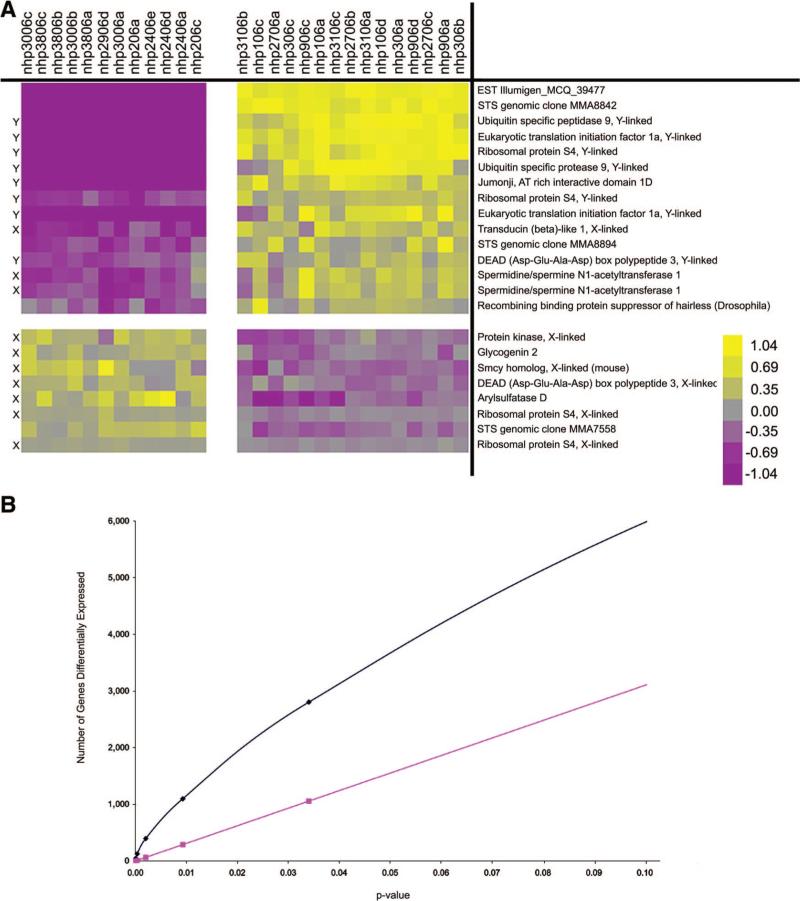
Figure 3 shows that the male and female lines cluster with themselves within the analysis. To determine whether gene expression differed significantly between male and female nhpESC lines and to identify which, if any, genes were over- expressed in male versus female lines, we defined the male and female lines for cluster analysis. Twenty-three transcripts were identified that had a statistically significant expression difference between male and female cell lines according to Bonferroni correction for multiple testing (Fig. 5A; p < .000001). Fifteen were overexpressed in male lines relative to female lines and the remaining eight overexpressed in female lines relative to male lines. Many but not all of the genes were sex chromosome linked (noted in Fig. 5A). When more relaxed statistical conditions were applied (FDR 5%), 94 differentially expressed transcripts were observed (supplemental online Fig. 2 [male overexpressed] and supplemental online Fig. 3 [female overexpressed]).
Figure 5.
Differential gene expression between male and female nonhuman primate embryonic stem cells. When individual samples were grouped according to their sex, we identified 15 transcripts, which were overexpressed in males relative to females, and 8 transcripts, which were overexpressed in female lines relative to male lines ([A], p < .000001). Using the chromosome locations of the human homologs to these monkey genes, we identified that 8 of the 15 transcripts overexpressed in males localized to the Y chromosome and 3 localized to the X chromosome. Of the eight transcripts overexpressed in females relative to males, seven localized to the X chromosome. When we compare our resultant differential gene expression ([B], blue line) with the expected number of gene differences based on statistical probability ([B], pink line), we see that many more genes were observed to be differentially expressed than expected due to statistical variation; yellow = relatively overexpressed and purple = relatively underexpressed. Abbreviations: X, X chromosome; Y, Y chromosome.
To address multiple testing and confirm the statistical significance of the differential gene expression between male and female lines depicted in Figure 5A, we performed overabun-dance analysis as previously described [39, 40] (Fig. 5B). In this analysis, the expected number of genes at any given p value in a random comparison is plotted (red line: Fig. 5B and supplemental online Fig. 4) and compared with the actual number of genes with the calculated p value in the data (blue line: Fig. 5B and supplemental online Fig. 4).
