Abstract
The DNA damage response (DDR) is a protein kinase cascade that orchestrates DNA repair processes via transcriptional and post-translational mechanisms. Cell cycle arrest is a hallmark of the DDR. We performed a damage-induced cell cycle arrest screen and uncovered a critical role for Fanconi anemia (FA) and homologous recombination (HR) proteins in ATR signaling. HR was required to maintain prolonged cell cycle arrest and to prevent massive genomic instability. Over 100 high scoring DDR candidates were interrogated for their roles in the DDR. Three proteins, INTS7, CLOCK and a novel protein RHINO, are recruited to sites of DNA damage. RHINO independently binds the Rad9-Rad1-Hus1 complex (9-1-1) and TopBP1. RHINO is recruited to sites of DNA damage by the 9-1-1 complex and is necessary to promote Chk1 activation. We suggest that RHINO functions together with the 9-1-1 complex and TopBP1 to fully activate ATR.
The importance of the DNA damage response (DDR) is underscored by the prevalence of mutations in this pathway found in cancers and developmental syndromes (1). Historically, most DDR genes were identified genetically in yeast as mutants defective in the transcriptional or cell cycle arrest responses to DNA damage. However, many mammalian DDR components are absent in yeast. To identify novel DDR genes, we developed a high throughput (HTP) microscopy-based assay using U2OS cells following siRNA depletion to measure inappropriate cell cycle entry into mitosis 18h after 10Gy IR, employing nocodazole to trap cells in mitosis (Fig. 1A). Most cells entering mitosis during this prolonged assay incurred damage during S phase (see Supplemental Text and Figure S1 for further details on the assay). Hits were stratified based on the fold change in mitotic index (MI) compared to negative control wells: Strong (>8 fold), Medium (4–8 fold) and Weak (2–4 fold) (Fig. 1B, Table S1). Since Chk1 did not score due to toxicity (Fig. S2), we rescreened the toxic subset of genes at a lower siRNA concentration resulting in an additional 98 pools scoring that included Chk1, PALB2, Wee1 and FANCM (Fig. S2D and Tables S1 and S2).
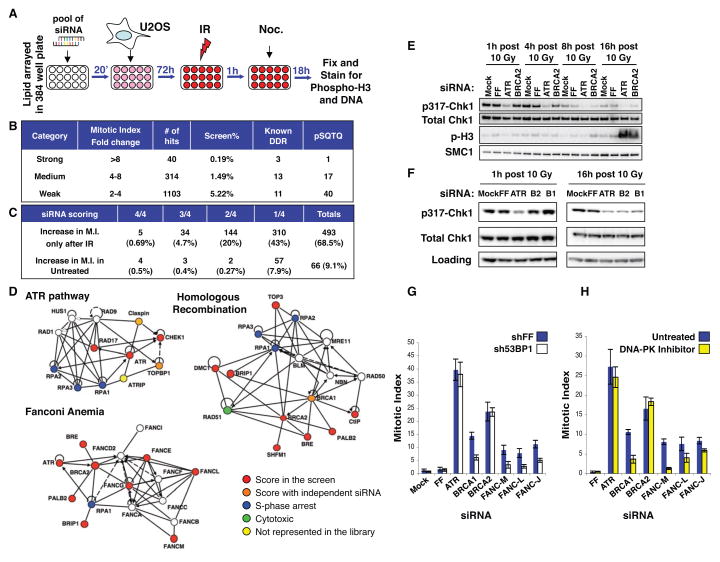
Figure 1. A screen for regulators of DDR signaling.
(A) Schematic of the screen.
(B) Primary screen statistics. The number of known DDR proteins and potential ATM/ATR substrates (pSQTQ) are listed.
(C) Secondary screen statistics for 720 candidate genes with and without DNA damage. For each gene, the fraction of siRNAs scoring and the total number of genes scoring is listed.
(D) DDR networks identified in primary screen using Ingenuity pathway analysis.
(E) ATR pathway signaling integrity after ATR and BRCA2 depletion. Cells collected at the indicated times after IR (10 Gy) were examined for Chk1 phosphorylation. Smc1 was used as loading control.
(F) ATR pathway signaling integrity after ATR, BRCA2 (B2) and BRCA1 (B1) depletion. Cells were collected 1 and 16 h after IR (10 Gy). Cyclin B1 and PCNA were used as loading controls for the left and right panels respectively.
(G) Depletion of 53BP1 with shRNAs restores cell cycle arrest in BRCA1, FANCM, FANCL and FANCJ depleted cells but not in ATR or BRCA2 depleted cells. MI calculated 18h after 10 Gy.
