STIMs (STIM1 and STIM2 in mammals) are transmembrane proteins that reside in the endoplasmic reticulum and regulate store-operated Ca2+ entry. STIM2 mediates cAMP/PKA-dependent phosphorylation of the AMPA receptor subunit GluA1 in excitatory neurons. In addition, STIM2 promotes cAMP-dependent surface delivery of GluA1.
Abstract
STIMs (STIM1 and STIM2 in mammals) are transmembrane proteins that reside in the endoplasmic reticulum (ER) and regulate store-operated Ca2+ entry (SOCE). The function of STIMs in the brain is only beginning to be explored, and the relevance of SOCE in nerve cells is being debated. Here we identify STIM2 as a central organizer of excitatory synapses. STIM2, but not its paralogue STIM1, influences the formation of dendritic spines and shapes basal synaptic transmission in excitatory neurons. We further demonstrate that STIM2 is essential for cAMP/PKA-dependent phosphorylation of the AMPA receptor (AMPAR) subunit GluA1. cAMP triggers rapid migration of STIM2 to ER–plasma membrane (PM) contact sites, enhances recruitment of GluA1 to these ER-PM junctions, and promotes localization of STIM2 in dendritic spines. Both biochemical and imaging data suggest that STIM2 regulates GluA1 phosphorylation by coupling PKA to the AMPAR in a SOCE-independent manner. Consistent with a central role of STIM2 in regulating AMPAR phosphorylation, STIM2 promotes cAMP-dependent surface delivery of GluA1 through combined effects on exocytosis and endocytosis. Collectively our results point to a unique mechanism of synaptic plasticity driven by dynamic assembly of a STIM2 signaling complex at ER-PM contact sites.
INTRODUCTION
The endoplasmic reticulum (ER) regulates structural and functional changes in neural circuits in both the developing and adult nervous systems (Mattson et al., 2000; Bardo et al., 2006). The ER is a continuous dynamic network of tubular membranes that connect the soma to the neuron's dendritic and axonal arbors and protrudes into large dendritic spines (Bourne and Harris, 2012). The organization of the ER is thus particularly suited to process synaptic inputs locally and integrate information over long distances. The ability of the ER to release Ca2+ in response to synaptic or other signaling inputs is an important mechanism by which this organelle fine-tunes synaptic Ca2+ transients and mediates synaptic plasticity (Garaschuk et al., 1997; Finch and Augustine, 1998; Lauri et al., 2003). Abnormal ER Ca2+ and protein homeostasis has been implicated in several major neurodegenerative disorders, including Alzheimer's and Parkinson's diseases (Mattson et al., 2000). Despite evidence linking the ER to normal and pathological synaptic functions, surprisingly little is known about how this organelle communicates with synapses.
Ultrastructural studies in the late 1950s first reported the existence of contact sites between the ER and the plasma membrane (PM) in muscles cells (Porter and Palade, 1957) and neurons (Rosenbluth, 1962). Stable ER-PM junctions have since been observed in most eukaryotic cells and consist of closely apposed membranes separated by a thin (10–30 nm) intermembrane space. The composition and function of neuronal ER-PM junctions are largely unknown, but ER-PM contact sites in nonexcitable cells have received considerable attention since the discovery of the stromal interaction molecule (STIM) proteins (Liou et al., 2005; Roos et al., 2005). STIMs (STIM1 and STIM2 in mammals) are single-pass transmembrane proteins that reside in the ER and sense changes in luminal ER Ca2+ concentration. After store depletion, STIMs oligomerize and migrate to ER-PM junctions, where they bind to and activate Orai1 channels at the plasma membrane, thereby allowing Ca2+ to flow into the cytoplasm (Feske et al., 2006; Vig et al., 2006; Zhang et al., 2006). The Ca2+ sensors STIMs and Orai1 are the main molecular components of store-operated Ca2+ entry (SOCE), also referred to as capacitative Ca2+ entry (Putney, 1986). Of importance, recent studies show that, in addition to Orai, STIM1 also modulates the activity of voltage-gated calcium channels (VGCCs; Park et al., 2010), adenylate cyclase (AC) (Lefkimmiatis et al., 2009), and the Ca2+ pump PMCA (Krapivinsky et al., 2011) and can operate as a sensor of heat (Xiao et al., 2011) and oxidative stress (Hawkins et al., 2010), suggesting that STIM proteins may affect multiple signal transduction pathways.
Whereas the role of STIM1 in regulating SOCE is undisputed, less is known about the function of STIM2. STIM2 has a lower affinity for Ca2+ than STIM1 and thus migrates to ER-PM contact sites in response to relatively small decreases in ER Ca2+ concentration (Brandman et al., 2007). However, coupling of STIM2 to Orai1 is weak, and STIM2 is a poor activator of Orai1 compared with STIM1 (Bird et al., 2009; Wang et al., 2014). This led to the idea that STIM2 is a homeostatic regulator of basal Ca2+ levels (Brandman et al., 2007). Whereas in most tissues, STIM1 is expressed at higher levels than STIM2, relative expression of these two isoforms is inverted in most parts of the nervous system (Skibinska-Kijek et al., 2009), suggesting brain-specific functions of STIM2.
The relevance of SOCE in the nervous system is being debated. In contrast to nonexcitable cells, for which SOCE serves as the predominant Ca2+ entry pathway, neurons display on their surface abundant ligand- and voltage-gated Ca2+ channels with much higher Ca2+ conductance than SOC channels. Indeed, store depletion leads to little if any Ca2+ entry in neurons compared with Ca2+ influx through VGCCs (Park et al., 2010). The controversial nature of SOCE in neurons is further fueled by a series of contradictory reports on the magnitude and properties of SOCE in these cells (Bouron et al., 2005; Berna-Erro et al., 2009; Park et al., 2010; Gruszczynska-Biegala et al., 2011; Lalonde et al., 2014; Sun et al., 2014) and by the fact that SOC channels have not been molecularly defined in nerve cells. Nevertheless, neuronal SOCE has been implicated in hypoxic neuronal cell death (Berna-Erro et al., 2009) and recently in dendritic spine maturation (Sun et al., 2014) and degradation of the transcription factor Sp4 (Lalonde et al., 2014).
The STIM1 and STIM2 genes are evolutionarily conserved and probably descend from duplication of an ancestral STIM gene ∼500 million yr ago (Collins and Meyer, 2011). Of interest, this gene duplication coincides with the emergence of dendritic spines and the explosion of brain complexity in higher vertebrates (Garcia-Lopez et al., 2010). Taken together, these data suggest the possibility that STIM2 regulates neuronal and synaptic functions through mechanisms other than SOCE.
Here we show that STIM2 regulates the formation and remodeling of dendritic spines in excitatory neurons. STIM2 signals through a cAMP/protein kinase A (PKA) pathway, rather than through SOCE, to promote phosphorylation and surface delivery of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors (AMPARs). Our data reveal an unsuspected connection between the ER and excitatory synapses and suggest a critical role of STIM2 signaling in AMPAR dynamics and synaptic plasticity.
RESULTS
STIM2 localizes to large dendritic spines and is enriched in the postsynaptic density
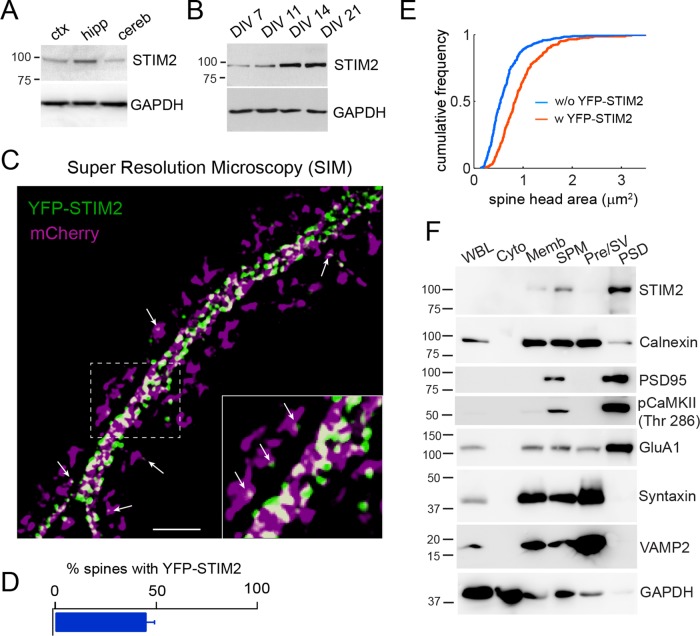
STIM2 is highly expressed in the hippocampus (Figure 1A; Skibinska-Kijek et al., 2009), and its expression in dissociated rat hippocampal neurons progressively increases until day in vitro (DIV) 14 (Figure 1B), around the time of synaptogenesis. Superresolution microscopy of mature hippocampal neurons showed that yellow fluorescent protein (YFP)–STIM2 localizes to patches in the dendritic shaft that appear connected by a network of ER-like tubules (Figure 1C). YFP-STIM2 puncta are also detected in ∼40% of dendritic spines (Figure 1D), in good agreement with a recent study (Sun et al., 2014) and the reported presence of the ER in ∼50% of spines in CA1 hippocampal neurons (Spacek and Harris, 1997). Dendritic spines receive most excitatory inputs in the brain (Yuste, 2011), and their size correlates with synaptic strength (Matsuzaki et al., 2004). We found that YFP-STIM2 is preferentially associated with large spine heads (Figure 1E), suggesting that STIM2 may regulate spine morphogenesis. To determine whether STIM2 associates with the postsynaptic density (PSD), a protein-dense specialization located at the tip of dendritic spines, we isolated synaptosomes (SPMs) from adult rat brains and separated presynaptic membranes/synaptic vesicles (Pre/SVs) from PSDs by fractionation on a sucrose gradient and detergent extraction. This procedure led to a clean separation of presynaptic and postsynaptic markers (Figure 1F) and showed clear enrichment of STIM2 in the PSD fraction, with no STIM2 detected in the Pre/SV fraction. These observations thus indicate that STIM2 is associated with the postsynaptic compartment in both hippocampal neuron cultures and adult brains.
FIGURE 1:
STIM2 localizes to large dendritic spines. (A) Immunoblots of STIM2 from adult rat cortex (ctx), hippocampus (hipp), and cerebellum (cereb). (B) Developmental expression pattern of STIM2 in dissociated hippocampal neurons. (C) Superresolution microscopy (SIM) of hippocampal neurons (DIV 21) cotransfected with YFP-STIM2 (green) and mCherry (magenta). Scale bar, 5 μm. Arrows point to YFP-STIM2 puncta inside spine heads. Note that spine necks connecting spine heads to the dendritic shaft are not always visible on these high-resolution images. (D) Percentage of spines containing at least one STIM2 punctum. n = 711 spines from two independent experiments. (E) Cumulative distribution of spine size (area) with (n = 282 spines) or without (n = 415 spines) YFP-STIM2. (F) Fractionation and immunoblot analysis of adult rat brains. Equal amounts of protein were loaded in each lane. Cyto, cytosol; Memb, total membranes; Pre/SV, presynaptic membranes and synaptic vesicles; PSD, postsynaptic density; SPM, synaptosomes; WBL, whole-brain lysate.
STIM2 regulates dendritic spine morphogenesis
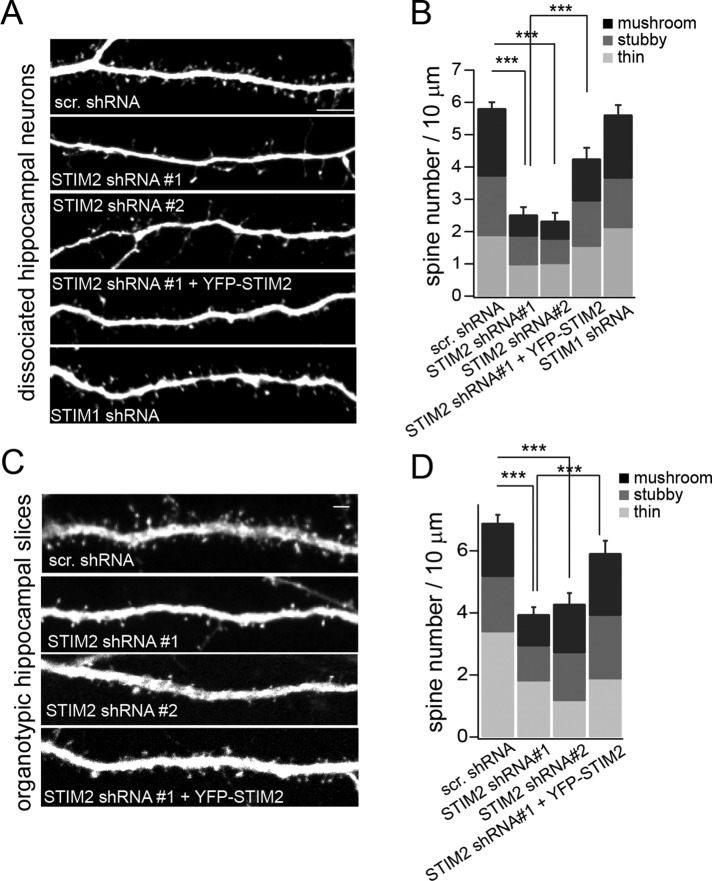
The presence of STIM2 in dendritic spines prompted us to examine whether STIM2 regulates spinogenesis. Silencing of STIM2 in dissociated hippocampal neurons (DIV 21–23) using two independent short hairpin RNA (shRNA) sequences (see Figure 4 later in this article for validation of these STIM2-targeting shRNAs) led to a decrease in dendritic spine density. This spine phenotype was partially rescued by introducing the human (RNA interference resistant) YFP-STIM2 variant (Figure 2, A and B). Although the fraction of mature (mushroom) spines tends to be smaller in STIM2-silenced neurons, there was no significant difference in the distribution of spine type (Figure 2, A and B). STIM1 silencing, on the other hand, had no detectable effect on spine density or shape (Figure 2, A and B; see Supplemental Figure S1 for validation of the STIM1 shRNA).
