Regulated turnover of miRNAs—tiny posttranscriptional regulators of gene expression in metazoan cells—is important to optimize cellular response to external stimuli. Cell–cell contact alters miRNA stability by restricting its exosomal export.
Abstract
MicroRNAs (miRNAs) are tiny posttranscriptional regulators of gene expression in metazoan cells, where activity and abundance of miRNAs are tightly controlled. Regulated turnover of these regulatory RNAs is important to optimize cellular response to external stimuli. We report that the stability of mature miRNAs increases inversely with cell proliferation, and the increased number of microribonucleoproteins (miRNPs) in growth-restricted mammalian cells are in turn associated with polysomes. This heightened association of miRNA with polysomes also elicits reduced degradation of target mRNAs and impaired extracellular export of miRNA via exosomes. Overall polysome sequestration contributes to an increase of cellular miRNA levels but without an increase in miRNA activity. Therefore miRNA activity and turnover can be controlled by subcellular distribution of miRNPs that may get differentially regulated as a function of cell growth in mammalian cells.
INTRODUCTION
MicroRNAs (miRNAs) are widely considered to be a key component of the gene regulatory circuit in metazoan cells. Essentially, miRNAs are 20- to 22-nucleotide (nt) noncoding RNAs that are reported to regulate a diverse array of genes, and perturbations of their levels and activities underlie several human diseases, including cancers (Lu et al., 2005; Bartel, 2009; Iorio and Croce, 2012; Mendell and Olson, 2012).
One of the strands of the mature miRNA gets incorporated into a multicomponent ribonucleoprotein (RNP) complex referred to commonly as miRNA-mediated silencing complex (miRISC) or microribonucleoproteins (miRNPs) and induces repression of the target mRNA by inhibiting the translation and/or affecting its stability (Filipowicz et al., 2008; Fabian et al., 2010; Huntzinger and Izaurralde, 2011). Argonaute (AGO) proteins form the essential component of a “minimal” miRNP that interacts with the GW182 family of proteins to induce clearance of miRNA-targeted mRNAs by promoting target mRNA deadenylation and degradation. In mammalian cells, miRNA-repressed mRNAs and miRNPs are reversibly targeted to P-bodies, the specialized cytoplasmic structures for storage or degradation of miRNA-repressed mRNAs in animal cells (Liu et al., 2005; Pillai et al., 2005).
In mammalian cells, of the four isoforms of Argonaute proteins, AGO2 has RNA endonuclease activity and is involved in cleavage of the target with perfect complementarity to the bound miRNA. In addition, it is also involved in the deadenylation and repression of translated messages bearing imperfect complementarity in their 3′ untranslated region (UTR; Liu et al., 2004; Rivas et al., 2005). A large fraction of cellular AGO2 is associated with the endoplasmic reticulum (ER) and Golgi membrane in animal cells (Cikaluk et al., 1999). Biochemical fractionation studies have confirmed the presence of miRNA and repressed messages with the detergent-insoluble, ER-enriched membrane fraction (Pillai et al., 2005). ER is also shown to be the site where the nucleation of small interfering RNA (siRNA) into functional RISC complex occurs (Stalder et al., 2013). The multivesicular body (MVB) is the other subcellular compartment that has recently received attention for its role in miRNA-mediated repression in animal cells and has been found to be important in restricting miRNP recycling (Gibbings et al., 2009; Lee et al., 2009). MVBs can potentially merge with cell membranes to form exosomes (Keller et al., 2006), which are small, 30- to 100-nm vesicles considered as means of intercellular communication and are secreted by animal cells to the extracellular space (Simons and Raposo, 2009). Several miRNAs and their associated proteins, such as AGO2 and GW182, are present in exosomes (Valadi et al., 2007). However, the significance of miRNA export via exosomes as a regulator of cellular miRNA activity is underexplored.
In contrast to the substantial amount of research on miRNA biogenesis, little work has been carried out on miRNA localization and turnover in mammalian cells. It has been observed that the half-lives of miRNAs in animal cells and tissues such as heart or liver are several hours or even days (van Rooij et al., 2007; Gatfield et al., 2009; Krol et al., 2010a). However, such a slow turnover rate is not likely to be the common feature of all miRNAs, as they play critical roles in developmental transitions that demand rapid turnover. Other investigations have indicated that both the cellular levels and activities of miRNAs are subjected to tight regulation. Tissue-specific expression and activity of hundreds of miRNAs are controlled at different levels to fine tune gene expression in metazoans (Bushati and Cohen, 2007), where Drosha-DGCR8 and Dicer-TRBP complexes act in tandem to generate mature 22-nt-long miRNAs from their precursors (Krol et al., 2010b). Expression and activity of miRNAs are also sensitive to cellular context (Hatfield et al., 2005; Sood et al., 2006), and cell–cell contact is reported to increase primary (pri-) miRNA processing in a majority of human cell lines tested (Hwang et al., 2009). Prototypes of such positive and negative feedback mechanisms to control miRNAs remain limited to specific miRNAs and are largely operative at miRNA synthesis and/or maturation steps (Krol et al., 2010a).
In this article, we document a global increase in miRNA stability in human cells grown to confluent conditions. Processed siRNA molecules that do not require an in vivo Dicer processing step were also found to be increasingly stable in growth-retarded human cells. Increased miRNAs that were found to be largely engaged with polysomal fractions were surprisingly inept at inducing degradation of target mRNAs. This is coupled with impaired export of miRNA via exosomes. On the contrary, in proliferating cells, miRNPs actively engaged in repression are predominantly nonpolysomal and can be efficiently exported via exosomes to the extracellular space. Therefore altered subcellular compartmentalization of miRNPs can result in a difference in activity and fate of miRNAs in mammalian cells.
RESULTS
Increased stability of mature miRNAs in human cells grown to high cell density in monolayer culture
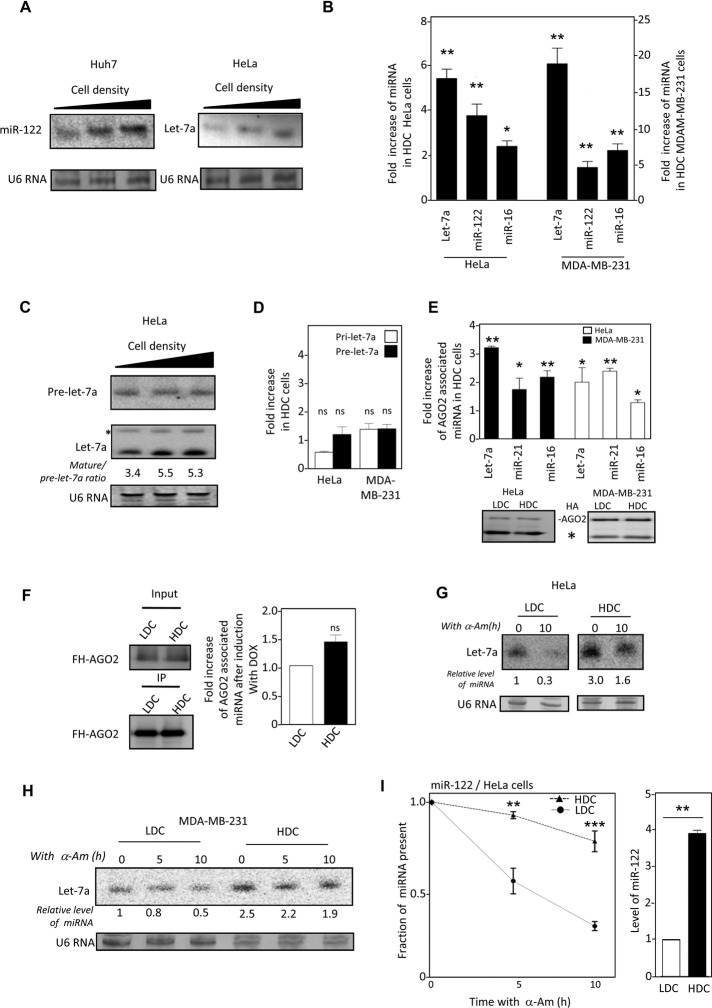
Increase in miRNA level with cellular confluency has been reported for the majority of miRNAs and was shown to be attributed to enhanced pri-miRNA processing by Drosha and higher loading of mature miRNAs to AGO2 in cells from confluent cultures (Hwang et al., 2009). To assess how cellular growth conditions may affect miRNA stability, we measured mature miRNA levels in human cells grown to different densities in monolayer culture. Toward that end, we incubated equal numbers of cells in culture plates of different sizes to achieve a gradient of cell density in culture. With changing cell densities from 25 to 100%, an increase in mature miR-122 level was observed in human Huh7 hepatoma cells. Similarly, mature let-7a level also increased with increasing cell density in the human cervical carcinoma cell line HeLa (Figure 1A). Similar increase of three independent miRNAs was detected in both HeLa and MDA-MB-231 human breast cancer cells grown to high-density culture (HDC; 90–100% cell density) in comparison to their levels in cells in low-density culture (LDC; 20–30% cell density; Figure 1B). Equal numbers of cells from HDC and LDC was used to avoid any variance during RNA extraction, and miRNA content was normalized against U6snRNA. miRNA levels in HDC and LDC cells were also compared and normalized against the cellular DNA content; they were found to be comparable between HDC and LDC states (Supplemental Figure S1A). miR-122 is a hepatocyte- specific miRNA that is not expressed in HeLa or MDA-MB-231 cells. The pre-miR-122 can be expressed exogenously from a plasmid under a constitutive U6 promoter driving pre-miR-122 expression. In mammalian cells, Drosha processes pri-miRNAs to generate pre-miRs. Therefore, mature miR-122 levels in HeLa and MDA-MB-231 cells expressing pre-miR-122 directly as a processed transcript could not be influenced by a change in Drosha activity. Of interest, as observed for other miRNAs, a comparable increase in mature miR-122 production in pre-miR-122–expressing HDC HeLa and MDA-MB-231cells was detected, which argues in favor of a Drosha-independent up-regulation of mature miR-122 level in these cells (Figure 1B). Consistent with this observation, the Drosha levels were found to be similar between HDC and LDC cells (Supplemental Figure S1B). In addition, only a fractional change in levels of miRNA precursors pri- and pre-let-7a, insufficient to explain the increase in mature let-7a content in HDC HeLa and MDAM-MB-231 cells, was noted with change in cell confluency (Figure 1, C and D).
