Abstract
Double-stranded DNA bacteriophages have motors that drive the genome into preformed capsids, using the energy releas ed by hydrolysis of ATP to overcome the forces opposing DNA packaging. Viral packaging motors are the strongest of all biological motors, but it is not known how they generate these forces. Several models for the process of mechanochemical force generation have been put forward, but there is no consensus on which, if any, of these is correct. All the existing models assume that protein-generated forces drive the DNA forward. The scrunchworm hypothesis proposes that the DNA molecule is the active force-generating core of the motor, not simply a substrate on which the motor operates. The protein components of the motor dehydrate a section of the DNA, converting it from the B form to the A form and shortening it by about 23%. The proteins then rehydrate the DNA, which converts back to the B form. Other regions of the motor grip and release the DNA to capture the shortening-lengthening motions of the B→A→B cycle (“scrunching”), so that DNA is pulled into the motor and pushed forward into the capsid. This DNA-centric mechanism provides a quantitative physical explanation for the magnitude of the forces generated by viral packaging motors. It also provides a simple explanation for the fact that each of the steps in the burst cycle advances the DNA by 2.5 base pairs. The scrunchworm hypothesis is consistent with a large body of published data, and it makes four experimentally testable predictions.
Keywords: DNA Bacteriophage, Viral Genome Packaging, Molecular Motors, Mechanochemistry, A-DNA, B-DNA
1. Introduction
Many bacteriophages have a double-stranded DNA genome that is packaged into a preformed capsid by an ATP-driven motor. The mechanism by which these motors convert the chemical energy of ATP hydrolysis into the mechanical energy of DNA motion remains unknown.
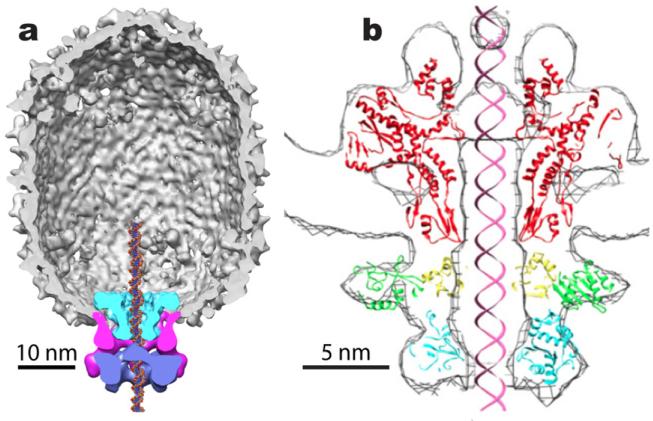
There is a growing body of structural information on the components of viral motors and their organization in procapsids, and in mature viruses (Morais, 2012). Two of these are shown in Fig. 1, 29 and T4. 29 is a particularly simple phage, so it is a common model for mechanochemical studies (Morais, 2012). 29 has two protein components, the dodecameric connector (gp10) that corresponds to similar portal proteins in other bacteriophages, and the pentameric ATPase (gp16). Between these is a pentameric ring of RNA (pRNA) that is unique to 29. T4 also has a pentameric ATPase (the terminase, gp1), and a much larger dodecameric portal. In the remainder of this paper, the word “motor” includes all components. It is generally believed that packaging mechanisms are similar among dsDNA bacteriophages, but it is not known how these molecules work together, capturing the energy from ATP hydrolysis to drive DNA into the capsid.
Fig. 1.
(a) Three dimensional reconstruction of the bacteriophage 29 capsid and motor (Morais, 2012; Morais et al., 2008). The capsid is shown cut away in grey, the connector in cyan, the pRNA in magenta, and the ATPase in purple. Note that the motor covers a footprint of about 4-5 complete turns of DNA. From (Morais et al., 2008), with permission. (b) A model for the T4 motor, consisting of the crystal structures of the SPP4 portal protein (red) and the T4 gp1 terminase (green, yellow and cyan), docked into the cryo-EM density (Sun et al., 2008). From (Sun et al., 2008), with permission.
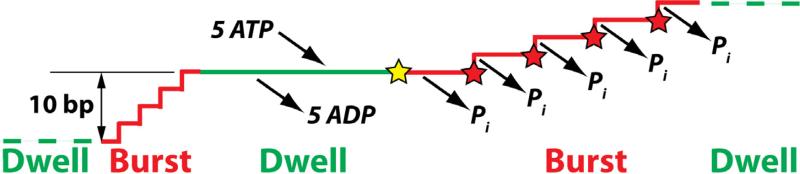
Much of what we know about the mechanochemisty of DNA packaging motors has come from elegant single-molecule experiments using laser tweezers; see (Chemla and Smith, 2012) for a recent review. These experiments have shown that viral packaging motors generate forces on the order of tens of pN (Chemla and Smith, 2012; Fuller et al., 2007b; Rickgauer et al., 2008; Smith et al., 2001), making them the strongest of all biological motors for which forces have been measured. When the DNA motions are not opposed by tension applied by the molecular tweezers, or by back-pressure when the capsid is nearly full, packaging proceeds in bursts of 10 base pairs (bp) followed by a dwell phase. Five ATPs are bound and five ADPs released during the dwell phase. The 10 bp burst contains four 2.5-bp steps, and each of these is accompanied by the hydrolysis of one molecule of ATP and the release of a single inorganic phosphate (Chistol et al., 2012; Liu et al., 2014b; Moffitt et al., 2009). The packaging motor resets after each burst, and the hydrolysis of the fifth ATP initiates the next burst (Chistol et al., 2012). The coordination of the mechanical and chemical cycles is shown in Fig. 2.
Fig. 2.
Relationship between the mechanical and chemical cycles. During the dwell phase, five molecules of ATP are bound, replacing five molecules of ADP. The burst phase contains four steps, during each of which one molecule of ATP is hydrolyzed (red stars), one molecule of inorganic phosphate is released, and the DNA advances by 8.5 Å (equivalent to 2.5 bp of B-DNA). Hydrolysis of the fifth ATP initiates the burst phase (yellow star). After (Liu et al., 2014b).