FIGURE 4:
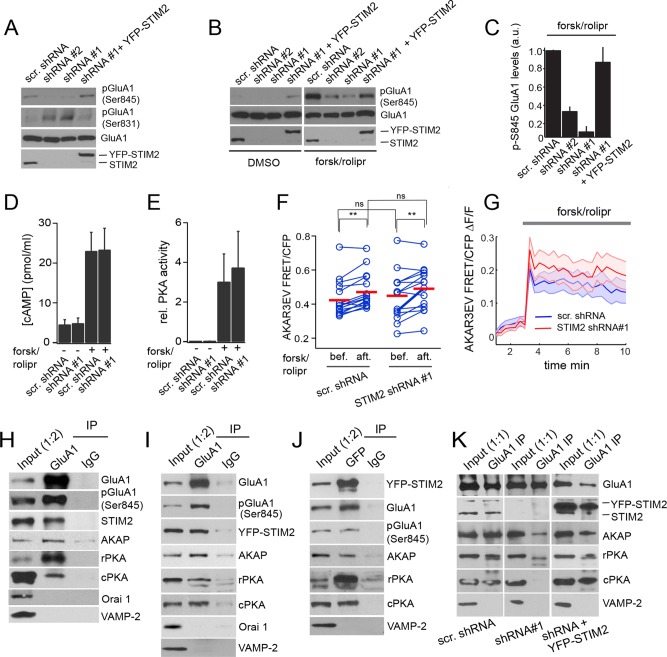
Reciprocal regulation of GluA1 Ser-831 and Ser-845 phosphorylation by STIM2. (A–C) Immunoblot analysis of hippocampal neurons (DIV 21) transduced with the indicated shRNAs and YFP-STIM2 for rescue experiments. (A) Decreased pSer-845 and increased pSer-831 in neurons expressing STIM2 shRNA#1 and #2. Both phosphorylation phenotypes are rescued by coexpression of YFP-STIM2. (B) Immunoblot analysis of GluA1 pSer-845 in cells treated with DMSO (vehicle) or 50 μM forskolin/0.1 μM rolipram for 30 min (same blot exposure for all conditions). (C) Quantification of GluA1 phospho–Ser-845 signals by densitometry after forsk/rolipr treatment (n = 4). (D, E) cAMP levels and PKA activity measured in DMSO or forsk/rolipr-treated neurons (DIV 20–21) transduced with the indicated shRNAs (n = 5 for each condition). (F, G) AKAR3EV FRET measurements in neurons (DIV 21) electroporated with the indicated shRNA constructs. (F) Pairwise analysis of AKAR3EV FRET in individual neurons before and 3 min after forsk/rolipr addition in control (n = 17) and STIM2-silenced cells (n = 15). Red bars indicate the mean. Cells were analyzed from three independent experiments. **p < 0.01, paired t tests. ns, nonsignificant (p > 0.05). (G) Fold change in AKAR3EV FRET induced by forsk/rolipr. The shaded areas represent SEM. The average fold increase in FRET in control and STIM2-silenced cells is not statistically different, p > 0.05, t test. (H–K) Co-IPs from adult rat brains (H) or DIV 21 hippocampal neurons (I–K) transduced with YFP-STIM2 (I, J) and the indicated shRNAs (K). (H, I, K) IPs with a GluA1 Ab or a control immunoglobulin G (IgG). (J) IP using anti-GFP Ab or a control IgG. Fractions were immunoblotted with the indicated Abs. The ratio indicated in the input lane reflects the fraction of input loaded relative to the IP fraction. See also Supplemental Figures S3 and S4 and Supplemental Movies S1 and S2.
FIGURE 2:
STIM2 regulates spinogenesis. Spine analysis in dissociated hippocampal neurons (A, B) or hippocampal organotypic slices (C, D). (A) Confocal images of hippocampal neurons (DIV 21) coexpressing mCherry and the indicated shRNAs. For rescue experiments, STIM2 shRNA#1 was coexpressed with YFP-STIM2. (B) Quantification of spine density and spine type for conditions shown in A, using NeuronStudio software. At least 50 dendritic segments comprising >850 spines from three independent experiments were scored for each condition. (C) CA1 neurons were biolistically transfected with the indicated shRNAs or the STIM2 shRNA#1 together with YFP-STIM2 for rescue experiments. Spines were imaged in distal apical primary and secondary dendrites (see also Supplemental Figure S2). (D) Quantification of spine size and type. At least 45 dendritic segments comprising >1000 spines in three independent experiments were analyzed for each condition. STIM2 silencing decreased spine density in both dissociated cultures and slices. ***p < 0.001, ANOVA. The percentages of thin, stubby, and mushroom spines were not significantly affected by STIM2 shRNA#1 or shRNA#2; p > 0.05, ANOVA. Scale bar, 5 μm. See also Supplemental Figures S1 and S2.
To determine whether STIM2 regulates spinogenesis in hippocampal neural circuits, we biolistically transfected organotypic slice cultures with shRNA constructs (Figure 2, C and D, and Supplemental Figure S2, A and B). Expression of two independent STIM2 shRNAs resulted in a reduction in spine density in both basal (unpublished data) and apical dendrites in mature (DIV 14–16) CA1 pyramidal neurons (Figure 2, C and D). This decrease in spine density could be partially rescued by expression of YFP-STIM2. STIM2 silencing had no significant effect on the distribution of spine type (Figure 2D) but resulted in a small decrease in apical dendrite complexity (Figure S2C). Together these results indicate that STIM2 influences the formation and/or maintenance of dendritic spines and fine-tunes the development of dendritic arbors.
STIM2 shapes spontaneous synaptic transmission
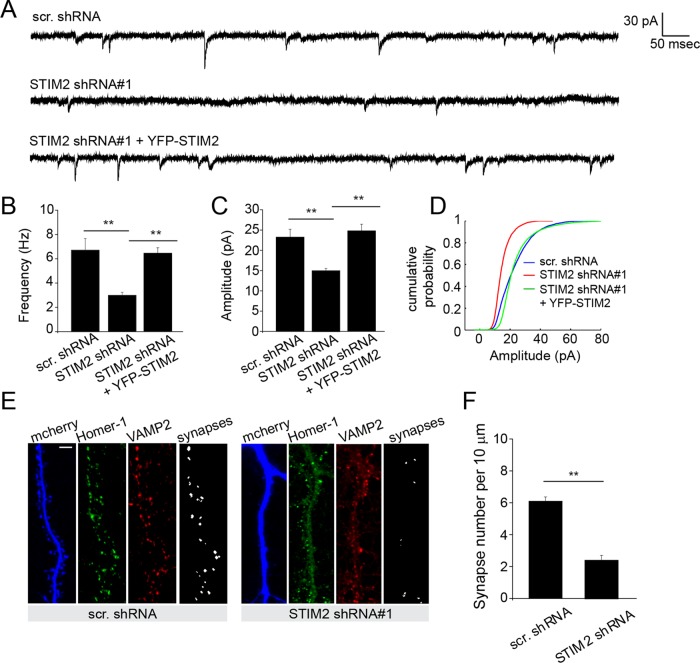
The spine phenotype observed in STIM2-silenced neurons led us to investigate whether STIM2 regulates synaptic transmission. Whole-cell patch-clamp recordings showed a clear reduction in both the frequency and amplitude of miniature excitatory postsynaptic currents (mEPSCs) in STIM2-silenced hippocampal neurons. These defects were again corrected by the introduction of YFP-STIM2 (Figure 3, A–D). In these sparsely transfected neuron cultures, presynaptic inputs originate primarily from nontransfected cells, suggesting that reduced mEPSC frequency likely results from a postsynaptic defect. To test whether a reduction in synapse density underlies this decrease in mEPSC frequency, we measured synapse density by scoring puncta costained by both presynaptic (vesicle-associated membrane protein 2 [VAMP2]) and postsynaptic (Homer-1) markers. STIM2-silenced neurons display fewer synapses than control cells (Figure 3, E and F), in accordance with the observed decrease in spine density. Reduced mEPSC amplitude, on the other hand, suggests a defect in AMPAR synaptic localization and/or channel properties.
FIGURE 3:
STIM2 shapes functional synaptic inputs. (A–D) whole-cell patch-clamp recordings of hippocampal neurons (DIV 15–18) sparsely transfected with the indicated shRNA constructs. (A) Individual mEPSC traces of cells expressing the indicated shRNAs. STIM2 shRNA#1 was coexpressed with YFP-STIM2 for rescue experiments. (B–D) Quantification of mEPSC frequency and amplitude (Scr, 23.2 ± 2 pA, 6.7 ± 1.0 Hz, n = 11; STIM2 shRNA#1, 15.0 ± 0.6 pA, 2.9 ± 0.3 Hz, n = 9; STIM2 shRNA#1 + YFP-STIM2, 24.7 ± 1.6 pA, 6.4 ± 0.5 Hz, n = 11; **p < 0.01, ANOVA). (D) Cumulative distribution of mEPSC amplitude. (E) Synaptic density measured in control and STIM2-silenced hippocampal neurons (DIV 21) by costaining with presynaptic (VAMP2) and postsynaptic (Homer1) markers. Scale bar, 5 μm. Overlapping VAMP2/Homer1 puncta are detected and scored (F) by a MATLAB script (see Materials and Methods). Synaptic density per 10 μm: scr. shRNA, 6.1 ± 0.27, n = 15; STIM2 shRNA #1, 2.4 ± 0.3, n = 17; p < 0.01, t test.
STIM2 reciprocally regulates phosphorylation of GluA1 on Ser-831 and Ser-845
AMPARs are the main mediators of excitatory neurotransmission in the brain (Huganir and Nicoll, 2013). They consist of four subunits (GluA1–4), which assemble in different combinations to form tetrameric channels. GluA1 has an unusually long C-terminal cytoplasmic tail, the phosphorylation of which regulates activity-dependent synaptic delivery and channel properties of the AMPAR (Derkach et al., 1999; Banke et al., 2000; Esteban et al., 2003). Two phosphorylation sites in the GluA1 cytoplasmic tail, Ser-831 and Ser-845, have been particularly well characterized. Phosphorylation of Ser-831 by Ca2+/calmodulin-dependent protein kinase II (CaMKII) and PKC regulates single-channel conductance of the AMPAR (Derkach et al., 1999), whereas cAMP/PKA-dependent phosphorylation of Ser-845 increases channel open probability (Banke et al., 2000) and promotes surface expression of AMPARs (Oh et al., 2006). Both pSer-845 and pSer-831 residues have been implicated in long-term potentiation (LTP) and long-term depression in the hippocampus, two forms of synaptic plasticity that have been associated with learning and memory (Esteban et al., 2003; Lee et al., 2003, 2010; Makino et al., 2011).
We thus probed the effect of STIM2 on GluA1 phosphorylation. Lentiviral transduction of hippocampal neurons with two different STIM2 shRNAs led to a marked decrease in the steady-state phosphorylation of GluA1 on Ser-845. Conversely, STIM2 silencing resulted in an increase in pSer-831 (Figure 4A). Both the Ser-845 and Ser-831 phosphorylation phenotypes were efficiently rescued by coexpression of YFP-STIM2 at a level comparable to endogenous STIM2 (Figure 4A). Expression levels of GluA1 (Figure 4A), PKA (Figure 4K), CaMKII, GluA2, or the N-methyl-d-aspartate (NMDA) receptor subunit GluN1 (Supplemental Figure S3) were not altered by STIM2 knockdown. Thus STIM2 reciprocally regulates GluA1 phosphorylation on Ser-845 and Ser-831.
Because STIM2 silencing results in a loss of function in Ser-845 phosphorylation and PKA-mediated phosphorylation of Ser-845 is required for synaptic recruitment of GluA1 (Esteban et al., 2003), we focused on the mechanisms underlying STIM2-dependent phosphorylation of GluA1 on Ser-845. PKA activity depends on cAMP-mediated dissociation of the regulatory subunit (rPKA) from the catalytic subunit (cPKA; Skroblin et al., 2010). We tested whether STIM2 also regulates GluA1 Ser-845 phosphorylation in response to acute PKA activation by treatment with forskolin (an AC agonist) and rolipram (a phosphodiesterase inhibitor). Forskolin/rolipram (forsk/rolipr) induced robust GluA1 Ser-845 phosphorylation (Figure 4, B and C), which was strongly inhibited (up to 80%) by two shRNAs against STIM2 and rescued by cotransduction of these cells with YFP-STIM2 (Figure 4, B and C) but not YFP-STIM1 (Supplemental Figure S4). The requirement of STIM2 in forsk/rolipr-induced GluA1 phosphorylation suggests that STIM2 operates downstream of AC. Indeed, STIM2 silencing had no detectable effect on cAMP levels at steady state or after forsk/rolipr treatment (Figure 4D).