(H) Chemical inhibition of DNA-PKcs restores cell cycle arrest in BRCA1, FANCM, FANCL and FANCJ depleted cells but not in ATR or BRCA2 depleted cells. The DNA-PK inhibitor was added 2h after IR (10Gy) at a final concentration of 1 μM. MI was calculated as above.
All strong and medium candidates and a subset of prioritized weak candidates, 720 in total, chosen for their degree of bypass or DDR phosphorylation status (2, 3) (pSQTQ, Fig. 1C, Table S1) were chosen for secondary screening. Pools of siRNAs were deconvoluted into 4 individual siRNAs and retested (Fig. 1C). More then 75% recapitulated with at least 1 siRNA (Fig. 1C, Table S3), 12% of these were eliminated because they increase MI in the absence of damage (Fig. 1C, Fig. S3B and Table S3).
DDR mutations often cause sensitivity to DNA damage, therefore sensitivity to mitomycin C (MMC) was assessed after gene depletion by siRNAs (Fig. S3C). Of these genes, 53% that scored with at least 2 siRNAs in the checkpoint assay also scored with two or more siRNAs in the MMC-sensitivity assay (97 genes). These genes were further interrogated using Dharmacon’s On target plus (OTP) technology and tested for checkpoint function, MMC-sensitivity and HR efficiency (4) (Fig. S4A, Table S4, see Supplemental Text for details). This high confidence list is enriched in the biological categories of DNA replication, recombination and repair as well as nucleic acid metabolism and cancer relevance (Fig. S4B).
Bioinformatic analysis revealed a strong enrichment for the ATR, Fanconi anemia (FA) and HR pathways (Fig. 1D and Fig. S4C). This seems counterintuitive since DSBs remain unrepaired in the absence of HR and signaling should persist until the repair process is complete. However, examination of Chk1 phosphorylation kinetics indicates that, in the absence of BRCA1 and BRCA2, ATR signaling is not sustained (Fig. 1E, F). While BRCA1 is known to regulate Chk1 phosphorylation (5), we did not observe a defect in Chk1 activation 1h post IR, but we observed a defect in maintenance of the activation (Fig. 1F). BRCA1 acts upstream of BRCA2, which has a direct role in promoting Rad51 filament formation (1). Although HR is the major DSB repair pathway in S/G2, other competing repair pathways such as non-homologous end-joining (NHEJ) also play a role. To examine whether the removal of signaling ends by NHEJ repair blocks HR in BRCA-depleted cells, we inhibited NHEJ by depleting 53BP1 with shRNA or using a DNA-PKcs inhibitor (NU7441). 53BP1 was chosen because of its role in promoting ligation of ends that are not in close proximity (1), a characteristic of ends that remain unrepaired for a significant amount of time. We observed a decrease in MI in BRCA1-depleted cells in both conditions (Fig. 1G, H), suggesting that NHEJ processes the breaks and this allows restart of the cell cycle. Interestingly, we did not observe a rescue of the BRCA2 defect, suggesting that either the processed DSB intermediates that accumulate in these cell lines are not channeled into NHEJ or that BRCA2 has an additional role in the DDR to promote arrest. Inhibiting NHEJ also suppressed the signaling defects in FANCM, L and J (Fig. 1G, H). These results are consistent with the recent findings that loss of 53BP1 partially rescues the HR defect and damage sensitivity of BRCA1 mutant cells, but not BRCA2 mutants, and sensitivity to damage in FA deficient cells (6–9).
Twenty-four genes were selected based on these secondary assays to assess their localization to sites of damage, a hallmark of DDR proteins (10). U2OS cells stably expressing GFP-fusions were created and subjected to laser-induced DNA damage, marked by γ-H2AX co-staining. Three proteins (INTS7, Clock and MGC13204 – an orphan protein) showed co-localization with γ-H2AX in the form of laser stripes (Fig. 2A), but did so independently of H2AX (Fig. S5A).
Figure 2. Characterization of several DDR candidates.
(A) Analysis of DNA damage localization. U2OS cells expressing GFP fusions to INTS7, Clock and RHINO were striped with a UV laser and co-stained with γ-H2AX to identify damaged cells.
(B) Cell cycle arrest of RHINO with individual siRNAs at 30 nM, 18 h post 10 Gy is shown. SiRNAs 9 and 11 result in a statistically significant increases in MI with P-values of 8×10−5 and 0.05 respectively determined by the two t-test.
(C) HR results using individual RHINO siRNAs normalized to the negative control (FF).
(D) The multi-color competition assay (MCA) was used to assess DNA damage sensitivity of the indicated depleted genes in response to the indicated damaging agents.