To date, there is no general agreement on how the motors propel DNA into the capsid. Several different mechanisms have been proposed. All of them involve some sort of grip/release cycle, coupled with conformational changes in the motor that drive the DNA forward. It has been shown that the motor does not rotate relative to the capsid (Baumann et al., 2006; Hugel et al., 2007), so, in all recent models, the DNA is pushed into the capsid by longitudinal conformational changes in the motor proteins (i.e., parallel to the direction of the required DNA motion). That is, the proteins grip the DNA, and ATP hydrolysis generates protein motions that pull or push the DNA along, much as the muscles in our arms move our hands as we pull a rope. With one exception, all of those models treat DNA as a passive substrate, which is fundamentally different from what is proposed here. The exception is the model of DNA “crunching” or torsional compression, put forward by Lindsay Black and his collaborators (Dixit et al., 2012; Oram et al., 2008; Ray et al., 2010); I will discuss it in more detail below. For a discussion of other models, I refer the reader to recent reviews (Casjens, 2011; Chemla and Smith, 2012; De-Donatis et al., 2014; Liu et al., 2014a; Morais, 2012; Rossmann and Rao, 2012; Serwer and Jiang, 2012; Sun et al., 2010).
2. The Model and Correlations with Existing Data
2.1 A-DNA and B-DNA
The Watson-Crick model of the DNA double helix (Watson and Crick, 1953) was based on x-ray fiber diffraction data obtained by Rosalind Franklin, when she was working in Maurice Wilkins's lab at Kings College, London. Franklin varied the humidity and obtained two diffraction patterns, which she designated “A” and “B” (Franklin and Gosling, 1953a; Franklin and Gosling, 1953b). The Watson-Crick model was for the high humidity form, B-DNA, which is considered the normal form in biological systems, because of the high water activity in vivo. That model has 10 base pairs per turn of the double helix, with a rise of 3.4 Å/bp (Watson and Crick, 1953), while the standard model for A-DNA has 11 base pairs per turn, and a rise of 2.6 Å/bp (Franklin and Gosling, 1953b).
For the purpose of simplicity in the remainder of this paper, we will use these idealized parameters for the structures of A- and B-DNA. The structures of DNA molecules do not match these ideal values; it has long been known that B-DNA in solution has an average repeat of about 10.4 bp/turn (Wang, 1979), and that the details of DNA conformation depend on the sequence (Bolshoy et al., 1991). Furthermore, we know almost nothing about the geometry of the junction between A-DNA and B-DNA, particularly inside a channel. Fortunately, the hypothesis presented here does not depend on these subtleties, so ideal conformations are used for simplicity.
2.2 The Scrunchworm Hypothesis
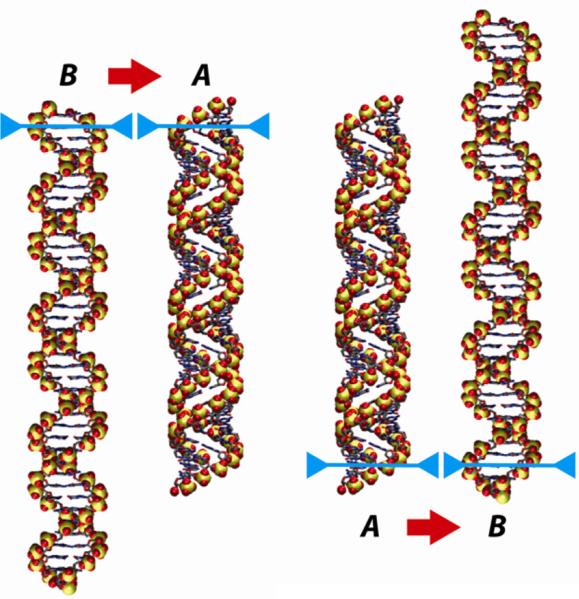
I propose that DNA packaging is driven by conformational changes within the DNA due to successive cycles of dehydration, which drives the DNA to the A form, and rehydration, which drives it back to the B form. A-DNA is more compact than B-DNA, so, as it passes through the B→A→B cycle, the DNA “scrunches up”, and then reelongates. If these conformational changes can be coordinated with an appropriate grip-release cycle, the cycle of DNA shortening and lengthening can move the DNA forward into the capsid (Fig. 3).
Fig. 3.
A cycle that takes DNA from the B form to the A form and back can be used to generate net motion of the molecule, because B-DNA is longer than A-DNA. If the molecule is anchored at the upper end (blue bars) when it undergoes the B→A transition, the other end will be drawn upward (left pair of images). If the molecule is anchored at the lower end during the transition back to the B form (right-hand pair of images), the entire molecule will have moved upward as a result of a complete B→A→B cycle. This figure shows idealized models for 40 base pairs of B-DNA (10 base pairs per turn, with a rise of 3.4 Å/bp) and A-DNA (11 base pairs per turn, rise = 2.6 Å/bp).