We next evaluated the impact of STIM2 on bulk PKA activity by enzyme-linked immunosorbent assay (ELISA) and found no significant changes between control and STIM2-silenced neurons in resting cells or after forsk/rolipr treatment (Figure 4E). We then turned to fluorescence resonance energy transfer (FRET) imaging as an alternative approach to measure PKA activity. We used an optimized FRET-based sensor of PKA activity, AKAR3EV (Komatsu et al., 2011), which we introduced in hippocampal neurons together with scramble or STIM2 shRNA–expressing vectors. Forsk/rolipr induced a similar increase in AKAR3EV FRET signals in control and STIM2-silenced cells (Figure 4, F and G, and Supplemental Movies S1 and S2). Analysis of time series showed that the kinetics of PKA activation was unaltered by STIM2 knockdown and that PKA activity reached slightly higher levels in STIM2-silenced cells, although this difference did not reach statistical significance (Figure 4G). Together these results show that STIM2 is essential for GluA1 Ser-845 phosphorylation and appears to function downstream of cAMP/PKA.
STIM2 couples GluA1 to PKA
PKA-dependent phosphorylation of GluA1 depends on the PKA scaffold AKAP150 (also called AKAP5 or AKAP79 in humans), which positions PKA in close proximity to its synaptic targets (Colledge et al., 2000; Hoshi et al., 2005). We thus asked whether STIM2 regulates the interaction of GluA1 with AKAP/PKA, by performing a series of coimmunoprecipitation (co-IP) experiments. IP of GluA1 from adult rat brain lysates efficiently pulled down endogenous STIM2 (Figure 4H). AKAP150 and both rPKA and cPKA were also coimmunopurified. Similarly, YFP-STIM2, AKAP, rPKA, and cPKA were copurified in GluA1 IPs prepared from hippocampal neurons transduced with YFP-STIM2 (Figure 4I). In the converse IP experiment, YFP-STIM2 efficiently pulled down GluA1, AKAP, and the two PKA subunits (Figure 4J). Of interest, however, Orai1 was not pulled down in GluA1 IPs derived from adult brain (Figure 4H) or YFP-STIM2–expressing neurons in culture (Figure 4I), suggesting that this STIM2/GluA1-containing protein complex differs from the classical STIM/SOCE machinery.
To determine whether GluA1 binding to AKAP/PKA is STIM2 dependent, we immunoprecipitated GluA1 from neurons transduced with scramble or STIM2 shRNAs. STIM2 silencing strongly reduced co-IP of AKAP and rPKA and completely disrupted binding of cPKA to GluA1 (Figure 4K). Interaction of both PKA subunits and AKAP150 with GluA1 was restored, however, by expression of YFP-STIM2 (Figure 4K). Loss of interaction between GluA1 and AKAP/PKA in STIM2-silenced cells does not appear to result from global disruption of synaptic organization, because interaction of the NMDA receptor with PSD95 is preserved (Supplemental Figure S5). Together these results indicate that STIM2 forms a complex with the AMPAR and AKAP/PKA and mediates GluA1 Ser-845 phosphorylation by coupling GluA1 to PKA.
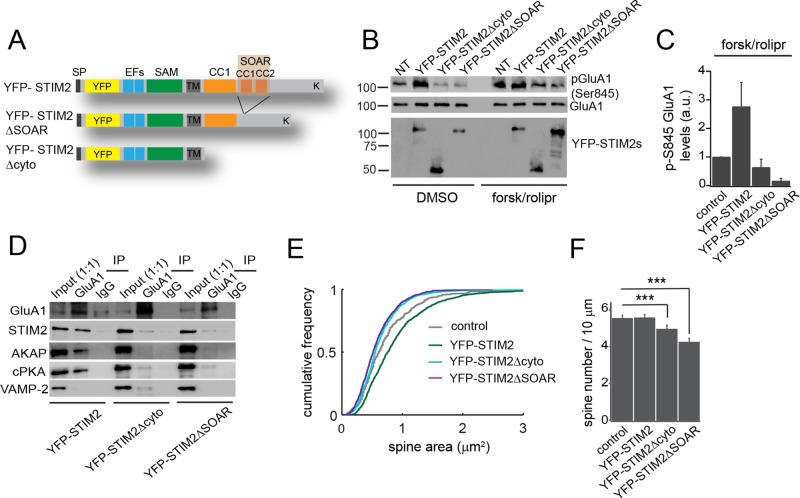
STIM2 mediates PKA-dependent phosphorylation of GluA1 through its SOAR domain
To identify the domain of STIM2 involved in GluA1 phosphorylation, we made two STIM2 mutants lacking the entire cytoplasmic moiety (YFP-STIM2Δcyto) or the SOAR (STIM-Orai-activating-region) domain (YFP-STIM2ΔSOAR), which was previously implicated in SOCE activation (Wang et al., 2014) and Cav1.2 inhibition (Park et al., 2010; Wang et al., 2010; Figure 5A). Both YFP-STIM2Δcyto and YFP-STIM2ΔSOAR retain their ER localization (Supplemental Figure S6A). These STIM2 mutants were introduced in hippocampal neurons by viral delivery alongside YFP-STIM2, and tested for their ability to drive GluA1 Ser-845 phosphorylation. Overexpression of YFP-STIM2 led to an increase in Ser-845 phosphorylation compared with nontransfected cells. In marked contrast, YFP-STIM2Δcyto and YFP-STIM2ΔSOAR failed to do so, indicating involvement of the SOAR domain (Figure 5, B and C). In fact, these two mutants appear to suppress GluA1 Ser-845 phosphorylation in resting cells or after forsk/rolipr treatment (Figure 5, B and C), suggesting that they act in a dominant-negative manner. Moreover, YFP-STIM2Δcyto and YFP-STIM2ΔSOAR disrupted the interaction of GluA1 with endogenous STIM2, AKAP, and cPKA (Figure 5D). Finally, expression of these mutants led to a reduction of spine head size and spine density, whereas YFP-STIM2 augmented dendritic spine size compared with control cells (Figure 5, E and F, and Supplemental Figure S6B). Thus the SOAR domain mediates STIM2-dependent phosphorylation of GluA1 and spine morphogenesis.
FIGURE 5:
The STIM2 SOAR domain mediates phosphorylation of and interaction with GluA1. (A) Cartoon showing the primary sequence of YFP-STIM2 WT, ΔSOAR, and Δcyto. (B–D) Cortical neurons (DIV 21) transduced with YFP-STIM2 WT and mutants. (B) Immunoblot analysis of GluA1 pSer-845 in cells expressing the indicated constructs and treated with DMSO (vehicle) or forsk/rolipr for 30 min. NT, nontransduced cells. (C) Densitometry analysis of pSer-845 after forsk/rolipr treatment from four independent experiments. (D) IPs from cells overexpressing YFP-STIM2 WT, Δcyto, or ΔSOAR with GluA1 Ab or control IgG. Lysates are the same used in B under DMSO condition. Note the marked decrease in endogenous STIM2, AKAP, and cPKA pulled down from cells expressing the STIM2 mutants. (E, F) Distribution of spine size (area) (E) and spine density (F) scored in neurons transduced with the indicated constructs or mock transduced. More than 400 spines from three independent experiments were analyzed for each condition. ***p < 0.001, ANOVA. See also Supplemental Figure S5.
cAMP triggers translocation of STIM2 to ER-PM junctions and promotes assembly of a STIM2/PKA/GluA1 complex
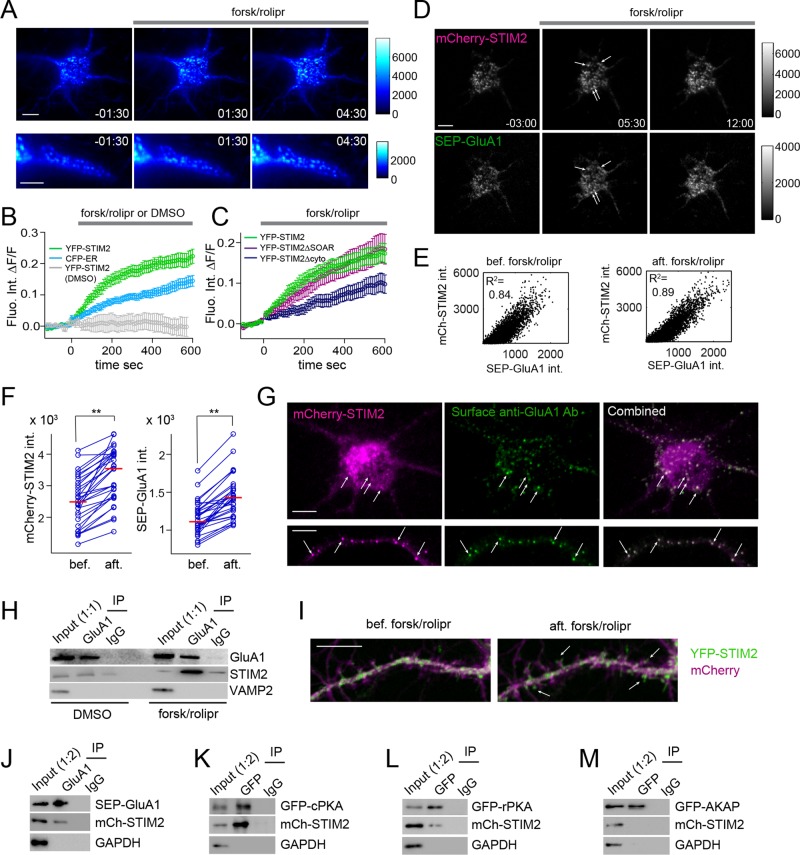
In nonexcitable cells, STIM1 regulates SOCE by interacting with Orai1 at ER-PM contact sites. To begin to address where STIM2-mediated phosphorylation of GluA1 occurs, we performed live-cell imaging of YFP-STIM2 in hippocampal neurons during cAMP elevation and opted for total internal reflection fluorescence (TIRF) microscopy to selectively image STIM2 near the PM. TIRF imaging revealed that a fraction of YFP-STIM2 localizes to puncta near the PM before stimulation. Elevation in cAMP levels by forsk/rolipr resulted in a rapid increase in the number and intensity of these puncta in both the cell body and proximal dendrites (Figure 6A and Supplemental Movie S3), suggesting redistribution of YFP-STIM2 to ER-PM contact sites.
FIGURE 6:
cAMP-induced recruitment of STIM2 and GluA1 to ER-PM contacts. (A–C) TIRF imaging of hippocampal neurons (DIV 6–8) showing cAMP-induced redistribution of YFP-STIM2 to puncta near the PM in the soma (top; see Supplemental Movie S3) and in a proximal dendrite (bottom). (B) cAMP-induced changes in fluorescence intensity in the soma of neurons coexpressing YFP-STIM2 (green, n = 10) and CFP-ER (cyan, n = 10). The vehicle (DMSO) has no effect on YFP-STIM2 intensity (gray, n = 5). (C) Quantification of cAMP-dependent increase in fluorescence intensity in the cell soma for YFP-STIM2 (green, n = 13), ∆SOAR (purple, n = 9), and ∆cyto (dark blue, n = 16). (D–F) TIRF imaging of hippocampal neurons (DIV 6–8) electroporated with mCherry-STIM2 and SEP-GluA1, showing colocalization of mCherry-STIM2 and SEP-GluA1 in puncta both before and after forsk/rolipr addition (see Supplemental Movies S4 and S5). (E) Scatterplots of mCherry-STIM2 against SEP-GluA1 fluorescence intensity (each dot represents a single pixel within the neuron) before and 10 min after forsk/rolipr addition, showing extensive colocalization of mCherry-STIM2 and SEP-GluA1. (F) Pairwise analysis of fluorescence intensity in individual puncta (derived from three neurons) for mCherry-STIM2 and SEP-GluA1 before and 10 min after forsk/rolipr addition. Red bars indicate the median. **p < 0.01, paired t test. (G) TIRF imaging of fixed hippocampal neurons (DIV 7) coexpressing mCherry-STIM2 and GFP-GluA1, treated with forsk/rolipr for 30 min, and stained with an N-terminal GluA1 Ab (Alexa 633) in nonpermeabilizing conditions. Only the mCherry-STIM2 and the surface Ab staining are shown. Arrows point to puncta in the soma (top) and dendritic segment (bottom) that contain both mCherry-STIM2 and surface GluA1. (H) Co-IPs of cortical neurons (DIV 21) treated with DMSO (vehicle) or forsk/rolipr for 30 min with a GluA1 Ab or control IgGs. (I) Dual-color confocal imaging of hippocampal neurons (DIV 21) electroporated with YFP-STIM2 and mCherry, showing cAMP-induced redistribution of YFP-STIM2 (green) in dendritic spines labeled with mCherry (magenta). Snapshots are shown before and 30 min after forsk/rolipr addition. Arrows indicate spines with increased YFP-STIM2 intensity after cAMP elevation (see Supplemental Movie S6). (J–M) Co-IPs of HeLa cells coexpressing mCherry-STIM2 with SEP-GluA1 (J), GFP-cPKA (K), GFP-rPKA (L), and GFP-AKAP (M) treated with forsk/rolipr for 30 min. IPs were carried out with a GluA1 Ab (J), GFP Ab (K–M), or control IgGs. Scale bar, 5 µm.