(E) Mass spec analysis of Flag-HA RHINO after IR. Genes are ranked according to the ratio of the number of peptides versus the molecular weight of total protein.
(F) Interaction of 9-1-1, TopBP1, Rad18 and Ubc13 with RHINO. 293 cells expressing GFP alone or Flag-HA-RHINO were treated with 10 Gy and harvested 4 h later and analyses for association by immunoblotting. The asterisk in the Rad9 panel represents the IgG heavy chain migrating above endogenous Rad9.
Clock (checkpoint bypass −7/8 siRNAs and MMC sensitivity −6/8 siRNAs) is a basic helix-loop-helix transcription factor that participates in circadian rhythm regulation. Numerous examples of crosstalk between DNA damage, cell cycle regulation and circadian rhythms have come to light in recent years, e.g. Timeless, Tipin, Chk2 and Period (11). Loss of Clock or its partner, BMAL1 in mice results in chronic enhanced sensitivity to DNA crosslinking agents, previously presumed to be due to misregulation of transcriptional targets (12). However, localization of Clock to sites of DNA damage suggests that it plays a more direct role in the DDR.
INTS7 is a member of the Integrator complex that interacts with RNA polymerase II to assist in snRNA processing (13). INTS7 depletion resulted in cell cycle arrest bypass (6/7 siRNAs) and MMC sensitivity (3/7 siRNAs). Interestingly, one component of the Integrator complex, INTS3, has been found associated with two related OB-fold proteins, SSB1 and SSB2, to form a complex recruited to damage sites and required for checkpoint function (14, 15). We also found INTS7 interacts with SSB1 (Fig. S5B).
MGC13204, referred to hereafter as RHINO, depletion led to checkpoint bypass and sensitivity to MMC, IR and camptothecin (Fig. 2D). Moreover, RHINO localizes to foci in response to IR (Fig. S6A), MMC and UV laser stripes (Fig. 2A). RHINO depletion levels strongly correlate with both checkpoint and HR defects (Fig. 2B, C and Fig. S6B and S8), which are suppressed by expression of a siRNA (#9)-resistant RHINO cDNA and partially suppressed by the WT cDNA (Fig. S7).
Mass spec analysis of Flag-HA RHINO immunoprecipitates identified peptides for each subunit of the damage specific clamp 9-1-1 (Rad9, Rad1 and Hus1), TopBP1, the E3 ubiquitin ligase Rad18 and the E2 conjugating enzyme Ubc13 (Fig. 2E). Western blotting independently verified these interactions (Fig. 2F). To determine if these components controlled RHINO’s damage recruitment, we depleted TopBP1, Rad18 and Rad17 (which loads 9-1-1) and quantified RHINO localization to UV-induced DSBs. Rad17 depletion caused a 40% reduction in RHINO recruitment (Fig. S9A). UV laser striping analysis of WT and mutant MEFs expressing GFP-RHINO directly demonstrated the requirement of the 9-1-1 complex in RHINO recruitment (Fig. 3A and Fig. S9B). For this reason, we named the orphan protein MGC13204 as RHINO (Rad9, Rad1, Hus1 Interacting Nuclear Orphan).
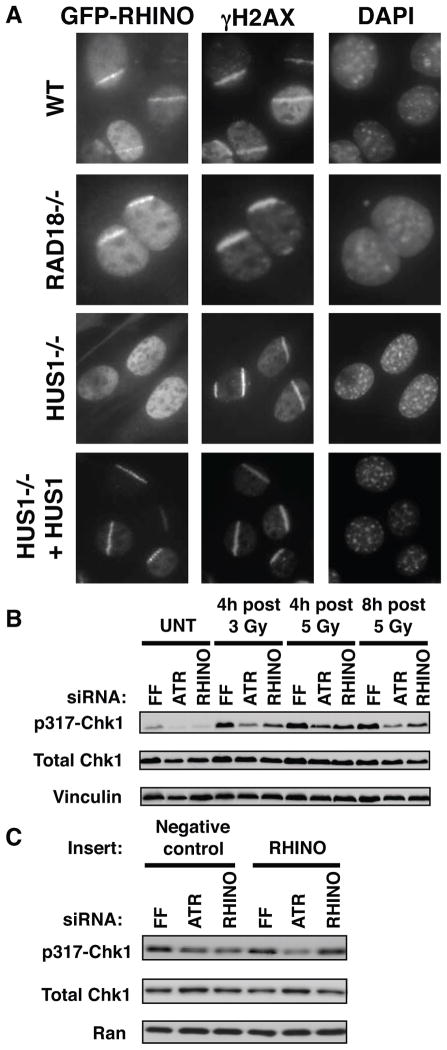
Figure 3. RHINO is required for DDR signaling.