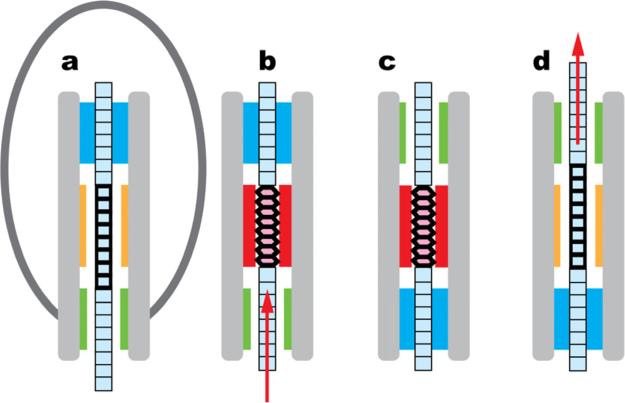
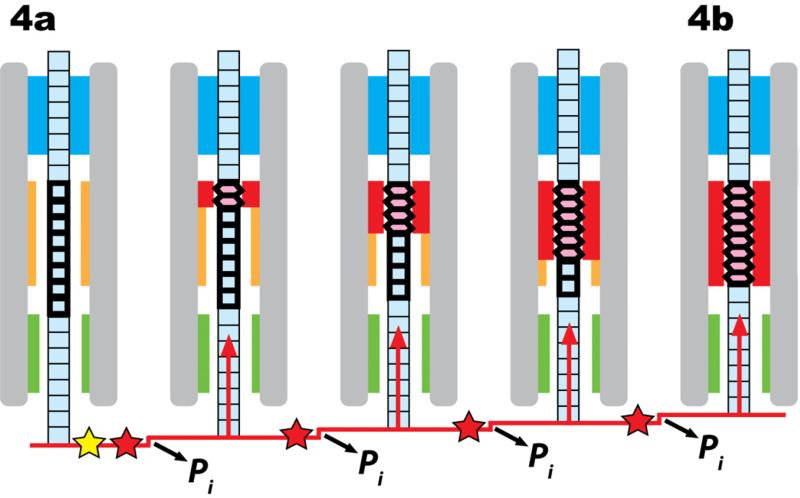
In this model, the DNA molecule must contain a “switch” region, so-called because it switches between the A and B conformations. In addition, the motor must have three components: a “dehydrator” that controls the water activity around the DNA switch region, and two “grips” that anchor the DNA – one ahead of the switch region (the distal grip) and the other behind it (the proximal grip). Fig. 4 shows a schematic representation of the mechanism whereby the cyclic motions of these components are coupled to the DNA conformational changes, leading to translocation.
Fig. 4.
Proposed mechanism for the scrunchworm translocation cycle. B-DNA is shown in pale blue, and A-DNA in pink. Heavy horizontal lines identify the conformationally active switch region. (Note that no part of this figure is drawn to scale, and the horizontal lines do not represent individual base pairs: if Scenario 1 is true, the switch region contains one turn of the double helix, while if Scenario 2 is true, the switch region contains four turns.) The capsid is shown only in panel (a) to clarify the orientation of the diagram. The motor contains two regions for gripping the DNA (blue) or letting it pass freely (green). It also contains a region for dehydrating and rehydrating the DNA (red and orange, respectively), and other regions (grey). (a) The distal (upper) grip grabs the DNA. (b) The dehydrator drives the DNA switch region from the B form to the A form, drawing the DNA tail into the proximal end of the motor. (c) The lower grip closes on the DNA molecule, and the upper grip opens. (d) The dehydrator moves away from the DNA, allowing rehydration; this drives DNA back into the B form, pushing DNA out of the distal end of the motor and into the capsid. The upper grip then closes on the DNA, and the lower grip opens, returning to state (a) and the beginning of a new translocation cycle.
The B→A transition shortens the DNA tail hanging out of the capsid, so the formation of A-DNA (the transition between Figs. 4a and 4b) coincides with the DNA motions seen in the burst phase in laser tweezer experiments (Chistol et al., 2012; Liu et al., 2014b; Moffitt et al., 2009). The energy released by ATP hydrolysis during each of these steps is used to dehydrate the DNA and drive it into the A form. It is released later in the form of mechanical work as the DNA is rehydrated and converts back to the lower energy B form (the transition between the states in Figs. 4c-d). The motor resets during the dwell, returning the system to the state in Fig. 4a.
To quantitatively connect the burst phase with the crunchworm hypothesis, we note that the experimentally measured distances were “obtained by ... assuming the average 3.4 Å/bp rise of B-form DNA”, and that “this conversion is correct irrespective of any distortions in the DNA structure inside the phage capsid or near the motor-DNA contacts, and relies solely on the DNA tether between the trapped beads being predominantly B-form.” (Both quotes come from the Supplementary Material to (Moffitt et al., 2009)). This means that each step of the burst, reported as 2.5 bp, moves the molecule forward by 2.5 × 3.4Å = 8.5Å. If the scrunchworm hypothesis is true, this requires the conversion of about 10.6 base pairs of DNA from the B form to the A form, because 10.6 × (3.4Å - 2.6Å) = 8.5Å. Interestingly, 10.6 bp is within a few percent of the number of base pairs in one turn of idealized B-DNA (10 bp), idealized A-DNA (11 bp), and the average repeat of DNA in solution (10.4 bp (Wang, 1979)). With the exception of the idealized A- and B-DNA models, all of these distances are approximations, with errors on the order of a few percent, so we will refer to the 2.5 bp step as corresponding to the conversion of approximately one turn of DNA. This suggests that the B→A transition is cooperative, and it raises the question, why is the cooperative unit equal to one turn of DNA? As will be seen below, this question has a simple mechanistic answer, and it provides support for the scrunchworm hypothesis.
If the scrunchworm hypothesis is true, there are two possible ways to connect the chemical cycle (Fig. 2) and the mechanical cycle (Fig. 4).
Scrunchworm Model, Scenario 1
One possibility is that the transition in Fig. 4a-4b represents one step of the burst. In this case, the cycle of Fig. 4 would be repeated four times for the complete burst. The switch region would contain about one turn of DNA, and the dehydrator would be about that length, too. (Note: Fig. 4 is not drawn to scale, and heavy lines in the DNA switch region do not represent individual base pairs.).
Scrunchworm Model, Scenario 2
Alternatively, the transition in Fig. 4a-4b could represent the complete burst. Since the burst has four 2.5 bp steps, there would have to be four hidden steps within the transition shown in Fig. 4a-4b. This could be explained if the dehydrator dehydrates four successive segments of about one turn of DNA at a time. If Scenario 2 is correct, then the switch region in Fig. 4 represents about four turns of DNA, and the dehydrator is long enough to cover four turns. (I again emphasize that Fig. 4 is not to scale.) The four steps within the burst for Scenario 2 are shown in Fig. 5.
Fig. 5.
