The retromer complex regulates vesicle transport at endosomes. Different members of the VPS9 domain–containing Rab5-family guanine nucleotide exchange factors interact with the yeast retromer complex and mediate its endosomal localization.
Abstract
The retromer complex facilitates the sorting of integral membrane proteins from the endosome to the late Golgi. In mammalian cells, the efficient recruitment of retromer to endosomes requires the lipid phosphatidylinositol 3-phosphate (PI3P) as well as Rab5 and Rab7 GTPases. However, in yeast, the role of Rabs in recruiting retromer to endosomes is less clear. We identified novel physical interactions between retromer and the Saccharomyces cerevisiae VPS9-domain Rab5-family guanine nucleotide exchange factors (GEFs) Muk1 and Vps9. Furthermore, we identified a new yeast VPS9 domain-containing protein, VARP-like 1 (Vrl1), which is related to the human VARP protein. All three VPS9 domain–containing proteins show localization to endosomes, and the presence of any one of them is necessary for the endosomal recruitment of retromer. We find that expression of an active VPS9-domain protein is required for correct localization of the phosphatidylinositol 3-kinase Vps34 and the production of endosomal PI3P. These results suggest that VPS9 GEFs promote retromer recruitment by establishing PI3P-enriched domains at the endosomal membrane. The interaction of retromer with distinct VPS9 GEFs could thus link GEF-dependent regulatory inputs to the temporal or spatial coordination of retromer assembly or function.
INTRODUCTION
Retromer is a retrograde endosomal trafficking complex that facilitates recycling of integral membrane proteins to the late Golgi and the plasma membrane (Seaman, 2005, 2012; Bonifacino and Hurley, 2008; Attar and Cullen, 2010). It was first identified in yeast as a complex required to recycle the acid hydrolase receptor Vps10 and maintain the Golgi localization of Kex2 (Seaman et al., 1998). Since then, it has been linked to many processes in higher organisms, including the endosome-to-Golgi recycling of the cation-independent mannose 6-phosphate receptor and the iron transporter DMT1 and the direct endosome-to-plasma membrane trafficking of the β2-adrenergic receptor (Seaman, 2004; Tabuchi et al., 2010; Temkin et al., 2011). In addition, deficiencies in the retromer complex and its associated factors have been linked to Parkinson's (Vilariño-Güell et al., 2011; Zavodszky et al., 2014) and Alzheimer's diseases (Fjorback et al., 2012).
Yeast retromer is composed of a structural subcomplex containing the sorting nexins Vps5/Vps17 and a cargo-selective subcomplex (CSC) comprising Vps26/Vps29/Vps35 (Seaman et al., 1998). The sorting nexins bind phosphatidylinositol 3-phosphate (PI3P) at endosomes and deform the membrane (Burda et al., 2002), whereas the CSC recruits cargo into the retromer tubule (Nothwehr et al., 1999; Seaman, 2005). Although these two subcomplexes form a stable pentamer in yeast, the CSC and sorting nexins are not tightly associated in mammalian cells (McGough and Cullen, 2011). The mammalian CSC assembles with Vps5/Vps17 homologues and with other sorting nexins to form spatially distinct classes of retromer tubules that engage different cargo (Harterink et al., 2011). Whereas the sorting nexin SNX3 is enriched at early endosomes, retromer tubules formed by the Vps5 and Vps17 homologues SNX1/2 and SNX5/6, respectively (Wassmer et al., 2009), are most abundant on endosomes undergoing the early-to-late transition (Rojas et al., 2008; Cullen and Korswagen, 2012; van Weering et al., 2012).
Rab GTPases are important for membrane identity, vesicle budding, and membrane fusion (Stenmark, 2009). Rabs are converted to their active, membrane-bound form by guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP and are inactivated by GTPase-activating proteins, which increase the rate of GTP hydrolysis by the Rabs. As early endosomes mature into late endosomes, they undergo a Rab conversion in which active Rab7-family GTPases are recruited and Rab5-family GTPases are inactivated and extracted from the membrane (Rink et al., 2005).
Both Rab5- and Rab7-family GTPases are implicated in the recruitment of retromer to endosomes in mammalian cells (Rojas et al., 2008; Liu et al., 2012). GTP-bound Rab5 does not bind retromer directly but instead recruits a complex containing VPS34, a phosphatidylinositol 3-kinase (PI3K; Christoforidis et al., 1999). This catalyzes the production of PI3P, which is recognized by the Phox homology (PX) domains of the sorting nexins (Xu et al., 2001; Yu and Lemmon, 2001; Cozier et al., 2002). In contrast, direct binding of Rab7 to Vps35 is essential for the endosomal recruitment of mammalian CSC (Harrison et al., 2014). The CSC also interacts with a number of regulatory factors, including the putative Rab7 GTPase-activating protein TBC1D5 (Seaman et al., 2009; Harbour et al., 2010), and retromer tubule formation is reported to be maximal at the time of Rab5-to-Rab7 conversion (Cullen and Korswagen, 2012; van Weering et al., 2012). Thus retromer assembly at endosomes may be tightly coupled to the regulation of Rab activation.
In yeast, the regulation of retromer function by Rab proteins is less well understood. Vps35 binds to the yeast Rab7 homologue Ypt7, but although this interaction directs it to the vacuole, it is not required for recruitment to endosomes (Balderhaar et al., 2010; Liu et al., 2012). Moreover, many of the retromer-associated regulatory proteins identified in mammalian cells are not conserved in yeast. Thus the extent to which retromer assembly at endosomes is subject to Rab-dependent regulation is unknown.
Here we show that retromer physically interacts with the VPS9-domain GEFs Muk1 and Vps9 and that at least one of these must normally be present for retromer recruitment at endosomes. We further identify a new yeast VPS9-domain protein related to human VARP that is present in wild but not laboratory strains of Saccharomyces cerevisiae. All three proteins can act through Rab5-family GTPases and the PI3K Vps34 to localize retromer to endosomal membranes. This suggests that association with Rab5 family GEFs could provide a mechanism to regulate the location or extent of retromer assembly.
RESULTS
Retromer physically interacts with Rab5-family GEFs
Previous high-throughput mass spectrometry studies of yeast protein complexes suggested an interaction between several subunits of the retromer complex and the VPS9-domain GEF Muk1 (Krogan et al., 2006; Babu et al., 2012). This interaction was surprising, as VPS9-domain GEFs activate Rab5-family GTPases, whereas yeast retromer has been shown to interact only with the Rab7-like GTPase, Ypt7.
To validate the retromer–Muk1 interaction, we expressed Muk1-hemagglutinin (HA) from a GAL1 promoter in strains containing different tandem affinity purification (TAP)–tagged retromer subunits and carried out pull downs using calmodulin resin under batch purification conditions similar to those used for mass spectrometry. This showed that all five retromer subunits were able to copurify Muk1 (Figure 1A).