(A) Hus1-dependent RHINO recruitment to sites of DSBs. MEFs (WT, Rad18−/−, Hus1−/− and Hus1−/− reconstituted with Hus1) expressing GFP-RHINO had DSBs introduced by UV laser treatment and RHINO recruitment was assessed.
(B) RHINO is required for Chk1 phosphorylation. Cells after ATR and RHINO depletion were treated with the indicated IR doses, collected at the indicated times and immunoblotted.
(C) Restoration of RHINO expression rescues DDR signaling. Three days after siRNA transfection, cells were infected at a MOI of 2 with a retrovirus expressing the Flag-HA negative control or Flag-HA-RHINO. Eighteen hours later cells were treated with 5 Gy and harvested 4 h later. Ran was used as a loading control.
Rad18 and Ubc13 function in post replication repair which controls PCNA ubiquitination (16). Unlike Rad18, RHINO depletion did not affect UV-induced PCNA monoubiquitination (Fig. S10). As TopBP1 and 9-1-1 play critical roles in ATR activation and Chk1 phosphorylation (17), we tested whether RHINO controlled IR-induced Chk1 phosphorylation. Indeed, RHINO depletion led to a reduction of Chk1 phosphorylation (Fig. 3B) that was rescued by re-expression of RHINO (Fig. 3C). Together, these data suggest a role for RHINO in ATR-mediated Chk1 activation driven by TopBP1 and 9-1-1.
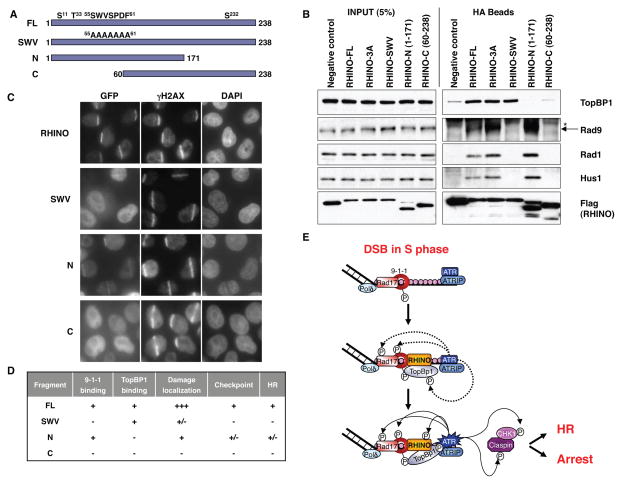
Clear RHINO orthologs exist in vertebrates, with high conservation in the N- and C-terminal regions (Fig. S11A). The N-terminus has similarity to the APSES domain (18) (Fig. S11B), a DNA binding domain homologous to the KilA-N domain found in eukaryotic viruses (K. Hofmann personal communication). To identify TopBP1 and 9-1-1 interaction regions, we mutated a conserved region in the APSES domain (referred to as SWV, Fig. S11), and made C- and N-terminal truncations (Fig. 4A). The SWV mutation specifically blocked the interaction with the 9-1-1 complex but not TopBP1 (Fig. 4B, D). While the N-terminal, but not the C-terminal, fragment bound 9-1-1, neither bound TopBP1 (Fig. 4B, D). The SVW mutant is completely defective in damage localization while the N-terminal fragment is partially impaired (Fig. 4C, D and Fig. S12A), suggesting that the C terminal region aids DNA damage localization. The interaction between RHINO and 9-1-1 is critical, as the SWV mutant RHINO failed to complement the checkpoint or HR defects (Fig. 4D, Fig. S12). The RHINO N-terminal fragment partially rescues both phenotypes, consistent with its partial localization phenotype.
Figure 4. RHINO interaction with 9-1-1 is critical for the DDR.
(A) Schematic representation of the different mutant and truncated versions of RHINO.
(B) RHINO binds 9-1-1 and TopBP1 independently. Extracts from 293 cells expressing either a Flag-HA negative control, Flag-HA-RHINO or the indicated mutants were immunoprecipitated and immunoblotted with the indicated antibodies. The asterisk represents the IgG heavy chain.
(C) RHINO localization requires 9-1-1. Hela cells expressing WT GFP-RHINO or the indicated mutants were treated with the UV laser to introduce DSBs and assessed for RHINO localization.
(D) Synopsis of the behavior of different mutants of RHINO.
(E) Model of the role of RHINO in DDR signaling. See text for further details.