Discussion
Studies investigating embryonic stem cells from nonhuman primates [21] have provided basic insights regarding hematopoiesis [45, 46] and cardiomyocyte differentiation [27] and imprinting [47], and encouraging preclinical results have been reported [48] after transplanting dopaminergic neurons differentiated in vitro from cynomolgus ESCs into nonhuman primate Parkinson disease models. Nonhuman primate ESCs continue to bridge the fundamental knowledge learned from mouse ESCs with the clinical information available by examining human ESCs. Here we generated 10 new pedigreed nhpESC lines including full siblings, half siblings, and an unrelated line. We demonstrate that nhpESCs are established more successfully from prime embryos versus developmentally delayed ones. We also found that transcriptomics analysis of these lines revealed striking homogeneity in gene expression among these lines. Despite this homogeneity, the nhpESC lines can be distinguished along familial patterns, with full siblings being the most closely related. This transcriptomic homogeneity enables subtle comparisons among lines including the identification of genes differentially expressed between male and female stem cell lines. These ten characterized lines are available for further study of the genetic relationships among familial ESCs.
Considerable heterogeneity among the available hESC lines including growth rate and genetic and epigenetic stability and gene expression profiles has been reported [2]. These gene expression comparisons have primarily focused on “stemness” genes, that is, those expressed in all pluripotent or multipotent lines but not in differentiated or somatic cells [3, 49–54]. The first two studies [51, 52] identified approximately 250 putative genes involved in mESC pluripotency, and similar genes were identified in hESCs. Abeyta et al. [1] compared two female lines, HSF-6 and H9 (derived in different laboratories), with one male line, HSF-1, derived in the same laboratory as HSF-6. HSF-6 expression was more similar to HSF-1 than to the female line derived elsewhere, suggesting that the variability among lines might be due to derivation and culture differences. However, these interpretations are complicated by the unknown pedigrees even when comparing cell lines isolated within the same laboratory as well as causes of the underlying infertility of the patient/couple.
More recent comprehensive expression studies confirm variability [4, 5, 55] among hESC lines, including among lines generated within the same lab by the same personnel. Li et al. [25] generated five new lines and found that ~10% of the probes were expressed in all five lines, although large variations among lines precluded direct line-by-line comparisons. Skottman et al. derived seven new hESC lines and, by comparing ~10,000 genes, found ~300 genes unique to each line. Interestingly, the gene expression profiles among four lines derived in Finland clustered more closely together than to their three lines derived in Sweden, raising questions regarding lab-to-lab variations as well as heredity. It will be interesting to determine whether similar relatedness in gene expression patterns is observed in related preimplantation embryos. Although not conducted in this report, methods exist to address this question in both humans [56, 57] and nonhuman primates [58]. This experiment would also be useful for determining whether the great similarity observed among these lines is related to extended in vitro culture under similar conditions. In mice, strain-specific differences in mESC gene profiles have been demonstrated [6, 7]. Taken together, these results suggest that knowledge of strain or family group origin might be essential for fully accurate interpretations. Access to infertility therapies in most countries is skewed by socioeconomic parameters, and it is likely that the donated embryos and resultant hESC lines are not representative of full demographic diversity. Consequently, even without direct knowledge of genetic origins, additional attention should be devoted to ensuring hESC research resources that are both well characterized and also inclusive of the population diversity.
Here, microarray comparisons within and between these nhpESC families show remarkably homogeneous gene expression profiles. It is important to highlight that these studies compared pluripotent ESCs with each other. Transcriptomic analysis of their progeny differentiated in vitro, in vivo in teratomas, and in utero in chimera is underway. The consistent and homogeneous gene expression of these nhpESCs (>96% identity) enables subtle analyses among ESCs that might otherwise be impossible, since the intrinsic variability described in hESCs would swamp minor differences. Recent transcriptional profiles of rhesus ESCs analyzed against human annotations reported an 85% [59] concordance. Our results achieved with rhesus ESCs are analyzed using the recently available rhesus gene annotations (http://www.affymetrix.com/products/arrays/specific/rhesus_macaque.affx). We found similar results when using the human annotations (not shown), reinforcing the reliability of our rhesus/rhesus results as well as the previous human/rhesus transcriptomics analysis [59]. Furthermore, analysis of average expression produced nearly identical results to comparisons of each of the individual triplicate samples, demonstrating the consistency of the Affymetrix rhesus chips and of the sample preparation. Additionally, the optimal leaf order analysis clustering precisely conforms with the experimentally designed parental origins and sex of each ESC line, providing another separate assurance of experimental accuracy beyond the independent genetic testing by the Veterinary Genetics Laboratory, University of California, Davis, on the genetic fidelity of each line reported here.