FIGURE 1:
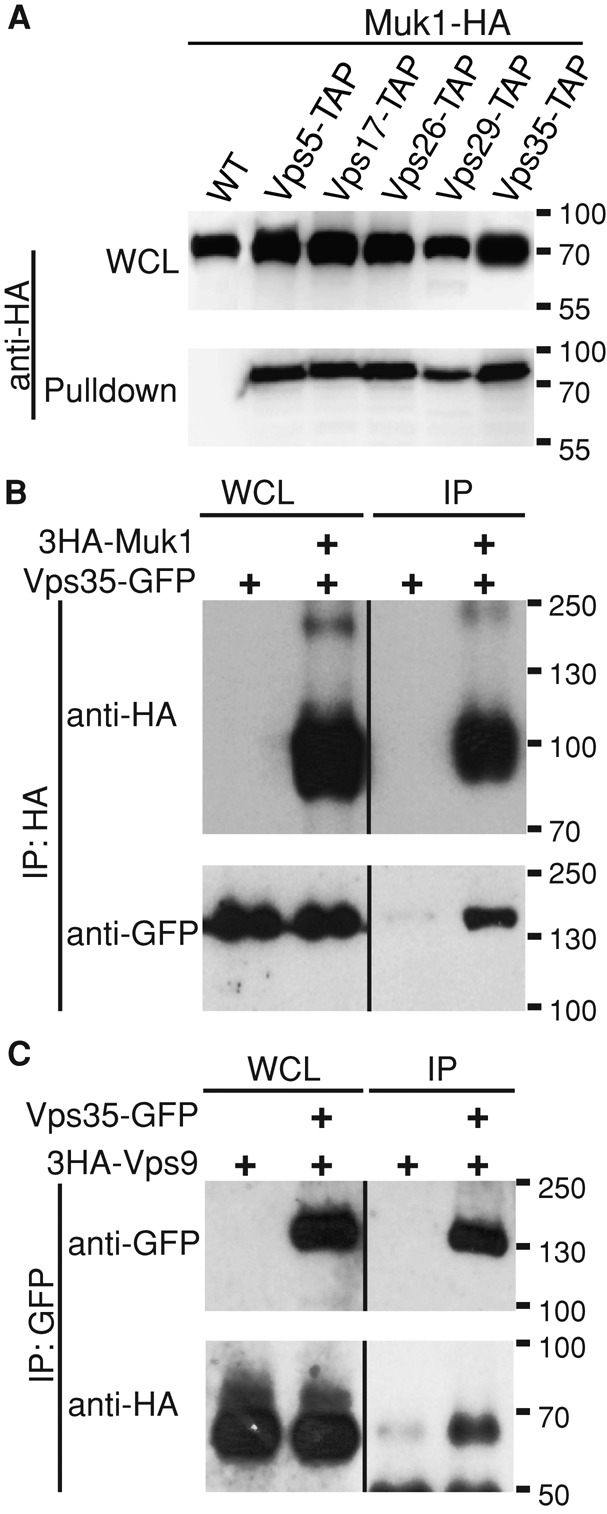
Retromer physically interacts with the Rab5-family GEFs Muk1 and Vps9. (A) TAP-tagged retromer subunits were pulled down using calmodulin beads from GAL1pr-MUK1-HA strains. Samples were resolved by 12% SDS–PAGE and detected by immunoblotting. Loading of lysates relative to the pull down was 1:25. (B) Endogenously tagged Vps35-GFP and ADH1pr-3HA-Muk1 were cross-linked with 1.6 mg/ml DSP, 3HA-Muk1 was immunoprecipitated, and copurifying Vps35-GFP was probed by immunoblotting. Loading of lysates relative to immunoprecipitate (IP) was 1:1.6 (anti-HA) and 1:388 (anti-GFP). (C) Cells expressing Vps35-GFP and ADH1pr-3HA-Vps9 were cross-linked and immunoprecipitated with anti-GFP. Copurification of 3HA-Vps9 was detected with anti-HA. Loading of lysates relative to IP was 1:1.6 (anti-GFP) and 1:388 (anti-HA).
To estimate the fraction of interacting proteins, we subsequently carried out small-scale coimmunoprecipitations in strains in which HA-Muk1 was expressed from the weaker ADH1 promoter and the retromer subunit Vps35 was green fluorescent protein (GFP) tagged at its endogenous locus. Interactions between Muk1 and retromer were reproducibly detected in the presence of the crosslinker dithiobis(succinimidyl propionate) (DSP). However, purification of ∼30% of cellular Muk1 coprecipitated <1% of the total pool of Vps35 (Figure 1B). The fact that interactions were detected only in the presence of the cross-linker and represented a minor fraction of the total Vps35 protein suggests that Vps35-Muk1 interactions are weak or disrupted on lysis and are likely to be substoichiometric. Furthermore, as retromer is a stable pentameric complex, we cannot exclude the possibility that Muk1 binds another subunit or interacts with retromer through a bridging protein.
Yeast express two known Rab5-family GEFs, Vps9 and Muk1, which share a catalytic VPS9 domain but have divergent C-termini (Carney et al., 2006; Balderhaar et al., 2010; Paulsel et al., 2013). Retromer–Vps9 interactions were not previously identified by large-scale mass spectrometry. Nevertheless, we found that HA-Vps9, when expressed from the ADH1 promoter, reproducibly copurified with Vps35-GFP from DSP-treated cell lysates (Figure 1C). These results indicate that retromer binds more than one VPS9-domain GEF.
Identification of Vrl1, a third member of the yeast VPS9-domain family related to mammalian VARP
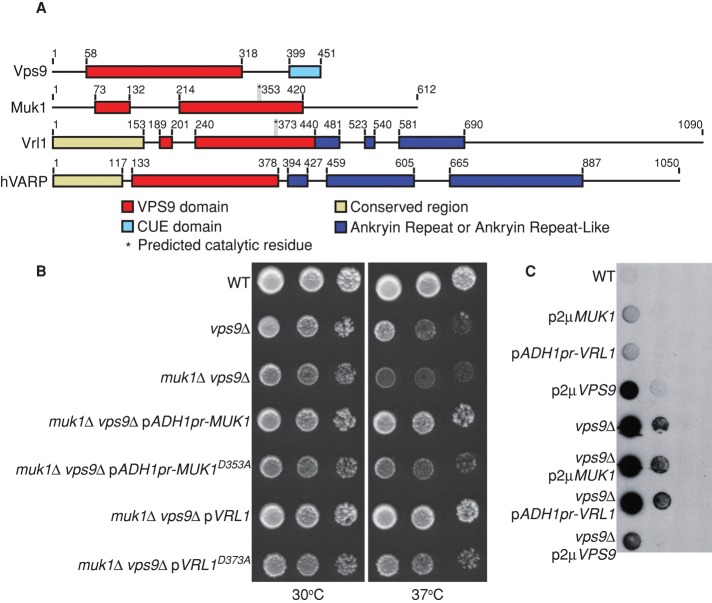
Muk1 and Vps9 are the only two VPS9-domain GEFs described in yeast to date. Of interest, the Superfamily database (Gough et al., 2001), which identifies domains using hidden Markov models, reported a partial VPS9 domain at the N-terminus of the uncharacterized open reading frame (ORF) YML002w. In wild strains of S. cerevisiae, YML002w is continuous with an upstream ORF (YML003w) and is predicted to encode a single protein of 1090 amino acids with a full-length VPS9 domain. Comparison of sequences from wild and laboratory strains of S. cerevisiae suggests that a single thymine residue at chrXIII:264337 is deleted in commonly studied strains, causing a frameshift and premature stop codon (Supplemental Figure S1A). Resequencing of the corresponding region in the BY4741 parental strain confirmed the presence of the mutation. Integration of a 3HA tag at the 5′ end of the upstream ORF, YML003w, under control of the ADH1 promoter, showed that the frameshift produces a truncated protein of the predicted size (Supplemental Figure S1B). Thus it appears that wild yeast strains encode a third VPS9-domain protein that has been mutated in laboratory strains of S. cerevisiae.
Sequence comparisons suggest that this new VPS9-domain protein is conserved in most species and is related to human VARP (hVARP), a Rab21 GEF (Zhang et al., 2006). Accordingly, we have named this ORF VRL1, for VARP-like1. hVARP is a multifunctional protein that binds the R–soluble N-ethylmaleimide–sensitive factor attachment protein receptor (R-SNARE) VAMP7 and is a key regulator of endosome-to–cell surface transport (Burgo et al., 2009, 2012; Ohbayashi et al., 2012). Whereas VARP GEF activity is essential for recycling to the cell surface in neurites, VARP has a separate role as a Rab32/38 effector in the trafficking of tyrosinase-related protein 1 to melanosomes, which does not require its GEF activity (Tamura et al., 2011; Ohbayashi et al., 2012). VARP and Vrl1 share extensive regions of conservation, including an N-terminal region not found in other GEFs, followed by the VPS9 domain and several ankyrin repeats (Figure 2A and Supplemental Figure S2). However, the yeast protein lacks the second of two sets of ankyrin repeats present in hVARP, and other hVARP features, including the Rab32/38-binding site and the VAMP7-interacting domain (Burgo et al., 2009; Ohbayashi et al., 2012; Schäfer et al., 2012), are only partially conserved, suggesting that the proteins may not be functionally identical (Supplemental Figure S2C).
FIGURE 2:
Vrl1 is a new yeast VPS9-domain protein. (A) Schematic of yeast VPS9 domain–containing proteins and human VARP based on Superfamily (Gough et al., 2001) and ClustalW alignments. (B) Yeast were spotted in 10× dilution series and grown 2 d at 37°C to assess temperature sensitivity of the indicated strains. VRL1 was expressed from its endogenous promoter. (C) A colony overlay assay was used to assess carboxypeptidase Y (CPY) secretion. Cells spotted in 10× dilution series were overlaid with nitrocellulose and incubated for 16 h. The nitrocellulose was then immunoblotted with anti-CPY antibodies.