FIGURE 1:
Effect of cell density on cellular levels, stability, and AGO2 association of miRNAs in monolayer culture of mammalian cells. (A) Northern blot detection of miR-122 and let-7a in Huh 7 (left) and HeLa cells (right) grown to increasing cell densities (25–100%). U6 RNA serves as a loading control. (B) Real-time-based quantitative estimation of the increase of miRNA let-7a, miR-16, and exogenously introduced miR-122 levels in HeLa and MDA-MB-231 cells grown in high-density (HDC) vs. low-density (LDC) cells. For comparison, individual miRNA levels in LDC conditions were taken as the unit. miRNA levels were normalized to U6 RNA levels for comparison. (C) Levels of pre-let-7a (top) detected with a pre-let-7a specific probe in HeLa cells vs. changing cell density. Membrane was also probed against the mature let-7a to detect both pre (denoted by an asterisk) and mature forms of this miRNA (bottom). U6 RNA serves as a loading control. (D) Relative change in the levels of pre-let-7a and pri-let-7a RNA in HDC HeLa and MDA-MB-231 cells estimated by real-time PCR. Levels of respective RNAs in LDC conditions were considered as the unit. (E) Quantitative estimation of associated miRNA in AGO2 immunoprecipitated materials from HeLa and MDA-MB-231 cells grown to low and high density. Top, relative increase of each miRNA in HDC conditions, considering LDC levels as the unit. Bottom, Western blot of HA-AGO2 immunoprecipitated using anti-HA antibody from HDC and LDC HeLa and MDAM-MB-231 cells expressing FH-AGO2. The heavy-chain band of the antibody used for precipitation is represented by a star. (F) Left, detection of FH-AGO2 in HA-immunoprecipitated materials from HDC and LDC HeLa cells expressing miR-122 from a Tet-ON inducible promoter and FH-AGO2. Right, real-time-based quantification of miR-122 in FH-AGO2 immunoprecipitated (IPed) materials after 4 h of induction with doxycycline (DOX). miR-122 levels in LDC condition was taken as the unit. (G) Half-life of let-7a in HeLa cells grown to low (LDC) or high (HDC) densities. Cells were treated for 10 h with α-Am before total RNA isolation and Northern blotting for let-7a. (H) let-7a detection in total RNA from LDC and HDC MDA-MB-231 cells subjected to α-Am treatment for 0, 5, or 10 h for let-7a. (I) Stability of exogenously expressed miR-122 in LDC and HDC HeLa cells. Cells expressing pre-miR-122 from a U6 promoter were grown to either HDC or LDC before α-Am was applied, and levels of mature miR-122 after 5 and 10 h of treatment were determined by real-time-based quantification and plotted. Initial level was considered as the unit in each case (left). The initial levels of miR-122 in pre-miR-122–expressing HeLa cells before addition of α-Am are plotted for comparison (right). ns, nonsignificant, *p < 0.05, **p < 0.01, ***p < 0.0001. The p values were determined by paired t test. All experiments were performed a minimum of three times. For densitometric quantification, the relative amounts of miRNAs were measured against the U6 RNA, which also served as loading control.
In HDC cells, there was also no gross change in AGO protein expression, whereas AGO2-associated miRNA levels increased (Figure 1E and Supplemental Figure S1, B and C). Moreover, the difference in amount of de novo synthesized miRNAs that get loaded to AGO2 in LDC and HDC cells expressing inducible pre-miR-122 was nonsignificant during the initial induction hours and therefore could not significantly account for the difference in mature miRNA content or its AGO2 association between LDC and HDC cells (Figure 1, B and F). Dicer1 immunoprecipitated from HDC or LDC cells also did not show any difference in pre-miR processing activity (Supplemental Figure S1D), and HDC and LDC cells showed a very similar level of Dicer1 expression (Supplemental Figure S1B). Therefore higher Dicer1 activity also could not be entirely responsible for the increased mature miRNAs found in HDC cells.
These results suggested that the enhanced miRNA levels in HDC cells are primarily due to changes in postmaturation stages of miRNAs in HDC. To verify this hypothesis, we measured the level of an exogenously administered siRNA in LDC and HDC cells and found it to be higher in HDC cells (Supplemental Figure S2, A–C). Note that siRNAs do not need any processing or maturation by Drosha, and their level in Dicer1-compromised HDC cells was similar to control HDC cells (Supplemental Figure S2B). Thus the higher level of miRNAs could not be solely due to either higher Drosha/Dicer1 activity or miRNA loading to AGO proteins in HDC cells.
We wanted to verify the stability of mature miRNAs in LDC and HDC mammalian cells. Application of the RNA polymerase II inhibitor α-amanitin (α-Am) was used to inhibit de novo production of let-7a transcripts (observed with reduction in pre-let-7a level with α-Am treatment; unpublished data). After 10 h of α-Am treatment, the residual amount of let-7a was higher in HDC than in LDC HeLa cells (Figure 1G). Increased cell density also reduced the depletion of let-7a in the presence of α-Am in human MDA-MB-231 breast cancer cells (Figure 1H). Similarly, quantitative estimation revealed a reduced half-life of ectopically expressed, liver- specific miR-122 in the presence of α-Am in LDC HeLa cells (Figure 1I). Similar to miRNAs, the half-life of a siRNA was also observed to be higher in HDC cells (Supplemental Figure S2C). The foregoing experiments show that the increased levels of miRNAs were primarily due to the increased half-life of the mature miRNAs in HDC cells.