Sex-specific differences between the male and female cell lines are observed as a subset of genes overexpressed in males and underexpressed in females and a different subset overex-pressed in female and underexpressed in male nhpESCs. Many but not all genes overexpressed in male lines are Y-chromosome linked, just as X-linked genes are overexpressed in the female lines. X inactivation is variable in pluripotent hESCs [60], perhaps explaining why not all sex linked genes are overex-pressed in lines of their respective sex. Additionally, we analyzed gene expression using Affymetrix monkey arrays, but the annotations analyzed are from human homologs and there probably does not exist a 1:1 ratio between these two species. Finally, we do not expect all genes to be expressed at this stage of development, and nonexpressed genes would be excluded from the analysis. However, the expression profile of nhp906 (trisomy 15) is not strikingly different from the euploid ones, suggesting that there are stringent mechanisms to compensate for gene dosages in ESCs. With the establishment of primate ESC lines that have consistent genetic expression comes the ability to use these lines for detailed studies of infertility and development. ESC lines established using pedigreed primates could be used as bioassays to probe pluripotency and developmental differences among blastocysts of varying qualities, those generated with differing ART methods (e.g., in vitro fertilization, ICSI, or spermatid injections), or after environmental exposures during gametogenesis or preimplantation development. These abilities to extract small but biologically meaningful differences in gene expression profiles are possible because of the commonality of the background profile. Parallel, complementary studies with hESCs might possibly uncover novel pathways for the donors’ underlying infertility, improved procedures for derivations, strategies to retain the fidelity of genomic imprints, and optimal growth and differentiation protocols.
Nearly identical expression profiles comparing different lines of either pluripotent mESCs or, as shown here, nhpESCs reinforce the reliability and biological utility of these research resources. Identifying the source or sources responsible for the newly recognized discordant profiling results with hESCs is important both for basic biology and also due to the clinical implications of hESC research findings. The existing federally approved hESC lines were derived from embryos discarded as suboptimal by the collaborating ART clinics and donated anonymously by the infertile patient/couple; therefore, no information is known regarding the relatedness of the existing lines. Also, the couple's infertility might have resulted from underlying genetic or epigenetic problems, further confounding the hESC line's status. Additionally, intrinsic defects might have resulted in the embryo's developmental delays, rendering them of limited clinical utility and therefore donated for fundamental studies. Finally, the lines were derived in labs distributed around the world and, thus, interlaboratory variability may play a role. hESC heterogeneity might be attributed to genetic variation due to parentage or sex, underlying causes of the couple's infertility or developmental delay of the embryo, variations in derivation and ESC establishment procedures and labs, and/or biological variations intrinsic to primate ESCs. Due to regulatory oversight policies, determination of these sources of hESC heterogeneity is not possible. In our study, embryos derived from rhesus monkeys of Indian origin more successfully produced ESC lines than did those from Chinese origin monkeys. These results suggest that hESC lines should be closely examined for ethnicity and utility parameters such as differentiation and transplantation, a set of experiments not possible with federal funding under the current guidelines.
Here, using embryos generated exclusively from gametes obtained from fertile rhesus, we find that nhpESC lines are more successfully generated from prime embryos versus subprime ones. This is somewhat surprising since the establishment of two-dimensional cell cultures would have been predicted to demand less stringency on the embryo versus the spatiotemporal challenges of implantation and pregnancy. Although neither pluripotency nor transcriptomic comparisons among the ESCs from the best embryos (lines 106, 206, 306, 2706, 2906, 3006, and 3106) and the subprime ones (lines 906, 2406, and 3806) show important differences, it is noteworthy that the one aneuploid line is from a subprime embryo (line 906). Furthermore, our most recent lines grow most consistently and reliably, suggesting that increased ESC experience improves the quality of the derived line and, by extrapolation, that newer lines are superior to older ones.