Vrl1, Muk1, and Vps9 have partially overlapping functions
Muk1 and Vps9 were previously shown to have redundant yet distinct functions. Muk1 overexpression rescues the temperature-sensitive (ts) growth phenotype of vps9 cells but cannot replace Vps9's role in the late endosomal sorting of carboxypeptidase Y (CPY; Paulsel et al., 2013). To determine whether Vrl1 can functionally substitute for Muk1 or Vps9, we cloned the genomic region that includes both YML003w and YML002w into a single-copy plasmid and inserted a nucleotide (T856) into YML003W to recreate the full-length ORF. We use VRL1 to refer to the full-length gene that results from the correction of the frameshift mutation; it is important to note that the laboratory yeast strains used here do not express functional VRL1. Introduction of a plasmid expressing VRL1 from its endogenous promoter fully restored growth of muk1 vps9 strains at high temperatures, suggesting that it can replace some function of Muk1 and/or Vps9 (Figure 2B).
The VPS9 domain of Rabex-5 contains a single invariant residue, D313, which is required for catalytic activity (Delprato and Lambright, 2007). We mutated the corresponding aspartate residues in Muk1 and Vrl1 and found that expression of Muk1D353A or Vrl1D373A failed to rescue the muk1 vps9 ts growth defect, suggesting that suppression requires an active VPS9 domain (Figure 2B). However, overexpression of Vrl1 did not rescue the CPY secretion of a vps9Δ strain. In addition, high levels of Vrl1 induced mild CPY secretion in a wild-type strain similar to overexpression of Muk1 (Paulsel et al., 2013; Figure 2C). These data suggest that although Vrl1, Muk1, and Vps9 show some degree of redundancy, they are not functionally equivalent.
Because both known VPS9-domain GEFs interact with retromer, we tested Vrl1–retromer binding by coimmunoprecipitation. No physical interaction between ADH1pr-driven HA-Vrl1 and the retromer subunit Vps35-GFP was observed, although HA-Vrl1 was expressed at a significantly lower level than Muk1 or Vps9 (∼5% of HA-Muk1 level; unpublished data), and thus any interaction may be below the detection threshold. Taken together, our data suggest that Vrl1 can partially substitute for Muk1/Vps9 but cannot replace all functions of these GEFs.
Retromer interacts with the VPS9 domain
Retromer physically interacts with both Muk1 and Vps9, and yet their sequence similarity outside the VPS9 domain is limited. To map the site of interaction, we used the integrated membrane yeast two-hybrid (iMYTH) system, which detects interactions of membrane-associated proteins at their normal organellar localization (Paumi et al., 2007; Snider et al., 2013). The N-terminal half of ubiquitin (NubG) was fused to full-length and truncated versions of Muk1 and coexpressed with retromer subunits fused to a ubiquitin C-terminus-transcription factor cassette (Figure 3A). Interactions that reconstitute ubiquitin result in the deubiquitinase-dependent release of the transcription factor and the activation of reporter genes. Full-length Muk1 and a truncated version (amino acids [aa] 1–417) corresponding to the VPS9 domain interacted with three of the five retromer subunits, and this interaction was not affected by the D353A mutation predicted to block Muk1's GEF activity (Figure 3B). Vps9 also interacted with Vps35–C-terminal ubiquitin fragment (Cub) in this assay, although no interaction was observed with the remaining retromer subunits. We were unable to detect an interaction between Vrl1–N-terminal ubiquitin fragment (Nub) and any of the retromer Cubs; however, Vrl1-Nub was expressed at relatively low levels compared with the other Nub fusions (Supplemental Figure S3A). Moreover, interactions will not be detected if the Nub and Cub fusions are not positioned in close proximity in the complex. Our results suggest that retromer interacts, directly or indirectly, with the Muk1 VPS9 domain. This is the only domain shared by Muk1 and Vps9, suggesting that retromer might recognize a conserved feature of this domain.
FIGURE 3:
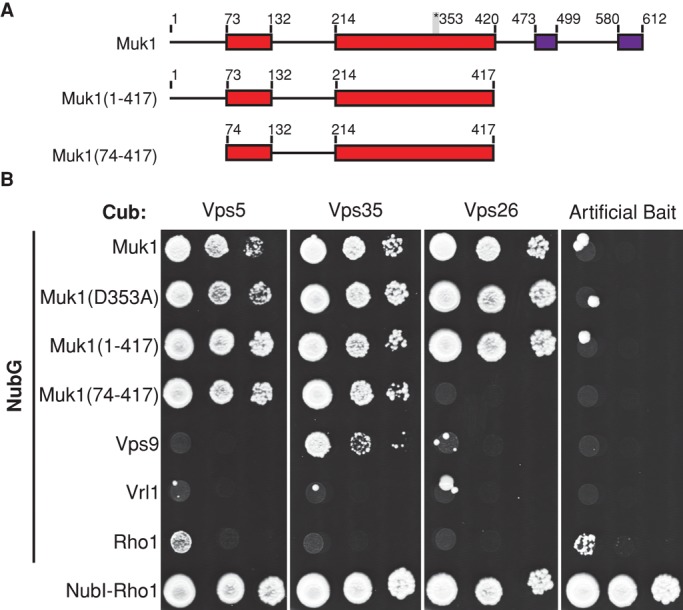
The VPS9 domain of Muk1 is sufficient for interaction with retromer subunits in the iMYTH assay. (A) Schematic of Muk1 truncations. The position of the invariant aspartate residue required for GEF activity and conserved C-terminal motifs are indicated. (B) Strains containing Cub-tagged retromer subunits and plasmids encoding Rab5-family GEFs tagged N-terminally with NubG were tested for activation of the HIS3 reporter on selective medium lacking tryptophan, adenine, and histidine. NubG-tagged Rho1 acted as a negative control, whereas the NubI tag, which binds tightly to Cub independently of other interactions, confirmed the expression of Cub fusions.
Muk1, Vps9, and Vrl1 can localize to endosomes
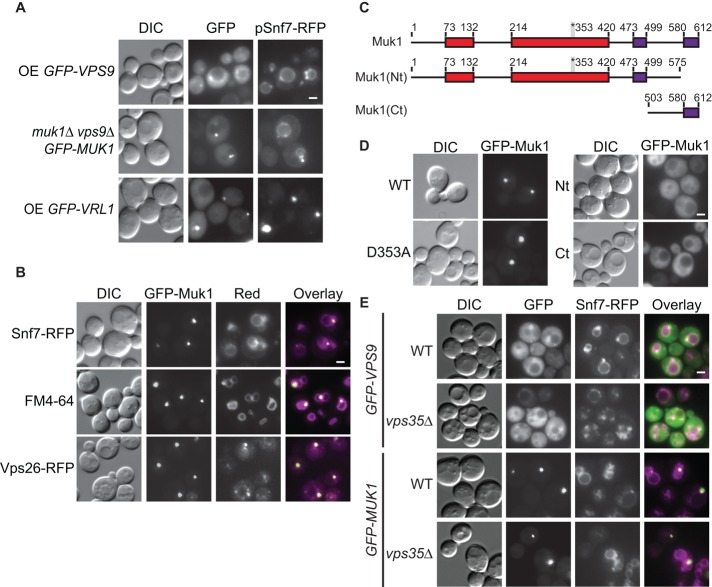
Both Muk1 and Vps9 have a predominantly cytosolic distribution in wild-type cells (Paulsel et al., 2013). Vps9 is believed to be recruited to ubiquitinated cargo via its C-terminal CUE domain (Carney et al., 2006), which is not present in Muk1. Accordingly, Vps9, but not Muk1, localizes to the aberrant endosome of endosomal sorting complex required for transport (ESCRT) mutants (Paulsel et al., 2013), where ubiquitinated proteins accumulate. Overexpression of the tagged ESCRT-III subunit Snf7–red fluorescent protein (RFP) disrupts ESCRT function and causes an endosomal maturation defect (Froissard et al., 2007). We confirmed that GFP-Vps9 colocalizes with Snf7-RFP at endosomes (21% of cells, SE = 6.5%; Figure 4A). Of interest, when expressed from a plasmid under its endogenous promoter, GFP-Muk1 could be detected at Snf7-RFP–marked endosomes in some cells (5%, SE = 1.5%; Figure 4A). Furthermore, GFP-Vrl1 formed puncta that colocalized with Snf7-RFP puncta in 16% (SE = 3.7%) of cells (Figure 4A). Taken together, these results suggest that all three VPS9-domain proteins are able to associate with endosomes to some extent.