Dual-color TIRF imaging of YFP-STIM2 and cyan fluorescent protein (CFP)–ER (an ER marker) showed that YFP-STIM2 redistribution to puncta in response to cAMP elevation is associated with bulk movement of the ER toward the PM, although the overall increase in CFP-ER fluorescence is significantly lower than that observed with YFP-STIM2 (Figure 6B and Supplemental Figure S7). ER Ca2+ store depletion in neurons induced similar behavior for both fluorescent proteins; the magnitude and kinetics of YFP-STIM2 migration toward the PM are comparable to those observed in response to cAMP and it is also accompanied by an increase in CFP-ER intensity. Thus cAMP and store depletion both trigger redistribution of STIM2 to ER-PM contact sites in neurons. The bulk movement of the ER toward the PM probably reflects the formation of new ER-PM junction sites and/or enlargement of preexisting contacts (see Discussion). Deletion of the SOAR domain had no effect on cAMP-induced redistribution of YFP-STIM2, whereas removal of the entire cytoplasmic domain significantly inhibited YFP-STIM2 migration toward the PM to levels comparable to CFP-ER (Figure 6C). Targeting of STIM2 to puncta and STIM2-mediated GluA1 phosphorylation are therefore mediated by different cytoplasmic motifs.
To learn whether cAMP-induced redistribution of STIM2 to puncta leads to corecruitment of GluA1, we imaged mCherry-STIM2 together with GluA1 tagged at the amino terminus with a pH-sensitive form of enhanced green fluorescent protein (eGFP; superecliptic pHluorin [SEP]; Miesenbock et al., 1998). SEP-GluA1 preferentially labels surface GluA1, as SEP fluorescence intensity is significantly quenched in acidic intracellular organelles (Miesenbock et al., 1998; Makino and Malinow, 2009). We observed substantial colocalization of mCherry-STIM2 and SEP-GluA1 in puncta before stimulation and further redistribution of both proteins to puncta after forsk/rolipr addition (Figure 6, D and E, and Supplemental Movies S4 and S5). Analysis of individual puncta from different cells shows cAMP-induced increase in fluorescence intensity for both mCherry-STIM2 and SEP-GluA1 (Figure 6F). To confirm that a surface-localized pool of GluA1 is recruited to ER-PM contacts, we cotransfected mCherry-STIM2 and GFP-GluA1 in neurons, stimulated these cells with forsk/rolipr, and labeled a surface-exposed epitope of GluA1 with an N-terminal GluA1 antibody (Ab) under nonpermeabilizing conditions. TIRF imaging showed a punctate distribution of surface GluA1, with many GluA1 puncta colocalizing with mCherry-STIM2 in both the cell body and proximal dendrites (Figure 6G), in accordance with the presence of a surface pool of GluA1 at ER-PM junctions.
Consistent with the assembly of a protein complex at the ER-PM interface, the amount of STIM2 coimmunopurified with GluA1 is increased after forsk/rolipr treatment (Figure 6H), indicating that the strength of this interaction correlates with the amount of STIM2 present at ER-PM junctions. Together these results suggest that cAMP triggers migration of STIM2 to ER-PM contact sites and corecruitment of a surface pool of GluA1, in a process similar to the assembly of the STIM1-Orai1 (Park et al., 2009) or STIM1-Cav1.2 (Park et al., 2010; Wang et al., 2010) complexes at the ER-PM interface. These data do not rule out the possibility, however, that a fraction of GluA1 interacts with STIM2 and is phosphorylated on Ser-845 intracellularly (see Discussion).
We then asked whether cAMP-induced relocalization of STIM2 toward the plasma membrane enhances its localization to dendritic spines, where a large fraction of the AMPAR resides. Because distal dendrites and spines are difficult to image by TIRF (they rarely are in direct contact with the glass coverslip), we turned to confocal imaging. Dual-color imaging of YFP-STIM2 and mCherry showed a marked redistribution of YFP-STIM2 to spines after cAMP elevation (Figure 6I and Supplemental Movie S6), where it presumably enhances phosphorylation of a synaptic pool of GluA1 on Ser-845.
Finally, we probed the interaction of STIM2 with its potential binding partners in a heterologous system. Coexpression of mCherry-STIM2 and SEP-GluA1 in HeLa cells, followed by a GluA1 IP after forsk/rolipr stimulation, shows that these two proteins interact (Figure 6J). Pairwise expression of mCherry-STIM2 with GFP-cPKA or GFP-rPKA revealed strong interaction of STIM2 with cPKA (Figure 6K) and weak binding to rPKA (Figure 6L). Moreover, no interaction was detected between mCherry-STIM2 and GFP-AKAP (Figure 6M). These data provide further support for an interaction of STIM2 with GluA1 and cPKA and suggest that STIM2 interaction with AKAP150 and rPKA observed in neurons (Figure 4, H–K) may be indirect.
cAMP-induced redistribution of STIM2 does not trigger SOCE
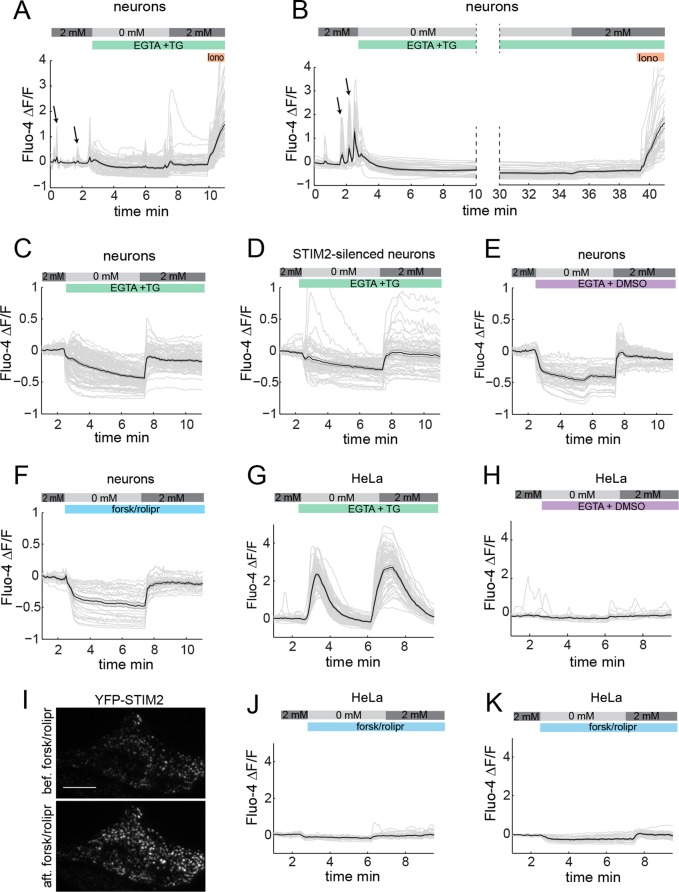
To determine whether cAMP-induced STIM2 translocation results in SOCE activation, we measured SOCE using a commonly used Ca2+ “addback” assay (Liou et al., 2005; Roos et al., 2005; Wong et al., 2010). In this assay, ER Ca2+ stores are depleted with thapsigargin in the absence of extracellular Ca2+, and SOCE is measured as the increase in cytoplasmic Ca2+ that follows addition of extracellular Ca2+. To measure accurately neuronal SOCE, we added a cocktail of channel inhibitors (see Materials and Methods) during Ca2+ addback (Figure 7, A and B) or throughout the experiment (Figure 7, C–F) to ensure that the increase in Ca2+ that occurs after Ca2+ addition is not mediated by Ca2+ channels that are not store operated.
FIGURE 7:
cAMP-induced redistribution of STIM2 does not trigger SOCE. (A, B) SOCE measured in Fluo4-loaded hippocampal neurons (DIV 14) after thapsigargin (TG)-induced short (A) or long (B) store depletion, with a cocktail of inhibitors (1 μM TTX, 10 μM AP-5, 10 μM CNQX, and 50 μM nifedipine) added during Ca2+ addback. Note the small magnitude of the Ca2+ influx upon Ca2+ readdition compared with that of spontaneous Ca2+ transients (arrows) or ionomycin (Iono)-induced Ca2+ influx. (C–F) SOCE measured in hippocampal neurons (DIV 14–15) with the inhibitor cocktail present throughout the time series and displayed at a different y-axis scale. (C, D) SOCE response in the presence of thapsigargin measured in control (C) or STIM2-silenced (D) neurons. (E, F) Ca2+ influx upon Ca2+ readdition in the absence of thapsigargin (E) or the presence of forsk/rolipr (F). (G, H) SOCE measured in Fluo4-loaded HeLa cells treated with thapsigargin (G) or DMSO (H). (I) cAMP-induced translocation of STIM2 to puncta in HeLa cells imaged by TIRF microscopy. Snapshots are shown before and 15 min after forsk/rolipr addition. Scale bar, 20 μm. (J, K) Forsk/rolipr does not trigger a SOCE response in HeLa cells expressing endogenous STIM2 (J) or mCherry-STIM2 (K). Ca2+ traces from individual cells are shown in light gray. The average trace is shown in black. Shaded error bar, SEM. The extracellular Ca2+ concentration (0 or 2 mM) is indicated on top of these traces. SOCE measurements in HeLa cells were also done in the presence of 1 μM TTX, 10 μM AP-5, 10 μM CNQX, and 50 μM nifedipine.
Store depletion in hippocampal neurons led to a small release of Ca2+ from the ER and little Ca2+ influx upon Ca2+ addition compared with spontaneous Ca2+ transients observed before Ca2+ depletion or to ionomycin-induced Ca2+ increase at the end of the experiment (Figure 7, A and B). Longer Ca2+ depletion protocols did not enhance neuronal SOCE (Figure 7B). Closer examination of Ca2+ responses in individual cells shows detectable ER Ca2+ release in roughly half of the cells (Figure 7C). Withdrawal of extracellular Ca2+ led to a rapid drop in cytoplasmic Ca2+ (Figure 7, A–F), as previously reported (Gemes et al., 2011; Gruszczynska-Biegala et al., 2011), which masks in part the Ca2+ increase induced by thapsigargin. We independently confirmed thapsigargin-induced lowering of ER Ca2+ by FRET measurements with an ER-targeted Ca2+ sensor (Supplemental Figure S8).
Ca2+ readdition restored baseline Ca2+ concentration to levels observed before Ca2+ depletion but failed to induce a sustained increase in intracellular Ca2+, although a SOCE-like response was occasionally detected in a few cells (Figure 7, A and C). This Ca2+-response profile is similar in STIM2-depleted neurons (Figure 7D) or in cells that have not been treated with thapsigargin before Ca2+ addition (Figure 7E). Finally, forsk/rolipr does not induce ER Ca2+ release, nor does it enhance Ca2+ influx after Ca2+ readdition (Figure 7F).
In marked contrast, thapsigargin-induced depletion of ER Ca2+ in HeLa cells triggered a robust SOCE response (Figure 7G), whereas no Ca2+ entry was detected when stores were full (Figure 7H). The lack of a significant SOCE response in primary neurons has been observed in some (Bouron et al., 2005; Park et al., 2010) but not all (Berna-Erro et al., 2009; Sun et al., 2014) studies (see Discussion) and prevented us from measuring the effect of cAMP on SOCE in these cells. We thus turned to HeLa cells because they display both robust SOCE activity (Figure 7G) and cAMP-induced redistribution of STIM2 to puncta (Figure 7I). Forsk/rolipr did not induce ER Ca2+ release or SOCE in these cells (Figure 7J) even when mCherry-STIM2 was overexpressed (Figure 7K). These findings, together with our data identifying cAMP as a novel cue that triggers redistribution of STIM2 to puncta near the PM, suggest that cAMP-induced translocation of STIM2 is functionally uncoupled from SOCE activation. Our data also indicate that a large majority of neurons do not exhibit a characteristic SOCE response.
STIM2 promotes surface expression of AMPARs
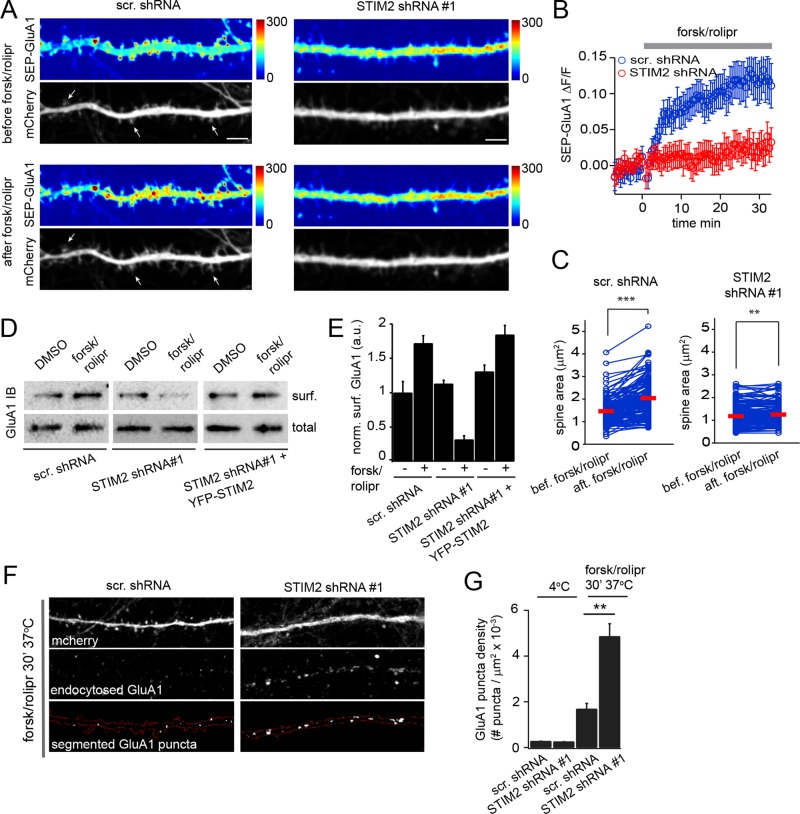
Phosphorylation of GluA1 on Ser-845 regulates activity-dependent exocytosis and endocytosis of AMPARs (Ehlers, 2000; Man et al., 2007), prompting us to evaluate the role of STIM2 in GluA1 trafficking. For this, we made use of SEP-GluA1 again, which has been employed by many groups to monitor AMPAR surface delivery in live cells (Makino and Malinow, 2009). GluA1 exocytosis was induced by a chemical LTP (cLTP) protocol based on PKA activation by forsk/rolipr, which has been reported to trigger AMPAR insertion, spine enlargement, and LTP in slices (Otmakhov et al., 2004). Of importance, cAMP-dependent PM insertion of GluA1 requires its phosphorylation on Ser-845 (Otmakhov et al., 2004). Time-lapse confocal imaging during cLTP shows a clear insertion of GluA1 to the dendritic plasma membrane in hippocampal neurons (DIV 18) coelectroporated with SEP-GluA1 and a control shRNA (mCherry; Figure 8, A and B, and Supplemental Movie S7). Insertion of SEP-GluA1 occurs mainly in the dendritic shaft and rarely in dendritic spines, consistent with earlier work reporting exocytosis of GluA1 to extrasynaptic sites after synaptic potentiation (Makino and Malinow, 2009). We also measured changes in spine size in the same cells using the mCherry fluorescence and found on average a 25% increase in spine surface area after cLTP (Figure 8, A and C, and Supplemental Movie S8). cLTP-induced SEP-GluA1 insertion was abolished upon STIM2 silencing (Figure 8, A and B, and Supplemental Movie S9), in line with STIM2-dependent GluA1 Ser-845 phosphorylation. cLTP-mediated spine enlargement in these cells was also strongly reduced (Figure 8, A and C, and Supplemental Movie S10), indicating that STIM2 is also required for cAMP-dependent structural plasticity.