Questions remain as to exactly how similar primate ESCs are to mESCs, and both limitations in the availability of primate embryos and ESC lines as well as appropriate ethical restrictions on certain types of experiments makes it challenging to answer these questions with certainly. However, nhpESC studies continue to offer opportunities for bridging the gap between mouse and responsible human ESC research in several ways, including the following: pluripotency and germ line transmission assayed in intra- and interspecific chimeras; derivations from prime versus delayed embryos; lines generated with deliberate and defined epigenetic and genetic characteristics including mitochondrial DNA pedigrees [61, 62]; families, including inbred ones, of related ESCs generated and derived simultaneously versus sibling ESCs generated at differing times to analyze environmental influences; aging or transformation of the cells in vitro as studied by changes between batches of varying passage numbers; gametogenesis potentials and functional tests of gametes produced in vitro or after intra- and interspecific chimeras; genetic manipulation of nhp gametes or embryos such as transgenic, knockout/knockin, or small interfering RNA to produce more meaningful animal models of human disease; and transplantation investigations in which differentiated progeny from these cells are transplanted into full-sibling offspring generated by the identical rhesus monkeys or other related primates. These familial-related nhpESCs now also afford opportunities to explore changes with passaging and other environmental factors as well as sex specificity. However, with only a single decade of hESC and nhpESC research, and with understandable research constraints on some types of hESC investigations, comprehensive and rigorous comparisons between and among the primate ESC lines is only now underway. We suggest that ready access to more homogeneous and reliable primate ESC lines—human and nonhuman—will accelerate reliable findings of biomedical importance.
Conclusion
Primate ESCs, established from prime-quality embryos generated by fertile pedigreed rhesus, display homogeneous gene expression. Unlike hESC heterogeneity, this family of primate ESCs is most similar among first-degree relatives and among females or males. Experimental comparisons among these primate ESCs may prove more reliable and interpretable, since, like inbred strains in which genetic variables are also reduced or eliminated, unknown genetic variations confound interpretations. Contrasting the similarities among these lines with the heterogeneous hESCs might suggest that additional, better hESC lines, with representation of fully inclusive demographic diversity, are justified.
Supplementary Material
Acknowledgments
We thank our veterinary team including Dr. Saverio (Buddy) Capuano, Dr. Mario Rodriquez, Jamie Tomko, Kevin Grund, and Thomas Richards for assistance in uploading expression data to GEO. Special thanks to Dr. Susanne Gollin (UPCI, Pittsburgh) for help in karyotyping the nhpESC lines and Dr. Cecilia Penedo (Veterinary Genetics Laboratory, University of California, Davis) for NHP and nhpESC genotyping as well as Diane Carlisle, Dave McFarland, Stacie Oliver, and Kari Panza. C.S.N. and G.S. conceived of the research and wrote the manuscript, the nhpESCs were derived and characterized by C.S.N., J.D.M.-B., and C.J.R. from embryos generated by C.R.S. and E.J. Transcriptomic and genomic analyses were performed by A.B.-Y., E.K.-N., and N.K., and teratomas were produced by M.S. and K.O. and analyzed by C.A.C. Funding by the National Institutes of Health (P01 HD 47675) is acknowledged gratefully.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Abeyta MJ, Clark AT, Rodriguez RT, et al. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- 2.Allegrucci C, Young LE. Differences between human embryonic stem cell lines. Hum Reprod Update. 2007;13:103–120. doi: 10.1093/humupd/dml041. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya B, Miura T, Brandenberger R, et al. Gene expression in human embryonic stem cell lines: Unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 4.Ware CB, Nelson AM, Blau CA. A comparison of NIH-approved human ESC lines. STEM CELLS. 2006;24:2677–2684. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- 5.Skottman H, Mikkola M, Lundin K, et al. Gene expression signatures of seven individual human embryonic stem cell lines. STEM CELLS. 2005;23:1343–1356. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- 6.Brambrink T, Hochedlinger K, Bell G, et al. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc Natl Acad Sci U S A. 2006;103:933–938. doi: 10.1073/pnas.0510485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakayama S, Jakt ML, Suzuki M, et al. Equivalency of nuclear transfer-derived embryonic stem cells to those derived from fertilized mouse blastocysts. STEM CELLS. 2006;24:2023–2033. doi: 10.1634/stemcells.2005-0537. [DOI] [PubMed] [Google Scholar]