FIGURE 4:
The yeast VPS9-domain proteins Muk1, Vps9, and Vrl1 localize to endosomes. (A) MUK1pr-GFP-Muk1, ADH1pr-GFP-Vps9, and ADH1pr-GFP-Vrl1 localize to Snf7-RFP–marked late endosomes by fluorescence microscopy. (B) Fluorescence microscopy of ADH1pr-GFP-Muk1 with Snf7-RFP, the lipophilic dye FM4-64, and the tagged retromer subunit RFP-Vps26. (C) Schematic of Muk1 truncations used in D, showing conserved C-terminal motifs identified by alignments (purple). (D) Fluorescence microscopy of full-length or mutated ADH1pr-GFP-Muk1. (E) Deletion of VPS35 does not disrupt the endosomal localization of overexpressed GFP-Vps9 or GFP-Muk1. Scale bars, 2 μm; OE, overexpressed.
When GFP-Muk1 was expressed from the stronger ADH1 promoter, bright puncta were seen in 37% (SE = 4.8%) of cells, and these overlapped with the endosomal markers FM4-64 and Snf7-RFP and with the retromer subunit Vps26-RFP (Figure 4B). Two regions in the Muk1 C-terminal domain, corresponding to residues 473–499 and 580–612, are highly conserved in different fungal species (Figure 4C). A truncation that removes the last 37 aa of Muk1 (GFP-Nt-Muk1) did not alter protein stability (Supplemental Figure S2B), and yet it abolished puncta formation (Figure 4D), suggesting that this region contributes to Muk1's localization. However, the Muk1 C-terminus alone (aa 503–612), although expressed at similar levels to the full-length GFP-Muk1, did not form puncta, indicating the Muk1 C-terminal domain is necessary but not sufficient for endosomal localization (Figure 4D). The localization determinant may not be correctly folded in the truncated protein. Alternatively, membrane localization of Muk1 may require multiple interacting domains.
To test whether retromer is involved in recruiting the Rab5 GEFs to endosomes, we deleted retromer subunits VPS26 or VPS35 from the strains expressing ADH1pr-driven GFP-Muk1 or GFP-Vps9. Both GEFs were found in puncta that colocalized with Snf7-RFP in the retromer deletion strains (Figure 4E), suggesting that Muk1 and Vps9 localize to endosomes independent of their interactions with retromer.
Retromer localization to endosomes is impaired by loss of Rab5 GEFs
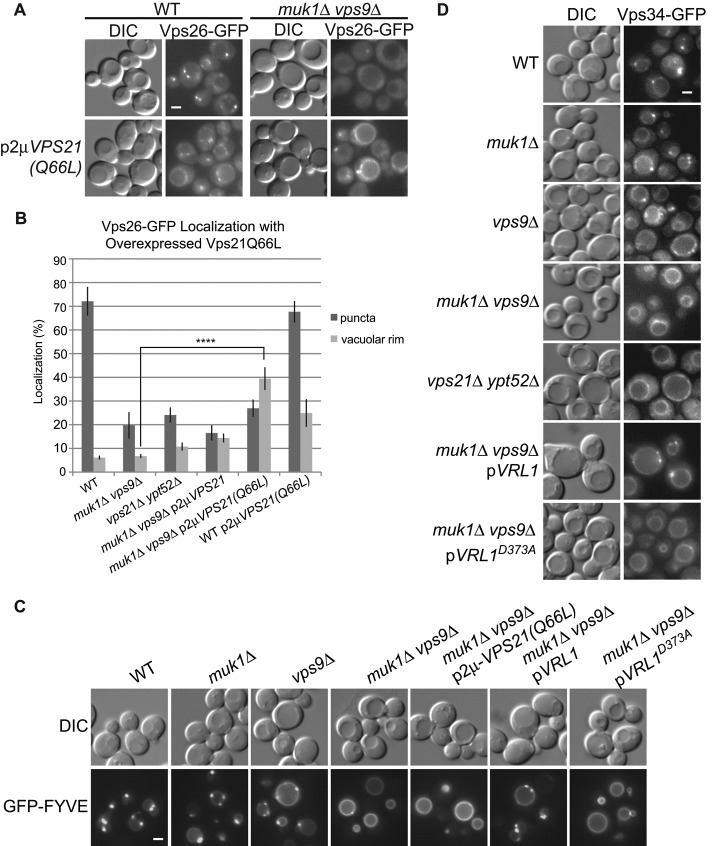
We showed that retromer interacts with two different VPS9-domain GEFs and that these GEFs can localize to endosomes. To test whether the GEFs are required for retromer recruitment, we deleted them singly and in combination (Figure 5A). Vps26-GFP localized to puncta in wild-type cells and muk1 and vps9 single mutants, although punctate localization was reduced in the vps9 strain. Strikingly, deletion of both MUK1 and VPS9 greatly reduced localization of Vps26-GFP and Vps5-GFP to endosomes. This was complemented by expression of either VPS9 or MUK1 from single-copy plasmids (Figure 5, A and B). Strikingly, the MUK1D353A GEF mutant failed to restore Vps26-GFP localization, demonstrating the importance of Muk1 GEF activity in vivo. Similarly, expression of Vrl1, but not Vrl1D373A, from its endogenous promoter also restored retromer localization to muk1 vps9 mutant cells. Thus any of the three active VPS9-domain proteins is sufficient to recruit retromer to endosomes.
FIGURE 5:
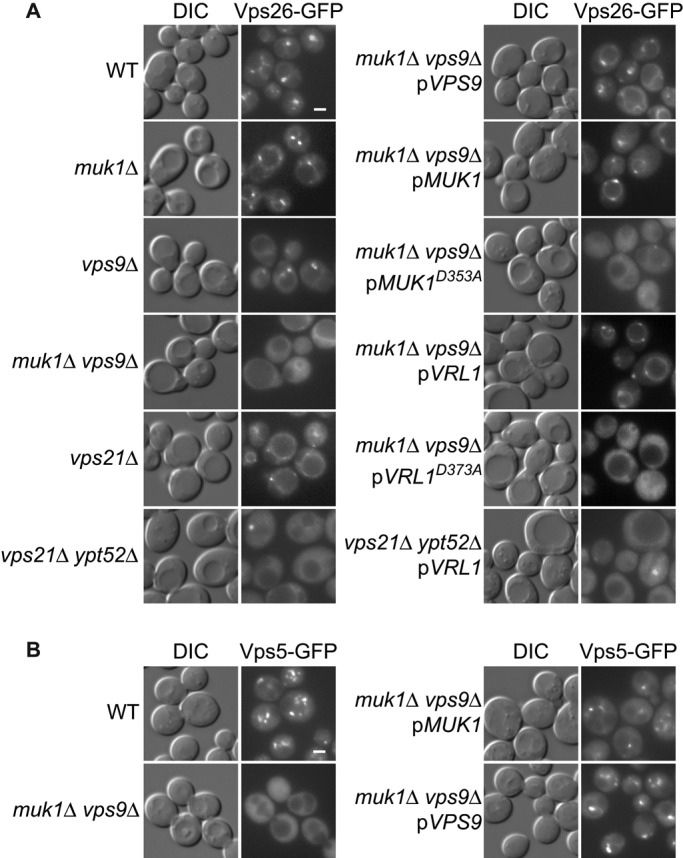
Rab5-family GEFs and GTPases are required for endosomal recruitment of retromer. (A) Fluorescence microscopy of Vps26-GFP in strains lacking Rab5-family GEFs and Rab5-family GTPases. Muk1 and Vrl1 are expressed from endogenous promoters. Muk1D353A and Vrl1D373A are predicted to be catalytically inactive. (B) Microscopy of Vps5-GFP in strains with deletions of Rab5-family GEFs. Scale bars, 2 μm.
The need for an intact VPS9 domain suggests that active Rabs are required for efficient retromer recruitment to endosomes. There are three Rab5-family GTPases in yeast: Vps21, Ypt52, and Ypt53. Ypt53 is normally expressed at very low levels and is activated only under conditions of stress (Nickerson et al., 2012). We found that loss of VPS21 alone caused a slight reduction in retromer recruitment (Figure 5A), consistent with other observations (Balderhaar et al., 2010), but deletion of both VPS21 and YPT52 strongly reduced retromer recruitment. This was not complemented by expression of VRL1, suggesting that Vrl1 acts upstream of the Rab5 GTPases, consistent with it role as a putative GEF. Together the results suggest the GEFs redundantly activate different Rab5 GTPases, and this in turn is needed for retromer's localization to endosomes.