Reduced miRISC activity in HDC cells
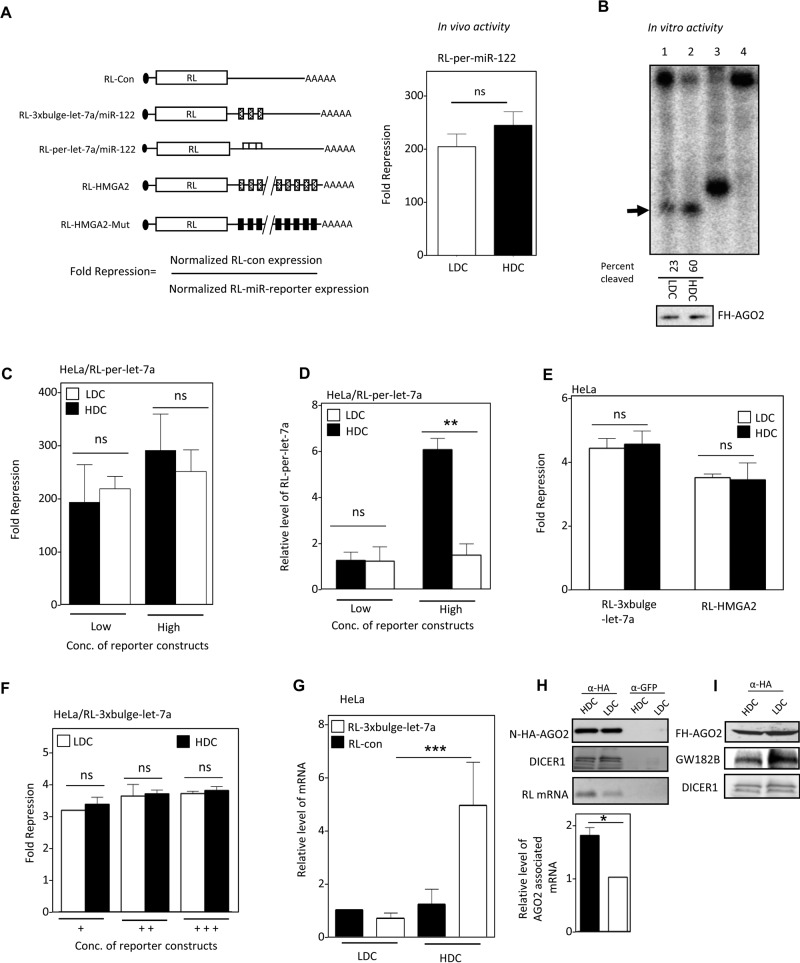
To ascertain whether the elevated miRNAs in HDC cells were part of “active” miRNPs, we expressed a Renilla reporter containing a perfect miR-122 binding site (Figure 2A, left) in HeLa cells coexpressing miR-122. Of interest, the in vivo activity of miR-122 RISC was almost equal between HDC and LDC cells under identical experimental parameters and was inconsistent with the higher miRNP present in HDC cells (Figure 2A, right, and Figure 1, B and E).
FIGURE 2:
Defective miRNA-mediated repression in HDC human cells. (A) Reporter mRNAs used to measure miRNA activity, showing how fold repression was measured (left). In vivo RISC activity of miR-122 in HDC and LDC HeLa cells expressing miR-122 and an RL reporter with one perfect miR-122 site (right). (B) RISC cleavage activity of miRISC-122 isolated from LDC and HDC HeLa cells cotransfected with FH-AGO2 construct and miR-122 expression plasmid. The activities were measured and quantified in an in vitro RISC cleavage reaction using 5′-32P-labeled miR-122 target RNA. Relative quantification of the RISC activity isolated from LDC and HDC HeLa cells (lanes 1 and 2) was done by densitometric analysis. Lane 4, no RISC; in lane 3, a 21-nt-long radiolabeled DNA oligo was used as size marker. The RISC-cleaved band is marked by an arrowhead. Western blot for the isolated RISC with anti-HA antibody confirmed equal levels of FH-AGO2 protein in each reaction (1 and 2). (C) Invariant repression levels of let-7a miRNA reporter in HDC and LDC cells. Cells transfected with low and high concentrations of plasmids encoding the RL reporter with one perfect let-7a binding site were grown to HDC and LDC, and the normalized repression values were measured in HeLa cells. (D) Reduced cleavage of mRNA targets with perfect miRNA site in HDC condition. Real-time quantification of RL mRNA level was performed in RNA isolated from RL-per-let-7a–transfected HeLa cells expressing low and high concentrations of reporter and grown to HDC and LDC conditions. The 18S rRNA normalized relative levels of RL mRNA for individual samples are plotted. Values in the LDC state are set to 1. (E) Relative repression level of two let-7a reporters with multiple imperfect let-7a binding sites in HeLa cells from HDC and LDC conditions. RL-HMGA2 has a 3′ UTR of let-7a target gene HMGA2. RL-HMGA2-Mut with mutated let-7a sites served as control. For RL-3xbulge-let-7a, RL-Con was used as control. (F) Relative fold repression of RL reporter with three imperfect let-7a binding sites in cells in which an increasing amount of reporter-encoding plasmid was used for transfection of HeLa cells. (G) Real-time PCR detection of RL mRNA in RL-3xbulge-let-7a– or RL-Con–transfected HeLa cells under similar experimental conditions as in E. (H) Association of target mRNA with AGO2 in HDC and LDC HeLa cells expressing RL-3xbulge-let-7a. N-HA-AGO2 was immunoprecipitated with anti-HA antibody from cells coexpressing it along with RL reporter. Control immunoprecipitation reaction was performed with anti-GFP antibody. Western blot analysis of the IPed AGO2 was performed, and associated DICER1 was detected (top). RNA extracted from the lysates was used to determine the amount of RL mRNA associated with AGO2 by PCR (top) and also quantified by real-time PCR (bottom). (I) Association of GW182B with AGO2 in HDC and LDC cells. Cells expressing FH-AGO2 were grown to HDC and LDC states, HA-AGO2 was immunoprecipitated with anti-FLAG antibody, and associated GW182B was detected. All experiments were performed a minimum of three times. Here Low and High refer, respectively, to 25 and 100 ng of plasmids used for transfection of cells present per well of a six-well plate. In F, +, ++, and +++ represent respectively 25, 100, and 500 ng of plasmid transfected per well of a six-well plate. For all relevant panels, ns, nonsignificant, *p < 0.05, **p < 0.01, and ***p < 0.0001. The p values were determined by paired t test, and results depict mean values from at least three experiments.
To ascertain whether the reduced in vivo activity is due to modification of miRNPs in HDC cells that might have rendered them nonfunctional, we purified the RISC complex from LDC and HDC cells. Consistent with the higher level of miR-122 in HDC cells, purified miRNP-122 from HDC HeLa cells showed 1.8-times-higher target cleavage activity in vitro for the same amount of AGO2 protein (Figure 2B). Reduced in vivo silencing by miRNPs in HDC cells could be due to the presence of limiting substrate RNA, which might lead to saturation of miRISC activity in LDC and HDC cells, or, alternatively, inefficiency of the increased number of miRNPs to bind their targets could make the repression partly defective in HDC cells. To test whether limiting substrate concentration could be the reason for identical in vivo repression levels observed in LDC and HDC cells, we used a higher concentration of reporter for expression in HeLa cells to rule out the target RNA unavailability issue and measured both repression and mRNA content in HDC and LDC cells. We did not detect any major difference in repression level between HDC and LDC cells with changing substrate concentration. Of interest, with a higher concentration of expression plasmid used for transfection, the target RNA level was much higher in HDC than in LDC cells (Figure 2, C and D). This observation ruled out limiting substrate concentration as a cause of comparable repression of miRNA reporter in LDC and HDC cells. Instead it suggested that the higher amount of miRNPs present in HDC cells showed impaired RISC-mediated target cleavage in HDC cells. Consistent with these arguments, we also observed identical in vivo repression levels with other miRNA-based reporters with three or more imperfect miRNA binding sites in their 3′ UTR by measuring the translation-repressive activity of let-7a and miR-122 miRNPs in HeLa and Huh7 cells grown to different cell densities (Figure 2. E and F, and Supplemental Figure S3A). The green fluorescent protein (GFP) reporter with let-7a sites also showed equal repression in LDC and HDC HeLa cells, ruling out any bias with Renilla-based reporters (Supplemental Figure S3B). miRNA repression is usually associated with target mRNA degradation in mammalian cells. Note that the RL-3xbulge-let-7a RNA exhibited unusual stability of transcripts in HDC conditions, analogous to what was obtained with high concentration of RL-per-let-7a, and the relative transcript level was severalfold higher in HDC cells than in LDC HeLa cells (Figure 2G). The mRNAs association with AGO2 was also enhanced in HDC cells (Figure 2H). No change in DICER1 association with hemagglutinin (HA)-AGO2 was noted. This was accompanied by a reduction in AGO2-GW182B protein interaction in HDC cells, although cellular GW182B level did not change with cell confluency (Figure 2I, and data not shown). GW182 proteins are known to play an important role in repression and degradation of target RNA, and therefore reduced AGO2-GW182B interaction in HDC cells correlates with reduced target RNA degradation in HDC cells. These observations imply the existence of miRNP-bound mRNAs that are translationally silent but are not degraded in HDC cells.