- 8.Rao MS, Civin CI. Translational research: Toward better characterization of human embryonic stem cell lines. STEM CELLS. 2005;23:1453. doi: 10.1634/stemcells.2005-ed.4. [DOI] [PubMed] [Google Scholar]
- 9.Civin CI, Rao MS. How many human embryonic stem cell lines are sufficient? A U.S. perspective. STEM CELLS. 2006;24:800–803. doi: 10.1634/stemcells.2006-0084. [DOI] [PubMed] [Google Scholar]
- 10.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 11.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 13.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 14.Egli D, Rosains J, Birkhoff G, et al. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- 15.Maherali N, Sridharan R, Xie W, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Wilmut I, Schnieke AE, McWhir J, et al. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 17.Wakayama T, Perry AC, Zuccotti M, et al. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 18.Ruhnke M, Ungefroren H, Zehle G, et al. Long-term culture and differentiation of rat embryonic stem cell-like cells into neuronal, glial, endothelial, and hepatic lineages. STEM CELLS. 2003;21:428–436. doi: 10.1634/stemcells.21-4-428. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Duan E, Sung LY, et al. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biol Reprod. 2005;73:149–155. doi: 10.1095/biolreprod.104.037150. [DOI] [PubMed] [Google Scholar]
- 20.Renfree MB, Shaw G. Diapause. Annu Rev Physiol. 2000;62:353–375. doi: 10.1146/annurev.physiol.62.1.353. [DOI] [PubMed] [Google Scholar]
- 21.Thomson JA, Kalishman J, Golos TG, et al. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 23.Akutsu H, Cowan CA, Melton D. Human embryonic stem cells. Methods Enzymol. 2006;418:78–92. doi: 10.1016/S0076-6879(06)18005-2. [DOI] [PubMed] [Google Scholar]
- 24.Cowan CA, Klimanskaya I, McMahon J, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 25.Li SS, Liu YH, Tseng CN, et al. Characterization and gene expression profiling of five new human embryonic stem cell lines derived in Taiwan. Stem Cells Dev. 2006;15:532–555. doi: 10.1089/scd.2006.15.532. [DOI] [PubMed] [Google Scholar]
- 26.Reubinoff BE, Pera MF, Fong CY, et al. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 27.Abdelalim EM, Takada T, Toyoda F, et al. In vitro expression of natriuretic peptides in cardiomyocytes differentiated from monkey embryonic stem cells. Biochem Biophys Res Commun. 2006;340:689–695. doi: 10.1016/j.bbrc.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 28.Gibbs RA, Rogers J, Katze MG, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki E, Hanazawa K, Kurita R, et al. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus). STEM CELLS. 2005;23:1304–1313. doi: 10.1634/stemcells.2004-0366. [DOI] [PubMed] [Google Scholar]
- 30.Suemori H, Tada T, Torii R, et al. Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Dev Dyn. 2001;222:273–279. doi: 10.1002/dvdy.1191. [DOI] [PubMed] [Google Scholar]
- 31.Thomson JA, Kalishman J, Golos TG, et al. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol Reprod. 1996;55:254–259. doi: 10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- 32.Cibelli JB, Grant KA, Chapman KB, et al. Parthenogenetic stem cells in nonhuman primates. Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- 33.Hewitson L, Takahashi D, Dominko T, et al. Fertilization and embryo development to blastocysts after intracytoplasmic sperm injection in the rhesus monkey. Hum Reprod. 1998;13:3449–3455. doi: 10.1093/humrep/13.12.3449. [DOI] [PubMed] [Google Scholar]
- 34.Simerly C, Navara C, Hwan Hyun S, et al. Embryogenesis and blastocyst development after somatic cell nuclear transfer in nonhuman primates: Overcoming defects caused by meiotic spindle extraction. Dev Biol. 2004;276:237–252. doi: 10.1016/j.ydbio.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Navara C, Redinger C, Mich-Basso J, et al. Derivation and characterization of nonhuman primate embryonic stem cells. In: Fisher S, editor. Current Protocols. John Wiley and Sons; Hoboken, NJ: 2007. [DOI] [PubMed] [Google Scholar]