Rab5 GEFs are required for normal PI3P localization
Overexpression of active Rabs can often overcome loss of their respective GEFs (Siniossoglou et al., 2000; Lynch-Day et al., 2010). Therefore we expressed different forms of Vps21 from high-copy plasmids to see whether this would rescue retromer recruitment (Figure 6, A and B). Surprisingly, expression of the constitutively active Vps21Q66L in wild-type (WT) and muk1 vps9 strains stimulated recruitment of retromer to the vacuolar membrane but not to endosomes.
FIGURE 6:
Rab5-family GEFs are needed for PI3P production at endosomes, and this cannot be bypassed by expressing the constitutively active Rab5-family GTPase Vps21(Q66L). (A) Fluorescence microscopy shows that Vps26-GFP is mislocalized to the vacuolar rim when Vps21(Q66L) is expressed, even in the absence of Rab5-family GEFs. (B) Quantification of Vps26-GFP fluorescence microscopy. Images of Vps26-GFP localization from four independent experiments were manually scored for localization to the vacuolar rim or puncta (N ≥ 140/strain per experiment). Unpaired one-way analysis of variance: p < 0.0001 overall; ****p < 0.0001. (C) Fluorescence microscopy of GFP-FYVE, a biomarker for PI3P, in strains with deletions of Rab5-family GEFs. (D) Vps34-GFP localization to puncta is dependent on expression of Rab5-family GEFs and GTPases. Vrl1 is expressed from the endogenous promoter in C and D. Scale bars, 2 μm.
Because retromer recruitment to early endosomes requires PI3P, its vacuolar localization might result from loss of endosomal PI3P or from elevated levels of PI3P at the vacuole in these mutant strains. Indeed, the PI3P-binding biosensor GFP-FYVE showed endosomal localization in WT strains but prominent vacuole rim staining in a muk1 vps9 strain (Figure 6C), similar to that of a vps21 ypt52 strain (Nickerson et al., 2012). This vacuolar pool of PI3P could originate at endosomes and accumulate at the vacuole due to a disruption in its turnover or metabolism. Alternatively, PI3P could be produced at the vacuolar membrane by a vacuole-localized pool of the PI3K Vps34 (Burda et al., 2002). We examined Vps34-GFP localization in each of the mutants and found it was strongly mislocalized to the vacuole in muk1 vps9 mutants and partially mislocalized in a vps9 mutant (Figure 6D). VPS34 is recruited by Rab5 GTPases in mammals (Christoforidis et al., 1999), and others have shown that, in yeast, constitutively active forms of Vps21, which cannot be extracted from membranes by Rab guanine dissociation inhibitors, are transported to the vacuolar membrane (Markgraf et al., 2009). Thus, overexpressed Vps21Q66L may activate Vps34 at the vacuole of muk1 vps9 strains, resulting in high levels of PI3P that drive retromer recruitment to the vacuolar membrane.
These results suggest that VPS9-domain GEFs are critical for maintaining an endosomal pool of Rab5-family GTPases that in turn recruit and activate the PI3K Vps34. The partial mislocalization of Vps34 in vps9 but not muk1 mutants suggests that the three VPS9-domain proteins do not contribute equally to this process. Nevertheless, expression of Vrl1, but not the VPS9-domain mutant Vrl1D373A, was found to rescue Vps34 localization (Figure 6D) and restore endosomal pools of PI3P in muk1 vps9 strains (Figure 6C). Taken together, these results suggest that Vps9-domain GEFs play a key role in promoting the enrichment of PI3P at endosomes.
DISCUSSION
Here we identify two VPS9-domain Rab5-family GEFs as new retromer-associated proteins and show that their activity is important for the recruitment of retromer to endosomal membranes. Although many of the retromer accessory factors that have been identified in higher cells are absent in yeast (Harbour et al., 2010; Seaman, 2012), VPS9-domain proteins constitute a broadly conserved family and may contribute to fundamental aspects of retromer assembly or function present in all eukaryotic cells.
Our coimmunoprecipitation experiments showed clear interactions between retromer and two different VPS9-domain GEFs, Muk1 and Vps9. Of interest, the interaction was mapped to the VPS9 domain of Muk1, suggesting that the binding of retromer to the VPS9 domain might regulate its GEF activity. Indeed, the human homologue of Vps9, Rabex-5, is autoinhibited by a conserved helix C-terminal to the VPS9 domain, and this is overcome by binding of the Rab5 effector Rabaptin-5 (Delprato and Lambright, 2007). Because Rabaptin-5 helps recruit Rabex-5 to endosomes, this creates a positive feedback loop that results in robust Rab5 activation (Horiuchi et al., 1997; Zhang et al., 2014). It is not known whether Muk1 is subject to autoinhibition or whether retromer binding could similarly affect GEF activity. However, the interaction with retromer is expected to increase the local concentration of the GEF, which would thus serve to enhance Rab5 activation at sites of retromer tubule formation.
Yeast retromer does not bind directly to Rab5-family GTPases (Liu et al., 2012). Instead, we found that Rab5-family GTPases and an active VPS9-domain GEF are required for correct localization of the PI3K Vps34 at endosomes and production of the endosomal pool of PI3P. Although Rab5 directly recruits the human homologue of Vps34, hVPS34, a similar relationship between yeast Rab5-family GTPases and Vps34 has not previously been reported (Christoforidis et al., 1999). Because the structural subcomplex of retromer binds PI3P through the PX domains of Vps5/17 (Burda et al., 2002), the loss of endosomal PI3P provides a likely explanation for the retromer localization defect in strains lacking active VPS9-domain GEFs.
The endosomal localization of yeast VPS9-domain GEFs does not depend on retromer, and thus a physical interaction between retromer and the GEF may not be absolutely required for endosomal PI3P production and retromer recruitment. Instead, we propose that retromer–GEF binding enhances the rate or extent of retromer assembly through a positive feedback loop like that described for Rabaptin-5 (Horiuchi et al., 1997; Figure 7A). In this model, the initial recruitment of retromer to PI3P-labeled endosomes allows it to bind and concentrate Muk1 and/or Vps9, which then recruit and activate Rab5-like GTPases in the vicinity of the forming retromer tubule. This, in turn, stimulates the recruitment and activation of PI3K, increasing local PI3P production and promoting further retromer assembly. Interactions with distinct VPS9-domain GEFs could conceivably enhance retromer assembly at different endosomal compartments or in response to different stimuli. A similar model has been proposed to explain the action of the Salmonella effector protein SopB, which causes overactivation of Rab5, promoting PI3P production that drives the formation of extensive Snx3 and Snx1-coated tubules at Salmonella-containing vacuoles (Braun et al., 2010).
FIGURE 7:
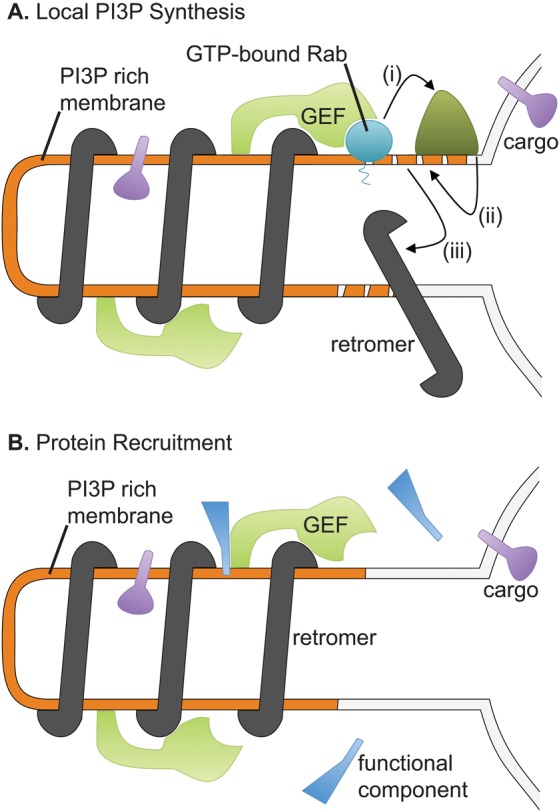
Models for the function of the interaction between retromer and Rab5-family GEFs at endosomes. (A) In the first model, the GEFs concentrate Rab5-family GTPases at the retromer tubules. The GTPases recruit the PI3K Vps34 (i), which locally increases the concentration of PI3P (ii), leading to further retromer recruitment (iii). (B) In the second model, GEFs physically recruit specific factors to the tubules.