Reduced polysomal disengagement is associated with low miRNA turnover and reduced RISC activity in human cells
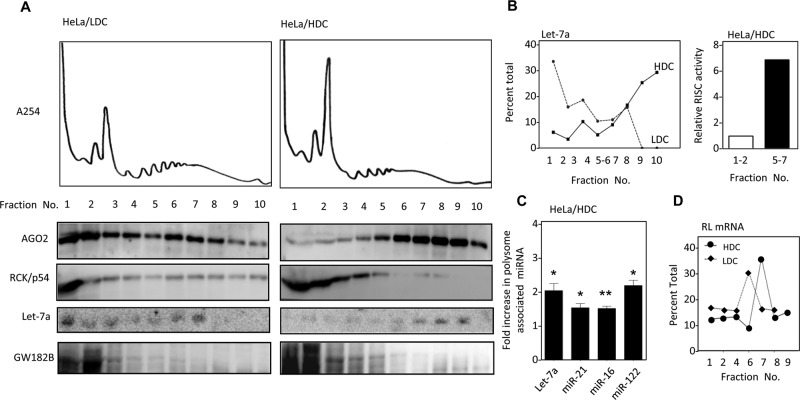
To resolve this apparent discrepancy between miRNA activity and its content in HDC cells and determine the biochemical fraction with which “inactive” miRNPs are associated in HDC cells, we analyzed whole-cell extracts of HDC and LDC HeLa cells on a 15–55% sucrose gradient. Apart from a relatively high 80S peak and comparatively lower peaks in the region corresponding to the polysomes, which implies reduced protein translation in HDC cells, the 254-nm absorption profiles of HDC and LDC extracts did not differ significantly (Figure 3A). Of interest, however, the gradient profiles of AGO2 protein were distinct for HDC and LDC samples. For HDC cell extracts, AGO2 and miRNA let-7a were both predominantly associated with the heavier fractions, whereas in LDC cells, AGO2, was present mostly in lighter fractions of the gradient, although some AGO2 was also found in the heavier fraction (Figure 3, A and B). Of interest, GW182 was found to primarily nonpolysomal in both LDC and HDC cells. This also supported the AGO2 immunoprecipitation data, which showed a reduction in AGO2-associated GW182B protein in HDC cells (Figures 3A and 2I). RCK/p54, another established component of the miRNA-mediated repression pathway, was considerably nonpolysomal in both HDC and LDC cells (Figure 3A). Polysome association of AGO2 in HDC cells was confirmed by treatment of the cell extract with puromycin to disrupt the elongating polysomes and associated AGO2 in HDC cell extract before it was analyzed on a sucrose density gradient (Supplemental Figure S3C). Polysome association of AGO2 was also found to be RNA dependent and sensitive to treatment with micrococcal nuclease (Supplemental Figure S3D). Therefore, in HDC cells, miRNPs remain associated with their target mRNA and are stabilized with polysomes.
FIGURE 3:
miRNPs in HDC human cells are retained with polysomes. (A, B) HDC and LDC HeLa cell extracts were analyzed on a 15–55% sucrose density gradient and collected in 10 different fractions while simultaneous absorption at 254 nm was monitored and plotted. The proteins isolated from individual fractions were Western blotted to obtain the distribution profile of AGO2, GW182, and RCK/p54 protein (A). Isolated RNAs from each fraction were analyzed by Northern blot for let-7a (A) and subsequently quantified as a percentage of total signals (B, left). FH-AGO2 immunoprecipitated from nonpolysomal fractions 1 and 2 and polysomal fractions 5–7 from transfected HeLa cells in a parallel experiment was assayed for RISC cleavage activity. (B) Right, representative graph of efficiency of RISC cleavage of the denoted fractions. (C) Polysomal miRNA content increase in HDC HeLa cells. RNA isolated from polysomes of LDC and HDC HeLa cells cotransfected with FH-AGO2 and miR-122 expression plasmid was quantified for endogenous miR-16, miR-21, let-7a, and exogenous miR-122 levels. (D) Association of let-7a reporter with different fractions of polysome gradient of HDC and LDC HeLa cells expressing RL-3xbulge-let-7a reporter mRNA. For all relevant panels, ns, nonsignificant, *p < 0.05, **p < 0.01, and ***p < 0.0001. The p values were determined by paired t test, and results depict mean values from at least three experiments.
Increase in cell density was marked by lower expression and reduced nuclear localization of proliferating cell nuclear antigen in mammalian cells (Supplemental Figure S4, A–C). Increased cellular density was also associated with a majority of the cells arrested in sub-G0/G1 phase of the cell cycle (Supplemental Figure S4D). In mammalian tissue such as liver, a majority of the cells are nonproliferative. Polysome profile analysis of mouse liver lysate confirmed polysome association of both AGO2 and miR-122 similar to what was documented in HDC cells, which are also nonproliferative (Supplemental Figure S3E). Therefore, in nonproliferative mammalian cells, miRNAs and AGO2 are primarily associated with polysomes.
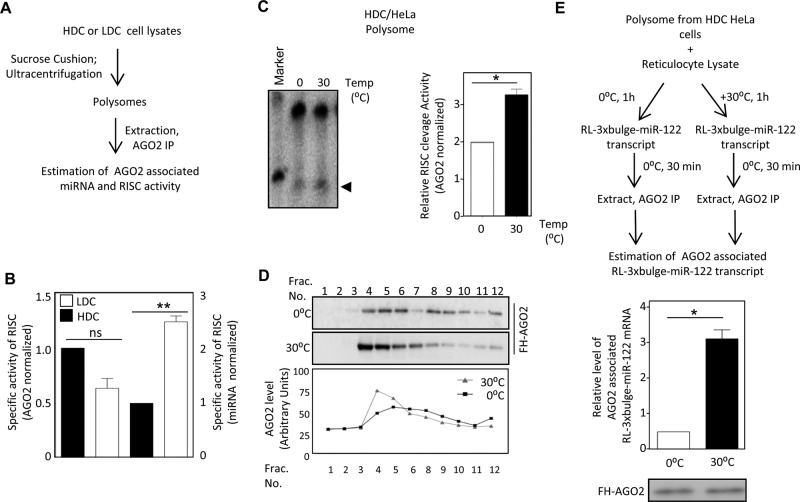
To determine whether the polysome-associated AGO2 in HDC cells was miRNA bound, we immunoprecipitated AGO2 extracted from polysomal fractions of HDC cells, which showed higher RISC cleavage activity than that isolated from nonpolysomal fraction. This indicated that miRNA-loaded AGO2 associated with polysomes in HDC cells (Figure 3B, right). This also rules out the existence of any specific modification of miRISC that could inhibit RISC activity of polysomal miRNPs in HDC cells. To reconcile this with the polysome association of miRNPs, we sought to investigate the cause and effect of this sequestration. Polysomes isolated from HDC HeLa cells showed twofold to threefold higher miRNA level than LDC polysomes, reconfirming that the excess miRNA in HDC cells is primarily polysomal (Figure 3C). This is also consistent with the increased target RNA association with polysomes in HDC cells evident from quantification of let-7a reporter mRNA RL-3xbulge-let-7a found with individual fractions of a sucrose density gradient (Figure 3D). However, polysome-associated AGO2 isolated from HDC cells, although enriched with miRNAs, showed relatively less RISC activity in vitro for the same amount of miRNA (Figure 4, A and B). From these data, we speculated that a fraction of AGO2- loaded miRNA that is associated with polysomes is ineffective for target cleavage in HDC cells. Is the activity of miRNPs inversely related to its polysome association? Boosting translation of isolated polysome-associated mRNA in vitro by incubating HDC polysomes with rabbit reticulocyte lysate released miRNP from polysomes and resulted in increased miRISC cleavage activity of polysome-associated miRNPs after treatment at 30°C as compared with control, where the ambient temperature for translation was not provided (Figure 4C). Higher translation of mRNAs with reticulocyte lysate at 30°C was verified by adding a firefly luciferase synthetic mRNA in the translation mixture under both conditions (unpublished data). This revival of polysomal miRNP activity isolated from HDC cells was accompanied by a concomitant shift of AGO2 to lighter fractions and was evident when the translation mixture after the reaction was resolved on a 30–55% sucrose gradient. This shift of AGO2 to lighter fractions indicated that the translational boost relieved AGO2 from polysomes and resulted in restoration of RISC activity (Figure 4D). Therefore, in HDC cells, low in vivo miRISC activity could be attributed to retention of a majority of miRISC with elongating polysomes. This in turn causes a relative reduction in accessibility of miRISC to de novo messages with a perfect site in HDC cells. We hypothesize that the retention of mRNPs with polysomes restricts its recruitment to new target mRNAs, which in turn reduces RISC activity in HDC cells. To explore this idea, we incubated the polysomes with reticulocyte lysates for 1 h at 0 or 30°C before incubating them with a target message for an additional 30 min at 0°C. The amount of AGO2-bound mRNA isolated under each condition was measured. We noted a higher level of target mRNA binding with AGO2 upon release of miRNP from polysome postincubation at 30°C (Figure 4E). Therefore, in HDC cells, polysomal miRNPs bound to target mRNAs showed reduced repressive activity, possibly due to impairment of subsequent steps necessary for clearing the miRNA-bound mRNAs from polysomes.
