Abstract
Mutations in the gene encoding profilin 1 (PFN1) have recently been shown to cause amyotrophic lateral sclerosis (ALS), a fatal neurodegenerative disorder. We sequenced the PFN1 gene in a cohort of ALS patients (n = 485) and detected 2 novel variants (A20T and Q139L), as well as 4 cases with the previously identified E117G rare variant (∼ 1.2%). A case-control meta-analysis of all published E117G ALS+/− frontotemporal dementia cases including those identified in this report was significant p = 0.001, odds ratio = 3.26 (95% confidence interval, 1.6–6.7), demonstrating this variant to be a susceptibility allele. Postmortem tissue from available patients displayed classic TAR DNA-binding protein 43 pathology. In both transient transfections and in fibroblasts from a patient with the A20T change, we showed that this novel PFN1 mutation causes protein aggregation and the formation of insoluble high molecular weight species which is a hallmark of ALS pathology. Our findings show that PFN1 is a rare cause of ALS and adds further weight to the underlying genetic heterogeneity of this disease.
Keywords: Amyotrophic lateral sclerosis, Profilin 1, TDP-43 proteinopathy
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a typically late onset, neurodegenerative disorder characterized by the degeneration of large motor neurons of the brain and spinal cord, resulting in death on average within 3 years of symptom onset. Five to 10 percent of patients have a family history of ALS and/or frontotemporal dementia. Mutations in 4 major genes: SOD1, TARDBP, FUS, and the intronic GGGGCC expansion within C9ORF72 account for approximately 60% of familial (FALS) and approximately 10% of sporadic ALS (SALS) cases (Al-Chalabi et al., 2012). Recently, missense mutations in profilin1 (PFN1) were identified in 2 large ALS families by exome capture and next generation sequencing, and screening of a larger cohort identified 7 further mutations in 274 FALS cases (approximately 2.5%) (Wu et al., 2012). An additional 10 reports of PFN1 gene screens have identified only 2 novel mutations: a T109M mutation in 1 German FALS case and a R136W mutation in a Chinese SALS case (Chen et al., 2013; Ingre et al., 2013). The previously described rare dinucleotide variant encoding E117G has been identified in 15 of 5601 cases in European ALS cohorts with or without frontotemporal dementia (FTD) (Daoud et al., 2013; Dillen et al., 2013; Fratta et al., 2013; Lattante et al., 2012; Tiloca et al., 2012; van Blitterswijk et al., 2013; Yang et al., 2013). See Supplementary Table 1 for a complete list of published PFN1 mutations.
In this study, we screened a cohort of 485 UK familial and sporadic ALS cases for PFN1 mutations and biologically characterized the cellular pathology of the novel variants.
2. Methods
2.1. Patient and DNA samples
Ninety-seven FALS and 215 SALS patient DNA samples from the research DNA resource within the Clinical Neuroscience Department at the Institute of Psychiatry, Psychology, and Neuroscience, King's College London, UK were analyzed. Consent was provided by patients after examination and diagnosis of definite or probable ALS based on revised El Escorial criteria (Brooks, 2000) by consultant neurologists at the Motor Neuron Disease clinic at King's College Hospital. DNA from an additional 3 FALS, 37 SALS/FTD, and 133 SALS cases were sourced from the Medical Research Council London Neurodegenerative Diseases Brain Bank. All cases were negative for mutations in SOD1, TDP-43, FUS, the C9ORF72 expansion, VCP, OPTN, and VAPB. DNA was extracted from white blood cells or frozen brain tissue using standard phenol and/or chloroform extraction procedures. Nine hundred and nine local control samples matched for sex and age were assayed using KASPar genotyping methods by LGC Genomics (LGC Genomics, Teddington, UK). No controls from Middle Eastern populations were available for screening.
2.2. Genetic screening
The 3 coding exons of PFN1 (Refseq ID NM_005022.3) were amplified using standard polymerase chain reaction procedures including at least 100 bp of flanking intronic sequence. See Supplementary Material for primers and cycling conditions. The amplicons were directly sequenced with Big-Dye Terminator v1.1 on an ABI3130 genetic analyzer (Applied Biosystems Pty Ltd, Warrington, UK). Sequence chromatograms were analyzed for mutations using Sequencher 4.10 (Gene Codes Corporation, Ann Arbor, Michigan, USA). Novel mutations were reconfirmed by amplifying a second aliquot of stock DNA sample.