Scenario 2, showing the details of the proposed burst steps comprising the transition between Figs. 4a-4b. The dehydrator converts four turns of DNA in the switch region from B-DNA to A-DNA in four steps of one turn each. Each step advances the DNA 2.5 bp and coincides with the hydrolysis of one molecule of ATP (red star) and the release of one inorganic phosphate group (Liu et al., 2014b). Five molecules of ATP are hydrolyzed in the complete cycle, with the fifth hydrolysis event either initiating the burst phase (yellow star). As in Fig. 4, nothing is drawn to scale.
The argument that the transition from B-DNA to A-DNA is cooperative, and that the length of the cooperative unit is about one turn of the double helix is true under either Scenario 1 or Scenario 2.
2.3 Comparison with Previous Models
In the scrunchworm model, the conformational changes in the DNA are driven by dehydration and rehydration. This contrasts sharply with models in which conformational changes in the protein motor generate forces that are applied to the DNA to push or pull it forward (Dixit et al., 2012; Draper and Rao, 2007; Fang et al., 2012; Lebedev et al., 2007; Moffitt et al., 2009; Oram et al., 2008; Ray et al., 2010; Schwartz et al., 2012; Sun et al., 2008; Yu et al., 2010). In all of those models but one, the DNA is a passive substrate, moved by forces arising from conformational changes in the proteins.
The conformational changes in the motor that produce the gripping/ungripping cycle described in Figs. 4-5 probably involve some sort of coupled twisting/untwisting and elongation/shortening motions, particularly in the connector. If so, they are similar to the intramolecular motions that were proposed in one of the earliest models for DNA packaging in 29 (Morais et al., 2001; Simpson et al., 2001; Simpson et al., 2000). As observed by Morais (2012), in that model “the connector essentially acts as an oscillating helical spring that pumps DNA into the prohead.” That model required that the connector rotate with respect to the capsid, but subsequent experiments have shown that the connector does not rotate (Baumann et al., 2006; Hugel et al., 2007). The scrunchworm model suggests similar conformational changes in the motor to attach and release the distal grip and to dehydrate and rehydrate the DNA, but without rotation of the proteins and not for pushing the DNA forward.
It has been recognized for some time that at least two valves or grips are required to alternately hold and release the DNA during translocation (Morais et al., 2008). The scrunchworm model has one feature in common with the previously proposed ratcheting model (Serwer, 2010) and the “push through a one-way valve” model from Guo's group (Fang et al., 2012; Schwartz et al., 2012), in that each of these invokes a rectification step to prevent the DNA from sliding backward after each forward step. But there are fundamental differences. Serwer's model assumes that the DNA motion is due to thermal fluctuations (longitudinal Brownian motion), and that rectification is achieved by coupling ATP hydrolysis to conformational changes in regions of the connector protein that grip the DNA. Guo's model uses the longitudinal motions of the ATPase to drive the DNA forward, and he proposes that the connector serves as a one-way valve, preventing backward motion. In contrast, DNA motions in the scrunchworm model are driven by the B→A→B cycle.
The scrunchworm model is also different from the “inchworm” models proposed for some phage motors (Draper and Rao, 2007; Moffitt et al., 2009), since these refer to longitudinal motions of specified regions of a motor protein, as they advance from one binding site on the DNA to the next. The protein is the “inchworm”, in contrast with the DNA “scrunchworm” proposed here.
As far as I am aware, only one other model has invoked an active role for DNA during translocation. Lindsay Black and his collaborators proposed that the DNA duplex “is translocated by a (DNA) compression and release mechanism that is also dependent upon storing torsional energy in the duplex” (Oram et al., 2008). This was later examined in a set of elegant fluorescence resonance energy transfer (FRET) experiments. Donor-acceptor pairs separated by 10 or 14 base pairs were placed in the stem of a branched (Y-shaped) DNA molecule being packaged in T4. The packaging motor loads the stem of the Y and translocates DNA until it reaches the junction, where it stalls. The efficiency of energy transfer increases when the stem of the Y is in the motor (Ray et al., 2010), indicating that the DNA is shortened by about 23% (Ray et al., 2010). The proposed “DNA crunching” by the motor received further support when they showed that intercalating agents are expell ed from duplex DNA during packaging (Dixit et al., 2012). Remarkably, A-DNA is 23% shorter than B-DNA ((3.4Å - 2.6Å)/3.4Å)). There are some questions about the explanation put forward for the results of the FRET experiments. It is not clear why one part of the motor would try to pull the DNA forward into the motor, while another part of the motor blocks that motion. It is also difficult to see how the T4 gp17 terminase (which constitutes the force-generating protein in Black's model) could reach beyond the junction of the Y as proposed (Fig. 4 of Ray et al., 2010), when the narrowest part of the channel through gp17 is at the entrance (Fig. 1b). It would seem that as the motor tries to pull a stalled Y-shaped DNA into the channel, the DNA should be under tension, not compression. The scrunchworm hypothesis suggests an alternative explanation for these experiments, namely that DNA is pulled into the capsid by a transition from the B form into A-DNA; apparently the motor can generate this transition even when the DNA is stalled by a Y-shaped junction at the entrance to the motor.
Black and his collaborators were the first to suggest an active role for the DNA in translocation. There are, however, subtle differences between their model and the model proposed here. Those authors propose “a transient linear grip-and-release motor mechanism that transiently compresses B-form DNA during translocation” (Dixit et al., 2012) (emphasis added). I assume that “linear” is equivalent to what I call “longitudinal”, i.e., along the direction of DNA motion. While it is possible that such motions do exist, the critical motions in the scrunchworm hypothesis are axial, i.e., perpendicular to the direction of DNA motion (Figs. 4-5). And, where Black's model suggests that B-DNA is compressed by the motor, the scrunchworm model proposed that it is dehydrated and converted into the A form. The two models agree that ATP drives conformational changes in the protein, which drive conformational changes in the DNA. They disagree on the details of the latter step.
2.4 Correlation of the Model with Experimental Data
The DNA packaging mechanism proposed here is consistent with a substantial body of experimental data.