FIGURE 4:
miRNP retention in polysomal pool precludes its RISC activity. (A) Assay to measure RISC cleavage activity of polysomal miRISC-122 isolated from LDC and HDC HeLa cells cotransfected with FLAG-HA-AGO2 and miR-122 expression plasmid. Normalization of RISC activity was done against the HA-AGO2 detected by Western blot analysis. (B) Relative specific activity of polysomal miRISC-122 isolated from HDC and LDC states with respect to AGO2 protein levels (left axis) or AGO2-associated miR-122 content (right axis). (C) In vitro translation activates polysomal miRISC. Polysomes isolated from HDC HeLa cells transfected with miR-122 expression plasmid was subjected to in vitro translation in rabbit reticulocyte lysates for 1 h at 30 and 0°C (control) before measuring the RISC activity at 30°C. Left, autoradiogram showing the end product of the reaction analyzed on a denaturing PAGE. Arrowhead marks the cleaved RNA bands. Right, relative RISC activity. (D) Distribution of FH-AGO2 on a 30–55% sucrose gradient after isolated polysomes from HDC HeLa cells were incubated with rabbit reticulocyte lysates at 30°C for 1 h. FH-AGO2 distribution at 0°C served as control. Top, FH-AGO2 detected by Western blot with anti-HA antibody. Bottom, quantification of the AGO2 levels in different fractions done by measuring the band intensity. (E) Binding of AGO2 to new target RNA is reciprocal to its association with polysomes. Top, scheme of the experiment. Bottom, relative amount of RL-3xbulge-miR-122 mRNA bound to FH-AGO2 after the polysomal FH-AGO2 was subjected to translation reaction at 37 or 0°C. ns, nonsignificant, *p < 0.05, **p < 0.01, and ***p < 0.0001. The p values were determined by paired t test. All experiments were performed a minimum of three times.
Translation slowdown down-regulates miRNA turnover in human cells
It seems from the previous experiments that retention of miRNPs with polysomes resulted in reduced recycling of miRNPs to bind new targets and also impairs the subsequent step of target RNA degradation by miRNPs. To validate the notion that slow translation-related retention of miRNPs with polysomes reduces miRNP turnover, we measured the AGO2-associated miRNA level in control and cycloheximide (CHX)-treated cells. Cycloheximide, when applied at a low concentration (100 ng/ml), affects the rate of protein translation in mammalian cells without freezing the elongating ribosomes (Kong et al., 2008). With CHX treatment, we observed an increase of miRNA content (Supplemental Figure S5A, left, bottom). As expected, this was also accompanied by an increased association of AGO2 with target mRNA (Supplemental Figure S5A, left, top). The repression level by increased numbers of miRNPs was also curtailed irrespective of the concentration of target mRNA expressed (Supplemental Figure S5A, right), whereas Western blot analysis showed no major change in levels of proteins associated with miRNA functions upon CHX treatment, except a relative decrease of AGO2 level possibly due to the higher turnover rate of this protein in mammalian cells (Supplemental Figure S5B). Gradient analysis also revealed that CHX at the used concentration did not grossly affect the polysome profile of LDC HeLa cells (Supplemental Figure S5C). Of interest, the miRNA content of the polysomes was significantly enhanced upon CHX treatment of LDC HeLa cells (Supplemental Figure S5D). Therefore slow translation in the presence of CHX resulted in increased retention of miRNA with polysomes, similar to what was observed with HDC cells.
Reduced exosomal export of miRNAs causes increased miRNA stability in nonproliferative cells
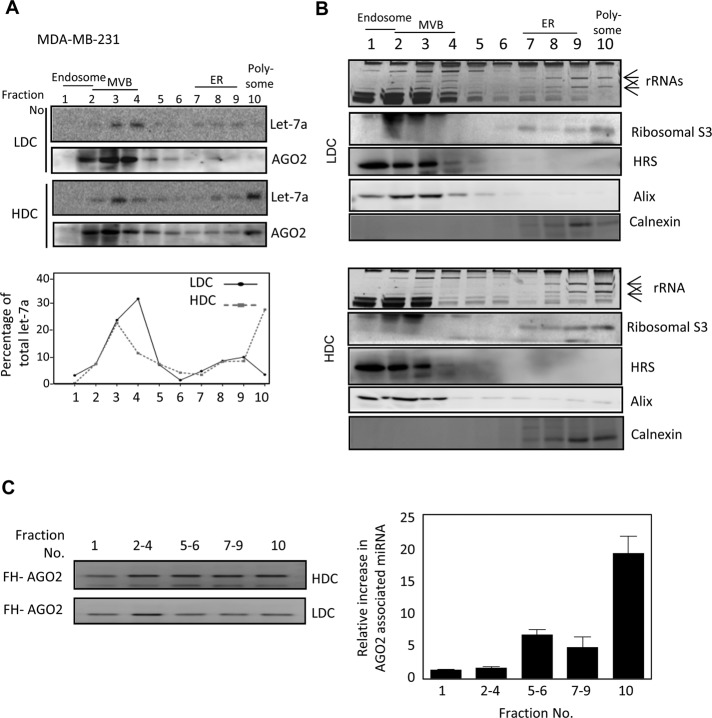
To identify the subcellular location of increased miRNAs in HDC human cells, we analyzed HDC and LDC cell lysates on an OptiPrep density gradient (Augustin et al., 2005) and found that a majority of let-7a miRNPs were associated with polysome-enriched/endoplasmic reticulum (ER) fractions in HDC cells, whereas in LDC cells, miRNA are primarily associated with MVBs, precursor organelles for exosome formation (Figure 5, A and B). AGO2-associated miRNAs are primarily located with polysomes and ER fractions in HDC cells (Figure 5C).