2.3. Mutation modeling and prediction of protein destabilization
Sequence-based assessment of the potential pathogenicity of each coding variant identified was assessed by submitting the genomic coordinates and alternate alleles to the SIFT (Ng and Henikoff, 2001) (http://sift.jcvi.org/www/SIFT_chr_coords_submit.html) and Provean web servers (Choi et al., 2012) (http://provean.jcvi.org/genome_submit_2.php) and the amino acid position and alternate residue to the PolyPhen-2 web server (Adzhubei et al., 2010) (http://genetics.bwh.harvard.edu/pph2/). The presence of multiple entries for the PFN1 protein in the Protein Data Bank (PDB, www.pdb.org) database of macromolecular structures also enabled the use of structure-based predictors of the likelihood of each mutation destabilizing the protein. The PDB structure 2PAV was chosen as being representative of PFN1 and submitted, along with the amino acid position and alternate residue, to the SDM (Worth et al., 2007) (http://mordred.bioc.cam.ac.uk/∼sdm/sdm.php), POPMUSIC-2.1 (Dehouck et al., 2011) (http://dezyme.com/), and I-Mutant-3.0 (Capriotti et al., 2008) (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi) web servers.
In silico mutagenesis was performed for the 3 mutations identified in this study, using the SwissModel webserver (Arnold et al., 2006) (http://swissmodel.expasy.org/) and the PDB template structure 2PAV (http://www.pdb.org/pdb/explore/explore.do?structureId=2pav). All amino acid side chains in close proximity to the mutated residues were subsequently fixed by simulated annealing in DeepView (Johansson et al., 2012), and the final diagrams rendered with PyMol (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC). All variants identified in this study were also assessed for changes to splicing using Netgene2 (Hebsgaard et al., 1996) (http://www.cbs.dtu.dk/services/NetGene2/).
2.4. Meta-analysis study
A Pubmed literature search was conducted for all articles containing the keywords “(PFN1 or profilin) and (ALS or amyotrophic)”. Six reports describing mutation screening of PFN1 in European cohorts and controls (Dillen et al., 2013; Fratta et al., 2013; Tiloca et al., 2012; van Blitterswijk et al., 2013; Wu et al., 2012) and this study were included in a meta-analysis for E117G frequency. Three reports (Daoud et al., 2013; Lattante et al., 2012; Yang et al., 2013) were excluded as they did not include screening of local population controls, and two reports (Chen et.al., 2013; Zou et.al., 2013) were excluded as the E117G variant has not been observed in any East Asian population. The meta-analysis was conducted using the Comprehensive Meta-analysis Version 2 software (Biostat Inc. Engelwood, NJ, USA) under both a fixed effects (Mantel-Haenszel) and random effects model (DerSimonian Laird).
2.5. Plasmids and cloning
PFN1 expression vectors (Gateway pcDNA3.1/nV5-DEST, Invitrogen) encoding V5 epitope tagged PFN1 wild type (WT), C71G and E117G were used in this study (Wu et al., 2012). Site-directed mutagenesis was performed according to the manufacturer's protocol (Quickchange II Site-Directed Mutagenesis Kit, Stratagene) using a V5-PFN1WT tagged pDONR221 entry clone plasmid to produce constructs harboring the novel mutants identified in this study (c.58G > A, A20T, and c.416A > T, Q139L). The pDONR221-PFN1 mutant constructs were then recombined with pcDNA3.1/nV5-DEST to create the final mutant expression constructs. All constructs were verified by DNA sequencing.
2.6. Transfection of HEK293T cells
HEK293T cells were maintained in DMEM with high glucose plus Glutamax (Life Technologies, Paisley, UK) with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin in a water-jacketed incubator at 5% CO2. For solubility fractionation, transfections were performed in 12-well plates with 500 ng of plasmid DNA and 1.5 μL of Fugene HD (Promega, Southampton, UK) per well, as per the manufacturer's protocol. For immunofluorescence, HEK293T cells were plated at 25,000 cells/cm2 on 13 mm diameter, 1.5-mm-thickness coverslips coated with poly-D-lysine (Sigma-Aldrich, Dorset, UK) in 24-well plates and transfected with 250 ng of DNA and 0.75 μL of Fugene HD.