FIGURE 8:
STIM2 mediates cAMP-dependent surface delivery of GluA1. (A–C) Hippocampal neurons (DIV 17–19) coelectroporated with scramble or STIM2 shRNA#1 (mCherry) and SEP-GluA1 were imaged by dual-color confocal microscopy during stimulation with forsk/rolipr. (A) SEP-GluA1 and mCherry fluorescence shown before and 30 min after forsk/rolipr treatment in control (left) and STIM2-silenced cells (right). SEP-GluA1 intensity is color coded. The arrows show examples of spines that have undergone forsk/rolipr-induced enlargement. Scale bar, 5 μm. See Supplemental Movies S7–S10 (B) Average change in SEP-GluA1 intensity in control and STIM2-silenced cells measured in 16 dendritic segments from four independent experiments for each condition. Error bars, SEM. (C) Pairwise analysis of individual spine surface area before and after forsk/rolipr in control and STIM2-silenced neurons. Red bars indicate the mean. ***p < 0.001, **p < 0.01, paired t tests. (D) Streptavidin pull downs from control, STIM2-silenced, or STIM2-silenced cells rescued with YFP-STIM2 that were surface biotinylated after DMSO (vehicle) or forsk/rolipr treatment. (E) Densitometry analysis of surface GluA1 for conditions shown in D. Average and SD are shown for three independent experiments. (F, G) GluA1 endocytosis assay. Surface GluA1 in control or STIM2-silenced neurons (DIV 20) was bound to an Ab at 4°C and the Ab–receptor complex allowed to internalize for 30 min at 37°C in the presence of forsk/rolipr. Surface Abs were stripped after incubation at 4°C (G) or 37°C (F, G). Endocytic GluA1 puncta located in mCherry-expressing dendrites were then segmented and scored using a MATLAB-based script. Scale bar, 10 μm. (G) Average density of GluA1 endocytic puncta. More than 50 dendritic segments from three independent experiments were quantified for each condition. p < 0.01, t test. See also Supplemental Movies S6–S9.
To confirm the role of STIM2 in regulating GluA1 surface expression, we used a surface biotinylation technique. Cortical neurons (DIV 21) transduced with scrambled or STIM2 shRNAs were surface biotinylated after 30 min of vehicle (dimethyl sulfoxide [DMSO]) or cLTP treatment. Affinity purification of biotinylated proteins with streptavidin showed a ∼50% increase in surface GluA1 (Figure 8, D and E), in agreement with previous reports (Man et al., 2007). STIM2 knockdown had little effect on GluA1 basal surface expression but completely blocked cLTP-mediated GluA1 insertion (Figure 8, D and E). In fact, levels of surface GluA1 in STIM2-silenced cells were even lower after cLTP than in control conditions, suggesting that STIM2 also inhibits GluA1 endocytosis after cAMP/PKA activation. This defect in GluA1 PM insertion was efficiently rescued by YFP-STIM2 (Figure 8, D and E).
To determine directly the effect of STIM2 on GluA1 endocytosis, endogenous surface GluA1 was bound to an Ab at 4°C, and the Ab–receptor complex was allowed to endocytose for 30 min at 37°C in the presence of forsk/rolipr. Analysis of the density of endocytic GluA1 puncta showed ∼60% increase in GluA1 uptake in STIM2-silenced cells (Figure 8, F and G). Together these data show that STIM2 promotes PKA-dependent AMPAR surface expression by both stimulating exocytosis and inhibiting endocytosis of GluA1.
DISCUSSION
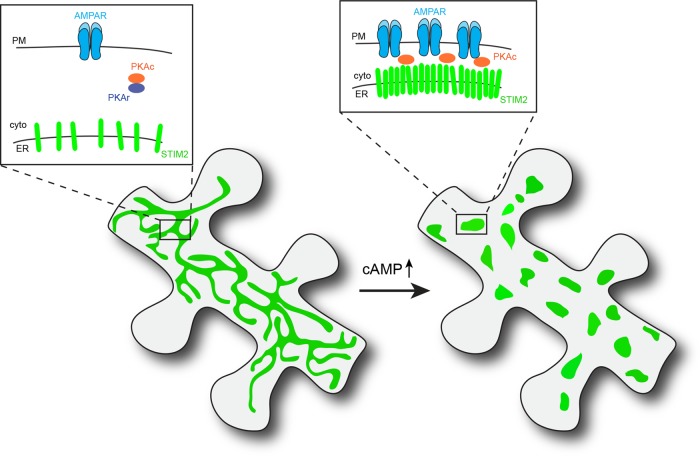
We report here a novel mechanism of synaptic remodeling that involves functional coupling between the ER and the dendritic PM. Our findings support a central role of the ER-resident protein STIM2 in regulating PKA-dependent AMPAR phosphorylation and trafficking at excitatory synapses. In addition, we show that STIM2 regulates dendritic spine morphogenesis and cAMP-dependent spine enlargement. These results led us to propose a model for STIM2 function in dendrites and spines (Figure 9) that is further discussed below.
FIGURE 9:
Model for STIM2-dependent regulation of AMPAR phosphorylation and trafficking at the interface between the ER and the PM. cAMP elevation resulting from synaptic or other signaling inputs triggers translocation of STIM2 to ER-PM contact sites and dynamic assembly of a PKA signaling complex that drives phosphorylation of GluA1 Ser-845 and surface delivery of the AMPAR.
cAMP: a novel cue that triggers translocation of STIM2 to ER-PM contact sites
We identify cAMP as a novel cue that triggers STIM2 translocation to ER-PM junctions, thus extending the repertoire of signals sensed by the STIM proteins. Dual-color TIRF imaging shows that cAMP- or thapsigargin-induced redistribution of STIM2 to puncta is associated with bulk movement of the ER toward to PM. Electron microscope data in HRP-STIM1–expressing Jurkat cells revealed that store depletion increases by ∼30% the number of ER-PM junctional contacts, as well as coverage of the cytoplasmic face of the PM by ER tubules (Wu et al., 2006). By analogy, it is possible that the global redistribution of the ER toward the PM observed in STIM2-overexpressing neurons reflects the formation of new contact sites and/or the expansion of preexisting ones. It should be mentioned, however, that Wu et al. (2006) did not observe bulk movement of the ER by TIRF microscopy. The reason for this discrepancy is not clear but could involve differences in cell shape (neurons are flatter than Jurkat cells), TIRF configuration, or STIM2-specific effects on ER dynamics.
Intriguingly, cAMP-induced migration of STIM2 to ER-PM contact sites does not result in any detectable SOCE activation, implying that these two processes are not necessarily coupled (Figure 7, E and F). This is in agreement with a recent study in pancreatic β cells showing cAMP-induced migration of STIM1 to puncta near the plasma membrane, without corecruitment of Orai1 (Tian et al., 2012). cAMP-dependent redistribution of STIM2 to ER-PM contact sites can also occur in the absence of any detectable release of Ca2+ from the ER (Figure 7, F and J). Store-independent translocation/activation of STIM1 has been reported (Hawkins et al., 2010; Xiao et al., 2011; Tian et al., 2012), suggesting that Ca2+ dissociation from the STIM sensors is not an absolute requirement for their oligomerization and migration to ER-PM contact sites. cAMP-induced redistribution of STIM2 to ER-PM contact sites depends on its cytoplasmic moiety but does not involve the SOAR domain (Figure 6C). How cAMP triggers redistribution of STIM2 to ER-PM junctions is not known.
STIM2 couples GluA1 to PKA and regulates phosphorylation of GluA1 on Ser-845
Our results show that STIM2 is essential for phosphorylation of GluA1 on Ser-845. How does STIM2 regulate GluA1 phosphorylation? By analogy with previous findings implicating STIM1 and SOCE in AC activation (Fagan et al., 1998; Lefkimmiatis et al., 2009), STIM2 could activate PKA by stimulating production of cAMP. This is unlikely, however, since STIM2 appears to function downstream of ACs (Figure 4, B–G) and because cAMP induces STIM2-dependent phosphorylation of GluA1 without activating SOCE (Figure 7, F, J, and K). Instead, we find that STIM2 is required for the assembly of a protein complex consisting of GluA1, PKA, and AKAP150. The absence of Orai1 in this complex provides further support for SOCE-independent STIM2 function.
STIM2 depletion results in uncoupling of GluA1 from PKA and AKAP150 (Figure 4K). Because AKAP150 anchors PKA in dendritic spines (Carr et al., 1992; Smith et al., 2006) and is responsible for PKA regulation of AMPAR activity (Lu et al., 2008), uncoupling of GluA1 from AKAP/PKA likely results in decreased phosphorylation of a synaptic pool of GluA1 and can explain to some degree the phosphorylation deficit detected in STIM2-silenced neurons. Of interest, change in GluA1-PKA coupling (and consequently GluA1 Ser-845 phosphorylation) has recently been proposed as a mechanism for homeostatic synaptic scaling (Diering et al., 2014). Based on these data and a large body of literature implicating AKAP/PKA in synaptic remodeling (Esteban et al., 2003; Lee et al., 2003, 2010; Hu et al., 2007; Makino et al., 2011), it is tempting to speculate that STIM2 may regulate various forms of synaptic plasticity by controlling GluA1-PKA coupling at excitatory synapses.
Surprisingly, we found that STIM2 negatively regulates phosphorylation of GluA1 on Ser-831 (Figure 4A), a CaMKII/PKC site implicated in synaptic plasticity and memory (Lee et al., 2003, 2010; Makino et al., 2011). Increased GluA1 pSer-831 in STIM2-silenced neurons appears to be CaMKII independent, since we observed reduced autophosphorylation of CaMKII on Thr286 in STIM2-silenced neurons (unpublished data, but see Sun et al., 2014). The mechanism by which STIM2 regulates GluA1 Ser-831 phosphorylation is under investigation. Together our results indicate that STIM2 regulates the phosphorylation state of two key serine residues in the cytoplasmic tail of GluA1.
Synaptic and nonsynaptic functions of STIM2
Where does STIM2-mediated phosphorylation of GluA1 take place? The presence of STIM2 in ∼40% of dendritic spines (Figure 1, C and D), the abundance of GluA1 in the postsynaptic membrane, and the role of STIM2 in coupling GluA1 to AKAP/PKA are consistent with a function at or near synapses. However, because STIM2 is an ER-resident protein, the bulk of it is clearly nonsynaptic. We find that in resting neurons, a significant fraction of STIM2 resides in puncta in the cell body and dendrites (Figures 1C and 6A). TIRF imaging shows that many of these puncta are located in close proximity to the PM, indicating that they likely correspond to ER-PM contact sites.
mCherry-STIM2 and SEP-GluA1 are corecruited to these puncta, and the presence of these two proteins at ER-PM junctions is further enhanced by cAMP elevation (Figure 6, D–F). This suggests that extrasynaptic ER-PM junctions are relevant sites of interaction for STIM2 and GluA1, where STIM2 presumably also couples PKA to GluA1 and promotes GluA1 Ser-845 phosphorylation. In line with this idea, AKAP150, a highly enriched PKA scaffold at the PSD, does not seem to be required for STIM2 to interact with PKA and GluA1 (Figure 6, J–L), suggesting that STIM2 is also able to promote GluA1 phosphorylation outside the context of a synapse. On the basis of these data, we speculate that synaptic and nonsynaptic functions of STIM2 may vary, depending on the type of stimulus that engages the STIM2/PKA signaling pathway. For example, local activation of PKA in response to synaptic inputs will likely recruit a pool of STIM2 present in spines, whereas cAMP production through neuromodulatory inputs such as norepinephrine (Hu et al., 2007) may, in contrast, engage STIM2/PKA signaling extrasynaptically (Figure 9).