FIGURE 5:
Reduced MVB localization and exosomal export of miRNA in HDC human cells. (A) Subcellular miRNA distribution in HeLa cells grown to LDC and HDC conditions. Cell homogenates from HDC and LDC cells were analyzed on a 3–30% OptiPrep gradient to separate the subcellular organelles, and Northern blot analysis for let-7a was performed with each fraction; relative distributions are plotted. AGO2 distribution was detected by Western blotting. (B) Subcellular distributions of different marker proteins and RNA to detect MVB, ER, and polysomal fractions in HDC and LDC cell extracts are shown to confirm effective fractionation. Positions of the 26S and 18S rRNAs are marked by arrows. Relative position of each organelle is shown in the top for both A and B. (C) Fold change in miRNA association of FH-AGO2 present in different OptiPrep fractions in HDC cells. FH-AGO2–expressing HeLa cells were grown to HDC and LDC states before lysis and separation of fractions on a gradient as described. Individual fractions were pooled, and FH-AGO2 was immunoprecipitated. From the IPed FH-AGO2, each fraction was used to measure associated let-7a by real time PCR. Left, Western blot of IPed FH-AGO2. **p < 0.01. The p value was determined by paired t test. Results depict mean values from at least three experiments.
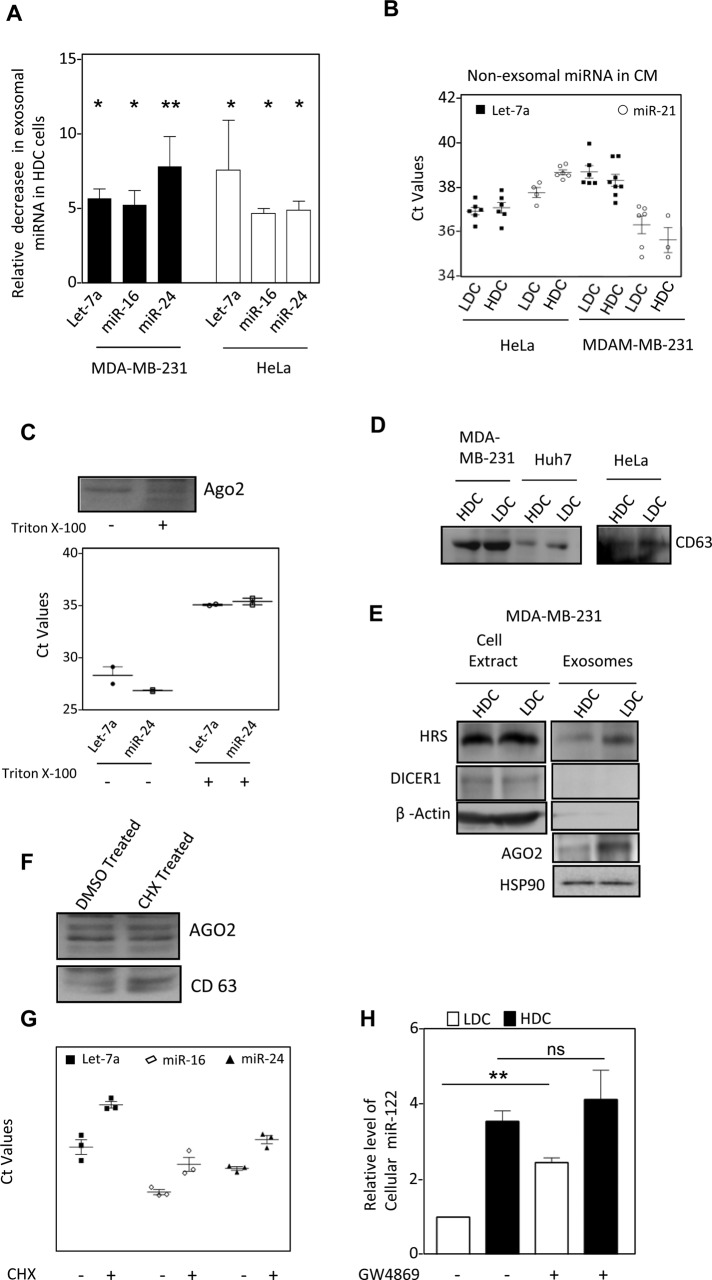
Because MVB association is a prerequisite for exosomal export of miRNAs (Gibbings et al., 2009), exosome-mediated export of miRNA in animal cells may also contribute to the clearance of miRNA from cells. This prompted us to explore how cellular confluency could in turn modulate cellular miRNA content by regulating the export process. Using isolated exosomes released by HDC and LDC in cell culture medium, we found that exosomal miRNA levels were notably lower for both HDC HeLa and MDA-MB-231 cells for all miRNAs tested without affecting extracellular, nonexosomal miRNA content (Figure 6, A and B). This occurs without a major change in exosomal content and character, apart from a reduction in exosomal AGO2 level in HDC cells, which was evident from biochemical or biophysical analysis of the isolated exosomes (Figure 6, C–E, and Supplemental Figure S6). Increased polysome association of miRNPs in HDC cells therefore could result in reduced exosomal export and increased miRNA stability in HDC HeLa and MDA-MB-231 cells. We earlier observed that retention of miRNPs with polysomes by application of a low concentration of CHX resulted in reduced miRNA turnover. To further assess whether polysome retention could restrict exosomal export of miRNA and account for enhanced cellular miRNA content, we measured the exosomal miRNA level upon application of CHX and observed that CHX treatment could also suppress exosomal export of miRNA in HeLa cells (Figure 6, F and G). From the foregoing experiments, we noted a good correlation of low miRNA export and high cellular miRNA content in HDC cells. We applied GW4869 as an inhibitor of SMPD2, a protein important for exosomal export of miRNA in animal cells (Kosaka et al., 2010). By preventing sphingomyelin synthesis, GW4869 restricts exosomal export of miRNA and enhances cellular miRNA content in LDC MDA-MB-231 cells. Of interest, GW4869-mediated increase of miRNA was moderate in HDC cells, arguing in favor of exosomal export of miRNA as a key mechanism of miRNA turnover that is already restricted in HDC cells. As a consequence, these cells are less responsive to GW4869 treatment and do not show a major increased miRNA level upon drug treatment (Figure 6H).
FIGURE 6:
Exosomal export of miRNA is curtailed in cells grown to high density (A) Levels of let-7a, miR-16 and miR-24 miRNAs in FACS sorted identical number of exosomes released by HeLa and MDA-MB-231 cells grown to LDC or HDC cells (mean+/– SEM, n = minimum 3). (B) Amount of miRNA present in cell culture supernatant of HDC and LDC cells after removal of exosomes. (C) miRNA and AGO2 present in cell culture supernatant of HeLa cells is membrane protected. Conditioned medium from LDC state HeLa cells were treated with Triton X-100 for 15 min before they were used for exosome isolation. The estimation of miRNA was done by real time quantification and Ct values are plotted. AGO2 levels were detected by Western blot analysis. (D) Levels of CD63 exosomal marker proteins, present in cell equivalent amount of exosomes isolated from HDC and LDC state cells. (E) Relative levels of exosomal AGO2, HRS and HSP90 in exosomes isolated from LDC and HDC MDA-MB-231 cells. Absence of Dicer and β-Actin was used to rule out any cellular contamination in isolated exosomes. (F, G) Effect of CHX treatment on AGO2 and miRNA export via exosomes. Western blot for AGO2 in exosomes from DMSO or CHX treated LDC HeLa cells. CD63 levels were used to negate the variance in the release of exosomes as a consequence of CHX treatment (F). miRNA levels were quantified by real-time based quantification and Ct values were plotted (G). (H) Effect of GW4869 treatment on miRNA content of HDC and LDC state. Real time PCR based estimation of miR-122 was done with RNA from miR-122 expressing HeLa cells grown to either LDC or HDC state in the presence or absence of GW4869. ns, nonsignificant, *p < 0.05, **p < 0.01, ***p < 0.0001. p values were determined by paired t test. Results depict mean values from at least three experiments. CM, conditioned medium.
DISCUSSION
With increasing cell density in monolayer culture, we documented an increase in mature miRNA levels in human cells due to stabilization of its mature form. According to Hwang et al. (2009), cell–cell contact increases miRNA production and activity in mammalian cells by increasing Drosha-DGCR8–mediated processing of pri-miRNAs and its incorporation to AGO proteins. This may be an alternative mechanism of increased miRNP production in HDC cells for specific miRNAs. However, siRNA and miRNA stability experiments confirmed that increased levels of these small RNAs in HDC cells are primarily caused by their higher stability in HDC cells.