2.7. Human dermal fibroblast isolation and culture
Punch biopsies were performed and collected in DMEM with high glucose plus Glutamax. After removal of fat tissue with a sterile scalpel blade, the biopsy was dissected into 0.5 mm explants. Explants were allowed to adhere to a dry flask, to which expansion medium (DMEM with high glucose plus Glutamax, with 20% fetal bovine serum, 1% non-essential amino acids, 100 U/mL penicillin, and 100 μg/mL streptomycin) was added. After 5–7 days, medium was changed to culture medium (DMEM with high glucose plus Glutamax, with 15% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin), cells were expanded once confluent and then banked. For solubility analyses, fibroblasts were harvested from confluent 6-well plates. Where used, 0.5 μM MG132 was added to the media 24 hours before cells were harvested.
2.8. Immunofluorescence
Immunofluorescence was performed as previously described (Vance et al., 2013). Briefly, cells were fixed with 4% paraformaldehyde at room temperature for 10 minutes and rinsed with phosphate-buffered saline (PBS). Cells were incubated with primary antibody overnight at 4 °C, then with fluorescent secondary antibodies for 3 hours at room temperature, all diluted in PBS with 0.2% Triton-X 100 and 1% goat serum. Mouse anti-V5 antibody was used at 1:500 (cat. no 46-0708, Life Technologies). Alexa Fluor goat secondary antibody was used at 1:500 (anti-Mouse 488, cat. no A11001, Life Technologies). After washing with PBS, the cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole), and the coverslips were mounted onto slides using fluorescence mounting medium (Dako UK Ltd, Ely, UK) and left to harden overnight. Images were acquired using a Zeiss Axiovert S100 (Carl Zeiss Ltd, Hertfordshire, UK) with a 63x/NA 1.25 Plan Neofluar oil immersion objective, fitted with a CoolSnap EZ digital camera (Photometrics, Tucson, AZ). The proportion of transfected cells containing cytoplasmic V5-PFN1 granules was determined by counting both the total number of transfected cells and those with granules. Counts were performed manually by an observer blinded to mutation status. A minimum of 15 images taken at 20× for each construct from 3 different experiments were counted. On average, over 700 cells were counted for each mutation in each experiment.
2.9. Fractionation of PFN1 by solubility
PFN1 was fractionated by solubility as described previously (Wu et al., 2012), but with several modifications. Briefly, cells were washed with cold PBS, then scraped into cold PBS containing protease inhibitors (Complete, Roche, UK) and phosphatase inhibitors (PhosSTOP, Roche). Cells were pelleted by centrifugation at 10,000g for 30 seconds at 4 °C, and the pellets resuspended in lysis buffer (1% NP-40, 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1 mM dithiothreitol) containing protease and phosphatase inhibitors. After 30 minutes rotating at 4 °C, an aliquot was taken representing whole cell lysate. The remaining lysate was centrifuged at 15,800g for 20 minutes at 4 °C. The supernatant was removed and kept as the soluble fraction. The insoluble pellet was washed with lysis buffer, recentrifuged, and then resuspended in a 4-fold smaller volume of urea-SDS buffer (NP-40 lysis buffer with 8 M urea, 3% SDS), then sonicated. This extract was then spun again at 15,800g for 20 minutes at room temperature and the supernatant kept as the insoluble fraction. Equal volumes of 2× Laemmli buffer (Laemmli, 1970) without bromophenol blue were added to all aliquots taken, and the lysate and soluble fractions (but not the urea-containing insoluble fraction) were boiled for 20 minutes. To enhance the detection of insoluble species in fibroblasts, the insoluble pellet was resuspended directly in a 4-fold smaller volume of 1× Laemmli buffer and boiled.