Discovery of a new yeast VPS9-domain protein
As this article was being prepared, it was reported that the human VPS9-domain GEF hVARP interacts with retromer and that this interaction is responsible for the normal trafficking of GLUT1 from endosomes to the plasma membrane (Hesketh et al., 2014). This, together with the work presented here, raises the possibility that retromer interacts with a variety of VPS9-domain GEFs and that retromer–GEF interactions may contribute to protein trafficking in yeast and mammals. The protein Vrl1, which is present in wild strains of S. cerevisiae but mutated in common lab strains, has many similarities to hVARP yet exhibits key differences. hVARP binds the retromer subunit VPS29 through two conserved cysteine-rich motifs, and this interaction recruits hVARP to endosomal tubules. These cysteine-rich motifs are not present in Vrl1, and we found that Vrl1 was not fully dependent on retromer for localization to endosomes. Although our coimmunoprecipitation and iMYTH experiments did not identify an interaction between retromer and Vrl1, such an interaction may be undetectable by these methods due to the relatively low level of Vrl1 expression.
A key role of hVARP in the trafficking of GLUT1 is to recruit the R-SNARE VAMP7 into retromer-derived vesicles and thus enable their fusion with the plasma membrane (Hesketh et al., 2014). hVARP has a VAMP7-interacting domain (Burgo et al., 2009; Ohbayashi et al., 2012; Schäfer et al., 2012), which is conserved in Vrl1 homologues from many fungal species, although it is less well conserved in S. cerevisiae. A retromer-mediated recycling pathway from endosomes to the plasma membrane has not been reported in yeast, but the laboratory strains used in most trafficking studies lack functional Vrl1. Thus it is intriguing to speculate that Vrl1, like hVARP, repurposes an R-SNARE to regulate a yet-undiscovered recycling pathway missing in lab strains but present in other yeast species.
The role of hVARP in VAMP7 transport suggests an alternative model for the function of retromer–GEF interactions: GEFs bind retromer to recruit specific cargo proteins or accessory factors to forming retromer tubules (Figure 7B). In fact, Vps9 has a ubiquitin-binding CUE domain that is important for the normal progression of ubiquitinated cargo through endosomes (Davies et al., 2003; Donaldson et al., 2003). Although Muk1 lacks a CUE domain or other recognizable motifs, it has conserved regions that could mediate interactions with additional factors that influence the composition or targeting of endosome-derived vesicles.
It is important to note that the two models presented in Figure 7 are not mutually exclusive. The retromer–GEF interaction could enhance retromer assembly by promoting local Rab5 activation and PI3P production while at the same time recruiting specific cargo or accessory factors. By providing a means of reinforcing local recruitment, this could explain why retromer coats subdomains of endosomes (Seaman et al., 1998; Temkin et al., 2011). Moreover, the positive feedback model could address puzzling aspects of retromer biology in higher cells. Because the mammalian CSC does not bind tightly to the PI3P-binding sorting nexins, it is unclear what prevents the sorting nexins from driving the formation of empty tubules devoid of cargo (Cullen and Korswagen, 2012). If recruitment of hVARP by the VPS29 subunit of the CSC also increases local PI3P production, this could enhance coassembly with the sorting nexin subcomplex and link membrane deformation to cargo recruitment.
The VPS9 family
There are three VPS9-domain proteins in yeast and at least nine in humans (Carney et al., 2006). Although the yeast VPS9-domain proteins share some overlapping functions, our results suggest they have unique functions that are as yet uncharacterized. Some mammalian Vps9-domain GEFs can preferentially activate a subset of Rab5-family GTPases (Delprato and Lambright, 2007) or contain domains that confer distinct localizations (Balaji et al., 2012). Although in vitro activity assays suggest that yeast VPS9-domain GEFs do not have differential specificity for Rab GTPases (Singer-Krüger et al., 1994; Cabrera et al., 2013; Paulsel et al., 2013), we and others have found that they show differential recruitment to endosomes. Further studies will be needed to determine whether the yeast GEFs localize to distinct endosome subpopulations or associate with membranes only in response to specific regulatory inputs. It will also be important to determine whether the retromer binding is a general feature of VPS9-domain GEFs in humans and to what extent the interaction serves to reinforce retromer recruitment or select other cargo.
MATERIALS AND METHODS
Yeast strains and plasmids
A list of strains and plasmids used can be found in Supplemental Tables S1 and S2. With the exception of the strains to be described here, all strains from this study were made by homologous recombination as described (Longtine et al., 1998; Janke et al., 2004; Sheff and Thorn, 2004). The iMYTH strains were made via integration of the L2 Cub cassette as previously described (Snider et al., 2013). VPS34 and VPS5 were tagged with a bright GFP variant, GFP+ (Scholz et al., 2000 ), by amplifying and transforming GFP+::NAT from pLC1318, a gift from R. Rachubinsky (University of Alberta).
Plasmids were made by homologous recombination in yeast (Scholz et al., 2000), rescued in Escherichia coli, and confirmed by sequencing. To make pGFP-FYVE(EEA1)::LEU2 (pBB21), pGFP-FYVE(EEA1)::TRP1 (#36096; Addgene, Cambridge, MA) was cut with Bsu36I and cotransformed with a LEU2 PCR fragment containing flanking homology to TRP1 5′ and 3′ regions. pNubG-HA-MUK1 FL (pAO470), pNubG-HA-MUK1 1-417 (pAO538), and pNubG-HA-MUK1 74-417 (pAO542) were made by cotransforming MUK1 gene regions amplified from yeast genomic DNA together with SmaI-digested pPR3-N MYTH prey vector, as described previously (Snider et al., 2013). The Muk1 D353A mutant (pA0531) was made via site-directed mutagenesis of pNubG-HA-MUK1 FL (pAO470) construct following the QuikChange II protocol (Agilent, Santa Clara, CA). pNubG-HA-VPS9 (pBB9) and pNubG-HA-VRL1 (pBB24) were made by EcoRI-HF/ClaI digestion of pPR3-N and its cotransformation with the respective genes as previously described (Snider et al., 2013).
Other MUK1 plasmids made in this study were based on pRS416 (URA3 CEN) as follows. We amplified 972 base pairs of the 5′ untranslated region of MUK1, referred to as MUK1pr, using primers with homology to pRS416 and two-thirds of a 3HA tag. A second product containing MUK1 was generated using primers with homology to two-thirds of the 3HA tag and a downstream pRS416 sequence. The two products were cotransformed with KpnI/SacII-digested pRS416 to generate pMUK1pr-3HA-MUK1 (pMD120). pMD120 was AatII/BamHI digested and cotransformed with a GFP PCR product with ends homologous to 5′ and 3′ of the 3HA tag to make pMUK1pr-GFP-MUK1 (pMD121). The pMD121 Muk1 promoter was KpnI/BamHI digested, and the cut plasmid was cotransformed with an ADH1 promoter PCR product that had homology outside of the cut region, forming pADHpr-GFP-MUK1 (pMT1). Both pMD120 and pMT1 were cut with HindIII/NruI and cotransformed with PCR products containing MUK1-D353A with terminal homology outside the cut sites to generate pMUK1pr-3HA-MUK1-D353A (pBB12) and pADH1pr-GFP-MUK1-D353A (pBB14). pMD120 and pBB12 were cut with KpnI and transformed with an ADH1 PCR product with flanking MUK1 homology to form pADH1pr-3HA-MUK1 (pBB33) and pADH1pr-3HA-MUK1-D353A (pBB34). pADH1pr-GFP-MUK1(1-575) (pBB15) was made by cutting out base pairs 544–1800 of MUK1 from pMT1 using BglII and cotransforming the cut plasmid with a PCR product containing base pairs 1–1725 of MUK1 with flanking homology to the plasmid. pADH1pr-GFP-MUK1(503-612) (pMT2) was made by excising the N-terminus of MUK1 in pMT1 with ClaI/HindIII and cotransforming the plasmid with a hybridized oligo with homology to GFP and MUK1 after base pair 1506.