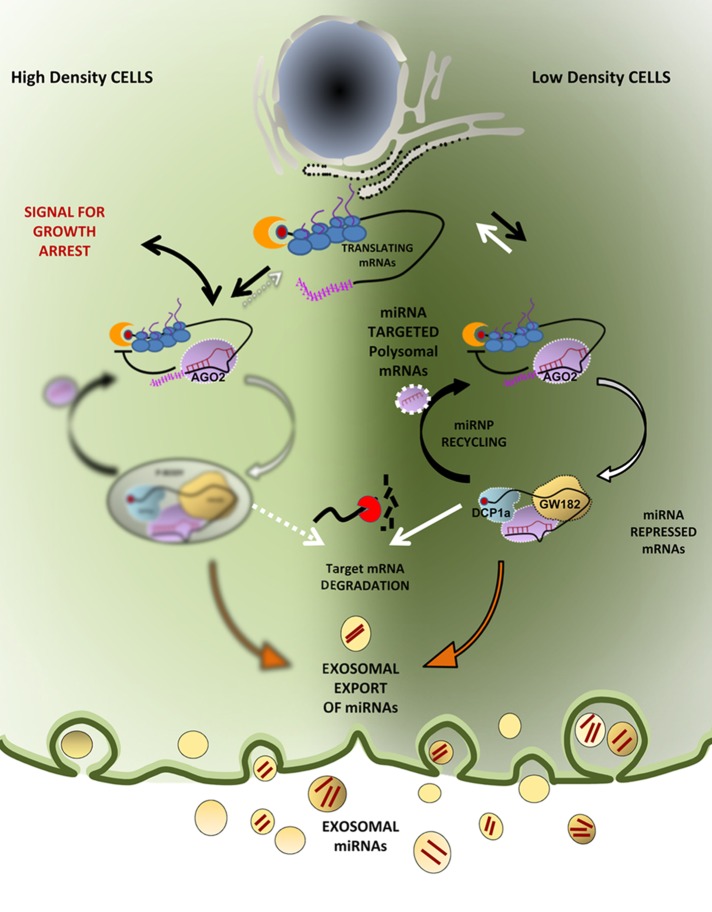
miRNP association with polysomes has long baffled miRNA researchers. For certain targets, repression is evident at protein levels only, without much effect on mRNA level of miRNA-targeted messages, and this too is a controversial aspect of miRNA research. In LDC cells, miRNAs are predominantly associated with lighter fractions of a sucrose density gradient, whereas miRNPs are primarily present with heavier polysomes in HDC cells. The GW182-CCR4-NOT1 complex recruits the ATPase RCK/p54 to AGO2 and induces translation repression and target RNA degradation in animal cells. Therefore relative loss of RCK/p54 from the polysomal fraction in HDC cells could be due to reduced interaction between AGO2 and GW182 (Chen et al., 2014; Mathys et al., 2014; Rouya et al., 2014). Ineffectiveness of polysome-attached miRNPs to induce degradation of target genes in HDC cells supports the model of miRNA-mediated repression at the initiation step followed by target RNA deadenylation and degradation (Djuranovic et al., 2012). Our data support a mechanism by which mRNAs attached to polysomes could be targeted by miRNPs but are not repressed or degraded before the elongating ribosomes complete their individual ongoing round of translation. Deployment of ribosomes from target mRNA is found to be essential for recycling or exosomal export of miRNAs. In HDC cells, where the translation rate is compromised (Azar et al., 2010), the polysome association time of miRNPs is prolonged and the miRNPs remain attached with translating mRNAs for a relatively longer time. This can explain why the increased numbers of miRNPs become less available for de novo target repression in HDC cells or in cells with a slow rate of translation. It suggests a processive mechanism of miRNA-mediated translation repression in which miRNPs, already engaged with target mRNAs, have to finish their repressive job before they can target another message. Thus arresting miRNPs with polysomal mRNAs will in turn increase stability of both miRNAs and target mRNAs with no change in the AGO2 protein level (Figure 7).
FIGURE 7:
Polysome retention precludes exosomal export of miRNAs in growth retarded human cells and modulate cellular miRNA level and activity. The polysomal miRNPs are found to be functionally inactive in processing of repressed mRNAs in HDC human cells. Our results support existence of miRNPs in mammalian cells in either active or inactive pools that is primarily determined by their polysome attachment. Reduced protein translation rate in HDC cells increases polysome association and alters subcellular distribution of miRNPs and hence extracellular secretion as well.
Our study showed that cell–cell contact has a strong effect on miRNA-mediated gene regulation pathways and plays a greater role than previously believed. It also satisfactorily answers and supports various proposed mechanisms of the mode of action of miRNAs.
Stalder et al. (2013) showed rough ER to be the site where miRNPs are enriched and interact with their targets. From this study, it seems that the majority of AGO2 is present with MVBs and/or polysomal fractions. How loaded miRNPs are transferred to MVB fractions is an unresolved issue. Regulated interorganellar transport to control miRNA activity in mammalian cells is a possibility that needs to be studied further.
Evidently, slow translation arrests miRNPs on polysomes but it is not clear why the miRNP retention with polysomes should restrict its MVB targeting. miRNA homeostasis is controlled by several factors that contribute to buffering cellular miRNA level and stability in animal cells. Although there are indications that RISC interacts with MVBs, very little is known as to why and under what circumstances this occurs. Exosomal export and delivery of several RNAs and proteins have received much attention, and they have been suggested to mediate changes in several physiological pathways often implicated in the regulation of cell growth. We speculate that compartmentalization of miRNAs to polysomes, thus affecting extracellular export, may have a key role in negotiating progrowth and antigrowth signals in cells in various environments.
MATERIALS AND METHODS
Cell culture and transfection
Human HeLa, MDA-MB-231, and Huh7 cells were all grown in DMEM supplemented with 2 mM l-glutamine and 10% heat-inactivated fetal calf serum (FCS). For transcription and translation inhibition experiments, α-Am (10 μg/ml) and CHX (100 ng/ml), both from Calbiochem (Billerica, MA), were dissolved in dimethyl sulfoxide (DMSO) and added to cells as described (Bhattacharyya et al., 2006). DMSO was added to controls (final concentration of 0.1%). For all experiments, cells were grown to 25–40% confluency (LDC) or were fully (100%) confluent (HDC) unless specified otherwise. Transfections were performed using Lipofectamine 2000 (Invitrogen, Bangalore, India) following the manufacturer's protocol. For miRNA repression assays, 25–500 ng of Renilla luciferase (RL) reporter plasmids with 500 ng of firefly luciferase (FL) plasmid were cotransfected per well of a six-well format (10-cm2 area), followed by splitting the cells to the desired HDC or LDC conditions and lysing them subsequently after at least 24 h of growth. siRNAs and miRVana mimics were transfected at 50 nM concentration. All transfections were followed by a cell split at 24 h posttransfection. Pre-miR-122 was cloned into pTRE-Tight-BI Vector from Clontech (Mountain View, CA) into NheI and NotI sites. Cells cotransfected with Tet on advanced plasmid and expressing FH-AGO2 was retransfected with the miR-122–expressing cassette, and miR-122 expression was induced with doxycycline (300 ng/ml) for 10 h during cell lysis and immunoprecipitation.
Luciferase assay and Northern and Western blots
RL and FL activities were measured using a Dual-Luciferase Assay Kit (Promega, Madison, WI) following the supplier's protocol on a VICTOR X3 Plate Reader with injectors (PerkinElmer, Waltham, MA). FL-normalized RL expression levels for reporter and control were used to calculate fold repression as the ratio of normalized control to reporter RL values. To avoid discrepancy in transfection efficiencies and avoid differential dilution of DNA cue to variable cell number, all transfections were performed in a single plate before splitting equal numbers of cells into plates of variable diameter to achieve LDC and HDC conditions.