One important feature of the scrunchworm hypothesis is that it offers a quantitative physical explanation for the magnitude of the forces that are generated by viral packaging motors. Consider first the elongation step, as DNA is rehydrated and advances into the capsid (the transition between Figs. 4c and 4d). There is a modest energy difference between the high-energy compact A form and the low-energy extended B form: G is on the order of 2.5 kJ/mol for a typical base pair step (Tolstorukov et al., 2001). On a single molecule basis, dividing by Avogadro's number gives ΔG ~4.2×10−21 J per base pair step. This energy difference is expended over a very short distance as the DNA is rehydrated and expands (Δx = 3.4Å - 2.6Å = 0.8Å = 8×10−11 m per base pair step). F = ΔG/Δx, so the resulting force is about 50 pN. (Since the motor is capable of packaging against pulling forces of this same magnitude, dehydration must make the A form favorable over the B form by a comparable value of G.) Earlier estimates had suggested that the maximum force exerted by the 29 motor are in the range of 70-80 pN (Chemla and Smith, 2012; Fuller et al., 2007b; Rickgauer et al., 2008; Smith et al., 2001), but recent work has shown that these are over-estimates, due to allosteric effects on packaging velocity, and that the maximum force resisting packaging is about 23 pN at high filling (Douglas E. Smith, personal communication). This is entirely consistent with forces generated under the scrunchworm hypothesis.
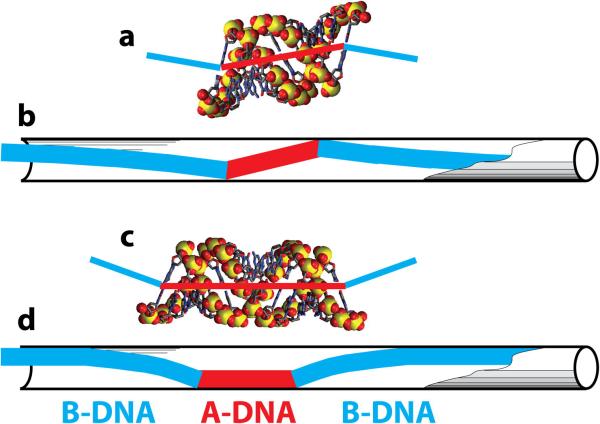
Another important feature of the scrunchworm hypothesis is that it offers an explanation for the experimentally observed size of each step in the burst. As explained earlier, a 2.5 bp step requires the B→A transition for about one turn of DNA. A simple argument can be made to show that the cooperative unit for the B→A transition in a confined space is approximately one turn of the double helix. Consider a narrow cylinder containing a segment of N base pairs of A-DNA within a long molecule in the B conformation. The junction between A-DNA and B-DNA necessarily produces a bend in the helix axis, because the base pairs in B-DNA are perpendicular to the helix axis, while those in A DNA are tilted (Fig. 3). Using the proper nomenclature, the base pair inclination is ~0-5° in B-DNA and ~15-20° in A-DNA (Olson et al., 2001). With bends at the two A/B junctions, there will be steric clashes of the B-DNA tails with the wall of the cylinder (Fig. 6), leading to an unfavorable elastic deformation energy, E(N) in the tails. If the segment of A-DNA is an integral number of turns, the helical axes of the two B-DNA tails will be parallel to one another where they connect to the A-DNA (Fig. 6a). When the B tails are in this trans configuration, they can bend gently to relieve the steric clashes with the walls (Fig. 6b). On the other hand, if the A-DNA segment contains a half-integral number of turns (1.5, 2.5, ...), the axes of the B-DNA tails will be cis to one another (Fig. 6c), so the tails must bend more sharply to relieve steric clashes with the wall of the cylinder (Fig. 6d). As a consequence, the least expensive B→A transition will involve approximately one turn of A-DNA – “approximately”, because we do not know the exact structure of the A/B junction and the exact periodicity of E(N). This simple argument explains why the cooperative unit in the B→A transition is about one turn, and why each step of the burst advances the DNA by 2.5 bp.
Fig. 6.
Elastic deformations of a B-DNA molecule containing inserts of A-DNA. (a) A model of one turn of A-DNA. The base pairs are inclined relative to the axis of the double helix (red line), so if this segment of DNA is inserted into B-DNA, the axes of the two B-DNA helices will be offset from one another, but parallel (blue lines). (b) A long BDNA double helix (blue) with an insert of one turn of A-DNA (red) can be fit into a long narrow cylinder with modest elastic deformations of the two B-DNA tails. (c) A model of one and a half turns of A-DNA. If this molecule is inserted into a long B-DNA duplex, the two axes of the B-DNA tails will be at a substantial angle to one another (blue). (d) When the construct in (c) is put into a narrow cylinder, the B-DNA tails will have a larger elastic deformation energy than when the A-DNA insert contains only one turn.
Perhaps the strongest direct support for the scrunchworm hypothesis is the experimental evidence that the DNA molecule in the motor is shortened during the generation of force (Dixit et al., 2012; Ray et al., 2010). As explained above, the FRET measurements of Ray et al. (2010) also provide quantitative support for the model, since the rise of A-DNA is 23% less than the rise of B-DNA, matching their results.
It has been argued that there are extensive nonspecific interactions between the DNA and the portal during the burst phase, so that the step size is not dictated by periodic repeats in the DNA structure, e.g., specific contacts with the phosphate groups (Aathavan et al., 2009; Moffitt et al., 2009). The 29 connector protein forms a dodecameric portal with a left-handed helical structure (Guasch et al., 2002), and left-handed structures are also found in the portals of SPP1 (Lebedev et al., 2007) and P22 (Olia et al., 2011). This mismatch between left-handed portals and right-handed DNA facilitates nonspecific interactions. Some of the nonspecific connector-DNA contacts are probably associated with the distal grip and the dehydrator.