Several lines of evidence suggest that STIM2 interacts with and promotes phosphorylation of a surface pool of GluA1. First, both SEP-GluA1 (which preferentially labels surface GluA1) and surface-exposed GFP-GluA1 detected by an Ab in nonpermeabilizing conditions colocalize with mCherry-STIM2 at ER-PM contact sites (Figure 6, D and G). Second, STIM2 inhibits GluA1 endocytosis, indicating that it likely operates on a PM pool of GluA1 (Figure 8, F and G). Third, the ability of STIM2 to drive GluA1 phosphorylation, interact with GluA1, and couple GluA1 to AKAP/PKA depends on its SOAR domain (Figure 5, A–D). This is analogous to STIM1 interaction with Orai1 and Cav1.2, where its SOAR domain binds to cytoplasmic domains of these PM effectors across the cytoplasmic space between the ER and the PM (Park et al., 2009, 2010; Wang et al., 2010). Finally, the amount of STIM2 coimmunopurified with GluA1 correlates with the presence of STIM2 at ER-PM junctions (Figure 6H).
Alternatively, it is also conceivable that STIM2 interacts with and regulates phosphorylation of an intracellular pool of GluA1. A significant fraction of GluA1 resides in early/recycling endosome, and recycling of GluA1 back to the PM is promoted by Ser-845 phosphorylation (Ehlers, 2000) and regulates LTP (Park et al., 2004). Another possible intracellular site of GluA1 phosphorylation is the ER itself. Newly synthesized AMPAR subunits dwell in the ER several hours before the assembled receptor exits the ER en route to the PM (Penn et al., 2008). CaMKII-dependent phosphorylation of GluA2 promotes its exit from the ER (Lu et al., 2014). Whether PKA phosphorylates an ER-localized pool of GluA1 and facilitates its exit from the ER is unknown.
STIM2 influences spine formation
Our results indicate that STIM2 regulates spinogenesis and cAMP-dependent spine enlargement. A recent study reported reduced levels of STIM2 and mature spines in a presenilin-1 (PS1) mouse model of familial Alzheimer's disease (Sun et al., 2014), and ectopic expression of STIM2 (but not STIM1) rescued mushroom spines in this PS1 (M146V) knock-in mouse (Sun et al., 2014). In addition, Cre-mediated excision of STIM2 in hippocampal neurons of floxed STIM2 mice resulted in a decrease in the fraction of mushroom spines (Sun et al., 2014), thereby providing independent evidence for a role of STIM2 in spine maturation. The authors attributed this spine maturation defect to reduced synaptic SOCE and CaMKII signaling (Sun et al., 2014), although they did not provide causal evidence for a role of SOCE in STIM2-dependent spine morphogenesis. Whether STIM2-dependent regulation of PKA signaling also contributes to spine morphogenesis remains to be established.
SOCE in neurons and nonexcitable cells
Although we monitored robust SOCE activity in HeLa cells, we were unable to detect a characteristic SOCE response in primary hippocampal neurons. The vast majority of store-depleted cells did not exhibit a sustained Ca2+ increase after addition of extracellular Ca2+ typical of SOCE (Figure 7). This is unlikely due to a cell health issue, as these neurons displayed spontaneous Ca2+ transients indicating network activity. Ca2+ readdition did restore, however, baseline Ca2+ levels, indicating that some form of Ca2+ entry took place. However, this also occurred in STIM2-silenced neurons or in neurons that were not store depleted before Ca2+ readdition. The small magnitude of that Ca2+ influx and its properties therefore argue against the presence of a classical SOCE response in hippocampal neurons.
The lack of a clear SOCE response in neurons was observed before (Bouron et al., 2005; Park et al., 2010), although neuronal SOCE was reported in other studies (Berna-Erro et al., 2009; Gemes et al., 2011; Gruszczynska-Biegala et al., 2011; Sun et al., 2014). Possible reasons for this discrepancy include 1) the use of various neuron cell types at different developmental stages (Bouron et al., 2005), 2) differences in the activation state of these cells (Lalonde et al., 2014), 3) cellular location of SOCE measurements (e.g., dendritic spines vs. cell body; Sun et al., 2014), and 4) variations in the Ca2+ addback protocol.
A recent report suggests that SOCE is constitutively active in resting cerebellar granule cells but appears to be attenuated when these neurons are active (depolarized; Lalonde et al., 2014). This study, if confirmed in other neuronal cell types, would imply that both ER Ca2+ content and SOCE strongly depend on neuronal activity, a property that could be responsible for some of the published differences on the magnitude of neuronal SOCE.
Finally, neurons are endowed with multiple Ca2+ channels that can be activated in response to changes in cytoplasmic/extracellular Ca2+ or, indirectly, by modulation of the network's excitability. The use of a cocktail of channel/receptor inhibitors during Ca2+ imaging is therefore recommended (see Materials and Methods) to exclude that Ca2+ enters through channels that are not store operated. Several but not all studies on neuronal SOCE report the use of these inhibitors, and those that do, use different sets of drugs. This could also potentially account for some of the inconsistencies in the literature. Overall, the lack of a standardized assay for SOCE in neurons, the presence of multiple Ca2+ channels in these cells with far greater Ca2+ conductance than SOC channels, the lack of specificity of current SOC channel inhibitors (particularly in neurons), and the absence of data on the identity of the neuronal SOC channel(s) complicate the analysis of SOCE in neurons. Clearly, more work is needed to understand the relevance of this Ca2+ entry pathway in excitable cells.
In conclusion, our findings highlight the existence of a novel mechanism that functionally couples the ER to excitatory synapses and provide evidence for a central role of ER-to-PM signaling in AMPAR function and synaptic remodeling. Our work, together with a series of recent reports (Giordano et al., 2013; Hartmann et al., 2014; Sun et al., 2014), also suggests that ER-PM coupling is a fundamental feature of cell and synapse physiology.
MATERIALS AND METHODS
DNA, shRNA constructs, lentiviruses, and antibodies
The lentiviral vectors pll3.7 (#11795) and FUGW (#14883) and the pCI-SEP-GluR1 plasmid (#24000) were from Addgene (Cambridge, MA). AKAR3EV was a gift from Miki Matsuda (Department of Bioimaging and Cell Signaling, Kyoto University, Kyoto, Japan; Komatsu et al., 2011). D1ER (Palmer et al., 2004) and GFP-AKAP150 were gifts from Roger Tsien (Department of Chemistry and Biochemistry, University of California, San Diego) and Mark Dell'Acqua (Department of Pharmacology, University of Colorado, Denver), respectively. CFP-ER was from Clontech (Mountain View, CA). A red version of pFUGW (pFUmCW) was made by replacing eGFP with mCherry using the BamHI and EcoRI sites. All shRNA constructs were cloned by introducing double-stranded DNA oligos in pll3.7 using the HpaI and Xho sites. PCR products consisting of the shRNA and the U6 promotor were then transferred from pll3.7 to pFUmCW or pFUGW using the PAC1 site. The following shRNA sequences were used: scramble, CGATACTGAACGAATCGAT; STIM2#1, ACCAAGAGCATGATCTTCA; STIM2#2, GGAACGAAAGATGATGGAT; STIM1, TCCAGGCAGGAAGAAGTTT. The human STIM2 gene was amplified by PCR from pDS-YFP-STIM2 (Brandman et al., 2007) and cloned into pFUGW (pFU-YFP-STIM2) using the BamH1 and EcoRI sites. The protein sequence encoded by pFU-YFP-STIM2 corresponds to human STIM2, accession number AAI71766. YFP-STIM2Δcyto and YFP-STIM2ΔSOAR constructs were made from pFU-YFP-STIM2 by deleting the cytoplasmic region (amino acids [aa] 329–841) and SOAR domain (aa 438–538) of the human STIM2 protein. Deletion of the SOAR domain was done by overlap extension PCR. All DNA and shRNA constructs used here were sequenced verified. pFUGW-based lentiviral particles were produced and purified according to Tiscornia et al. (2006). Multiplicity of infection (MOI) between 2 and 3 was used for all viral transduction experiments.
Table 1 lists the antibodies used in this work.
TABLE 1:
Antibodies used in this work.
| Antibody | Species/type | Clone | Application/dilution | Source |
|---|---|---|---|---|
| α-Stim2 | Rabbit, pAb | NA | WB 1:500 | Cell Signaling (Danvers, MA), 4917S |
| α-Stim1 | Rabbit, pAb | NA | WB 1:500 | ProSci (Loveland, CO), 4119 |
| α-GluA1-NT | Mouse, mAb | RH95 | WB 1:1000, IP 1:10, ICC 1:500 | Millipore, MAB2263 |
| α-pGluA1 Ser-845 | Mouse, mAb | EPR 2148 | WB 1:1000 | Millipore, 04-1073 |
| α-pGluA1 Ser-831 | Mouse, mAb | N453 | WB 1:1000 | Millipore, 04-823 |
| α-GluA2 | Rabbit, pAb | WB 1:1000 | Synaptic Systems (Goettingen, Germany), 182 103 | |
| α-GluN1 | Mouse, mAb | M68 | WB 1:1000 | Synaptic Systems, 114 011 |
| α-CaMKII | Rabbit, pAb | NA | WB 1:1000 | Cell Signaling, 3362 |
| α-syntaxin | Mouse, mAb | HPC1 | WB 1:20,000 | Sigma-Aldrich, S0664 |
| α-VAMP2 | Mouse, mAb | SP10 | WB 1:1000, ICC 1:1000 | Covance (Princeton, NJ), MMS-616R |
| α-Homer | Rabbit, pAb | NA | ICC 1:1000 | Synaptic Systems, 160003 |
| α-PSD95 | Mouse, mAb | 70-18 | WB 1:1000 | Millipore, MAB1596 |
| α-PKA-R1-α/β | Rabbit, pAb | NA | WB 1:1000 | Cell Signaling, 3927 |
| α-PKA-C-α | Rabbit, mAb | EP2102Y | WB 1:1000 | Abgent (San Diego, CA), AJ1612a |
| α-PKA-C-α | Mouse, mAb | 5B | ICC 1:500 | BD Transduction Laboratories (San Jose, CA), 610980 |
| α-AKAP150 | Goat, pAb | NA | IP 1:10 | Santa Cruz Biotechnology, sc-6445 |
| α-AKAP150 | Rabbit, pAb | NA | WB 1:1000 | Millipore, 07-210 |
| α-calnexin | Rabbit, pAb | NA | 1:2000 | Abcam (Cambridge, UK), 22595 |
| α-actin | Mouse, mAb | AC15 | WB 1:20,000 | Sigma-Aldrich, A1978 |
| α-GAPDH | Mouse, mAb | 6C5 | WB 1:20,000 | Millipore, MAB374 |
| α−NR1 | Mouse, mAb | NA | WB 1:1000 | Millipore, 05-432 |
| α−NR2A | Rabbit, pAb | NA | WB 1:1000, IP 1:10 | Millipore, 07-632 |
| α−Orai1 | Rabbit, pAb | NA | WB 1:1000 | ProScience, 4281 |
| α-rIgG HRP | Goat | NA | WB 1:10,000 | Jackson, 111-001-003 |
| α-mIgG HRP | Goat | NA | WB 1:10,000 | Jackson, 115-001-003 |
| Alexa Fluor 488 Anti-mIgG | Goat | NA | ICC 1:1000 | Invitrogen, A110011 |
| Alexa Fluor 568 anti-mIgG | Goat | NA | ICC 1:1000 | Invitrogen, A11004 |
| Alexa Fluor 488 anti-rIgG | Goat | NA | ICC 1:1000 | Invitrogen, A11008 |
| Alexa Fluor 568 anti-mIgG | Goat | NA | ICC 1:1000 | Invitrogen, A11011 |
| Alexa Fluor 633 anti-mIgG | Goat | NA | ICC 1:500 | Invitrogen, A21050 |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ICC, immunocytochemistry; mAb, monoclonal antibody; pAb, polyclonal antibody; WB, Western blot.
Primary neuron cultures, organotypic slices, and transfection/transduction protocols
Primary hippocampal and cortical neurons were prepared from E18 and E16 rat embryos, respectively, as previously described (Fivaz and Meyer, 2005; Kaech and Banker, 2006). Most experiments were done using hippocampal neurons. We used cortical neurons for a few biochemistry experiments requiring large amount of cells. The type of neuron culture used is indicated in the figure legends. For imaging and electrophysiology experiments, hippocampal neurons were grown on glass coverslips on top of a glial feeder layer, strictly adhering to the Banker protocol (Kaech and Banker, 2006). For biochemical experiments, hippocampal and cortical neurons were grown at high density on poly-l-lysine–coated dishes. Neurons were maintained in Neurobasal/B27 medium. DNA and shRNA constructs were introduced in neurons by electroporation using the Rat Neuron Nucleofector kit II (Amaxa Biosystems, Lonza; for live imaging experiments) or transfection with Lipofectamine 2000 (Invitrogen, Carlsbad, CA; for imaging and electrophysiology experiments) 1 d after plating. For biochemical experiments, neurons were transduced with lentiviruses (MOI 2–3) the day of plating. Hippocampal organotypic slices were prepared from postnatal day 5–6 rats according to the following protocol (Gogolla et al., 2006). Hippocampal slices (300 μm) were prepared using a tissue chopper, transferred to cell culture inserts (Millicell, 0.4 μM pore size; Millipore, Billerica, MA) and maintained in slice medium (50% MEM, 25% Hank's balanced salt solution, 25% horse serum, 1× GlutaMAX, 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml sodium selenite, 2.64 mg/ml glucose, 0.8 μg/ml vitamin C, and 1× penicillin and streptomycin). Slices were transfected using biolistic gene delivery (Bio-Rad) after DIV 2–3.