Northern blotting of total cellular RNA (5–15 μg) was performed as described by Pillai et al., (2005). For detection, 32P-labeled 22-nt antisense DNA probes specific for respective miRNAs or siRL or U6 snRNA were used. Phosphorimaging of the blots was performed in the Cyclone Plus Storage Phosphor System (PerkinElmer), and Optiquant software (PerkinElmer) was used for quantification.
Western analyses of different miRNP components (AGO2, RCK/p54, and XRN1) were performed as described previously. Detailed list of antibodies used are given in Supplemental Table S3. Imaging of all Western blots was performed using an UVP BioImager 600 system equipped with VisionWorks Life Science software, version 6.80 (UVP, Cambridge, UK).
Polysome analysis
For polysome analysis, ∼2 × 107 HeLa cells grown to the desired level of confluency (LDC and HDC) were lysed in a buffer containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 8.0, 25 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 5 mM vanadyl ribonucleoside complex, 1% Triton X-100, 1% sodium deoxycholate, and 1× EDTA-free protease inhibitor cocktail (Roche) supplemented with CHX at 100 μg/ml (Calbiochem). Gradient analysis was performed as described previously (Pillai et al., 2005; Bhattacharyya et al., 2006). The absorbance profile was obtained by using an ISCO UA-6 absorbance monitor, and fractions were collected on a ISCO gradient fractionator. RNA and proteins from the individual fractions were isolated and analyzed following published procedures (Bhattacharyya et al., 2006).
Affinity purification of miRISC and in vitro RISC cleavage assay
FLAG-HA-AGO2 (FH-AGO2) expression plasmid was cotransfected in HeLa cells with plasmid expressing pre-miR122. Cell lysis and miRISC purification was done following published protocols (Kundu et al., 2012).
Affinity-purified miRISC-122 was assayed for target RNA cleavage using a 36-nt RNA, 5′AAAUUCAAACACCAUUGUCACACUCCACCAGAUUAA3′, bearing the sequence complementary to mature miR-122 (underlined). Target RNA cleavage assays were carried out in a total volume of 30 μl with 10 fmol of 5′ 32P-labeled RNA and protein-equivalent amounts of RISC at 30°C for 30 min, followed by RNA isolation, and cleaved products were analyzed on a 12% denaturing 8 M urea-PAGE before autoradiography.
In vitro translation with rabbit reticulocyte lysate
Total polysomes separated on a 30% sucrose cushion were resuspended in polysome buffer lacking one of 1% Triton X-100, 1% sodium deoxycholate, or CHX. Isolated polysomes corresponding to 4 × 106 HDC HeLa cells were used for in vitro translation as per manufacturer's protocol (Promega). Translation was carried out at 30°C for 1 h; control sets were incubated on ice for the same duration. The reaction was terminated with the addition of 100 μg/ml CHX, and RISC cleavage assay of the total reaction mixture was performed as described previously. To check the distribution of AGO2 in the polysomes upon translation, instead of performing the RISC cleavage assay, the whole translation mixture was loaded on a 30–55% sucrose cushion and centrifuged for 2 h at 36,000 rpm in a SW60 rotor in an ultracentrifuge.
Exosome isolation and characterization
Exosomes were isolated based on published protocols (Thery et al., 2006) in a two-step process using a sucrose cushion in an ultracentrifuge. For affinity-based purification of exosomes, Streptavidin magnetic beads coated with biotinylated capture antibodies were used as per manufacturer's protocol (EXOFLOW150A-1; System Biosciences). All of the precipitated MDA-MB-231 exosomes were added to the beads and kept at 4°C overnight for affinity interaction. Beads containing pulled-down exosomes were then washed (as per manufacturer's protocol) and stained with the Exo-FITC universal exosome stain provided in the kit. Stained exosomes were then visualized on a BD LSR Fortessa FACS machine (BD Biosciences). An identical number of affinity-purified exosomes was eluted from the beads (as per manufacturer's protocol) and proteins and RNA isolated for subsequent analysis. Biophysical analysis of exosomes from MDA-MB-231 cells was carried out as described (Basu and Bhattacharyya, 2014).
Fractionation and separation of endosomes and ER on OptiPrep gradients
OptiPrep (Sigma-Aldrich, St. Louis, MO) was used to prepare a 3–30% continuous gradient in a buffer containing 78 mM KCl, 4 mM MgCl2, 8.4 mM CaCl2, 10 mM ethylene glycol tetraacetic acid (EGTA), and 50 mM HEPES-NaOH (pH 7.0) for separation of subcellular organelles as described previously, with minor modifications (Gibbings et al., 2009). Cells were trypsinized, washed, and homogenized with a Dounce homogenizer in a buffer containing 0.25 M sucrose, 78 mM KCl, 4 mM MgCl2, 8.4 mM CaCl2, 10 mM EGTA, and 50 mM HEPES-NAOH, pH 7.0, supplemented with 100 μg/ml CHX, 5 mM vanadyl ribonucleoside complex (Sigma-Aldrich), 0.5 mM DTT, and 1× protease inhibitor. The lysate was clarified by centrifugation at 1000 × g for 5 min and layered on top of the prepared gradient. The tubes were centrifuged for separation of gradient using established protocols, and 10 fractions were collected for further analysis or pooled for immunoprecipitation studies.
Postimaging analysis and others
All Western blot and Northern blot images were processed with Photoshop CS4 (Adobe) for all linear adjustments and cropping. Image cropping was done using Photoshop CS4. Statistical analysis of data was done by performing paired/unpaired, nonparametric, two-tailed t tests with a confidence interval of 95%, and four significant digits were considered in all cases. ns, nonsignificant, *p < 0.01, **p < 0.001, and ***p < 0.0001 in all cases. Additional information on plasmids, siRNAs, oligos, and antibodies used is provided in Supplemental Tables S1 and S2. Additional information on methodologies is available in the Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank W. Filipowicz, G. Meister, R. Pillai, E. Bertrand, S. Bandhopadhyay, S. Feroj, and B. Barman for generous help with reagents and plasmids constructs. We are particularly indebted to Witold Filipowicz for his critical comments and suggestions. The work was primarily funded by the Human Frontier Science Program Organization Career Development Fund and the Wellcome Trust International Senior Research Fellowship Fund (to S.N.B.) and was also supported by a Council of Scientific and Industrial Research EMPOWER grant. All the authors except S.N.B. were supported by fellowships from the Council of Scientific and Industrial Research or the Wellcome Trust. For imaging, the live-cell imaging facility at the Indian Institute of Chemical Biology, funded by the Council of Scientific and Industrial Research and the Wellcome Trust, was used.
Abbreviations used:
- ER
endoplasmic reticulum
- HDC
high-density culture
- LDC
low-density culture
- MVB
multivesicular body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1521) on January 21, 2015.
The authors declare no conflict of interest.
REFERENCES
- Augustin R, Riley J, Moley KH. GLUT8 contains a [DE]XXXL[LI] sorting motif and localizes to a late endosomal/lysosomal compartment. Traffic. 2005;6:1196–1212. doi: 10.1111/j.1600-0854.2005.00354.x. [DOI] [PubMed] [Google Scholar]
- Azar R, Susini C, Bousquet C, Pyronnet S. Control of contact-inhibition by 4E-BP1 upregulation. Cell Cycle. 2010;9:1241–1245. doi: 10.4161/cc.9.7.11047. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Bhattacharyya SN. Insulin-like growth factor-1 prevents miR-122 production in neighbouring cells to curtail its intercellular transfer to ensure proliferation of human hepatoma cells. Nucleic Acids Res. 2014;42:7170–7185. doi: 10.1093/nar/gku346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Cikaluk DE, Tahbaz N, Hendricks LC, DiMattia GE, Hansen D, Pilgrim D, Hobman TC. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Martelot G Le, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci USA. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Kong YW, Cannell IG, Moor de CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci USA. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010a;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010b;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014;54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Rouya C, Siddiqui N, Morita M, Duchaine TF, Fabian MR, Sonenberg N. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA. 2014;20:1398–1409. doi: 10.1261/rna.045302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Raposo G. Exosomes—vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013;32:1115–1127. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.