There is also evidence supporting the proposed dehydration of DNA by proteins. It is commonly assumed that DNA in vivo has the B form associated with high water activity, but examination of a collection of high-resolution crystal structures revealed that a large fraction of base pair steps adopt the A conformation (Lu et al., 2000). Moreover, it has been known for years that very subtle environmental changes can drive a given DNA from one conformation to another. As just two examples, different fragments of DNA crystallized under virtually identical conditions can crystallize in either the A, B or Z form (Guschlbauer and Saenger, 1987), and one experiment reported a mixture of octamers in the A form and octamers in the B form in a single crystal (Doucet et al., 1989). These results all suggest that relatively modest motions of the motor proteins could alter the solvation state of DNA sufficiently to drive the B→A and A→B transitions, which is consistent with the scrunchworm hypothesis.
Finally, the scrunchworm hypothesis may offer an explanation for the surprising ability of viruses to package dsDNA molecules containing large gaps or regions with extensive chemical modifications (Aathavan et al., 2009; Moll and Guo, 2005; Oram et al., 2008). Under Scenario 2, the distance between the proximal and distal grips must be ~110Å or more if the dehydrator covers about four turns of A-DNA. This is more than enough to accommodate a substantial length of inactive DNA (gapped or chemically modified) along with sections of active DNA that move the molecule forward by cyclic B→A→B transitions. This substrate promiscuity is probably the strongest argument in support of Scenario 2.
3. Predictions and Open Questions
3.1 Predictions
One advantage of the scrunchworm hypothesis is that it makes four specific predictions that can be tested experimentally.
Prediction 1: Viral motors can't package long segments of double-stranded RNA. dsRNA is always in the A form, so it is incapable of undergoing the B→A→B transition, which is at the heart of the scrunchworm hypothesis. This prediction can be tested by engineering dsRNA inserts into dsDNA molecules and using them as substrates in laser tweezers experiments. These motors are known to be quite promiscuous with regard to packaging DNAs with nicks, gaps, abasic stretches and non-standard backbones (Aathavan et al., 2009; Oram et al., 2008), so dsRNA inserts of modest length can probably be tolerated. As these grow longer and longer, the motor should have more difficulty with them, and when they exceed the length of the dehydrator, packaging should cease completely. These experiments should also shed light on the size of the dehydrator.
Prediction 2: The pulling force generated by the B→A transition induced by dehydration is 50 pN or more, since viruses can package DNA when pulling forces of this magnitude are applied in laser tweezer experiments. There has been one published attempt at detecting the B→A transition in laser tweezer experiments, varying the concentration of ethanol in aqueous solution to dehydrate the DNA (Hormeno et al., 2011).
Unfortunately, instead of the anticipated shortening, the authors observed a total collapse of the DNA, which they attributed to aggregation. They did not try other dehydrating agents, so the experiment bears repeating with trifluoroethanol, which is reported to generate the B→A transition with less risk of aggregation than ethanol (Tolstorukov et al., 2001). Other organic liquids that are miscible with water might also be tried.
Prediction 3: The force generated by the packaging motor depends on the DNA sequence. Some DNA sequences are more resistant to conversion into the A form than others (Ivanov and Minchenkova, 1995; Tolstorukov et al., 2001). The B→A transition (Fig. 4a-4b) should generate larger pulling forces for “A-philic” sequences than for “A-phobic” sequences. This prediction can be tested in single molecule laser tweezer experiments, comparing molecules containing an abundance of A-philic regions against those containing many A-phobic regions. In contrast, the pushing forces of the A→B transition (Fig. 4c-4d) should show the opposite trend, and A-phobic sequences should be able to overcome larger back-pressures as the capsid fills than A-philic sequences. As one concrete prediction, according to the scale of (Tolstorukov et al., 2001), d(CAATT)n •d(AATTG)n should generate a pushing force of about 80 pN, vs. ~40 pN for d(TACCC)n•d(GGGTA)n.
Prediction 4: The cyclic B→A→B transitions that drive DNA into the capsid can be detected in laser tweezer experiments. The experiment would require beads on both ends of a dsDNA molecule threaded through the motor in a perforated capsid (one that has a hole in it) (Liu et al., 2014b). As ATP is hydrolyzed, the capsid should move forward along the DNA without resistance. If the DNA is held under light tension (~5 pN), there should be no serious opposition to the B→A transition, and this should appear as a shortening event (the burst), followed by a return to the longer B form during the dwell. It will be interesting to see whether the A→B transition occurs after each shortening event (Scenario 1), or whether it occurs after four shortening steps (Scenario 2). As in previous experiments, further information on the mechanochemical cycle described in Figs. 4-5 can be teased out by varying the tension in the DNA, by varying the concentrations of ATP and ADP, and by the addition of nonhydrolyzable ATP analogues to slow or stall events.
3.2 Open Questions
The scrunchworm hypothesis suggests that repeated transitions between B-DNA and ADNA generate force and motion in dsDNA viral packaging motors. This raises three questions. First, how does the motor alternatively dehydrate and rehydrate DNA in the switch region? Second, how does the motor coordinate the cycles of grip/release and dehydration/rehydration? Third, how are the binding and hydrolysis of ATP and the release of ADP and inorganic phosphate coupled to these mechanical steps?
If we focus on 29, the questions can be made more specific.
What are the sites of DNA-protein interactions during dehydration, and during the locking steps at each of the grips? Morais has suggested that, in 29, one of the grips is found in the ATPase, and the other might be in the connector (Morais et al., 2008). If this is true, then the dehydrator probably includes the pRNA and part of both proteins. This would be easily accommodated under Scenario 2, since the total length of the channel through the motor is about 150Å (Fig. 1), which is enough to cover four turns of A-DNA (104Å) and two grip regions. The interior surface of the 29 connector is mostly negatively charged, but there are two rings of lysine side chains about 20Å apart (Guasch et al., 2002). It remains to be seen what functional role these have.