Live-cell confocal microscopy and FRET imaging
Time-lapse confocal microscopy was performed on an inverted Nikon (Tokyo, Japan) Eclipse TE2000-E microscope equipped with a spinning-disk confocal scan head (CSU-10; Yokogawa), an autofocusing system (PFS; Nikon), and a temperature-controlled stage. Images were acquired with a CoolSNAP HQ2 charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) driven by MetaMorph 7.6 (Universal Imaging). Neurons were imaged in ACSF –Mg2+ (125 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 30 mM glucose, and 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4) at 36.5°C with a 60× (numerical aperture [NA] 1.4) objective and stimulated with 50 μM forskolin/0.1 μM rolipram. AKAR3EV FRET imaging was performed as previously described (Thevathasan et al., 2013). FRET between eCFP and YPet is displayed as the intensity in the FRET channel (corrected for bleedthrough) divided by the donor (eCFP) intensity. Because AKAR3EV FRET was imaged in cells coexpressing mCherry, we made sure that there was no contribution of mCherry to the FRET channel.
TIRF microscopy
Time-lapse TIRF imaging was performed on an inverted Nikon Ti-E microscope equipped with a TIRF illuminator, a 60×/1.49 NA objective lens, an autofocusing system (Perfect Focus), and a motorized XY stage. The sample was illuminated with the 442, 514, 561, and 647 lines of solid-state lasers with dichroic and filter sets designed for dual CFP/YFP or triple GFP/RFP/647 imaging. Images were acquired with an Andor EM CCD iXon3 897 camera driven by NIS-Elements (Nikon Imaging).
Superresolution microscopy
Structured illumination microscopy (SIM) was performed using a 63×/1.4 NA objective on a Zeiss (Jena, Germany) inverted SIM microscope (ELYRA). Three-dimensional (3D) stacks were maximally projected on a single plane or subjected to 3D surface rendering using Imaris software.
Immunofluorescence and confocal microscopy of cultured neurons and slices
For immunofluorescence (IF), neurons grown on glass coverslips were fixed in 4% paraformaldeyhyde (PFA) and 4% sucrose and permeabilized using 0.25% Triton X-100. Five percent goat serum was used to block nonspecific binding sites. Primary and secondary Abs were incubated at room temperature for 1 h each, and cells were mounted on a glass slide for imaging. Double or triple stains were done using Alexa 488, 568, and 633–conjugated secondary Abs. Organotypic hippocampal slices (300 μm) were fixed in 4% PFA and 4% sucrose and cleared using the Scale A2 solution (4 M urea, 10% glycerol, and 0.1% Triton X-100; Hama et al., 2011) before they were mounted on a glass slide. Fixed samples were imaged with an upright laser scanning confocal microscope (LSM710; Zeiss).
Measuring SOCE in primary neurons and HeLa cells
SOCE was measured in neurons and HeLa cells as described earlier (Wong et al., 2010). Briefly, Fluo-4 loaded cells were switched from a Ca2+-containing (2 mM) to a Ca2+-free buffer (2.1 mM ethylene glycol tetraacetic acid [EGTA]), and stores were depleted with 1 μm thapsigargin. SOCE was then triggered by the addition of 2 mM Ca2+. In control experiments, thapsigargin was replaced by DMSO or forsk/rolipr. Neurons (and HeLa cells for faithful comparison) were exposed to a cocktail of channel inhibitors (1 μM tetrodotoxin [TTX], 10 μM 2-amino-5-phosphonopentanoic acid [AP-5], 10 μM 6-cyano-7-nitroquinoxaline-2,3-dione [CNQX], and 50 μM nifedipine) throughout the experiment or during Ca2+ addback as stated. Cells were imaged by confocal microscopy using a 20× objective and Ca2+ traces measured for each individual cells using software described in Wong et al. (2010). For SOCE measurements in mCherry-expressing neurons, Ca2+ traces were measured in cells segmented based on their mCherry-STIM2 expression (Wong et al., 2010). At least 50 cells (from separate fields) were measured for each condition.
Image analysis
Spine detection and classification.
Spines were automatically detected and classified using the NeuronStudio software (Rodriguez et al., 2008). Z-stacks of dendritic segments imaged by confocal microscopy and covering the entire 3D volume of spines were used for analysis.
Scoring synaptic density.
A MATLAB script was written to analyze three-color images consisting of mCherry (shRNA), VAMP2 (633 nm; presynaptic marker), and Homer-1 (488 nm; postsynaptic). The script identifies puncta (synapses) that are positive for both VAMP2 and Homer-1 and overlap with mCherry-expressing dendrites. The same intensity thresholds were used to segment VAMP2 and Homer-1 puncta in control and STIM2-silenced neurons. Synapse density was obtained by computing the number of detected synapses per unit area of mCherry-expressing dendrites.
Sholl analysis.
Sholl analysis was done on apical dendrites of CA1 pyramidal neurons from organotypic slice cultures. Neurite traces were drawn using NeuroStudio, and the Sholl analysis was done using Simple Neurite Tracer Plugin in Fiji (Longair et al., 2011).
SEP-GluA1 insertion.
SEP-GluA1 insertion was quantified by measuring the fold change in SEP-GluA1 intensity in dendritic segments.
Detection of endocytic GluA1 puncta.
A MATLAB script was written to detect and score GluA1 endocytic puncta. GluA1 puncta were segmented based on intensity and inclusion in mCherry-expressing dendritic segments. The same intensity threshold was used for both control and STIM2-silenced cells. Puncta density was obtained by computing the number of detected puncta per unit area of mCherry-expressing dendrites.
All MATLAB scripts are available upon request.
Electrophysiology
Whole-cell patch-clamp recordings were performed in DIV 15–18 hippocampal neurons. Neurons were held at −70 mV using a multiClamp 700B amplifier (Axon Instruments, Sunnyvale, CA) driven by pClamp (Axon Instruments). Recording pipettes with resistances of 3–5 MΩ were filled with an internal solution containing (in mM) 120 K-gluconate, 9 KCl, 10 KOH, 3.48 MgCl2, 4 NaCl, 10 HEPES, 4 Na2ATP, 0.4 Na3GTP, 19.5 sucrose, and 5 EGTA. Neurons were bathed in external solution containing (in mM) 110 NaCl, 5 KCl, 2 CaCl2, 0.8 MgCl2, 10 HEPES, pH 7.4, and 10 d-glucose and supplemented with 0.5 μM TTX to prevent action potential-evoked EPSCs and 10 μM bicuculline methiodide to block GABAergic inhibitory postsynaptic potentials. Data were analyzed using pClamp. Only recording epochs in which series and input resistances varied by <10% were included in the analysis.
Measurement of cAMP levels and PKA activity
Hippocampal neurons (DIV 21) were treated with 50 μM forskolin (Sigma-Aldrich, St. Louis, MO)/0.1 μM rolipram (Sigma-Aldrich) or DMSO for 30 min before harvesting. Cells were harvested with HCl (0.1 N), and cAMP levels were measured using ELISA (cAMP Direct Immunoassay Kit; Calbiochem, San Diego, CA) according to the manufacturer's instructions. For PKA activity, cells were harvested and PKA activity measured by ELISA (PKA kinase activity kit, ADI-EKS-390A; ENZO Lifescience, Farmingdale, NY), according to the manufacturer's instructions. PKA activity was normalized against protein concentration.
Biochemical isolation of the PSD
Isolation of PSDs was adapted from described procedures (Cotman and Taylor, 1972; Cotman et al., 1974; Hahn et al., 2009). All steps were done at 4°C. Adult rat forebrains were dissected and homogenized using a glass/Teflon Dounce homogenizer. The homogenate was spun at 1000 × g to remove cell debris. Centrifugation of the supernatant at 16,000 × g for 20 min yielded membrane and cytoplasmic fractions. The membrane fraction was further fractionated on a sucrose gradient (1.2/1/0.85 M sucrose, 100,000 × g for 2 h) to isolate SPMs. Triton X-100 (1%) extraction and centrifugation (35,000 × g for 20 min) of SPMs yielded a supernatant and a pellet. The supernatant contains SVs. The pellet was further extracted in 1.5% Triton X-100 and spun at 140,000 × g for 30 min. The supernatant contains presynaptic membranes and was pooled with the SV fraction (Pre/SV). The pellet is the final PSD fraction. All of these fractions (equal amount of proteins) were then immunoblotted for several markers and the STIM2 proteins (Figure 1F).
Immunoblotting and immunoprecipitation
Cultured neurons or brain tissue were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed with immunoblotting or IP lysis buffer (25 mM Tris-HCl, pH 8, 27.5 mM NaCl, 20 mM KCl, 25 mM sucrose, 10 mM EDTA, 1 mM dithiothreitol, 10% [vol/vol] glycerol, 0.5% NP-40, complete protease inhibitors [Roche], and phosphatase inhibitors [Roche]) or RIPA buffer without SDS (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 1% NP-40, complete protease inhibitors [Roche], and phosphatase inhibitors [Roche]), respectively. Lysates were cleared by centrifugation. For immunoblotting, 20–40 μg of total protein was loaded. For IP, 200 μg to 1 mg of proteins was immunoprecipitated with 1 μg of primary Ab per 100 μg of input and protein A/G-Sepharose overnight at 4°C. IP and input fractions were then analyzed by SDS–PAGE and immunoblotting. Immunoblots were developed using horseradish peroxidase (HRP)–conjugated Abs (Jackson, West Grove, PA), followed by detection with enhanced chemiluminescence (ECL; Pierce).
Surface biotinylation, AMPAR exocytosis and endocytosis, and surface GluA1 staining
To quantify GluA1 insertion into the PM, cortical neurons (DIV 21) treated with DMSO or 50 μM forskolin/0.1 μM rolipram for 30 min at 37°C were rinsed twice with ice-cold PBS and incubated with sulfo-NHS-SS-biotin (Pierce) in ice-cold PBS for 30 min. Cells were then washed in PBS, lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.2, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS), and biotinylated proteins were isolated with NeutrAvidin Resin (Pierce). Input, flowthrough, and eluate (biotinylated fraction) were immunoblotted with GluA1 Ab. To measure GluA1 internalization, hippocampal neurons (DIV 21) were incubated with N-terminal GluA1 Ab (1:100) in ACSF at 4°C for 30 min. DIV 21 hippocampal neurons were then washed and incubated at 37°C for 30 min in the presence of 50 μM forskolin/0.1 μM rolipram. Surface Abs were stripped (0.5 M NaCl and 0.2 M acetic acid) at 4°C, and cells were then fixed, permeabilized, and immunostained for GluA1. To control for efficient stripping of surface Abs, cells were incubated with N-GluA1 Abs at 4°C, acid stripped, fixed, and immunostained for GluA1. To probe colocalization of STIM2 with a surface pool of GluA1, hippocampal neurons (DIV 7) cotransfected with mCherry-STIM2 and GFP-GluA1 were incubated at 37oC for 30 min in the presence of 50 μM forskolin/0.1 μM rolipram. Neurons were then incubated with N-terminal GluA1 Ab (1:100) in culture medium at 37°C for 60 min after which cells were fixed in 4% PFA and 4% sucrose and immunostained for GluA1 with a secondary antibody coupled to Alexa 633.
Statistics
Average data are represented as means ± SEM unless indicated otherwise. Statistical significance was determined using two-tailed unpaired or paired t tests on data sets obtained from cell populations or individual cells, respectively. One-way analysis of variance (ANOVA) was used when simultaneously comparing three or more data sets. In this case, p values were derived from a posthoc Bonferroni test. ANOVA and posthoc Bonferroni tests were done using the ANOVA1 and multcompare functions in MATLAB.
Supplementary Material
Acknowledgments
We thank Dale Purves, David Virshup, and Gisou van der Goot for critical reading of the manuscript. We are grateful to Zeiss Singapore for use of their SIM system and Nikon Singapore for their help with TIRF microscopy. This work was supported by the Singapore Ministry of Education Academic Research Fund (MOE2011-T2-1-107), the Duke-NUS Signature Research Program funded by the Agency for Science, Technology and Research and the Ministry of Health, Singapore, a CRP (Competitive Research Programme) grant from the National Research Foundation of Singapore, and the World Class Institute (Program of the National Research Foundation of Korea) funded by the Ministry of Education, Science and Technology of Korea (NRF Grant WCI 2009-003).
Abbreviations used:
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- FRET
Forster resonance energy transfer
- SIM
structured illumination microscopy
- SOAR
STIM-Orai-activating-region
- STIM
stromal interaction molecule
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1222) on January 21, 2015.
*These authors contributed equally to this work.
Present addresses: †MRC Centre for Developmental Neurobiology, King's College London, London SE1 1UL, United Kingdom;
‡Department of Biochemistry, Oslo Universitetssykehus, 0424 Oslo, Norway;
§Department of Systems Medicine and Bioengineering, Houston Methodist Research Institute, Houston, TX 77030.