Rad17, RHINO or TopBp1 depletion results in a partial HR defect, although not as strong as ATR depletion (Fig. S4A and Fig. S13). Notably, the double depletion of Rad17 and RHINO leads to a similar HR defect as each individual depletion, suggesting that RHINO mediates the 9-1-1’s role in HR (Fig. S13). Together, these data suggest that the 9-1-1/RHINO/TopBp1 axis promotes DDR activation in response to lesions occurring during S phase (Fig. 4E).
Through genetic analysis we have uncovered a large number of genes and pathways required to maintain prolonged cell cycle arrest in response to DNA damage occurring in S phase, the cell cycle phase in which DNA damage is the most likely to contribute to genomic instability. We uncovered an unexpected role for the HR and FA pathways in promoting cell cycle arrest. While this may in part reflect differential funneling of repair intermediates into NHEJ, the inability of NHEJ inhibition to rescue the signaling defect from BRCA2-depletion suggests the possibility that cells may retain the capacity to signal if they process lesions through the HR pathway. Currently, there are two ssDNA binding complexes required for DDR signaling, RPA and the SSB1 and SSB2 complexes. The HR pathway, which replaces RPA with another ssDNA binding protein, Rad51, may represent a third (Fig. S14 and Supplemental Text). We were unable to further test this possibility due to extreme toxicity of Rad51 depletion. However, it makes biological sense that cells would evolve the capacity to sense ongoing repair, as it is the completion of repair that is the relevant biological event with respect to turning off the DDR.
We also defined a new component of the Rad17/911/TopBP1 pathway, RHINO. TopBP1 recruitment to sites of damage through the Rad17/911 complex is important for activation of ATR (19). This recruitment has been suggested to occur through association of the BRCT-repeats of TopBP1 with constitutively phosphorylated Rad9 (20). However, recent experiments in Xenopus have indicated that there is a 911-dependent but Rad9-phosphorylation independent mechanism that also operates to recruit TopBP1 (21). The association of RHINO with both 911 and TopBP1 through independent domains on RHINO suggests that it may bridge these to help recruit TopBP1 to the 911 complex. Whether RHINO is involved in this second mechanism and why two different mechanisms operate remains to be determined but it is possible that they are each used in response to different classes of lesions or to bolster long-term signaling. Alternatively RHINO could allosterically regulate TopBP1 to promote its association with, or activation of ATR/ATRIP. These and other issues raised by the discovery of this new group of genes involved in maintaining DDR signaling during prolonged arrest initiated by S phase DNA damage should provide fertile grounds for future exploration.
Supplementary Material
Acknowledgments
We thank the ICCB-Longwood for screen assistance. We also thank A. Ciccia, A. Bredemeyer, B. Adamson, A. Burrows, A. Smogorzewska, and A. Brass for helpful discussions. This work was supported by NIH grants to S.J.E. and J.W.H., C.C.R. is the recipient of a long term EMBO fellowship. S.J.E. is a Howard Hughes Medical Institute investigator.
References and Notes
- 1.Ciccia A, Elledge SJ. Mol Cell. 2010;40:179. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuoka S, et al. Science. 2007;316:1160. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 3.Stokes MP, et al. Proc Natl Acad Sci U S A. 2007;104:19855. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierce AJ, Johnson RD, Thompson LH, Jasin M. Genes Dev. 1999;13:2633. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. Nat Genet. 2002;30:285. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 6.Adamo A, et al. Mol Cell. 2010;39:25. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Bouwman P, et al. Nat Struct Mol Biol. 2010;17:688. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunting SF, et al. Cell. 2010;141:243. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pace P, et al. Science. 2010;329:219. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 10.Bartek J, Lukas J. Curr Opin Cell Biol. 2007;19:238. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Collis SJ, Boulton SJ. Chromosoma. 2007;116:331. doi: 10.1007/s00412-007-0108-6. [DOI] [PubMed] [Google Scholar]
- 12.Gorbacheva VY, et al. Proc Natl Acad Sci U S A. 2005;102:3407. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baillat D, et al. Cell. 2005;123:265. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Gong Z, Ghosal G, Chen J. Mol Cell. 2009;35:384. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, et al. J Biol Chem. 2009;284:23525. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moldovan GL, Pfander B, Jentsch S. Cell. 2007;129:665. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Cimprich KA, Cortez D. Nat Rev Mol Cell Biol. 2008;9:616. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer LM, Koonin EV, Aravind L. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-research0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumagai A, Lee J, Yoo HY, Dunphy WG. Cell. 2006;124:943. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 20.Delacroix S, et al. Genes Dev. 2007;21:1472. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Dunphy WG. Mol Biol Cell. 2010;21:926. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.