What conformational changes in the ATPase (gp16) accompany each step of the chemical cycle, and how are these propagated to the conformational changes in the connector? What role does the pRNA play in these processes? One major puzzle is that there are only four bursts of DNA motion while five ATP molecules are hydrolyzed in each cycle (Moffitt et al., 2009). Four ATPs are hydrolyzed during the burst, one with each 2.5 bp step, and the fifth hydrolysis event “either initiates or concludes the hydrolysis cascade (of the burst phase)” (Chistol et al., 2012). This observation, coupled with an observed DNA rotation of −15° per 10 bp burst (Liu et al., 2014b) led to the suggestion that load-bearing interactions between the DNA and one specific subunit in the pentameric ATPAse (the “coordinating subunit”) accompany the dwell phase, and that these must be reset after each four-step burst (Chistol et al., 2012; Liu et al., 2014b). There is nothing in the scrunchworm hypothesis that conflicts with or supports this suggestion.
When we consider other dsDNA viral motors, one of the most striking features is the variation in the sizes of the portal proteins (Fig. 1). What is the role of channel length in the mechanochemical cycle? The 29 motor is smaller than those of some other dsDNA bacteriophages such as P22 (Olia et al., 2011), T4 (Sun et al., 2008), T7 (Guo et al., 2013) and ε15 (Jiang et al., 2006), whose cores extend 200Å or more into the capsid's interior, and where the total length of the channel through the portal/core complex can be as much as 300Å. It has been suggested that such core structures facilitate the organization of the packaged genome (Jiang et al., 2006; Olia et al., 2011; Petrov et al., 2007b). An alternative possibility is that different core structures have different mechanisms for gripping and dehydrating the DNA. In any case, the effects of channel length and structure on packaging kinetics, motor strength, and efficiency remain to be determined.
The characteristics of other motors will certainly inform further development of the scrunchworm hypothesis, especially those for which laser tweezer data are available, such as T4 (Fuller et al., 2007a) and lambda (Fuller et al., 2007c). Of particular interest is the question of how lambda generates packaging velocities that are more than three times as great as those of 29 (Fuller et al., 2007c). It would also be interesting to test the prediction that ε15 generates higher forces than 29 (Petrov et al., 2007a) and, if that prediction is sustained, to determine how the ε15 motor does this.
4. Future Directions
Several experiments have examined how the packaging process is affected by the concentrations of ATP and ADP, by external forces, and by increasing internal pressure as the capsid fills with DNA (Chistol et al., 2012; Fuller et al., 2007a; Fuller et al., 2007c; Liu et al., 2014b; Moffitt et al., 2009; Rickgauer et al., 2008; Smith et al., 2001). Recent work has also shown that some of the effects of filling are due to allosteric effects (Douglas E. Smith, personal communication). Table 1 summarizes these data.
Table 1.
Effects of External Force, Filling, and ATP Concentration on DNA Packaging
| Effects | No Effect On | |
|---|---|---|
| External Force (pulling) | Induces slips | Motor-ATP interactions |
| Slows burst | ||
| Internal Force (filling) | Induces slips | |
| Slows burst | ||
| Reduces burst size | ||
| Lengthens dwell | ||
| Creates long-lived pauses | ||
| ATP Concentration | Duration of dwell | Duration of burst |
High-resolution structural data on the DNA-motor complex at various steps in the packaging process are needed to explain these effects. These data should also facilitate examination of the scrunchworm hypothesis. In the absence of such data, detailed molecular modeling studies on the model presented in Figs. 4-5 offer the best hope for moving forward.
One existing model is particularly promising for modeling studies, the hybrid model for the T4 motor (Fig. 1b). For 29, modeling will be facilitated by the availability of crystal structures of the connector (Simpson et al., 2000), and of the connector-binding domain of the pRNA (Ding et al., 2011). There is also a sub-nanometer asymmetric reconstruction of the complex between these molecules, based on data from cryo-electron microscopy (Cao et al., 2014). Unfortunately, there is no crystal structure of the 29 ATPase (gp16), although a homology model for the gp16 pentamer is available (Yu et al., 2010). It was based on the crystal structure of the hexameric P4 ATPase of bacteriophage 12 (Mancini et al., 2004). There are three caveats in using P4 as a homology model. First, the genome of 12 is double-stranded RNA, not DNA. RNA adopts the A form, and there is no reason to believe dsDNA motors have the same mechanism as the motors of dsRNA viruses. Second, phages of the 12 class have very simple motors that contain only the ATPase. They do not have molecules corresponding to pRNA and the 29 connector (the portal protein in other dsDNA viruses). There is no reason to assume that portal proteins are passive elements in translocation. Third, those authors proposed a “push and roll” mechanism for packaging and predicted that DNA would rotate through an angle of about 48° for a complete ten base pair burst (Yu et al., 2010). This is more than three times the observed rotation of 15° (Liu et al., 2014b).
Beyond the issue of viral packaging motors, it is possible that the scrunchworm hypothesis may be relevant to other molecular motors that translocate along double-stranded DNA. As just one example, it is interesting to note that the replicative DnaB helicase, whose hexamer wraps around single-stranded A-form DNA (Itsathitphaisarn et al., 2012), is also capable of translocating along dsDNA (Kaplan and O'Donnell, 2002).
5. Summary
The scrunchworm hypothesis suggests that DNA is not simply a passive element driven into the capsid by the motor. Instead, repeated B→A→B transitions make DNA the heart of the motor that actually generates force and longitudinal motion.
This model can explain how dsDNA packaging motors generate the highest known forces of any molecular motors, because the energy difference between the high-energy compact A form and the low-energy extended B form is expended over a short distance as the DNA is rehydrated and converts from A-DNA to B-DNA. In addition, it suggests that the cooperative unit for the A→B transition is one turn of the double helix, providing a structural basis for the experimental observation that packaging proceeds in bursts comprised of 2.5 bp steps.
Further research is needed to determine the validity of the scrunchworm hypothesis. Four predictions to test it have been presented here. The proposed experiments should deepen our understanding of the mechanochemistry of dsDNA viral motors, regardless of their outcomes.