REFERENCES
- Banke TG, Bowie D, Lee H, Huganir RL, Schousboe A, Traynelis SF. Control of GluR1 AMPA receptor function by cAMP-dependent protein kinase. J Neurosci. 2000;20:89–102. doi: 10.1523/JNEUROSCI.20-01-00089.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardo S, Cavazzini MG, Emptage N. The role of the endoplasmic reticulum Ca2+ store in the plasticity of central neurons. Trends Pharmacol Sci. 2006;27:78–84. doi: 10.1016/j.tips.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Berna-Erro A, Braun A, Kraft R, Kleinschnitz C, Schuhmann MK, Stegner D, Wultsch T, Eilers J, Meuth SG, Stoll G, Nieswandt B. STIM2 regulates capacitive Ca2 +entry in neurons and plays a key role in hypoxic neuronal cell death. Sci Signal. 2009;2:ra67. doi: 10.1126/scisignal.2000522. [DOI] [PubMed] [Google Scholar]
- Bird GS, Hwang SY, Smyth JT, Fukushima M, Boyles RR, Putney JW., Jr STIM1 is a calcium sensor specialized for digital signaling. Curr Biol. 2009;19:1724–1729. doi: 10.1016/j.cub.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Nanoscale analysis of structural synaptic plasticity. Curr Opin Neurobiol. 2012;22:372–382. doi: 10.1016/j.conb.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouron A, Altafaj X, Boisseau S, De Waard M. A store-operated Ca2+ influx activated in response to the depletion of thapsigargin-sensitive Ca2+ stores is developmentally regulated in embryonic cortical neurons from mice. Brain Res Dev Brain Res. 2005;159:64–71. doi: 10.1016/j.devbrainres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr DW, Stofko-Hahn RE, Fraser ID, Cone RD, Scott JD. Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J Biol Chem. 1992;267:16816–16823. [PubMed] [Google Scholar]
- Colledge M, Dean RA, Scott GK, Langeberg LK, Huganir RL, Scott JD. Targeting of PKA to glutamate receptors through a MAGUK-AKAP complex. Neuron. 2000;27:107–119. doi: 10.1016/s0896-6273(00)00013-1. [DOI] [PubMed] [Google Scholar]
- Collins SR, Meyer T. Evolutionary origins of STIM1 and STIM2 within ancient Ca2+signaling systems. Trends Cell Biol. 2011;21:202–211. doi: 10.1016/j.tcb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman CW, Banker G, Churchill L, Taylor D. Isolation of postsynaptic densities from rat brain. J Cell Biol. 1974;63:441–455. doi: 10.1083/jcb.63.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman CW, Taylor D. Isolation and structural studies on synaptic complexes from rat brain. J Cell Biol. 1972;55:696–711. doi: 10.1083/jcb.55.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkach V, Barria A, Soderling TR. Ca2+/calmodulin-kinase II enhances channel conductance of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate type glutamate receptors. Proc Natl Acad Sci USA. 1999;96:3269–3274. doi: 10.1073/pnas.96.6.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diering GH, Gustina AS, Huganir RL. PKA-GluA1 coupling via AKAP5 controls AMPA receptor phosphorylation and cell-surface targeting during bidirectional homeostatic plasticity. Neuron. 2014;84:790–805. doi: 10.1016/j.neuron.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- Esteban JA, Shi SH, Wilson C, Nuriya M, Huganir RL, Malinow R. PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat Neurosci. 2003;6:136–143. doi: 10.1038/nn997. [DOI] [PubMed] [Google Scholar]
- Fagan KA, Mons N, Cooper DM. Dependence of the Ca2+-inhibitable adenylyl cyclase of C6–2B glioma cells on capacitative Ca2+ entry. J Biol Chem. 1998;273:9297–9305. doi: 10.1074/jbc.273.15.9297. [DOI] [PubMed] [Google Scholar]
- Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- Finch EA, Augustine GJ. Local calcium signalling by inositol-1,4,5-trisphosphate in Purkinje cell dendrites. Nature. 1998;396:753–756. doi: 10.1038/25541. [DOI] [PubMed] [Google Scholar]
- Fivaz M, Meyer T. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J Cell Biol. 2005;170:429–441. doi: 10.1083/jcb.200409157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaschuk O, Yaari Y, Konnerth A. Release and sequestration of calcium by ryanodine-sensitive stores in rat hippocampal neurones. J Physiol. 1997;502:13–30. doi: 10.1111/j.1469-7793.1997.013bl.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez P, Garcia-Marin V, Freire M. Dendritic spines and development: towards a unifying model of spinogenesis—a present day review of Cajal's histological slides and drawings. Neural Plast. 2010;2010:769207. doi: 10.1155/2010/769207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemes G, Bangaru ML, Wu HE, Tang Q, Weihrauch D, Koopmeiners AS, Cruikshank JM, Kwok WM, Hogan QH. Store-operated Ca2+ entry in sensory neurons: functional role and the effect of painful nerve injury. J Neurosci. 2011;31:3536–3549. doi: 10.1523/JNEUROSCI.5053-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolla N, Galimberti I, DePaola V, Caroni P. Preparation of organotypic hippocampal slice cultures for long-term live imaging. Nat Protoc. 2006;1:1165–1171. doi: 10.1038/nprot.2006.168. [DOI] [PubMed] [Google Scholar]
- Gruszczynska-Biegala J, Pomorski P, Wisniewska MB, Kuznicki J. Differential roles for STIM1 and STIM2 in store-operated calcium entry in rat neurons. PLoS One. 2011;6:e19285. doi: 10.1371/journal.pone.0019285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CG, Banerjee A, Macdonald ML, Cho DS, Kamins J, Nie Z, Borgmann-Winter KE, Grosser T, Pizarro A, Ciccimaro E, et al. The post-synaptic density of human postmortem brain tissues: an experimental study paradigm for neuropsychiatric illnesses. PLoS One. 2009;4:e5251. doi: 10.1371/journal.pone.0005251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hartmann J, Karl RM, Alexander RP, Adelsberger H, Brill MS, Ruhlmann C, Ansel A, Sakimura K, Baba Y, Kurosaki T, et al. STIM1 controls neuronal Ca(2+) signaling, mGluR1-dependent synaptic transmission, and cerebellar motor behavior. Neuron. 2014;82:635–644. doi: 10.1016/j.neuron.2014.03.027. [DOI] [PubMed] [Google Scholar]
- Hawkins BJ, Irrinki KM, Mallilankaraman K, Lien YC, Wang Y, Bhanumathy CD, Subbiah R, Ritchie MF, Soboloff J, Baba Y, et al. S-glutathionylation activates STIM1 and alters mitochondrial homeostasis. J Cell Biol. 2010;190:391–405. doi: 10.1083/jcb.201004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi N, Langeberg LK, Scott JD. Distinct enzyme combinations in AKAP signalling complexes permit functional diversity. Nat Cell Biol. 2005;7:1066–1073. doi: 10.1038/ncb1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Real E, Takamiya K, Kang MG, Ledoux J, Huganir RL, Malinow R. Emotion enhances learning via norepinephrine regulation of AMPA-receptor trafficking. Cell. 2007;131:160–173. doi: 10.1016/j.cell.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80:704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Krapivinsky L, Stotz SC, Manasian Y, Clapham DE. POST, partner of stromal interaction molecule 1 (STIM1), targets STIM1 to multiple transporters. Proc Natl Acad Sci USA. 2011;108:19234–19239. doi: 10.1073/pnas.1117231108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde J, Saia G, Gill G. Store-operated calcium entry promotes the degradation of the transcription factor Sp4 in resting neurons. Sci Signal. 2014;7:ra51. doi: 10.1126/scisignal.2005242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauri SE, Bortolotto ZA, Nistico R, Bleakman D, Ornstein PL, Lodge D, Isaac JT, Collingridge GL. A role for Ca2+ stores in kainate receptor-dependent synaptic facilitation and LTP at mossy fiber synapses in the hippocampus. Neuron. 2003;39:327–341. doi: 10.1016/s0896-6273(03)00369-6. [DOI] [PubMed] [Google Scholar]
- Lee HK, Takamiya K, Han JS, Man H, Kim CH, Rumbaugh G, Yu S, Ding L, He C, Petralia RS, et al. Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell. 2003;112:631–643. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- Lee HK, Takamiya K, He K, Song L, Huganir RL. Specific roles of AMPA receptor subunit GluR1 (GluA1) phosphorylation sites in regulating synaptic plasticity in the CA1 region of hippocampus. J Neurophysiol. 2010;103:479–489. doi: 10.1152/jn.00835.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkimmiatis K, Srikanthan M, Maiellaro I, Moyer MP, Curci S, Hofer AM. Store-operated cyclic AMP signalling mediated by STIM1. Nat Cell Biol. 2009;11:433–442. doi: 10.1038/ncb1850. [DOI] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Lu W, Khatri L, Ziff EB. Trafficking of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA) receptor subunit GluA2 from the endoplasmic reticulum is stimulated by a complex containing Ca2+/calmodulin-activated kinase II (CaMKII) and PICK1 protein and by release of Ca2+ from internal stores. J Biol Chem. 2014;289:19218–19230. doi: 10.1074/jbc.M113.511246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhang M, Lim IA, Hall DD, Allen M, Medvedeva Y, McKnight GS, Usachev YM, Hell JW. AKAP150-anchored PKA activity is important for LTD during its induction phase. J Physiol. 2008;586:4155–4164. doi: 10.1113/jphysiol.2008.151662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y, Johnson RC, Yu Y, Takamiya K, Huganir RL. Enhanced synaptic plasticity in mice with phosphomimetic mutation of the GluA1 AMPA receptor. Proc Natl Acad Sci USA. 2011;108:8450–8455. doi: 10.1073/pnas.1105261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Malinow R. AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron. 2009;64:381–390. doi: 10.1016/j.neuron.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man HY, Sekine-Aizawa Y, Huganir RL. Regulation of {alpha}-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking through PKA phosphorylation of the Glu receptor 1 subunit. Proc Natl Acad Sci USA. 2007;104:3579–3584. doi: 10.1073/pnas.0611698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, LaFerla FM, Chan SL, Leissring MA, Shepel PN, Geiger JD. Calcium signaling in the ER: its role in neuronal plasticity and neurodegenerative disorders. Trends Neurosci. 2000;23:222–229. doi: 10.1016/s0166-2236(00)01548-4. [DOI] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- Oh MC, Derkach VA, Guire ES, Soderling TR. Extrasynaptic membrane trafficking regulated by GluR1 serine 845 phosphorylation primes AMPA receptors for long-term potentiation. J Biol Chem. 2006;281:752–758. doi: 10.1074/jbc.M509677200. [DOI] [PubMed] [Google Scholar]
- Otmakhov N, Khibnik L, Otmakhova N, Carpenter S, Riahi S, Asrican B, Lisman J. Forskolin-induced LTP in the CA1 hippocampal region is NMDA receptor dependent. J Neurophysiol. 2004;91:1955–1962. doi: 10.1152/jn.00941.2003. [DOI] [PubMed] [Google Scholar]
- Palmer AE, Jin C, Reed JC, Tsien RY. Bcl-2-mediated alterations in endoplasmic reticulum Ca2 +analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci USA. 2004;101:17404–17409. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Shcheglovitov A, Dolmetsch R. The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science. 2010;330:101–105. doi: 10.1126/science.1191027. [DOI] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH. Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J. 2008;27:3056–3068. doi: 10.1038/emboj.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter KR, Palade GE. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J Biophys Biochem Cytol. 1957;3:269–300. doi: 10.1083/jcb.3.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J. Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol. 1962;13:405–421. doi: 10.1083/jcb.13.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinska-Kijek A, Wisniewska MB, Gruszczynska-Biegala J, Methner A, Kuznicki J. Immunolocalization of STIM1 in the mouse brain. Acta Neurobiol Exp (Wars) 2009;69:413–428. doi: 10.55782/ane-2009-1753. [DOI] [PubMed] [Google Scholar]
- Skroblin P, Grossmann S, Schafer G, Rosenthal W, Klussmann E. Mechanisms of protein kinase A anchoring. Int Rev Cell Mol Biol. 2010;283:235–330. doi: 10.1016/S1937-6448(10)83005-9. [DOI] [PubMed] [Google Scholar]
- Smith KE, Gibson ES, Dell'Acqua ML. cAMP-dependent protein kinase postsynaptic localization regulated by NMDA receptor activation through translocation of an A-kinase anchoring protein scaffold protein. J Neurosci. 2006;26:2391–2402. doi: 10.1523/JNEUROSCI.3092-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spacek J, Harris KM. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J Neurosci. 1997;17:190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang H, Liu J, Popugaeva E, Xu NJ, Feske S, White CL, 3rd, Bezprozvanny I. Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice. Neuron. 2014;82:79–93. doi: 10.1016/j.neuron.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevathasan JV, Tan E, Zheng H, Lin YC, Li Y, Inoue T, Fivaz M. The small GTPase HRas shapes local PI3K signals through positive feedback and regulates persistent membrane extension in migrating fibroblasts. Mol Biol Cell. 2013;24:2228–2237. doi: 10.1091/mbc.E12-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Tepikin AV, Tengholm A, Gylfe E. cAMP induces stromal interaction molecule 1 (STIM1) puncta but neither Orai1 protein clustering nor store-operated Ca2 +entry (SOCE) in islet cells. J Biol Chem. 2012;287:9862–9872. doi: 10.1074/jbc.M111.292854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Zhou Y, Hendron E, Mancarella S, Andrake MD, Rothberg BS, Soboloff J, Gill DL. Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nat Commun. 2014;5:3183. doi: 10.1038/ncomms4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Deng X, Mancarella S, Hendron E, Eguchi S, Soboloff J, Tang XD, Gill DL. The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science. 2010;330:105–109. doi: 10.1126/science.1191086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LC, Lu B, Tan KW, Fivaz M. Fully-automated image processing software to analyze calcium traces in populations of single cells. Cell Calcium. 2010;48:270–274. doi: 10.1016/j.ceca.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Coste B, Mathur J, Patapoutian A. Temperature-dependent STIM1 activation induces Ca(2)+ influx and modulates gene expression. Nat Chem Biol. 2011;7:351–358. doi: 10.1038/nchembio.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.