VRL1 plasmids were based on pADH1pr-GFP-MUK1 (pMT1). Genomic DNA was used as a template for VRL1, and primers were used to correct the thymine deletion in YML003W of the parental yeast strain. The DNA upstream of the deletion was amplified with the reverse oligo (5′ ATATTTATATTTTTCAGTGTCTACTTCGTGGCCTTTGAAATGTGTAGTAAGCCTAGACCA), and the region downstream was amplified with (5′ TGGTCTAGGCTTACTACACATTTCAAAGGCCACGAAGTAGACACTGAAAAATATAAATAT). The reverse oligo was used to amplify upstream YML003W with homology to either the ADH1 promoter or GFP of pMT1, and the forward oligo was used to amplify downstream YML003W and YML002W with homology to pMT1 after MUK1. The two sets of PCR products were cotransformed with HindIII-cut pMT1 to make plasmids pADH1pr-VRL1 (pBB23) and pADH1pr-GFP-VRL1 (pBB22). The GFP tag of pBB22 was cut with HpaI and cotransformed with an oligo containing 3HA and flanking homology to make pADH1pr-3HA-VRL1 (pBB25). Expression of N-terminally HA- or GFP-tagged forms of VRL1 resulted in a protein of the expected size. The VRL1 promoter was substituted for the ADH1 promoter by cutting pBB25 with SphI and cotransforming with a PCR product containing 366 base pairs upstream of YML003w and flanking homology to form pVRL1pr-3HA-VRL1 (pBB25). pBB25 was cut with MscI and BglII and cotransformed with two overlapping PCR products containing VRL1 with the D373A mutation, forming pVRL1pr-3HA-VRL1D373A (pBB32).
Growth and colony overlay assays
For the concentration-limiting growth assays, 4 μl of 1 OD600/ml log-phase yeast was spotted onto yeast extract/peptone/dextrose (YPD) medium in 10× serial dilutions and imaged using a CanoScan 4400F scanner after 2 d of growth at the indicated temperatures. For the colony overlay assay, yeast (containing pRS415 and pRS416 as required) were spotted onto synthetic amino acid dropout medium lacking histidine and uracil and overlaid with a nitrocellulose membrane. After 16 h, the membrane was blotted with mouse anti–carboxypeptidase Y (A6428; Molecular Probes, Carlsbad, CA) and then goat anti-mouse conjugated to horseradish peroxidase (HRP; 115035146; Jackson ImmunoResearch, West Grove, PA). The blot was developed with the enhanced chemiluminescent West Pico (34077; Pierce, Rockford, IL) and exposed to Amersham Hyperfilm (28906839; GE Healthcare, Little Chalfont, United Kingdom).
Western blotting and coimmunoprecipitation
For coimmunoprecipitation from batch cultures, 50-ml samples of the appropriate strains were grown in YPD medium to mid log phase, washed, and transferred to galactose medium for 1 h. Cells were harvested, resuspended in an equal volume of IPLB buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 7.4, 150 mM KOAc, 2 mM Mg(Ac)2, 1 mM ethylene glycol tetraacetic acid, 10% glycerol, protease inhibitor cocktail, and 1% Triton), and disrupted by glass beads for 5 min. Cell lysates were then cleared by centrifugation at 1500 × g for 3 min. Half of the cleared lysate was incubated with 50 μl of calmodulin beads for 2 h and washed with 10 volumes of fresh IPLB. Then10 μl of loading buffer (5% SDS, 50 mM Tris, pH 6.8, 0.4 mg/ml bromophenol blue, and 1% β-mercaptoethanol) was added to 20 μl of beads, and eluates were resolved on 12% SDS–PAGE gels. Gels were then transferred to nitrocellulose membrane using the iBlot transfer system following the manufacturer's instructions (Life Technologies, Carlsbad, CA). Membranes were probed using anti-HA (sc-805; Santa Cruz Biotechnology, Dallas, TX) or anti-TAP (#A01435; GenScript, Piscataway, NJ) rabbit primary antibodies and HRP-tagged goat anti-rabbit secondary antibody (#31462; Pierce) and visualized using a Kodak image station 4000.
Western lysates were prepared from log-phase cells by bead bashing, freezing, and resuspension in Thorner buffer (8 M urea, 5% SDS, 50 mM Tris, pH 6.8, 0.4 mg/ml bromophenol blue, and 1% β-mercaptoethanol). Lysates were heated to 70°C, and the equivalent of 0.5 OD600 of cells was loaded into 10% SDS–PAGE gels. In cross-linking coimmunoprecipitation experiments, fresh spheroplasts were prepared by digesting cell walls with Zymolyase (SK1204911; MJS BioLynx, Brockville, Canada). We cross-linked 20 OD600 of spheroplasts with 1.6 mg/ml DSP and lysed them as described (Cˇopicˇ et al., 2007), except that we used 1% n-dodecyl-β-d-maltoside in the lysis buffer instead of Triton X-100. The lysates were incubated at 4°C with rabbit anti-HA (sc-805; Santa Cruz Biotechnology) or rabbit anti-GFP (A6455; Molecular Probes), followed by protein A–Sepharose beads (17-5280-04; GE Healthcare). The beads were washed, and proteins were eluted by heating at 95°C for 5 min. Samples were loaded into 10% SDS–PAGE gels. For both Western blots and coimmunoprecipitations, proteins were transferred overnight to nitrocellulose membranes and blotted with either mouse anti-GFP (11814460001; Roche, Basel, Switzerland) or mouse anti-HA (MMS-101R; Covance, Princeton, NJ). Probing with secondary antibodies and exposure were done as in the colony overlay assay.
iMYTH
Log-phase yeast were serially diluted by a factor of 10 from 1 OD600, and 4 μl was spotted onto synthetic dextrose dropout plates lacking tryptophan, adenine, and histidine. Tryptophan selected for Nub plasmids and adenine and histidine for an interaction between the Cub bait and Nub prey constructs (Snider et al., 2013). Yeast were grown at 37°C for 4 d and then imaged using a CanoScan 4400F scanner.
Fluorescence microscopy
Log-phase yeast were imaged in minimal selective medium at room temperature with a Plan-Apochromat 100×/1.40 numerical aperture oil immersion objective lens on an Axioplan 2 fluorescence microscope (Carl Zeiss, Jena, Germany). Images were taken with a CoolSNAP camera (Roper Scientific; Tucson, AZ) using MetaMorph 7.7 software (MDS Analytical Technologies, Sunnyvale, CA) and adjusted using MetaMorph and Photoshop CS5 (Adobe, San Jose, CA). Exposure times varied from 100 ms to 3 s based on the protein tagged with GFP or RFP but were kept the same within a given experiment. When the FM4-64 (T-3166; Life Technologies) lipophilic dye was used, cells were incubated with the dye for 1 h in minimal medium, washed once, and then grown another hour in minimal medium before imaging. Cellular features were quantified by manually scoring images.
Supplementary Material
Acknowledgments
We thank Christian Ungermann for reagents and Nayomi Gomes, Amandeep Parhar, and Teresa Lee for technical assistance. This work was supported by funding from the Canadian Institutes of Health Research (MOP #64394 to E.C. and MOP #125952 and MOP #132191 to M.B.). The work in the Stagljar lab is supported by grants from the Canadian Foundation for Innovation, the Natural Sciences and Engineering Research Council of Canada, the Ontario Genomics Institute, the Canadian Cystic Fibrosis Foundation, the Canadian Cancer Society, Pancreatic Cancer Canada, and the University Health Network.
Abbreviations used:
- CPY
carboxypeptidase Y
- CSC
cargo-selective subcomplex (of retromer)
- Cub
C-terminal ubiquitin fragment
- DSP
dithiobis(succinimidyl propionate)
- ESCRT
endosomal sorting complex required for transport
- GEF
guanine nucleotide exchange factor
- iMYTH
integral membrane yeast two-hybrid
- Nub
N-terminal ubiquitin fragment
- ORF
open reading frame
- PI3K
phosphatidylinositol 3-kinase
- PI3P
phosphatidylinositol 3-phosphate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1281) on January 21, 2015.