- 36.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 37.Navara CS, Benyumov A, Vassilev A, et al. Vanadocenes as potent anti-proliferative agents disrupting mitotic spindle formation in cancer cells. Anticancer Drugs. 2001;12:369–376. doi: 10.1097/00001813-200104000-00010. [DOI] [PubMed] [Google Scholar]
- 38.Pardo A, Gibson K, Cisneros J, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2005;2:e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barash Y, Dehan E, Krupsky M, et al. Comparative analysis of algorithms for signal quantitation from oligonucleotide microarrays. Bioinformatics. 2004;20:839–846. doi: 10.1093/bioinformatics/btg487. [DOI] [PubMed] [Google Scholar]
- 40.Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am J Respir Cell Mol Biol. 2002;27:125–132. doi: 10.1165/ajrcmb.27.2.f247. [DOI] [PubMed] [Google Scholar]
- 41.Segal E, Friedman N, Koller D, et al. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Dor A, Bruhn L, Friedman N, et al. Tissue classification with gene expression profiles. J Comput Biol. 2000;7:559–583. doi: 10.1089/106652700750050943. [DOI] [PubMed] [Google Scholar]
- 43.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 44.Bar-Joseph Z, Demaine ED, Gifford DK, et al. K-ary clustering with optimal leaf ordering for gene expression data. Bioinformatics. 2003;19:1070–1078. doi: 10.1093/bioinformatics/btg030. [DOI] [PubMed] [Google Scholar]
- 45.Shibata H, Ageyama N, Tanaka Y, et al. Improved safety of hematopoietic transplantation with monkey embryonic stem cells in the allogeneic setting. STEM CELLS. 2006;24:1450–1457. doi: 10.1634/stemcells.2005-0391. [DOI] [PubMed] [Google Scholar]
- 46.Hematti P, Obrtlikova P, Kaufman DS. Nonhuman primate embryonic stem cells as a preclinical model for hematopoietic and vascular repair. Exp Hematol. 2005;33:980–986. doi: 10.1016/j.exphem.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Mitalipov S, Clepper L, Sritanaudomchai H, et al. Methylation status of imprinting centers for H19/IGF2 and SNURF/SNRPN in primate embryonic stem cells. STEM CELLS. 2007;25:581–588. doi: 10.1634/stemcells.2006-0120. [DOI] [PubMed] [Google Scholar]
- 48.Takagi Y, Takahashi J, Saiki H, et al. Dopaminergic neurons generated from monkey embryonic stem cells function in a Parkinson primate model. J Clin Invest. 2005;115:102–109. doi: 10.1172/JCI21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharya B, Cai J, Luo Y, et al. Comparison of the gene expression profile of undifferentiated human embryonic stem cell lines and differentiating embryoid bodies. BMC Dev Biol. 2005;5:5–22. doi: 10.1186/1471-213X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai J, Chen J, Liu Y, et al. Assessing self-renewal and differentiation in human embryonic stem cell lines. STEM CELLS. 2006;24:516–530. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanova NB, Dimos JT, Schaniel C, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 52.Ramalho-Santos M, Yoon S, Matsuzaki Y, et al. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 53.Sato N, Sanjuan IM, Heke M, et al. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 54.Sperger JM, Chen X, Draper JS, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, Shin S, Zeng X, et al. Genome wide profiling of human embryonic stem cells (hESCs), their derivatives and embryonal carcinoma cells to develop base profiles of U.S. Federal government approved hESC lines. BMC Dev Biol. 2006;6:20. doi: 10.1186/1471-213X-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobson AT, Raja R, Abeyta MJ, et al. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- 57.Kocabas AM, Crosby J, Ross PJ, et al. The transcriptome of human oocytes. Proc Natl Acad Sci U S A. 2006;103:14027–14032. doi: 10.1073/pnas.0603227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Latham KE. The primate embryo gene expression resource in embryology and stem cell biology. Reprod Fertil Dev. 2006;18:807–810. doi: 10.1071/rd06110. [DOI] [PubMed] [Google Scholar]
- 59.Byrne JA, Mitalipov SM, Clepper L, et al. Transcriptional profiling of rhesus monkey embryonic stem cells. Biol Reprod. 2006;75:908–915. doi: 10.1095/biolreprod.106.053868. [DOI] [PubMed] [Google Scholar]
- 60.Hoffman LM, Hall L, Batten JL, et al. X-inactivation status varies in human embryonic stem cell lines. STEM CELLS. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 61.St John JC, Ramalho-Santos J, Gray HL, et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–153. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- 62.St John JC, Schatten G. Paternal mitochondrial DNA transmission during nonhuman primate nuclear transfer. Genetics. 2004;167:897–905. doi: 10.1534/genetics.103.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.