Acknowledgments
Supported by grant R01 GM70785 from the US National Institutes of Health. I am indebted to Roger Wartell and Doug Smith for inciteful comments on the manuscript. Marc Morais kindly provided Fig. 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–73. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Bolshoy A, McNamara P, Harrington RE, Trifonov EN. Curved DNA Without A-A: Experimental Estimation of All 16 DNA Wedge Angles. Proc. Natl. Acad. Sci. USA. 1991;88:2313–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Saha M, Zhao W, Jardine PJ, Zhang W, Grimes S, Morais MC. Insights into the structure and assembly of the bacteriophage 29 double-stranded DNA packaging motor. J Virol. 2014;88:3986–96. doi: 10.1128/JVI.03203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–57. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Chemla YR, Smith DE. Single-molecule studies of viral DNA packaging. Adv Exp Med Biol. 2012;726:549–84. doi: 10.1007/978-1-4614-0980-9_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, Jardine PJ, Bustamante C. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151:1017–28. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Donatis GM, Zhao Z, Wang S, Huang LP, Schwartz C, Tsodikov OV, Zhang H, Haque F, Guo P. Finding of widespread viral and bacterial revolution dsDNA translocation motors distinct from rotation motors by channel chirality and size. Cell Biosci. 2014;4:30. doi: 10.1186/2045-3701-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc Natl Acad Sci U S A. 2011;108:7357–62. doi: 10.1073/pnas.1016690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit AB, Ray K, Black LW. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc Natl Acad Sci U S A. 2012;109:20419–24. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet J, Benoit JP, Cruse WB, Prange T, Kennard O. Coexistence of A- and B-form DNA in a single crystal lattice. Nature. 1989;337:190–2. doi: 10.1038/337190a0. [DOI] [PubMed] [Google Scholar]
- Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J Mol Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Fang H, Jing P, Haque F, Guo P. Role of channel lysines and the “push through a one-way valve” mechanism of the viral DNA packaging motor. Biophys J. 2012;102:127–35. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RE, Gosling RG. Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate. Nature. 1953a;172:156–7. doi: 10.1038/172156a0. [DOI] [PubMed] [Google Scholar]
- Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953b;171:740–1. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007a;104:16868–73. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi29. Proc Natl Acad Sci U S A. 2007b;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007c;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–76. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci U S A. 2013;110:6811–6. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschlbauer W, Saenger W. DNA-Ligand Interactions from Drugs to Proteins Plenum. New York: 1987. [Google Scholar]
- Hormeno S, Moreno-Herrero F, Ibarra B, Carrascosa JL, Valpuesta JM, Arias-Gonzalez JR. Condensation prevails over B-A transition in the structure of DNA at low humidity. Biophys J. 2011;100:2006–15. doi: 10.1016/j.bpj.2011.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–77. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov VI, Minchenkova LE. The A-form of DNA: in search of the biological role. Mol Biol. 1995;28:780–8. [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–6. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DL, O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell. 2002;10:647–57. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–94. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chistol G, Bustamante C. Mechanical operation and intersubunit coordination of ring-shaped molecular motors: insights from single-molecule studies. Biophys J. 2014a;106:1844–58. doi: 10.1016/j.bpj.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine PJ, Bustamante C. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014b;157:702–13. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Shakked Z, Olson WK. A-form Conformational Motifs in Ligand-bound DNA Structures. J Mol Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–55. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–50. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll WD, Guo P. Translocation of nicked but not gapped DNA by the packaging motor of bacteriophage phi29. J Mol Biol. 2005;351:100–7. doi: 10.1016/j.jmb.2005.05.038. [DOI] [PubMed] [Google Scholar]
- Morais MC. The dsDNA packaging motor in bacteriophage phi29. Adv Exp Med Biol. 2012;726:511–47. doi: 10.1007/978-1-4614-0980-9_23. [DOI] [PubMed] [Google Scholar]
- Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–74. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Tao Y, Olson NH, Grimes S, Jardine PJ, Anderson DL, Baker TS, Rossmann MG. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Prevelige PE, Jr., Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu XJ, Neidle S, Shakked Z, Sklenar H, Suzuki M, Tung C-S, Westhof E, Wolberger C, Berman HM. A standard reference frame for the description of nucleic acid base- pair geometry. J Mol Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Lim-Hing K, Harvey SC. Packaging of DNA by bacteriophage epsilon15: structure, forces, and thermodynamics. Structure. 2007a;15:807–12. doi: 10.1016/j.str.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Boz MB, Harvey SC. The conformation of double-stranded DNA inside bacteriophages depends on capsid size and shape. J Struct Biol. 2007b;160:241–8. doi: 10.1016/j.jsb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010;398:224–32. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29. Biophys J. 2008;94:159–67. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Rao VB. Viruses: sophisticated biological machines. Adv Exp Med Biol. 2012;726:1–3. doi: 10.1007/978-1-4614-0980-9_1. [DOI] [PubMed] [Google Scholar]
- Schwartz C, Fang H, Huang L, Guo P. Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging. Nucleic Acids Res. 2012;40:2577–86. doi: 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P. A hypothesis for bacteriophage DNA packaging motors. Viruses. 2010;2:1821–43. doi: 10.3390/v2091821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P, Jiang W. Dualities in the analysis of phage DNA packaging motors. Bacteriophage. 2012;2:239–255. doi: 10.4161/bact.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr. 2001;57:1260–9. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Sun S, Rao VB, Rossmann MG. Genome packaging in viruses. Curr Opin Struct Biol. 2010;20:114–20. doi: 10.1016/j.sbi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–62. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Tolstorukov MY, Ivanov VI, Malenkov GG, Jernigan RL, Zhurkin VB. Sequence-dependent BA transition in DNA evaluated with dimeric and trimeric scales. Biophys J. 2001;81:3409–21. doi: 10.1016/S0006-3495(01)75973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979;76:200–3. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FHC. A Structure for Deoxyribose Nucleic Acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. Mechanochemistry of a viral DNA packaging motor. J Mol Biol. 2010;400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]