REFERENCES
- Attar N, Cullen PJ. The retromer complex. Adv Enzyme Regul. 2010;50:216–236. doi: 10.1016/j.advenzreg.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BDM, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- Balaji K, Mooser C, Janson CM, Bliss JM, Hojjat H, Colicelli J. RIN1 orchestrates the activation of RAB5 GTPases and ABL tyrosine kinases to determine the fate of EGFR. J Cell Sci. 2012;125:5887–5896. doi: 10.1242/jcs.113688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar H, Arlt H, Ostrowicz C, Brocker C, Sundermann F, Brandt R, Babst M, Ungermann C. The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J Cell Sci. 2010;123:4085–4094. doi: 10.1242/jcs.071977. [DOI] [PubMed] [Google Scholar]
- Bonifacino J, Hurley J. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Wong A, Landekic M, Hong W, Grinstein S, Brumell J. Sorting nexin 3 (SNX3) is a component of a tubular endosomal network induced by Salmonella and involved in maturation of the Salmonella-containing vacuole. Cell Microbiol. 2010;12:1352–1367. doi: 10.1111/j.1462-5822.2010.01476.x. [DOI] [PubMed] [Google Scholar]
- Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Burgo A, Proux-Gillardeaux V, Sotirakis E, Bun P, Casano A, Verraes A, Liem RKH, Formstecher E, Coppey-Moisan M, Galli T. A molecular network for the transport of the TI-VAMP/VAMP7 vesicles from cell center to periphery. Dev Cell. 2012;23:166–180. doi: 10.1016/j.devcel.2012.04.019. [DOI] [PubMed] [Google Scholar]
- Burgo A, Sotirakis E, Simmler M-C, Verraes A, Chamot C, Simpson JC, Lanzetti L, Proux-Gillardeaux V, Galli T. Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 2009;10:1117–1124. doi: 10.1038/embor.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Arlt H, Epp N, Lachmann J, Griffith J, Perz A, Reggiori F, Ungermann C. Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J Biol Chem. 2013;288:5166–5175. doi: 10.1074/jbc.M112.431536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Cˇopicˇ A, Starr T, Schekman R. Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol Biol Cell. 2007;18:1803–1815. doi: 10.1091/mbc.E06-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozier GE, Carlton J, McGregor AH, Gleeson Pa, Teasdale RD, Mellor H, Cullen PJ. The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J Biol Chem. 2002;277:48730–48736. doi: 10.1074/jbc.M206986200. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2012;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BA, Topp JD, Sfeir AJ, Katzmann DJ, Carney DS, Tall GG, Friedberg AS, Deng L, Chen Z, Horazdovsky BF. Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. J Biol Chem. 2003;278:19826–19833. doi: 10.1074/jbc.M301059200. [DOI] [PubMed] [Google Scholar]
- Delprato A, Lambright D. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat Struct Mol Biol. 2007;14:406–412. doi: 10.1038/nsmb1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Yin H, Gekakis N, Supek F, Joazeiro CaP. Ubiquitin signals protein trafficking via interaction with a novel ubiquitin binding domain in the membrane fusion regulator, Vps9p. Curr Biol. 2003;13:258–262. doi: 10.1016/s0960-9822(03)00043-5. [DOI] [PubMed] [Google Scholar]
- Fjorback AW, Seaman M, Gustafsen C, Mehmedbasic A, Gokool S, Wu C, Militz D, Schmidt V, Madsen P, Nyengaard JR, et al. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci. 2012;32:1467–1480. doi: 10.1523/JNEUROSCI.2272-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froissard M, Belgareh-Touzé N, Dias M, Buisson N, Camadro J-M, Haguenauer-Tsapis R, Lesuissse E. Trafficking of siderophore transporters in Saccharomyces cerevisiae and intracellular fate of ferrioxamine B conjugates. Traffic. 2007;8:1601–1616. doi: 10.1111/j.1600-0854.2007.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Harbour M, Breusegem S, Antrobus R, Freeman C, Reid E, Seaman M. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J Cell Sci. 2010;123:3703–3717. doi: 10.1242/jcs.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MS, Hung C-S, Liu T, Christiano R, Walther TC, Burd CG. A mechanism for retromer endosomal coat complex assembly with cargo. Proc Natl Acad Sci USA. 2014;111:267–272. doi: 10.1073/pnas.1316482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M, Port F, Lorenowicz MJ, Mcgough IJ, Silhankova M, Betist MC, van Weering JRT, van Heesbeen RGHP, Middelkoop TC, Basler K, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh GG, Pérez-Dorado I, Jackson LP, Wartosch L, Schäfer IB, Gray SR, McCoy AJ, Zeldin OB, Garman EF, Harbour ME, et al. VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev Cell. 2014;29:1–16. doi: 10.1016/j.devcel.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera M, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Liu TT, Gomez TS, Sackey BK, Billadeau DD, Burd CG. Rab GTPase regulation of retromer-mediated cargo export during endosome maturation. Mol Biol Cell. 2012;23:2505–2515. doi: 10.1091/mbc.E11-11-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine S, Pringle M, RJ Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lynch-Day M, Bhandari D, Menon S, Huang J, Cai H, Bartholomew C, Brumell J, Ferro-Novick S, Klionsky D. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markgraf DF, Ahnert F, Arlt H, Mari M, Peplowska K, Epp N, Griffith J, Reggiori F, Ungermann C. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol Biol Cell. 2009;20:5276–5289. doi: 10.1091/mbc.E09-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I, Cullen P. Recent advances in retromer biology. Traffic. 2011;12:963–971. doi: 10.1111/j.1600-0854.2011.01201.x. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Russell MRG, Lo S-Y, Chapin HC, Milnes J, Merz AJ. Msb3/Gyp3 GAP selectively opposes Rab5 signaling at endolysosomal organelles. Traffic. 2012;13:1411–1428. doi: 10.1111/j.1600-0854.2012.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bruinsma P, Strawn LA. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol Biol Cell. 1999;10:875–890. doi: 10.1091/mbc.10.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi N, Yatsu A, Tamura K, Fukuda M. The Rab21-GEF activity of Varp, but not its Rab32/38 effector function, is required for dendrite formation in melanocytes. Mol Biol Cell. 2012;23:669–678. doi: 10.1091/mbc.E11-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsel AL, Merz AJ, Nickerson DP. Vps9 family protein Muk1 is the second Rab5 guanosine nucleotide exchange factor in budding yeast. J Biol Chem. 2013;288:18162–18171. doi: 10.1074/jbc.M113.457069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi CM, Menendez J, Arnoldo A, Engels K, Iyer KR, Thaminy S, Georgiev O, Barral Y, Michaelis S, Stagljar I. Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol Cell. 2007;26:15–25. doi: 10.1016/j.molcel.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJR, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer IB, Hesketh GG, Bright Na, Gray SR, Pryor PR, Evans PR, Luzio JP, Owen DJ. The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation. Nat Struct Mol Biol. 2012;19:1300–1309. doi: 10.1038/nsmb.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz O, Thiel A, Hillen W, Niederweis M. Quantitative analysis of gene expression with an improved green fluorescent protein. p6. Eur J Biochem. 2000;267:1565–1570. doi: 10.1046/j.1432-1327.2000.01170.x. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ. Recycle your receptors with retromer. Trends Cell Biol. 2005;15:68–75. doi: 10.1016/j.tcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Seaman MN. The retromer complex—endosomal protein recycling and beyond. J Cell Sci. 2012;125:4693–4702. doi: 10.1242/jcs.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122:2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M, Thorn K. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Singer-Krüger B, Stenmark H, Düsterhöft a, Philippsen P, Yoo JS, Gallwitz D, Zerial M. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J, Hanif A, Lee ME, Jin K, Yu AR, Graham C, Chuk M, Damjanovic D, Wierzbicka M, Tang P, et al. Mapping the functional yeast ABC transporter interactome. Nat Chem Biol. 2013;9:565–572. doi: 10.1038/nchembio.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Yanatori I, Kawai Y, Kishi F. Retromer-mediated direct sorting is required for proper endosomal recycling of the mammalian iron transporter DMT1. J Cell Sci. 2010;123:756–766. doi: 10.1242/jcs.060574. [DOI] [PubMed] [Google Scholar]
- Tamura K, Ohbayashi N, Ishibashi K, Fukuda M. Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes. J Biol Chem. 2011;286:7507–7521. doi: 10.1074/jbc.M110.191205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P, Lauffer B, Jäger S, Cimermancic P, Krogan NJ, Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13:715–721. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Weering JRT, Verkade P, Cullen PJ. SNX–BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic. 2012;13:94–107. doi: 10.1111/j.1600-0854.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- Vilariño-Güell C, Wider C, Ross O, Dachsel J, Kachergus J, Lincoln S, Soto-Ortolaza A, Cobb S, Wilhoite G, Bacon J, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer T, Attar N, Harterink M, van Weering JRT, Traer CJ, Oakley J, Goud B, Stephens DJ, Verkade P, Korswagen HC, et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell. 2009;17:110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Seet LF, Hanson B, Hong W. The Phox homology (PX) domain, a new player in phosphoinositide signalling. Biochem J. 2001;360:513–530. doi: 10.1042/0264-6021:3600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Lemmon Ma. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- Zavodszky E, Seaman MNJ, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC. Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. Nat Commun. 2014;5:3828. doi: 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, He X, Fu X-Y, Chang Z. Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J Cell Sci. 2006;119:1053–1062. doi: 10.1242/jcs.02810. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang T, Wang S, Gong Z, Tang C, Chen J, Ding J. Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5. Elife. 2014:e02687. doi: 10.7554/eLife.02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.