2.10. Western blotting and densitometry analysis
Western blotting was performed as previously described (Scotter et al., 2014). Whole cell lysate protein concentrations were quantified using the BioRad DC Protein Assay (BioRad, Hemel Hempstead, UK). A total of 5 μg (or equivalent liquid volume of the soluble and insoluble fractions) was loaded per well of a 10% NuPAGE Novex Bis-Tris gel (Life Technologies). Gels were transferred onto nitrocellulose using the iBlot (Life Technologies), stained with Ponceau S, then blocked in TBS with 0.05% Tween 20 (TBS-T) with 5% non-fat dried milk (Sigma-Aldrich) for 30 minutes. Blots were probed overnight at 4 °C with primary antibody, then for 3 hours at room temperature with secondary antibodies (all in TBS-T plus 1% non-fat dried milk). Antibodies used for blotting were V5 (mouse: cat. no. 46-0708, 1:5000, Life Technologies), PFN1 (mouse: cat. no. SAB4100041, 1:1000 Sigma), histone H3 (rabbit: cat. no. H0164, 1:10,000, Sigma-Aldrich), GAPDH (mouse: cat. no. G9545, 1:1000, Sigma-Aldrich), and Dylight fluorescent secondary antibodies (goat anti-mouse 680 nm, cat. no. 35521,1:5000; goat anti-rabbit 700 nm, cat. no. 35568,1:10,000, Fisher Scientific UK Ltd, Leicestershire, UK).
Blots were scanned on the Li-Cor Odyssey gel scanner (Li-Cor Biotechnology, Cambridge, UK). Blots were then reprobed (without stripping) for loading control proteins using the same antibody conditions and scanning protocol. Blot images from 3 separate experiments in TIF format were quantified using the ImageJ gel analyzer tool (Image J 1.45e, NIH, Bethesda, USA, http://rsb.info.nih.gov/ij/). Integrated band intensities for V5-PFN1 (transfected HEK293T cells) or PFN1 (fibroblasts) were normalized for loading (soluble fraction, GAPDH; insoluble fraction, H3), and total PFN1 protein (PFN1 in the lysate fraction), and then normalized to those for WT PFN1.
2.11. Brain tissue collection and neuropathologic assessment
Brain and spinal cord tissues in 10% formalin-fixed, paraffin-embedded tissue blocks were available from the Medical Research Council London Neurodegenerative Diseases Brain Bank (Institute of Psychiatry, King's College London, UK). Consent for autopsy, neuropathologic assessment, and research was obtained from all subjects. Block taking for histologic and immunohistochemical studies and neuropathologic assessment for neurodegenerative diseases was performed in accordance with institutional and national guidelines.
2.12. Immunohistochemistry
Immunohistochemistry was carried out as per previously published protocols (Maekawa et al., 2009). In brief, sections of 7 μm thickness were cut from the paraffin-embedded tissue blocks, deparaffinized in xylene, endogenous peroxidase was blocked by 2.5% H2O2 in methanol and immunohistochemistry performed. To enhance antigen retrieval, sections were kept in citrate buffer for 10 minutes following microwave treatment. After blocking in normal swine or rabbit serum (Dako UK Ltd), primary antibody was applied overnight at 4 °C. Following washes, sections were incubated with biotinylated secondary antibody (Dako UK Ltd), followed by avidin:biotinylated enzyme complex (Vectastain Elite ABC kit, Vector Laboratories, Peterborough, UK). Finally, sections were incubated for 10–15 minutes with 0.5 mg/mL 3,3′-diaminobenzidine chromogen (Sigma-Aldrich) in Tris-buffered saline (pH 7.6) containing 0.05% H2O2. Sections were counterstained with Harris hematoxylin and immunostaining analyzed using a Leica microscope (Leica, Wetzlar, Germany). Antibodies used were PFN1 (rabbit: cat. no. AV48269, 1:500; mouse: cat. no. SAB410041, 1:500; rabbit: cat. no. P7624, 1:500, all Sigma-Aldrich), phospho-TAR DNA-binding protein 43 (TDP-43) pS409/410-2 (rabbit: cat. no. TIP-PTD-P02, 1:3500) and p62 (mouse: cat. no. 610833, 1:200, BD Biosciences).
2.13. Statistical analyses
Data handling and graphical representations were performed using Microsoft Excel 2010 (Microsoft Corp, Redmond, WA, USA). Statistical analyses as described in the text were performed using SigmaPlot v. 12.5 (Systat Software Inc, GmbH, Germany). One-way analysis of variance (ANOVA) was performed after Shapiro-Wilk testing for normality and Bartlett testing for equal variance. Posttest comparisons were performed against control using Dunnett test, and pairwise comparisons were performed using Holm-Sidak test. When normality or equal variance conditions were not met, Kruskal-Wallis 1-way ANOVA on ranks was performed. Posttest comparisons were performed against control using Dunnett test, and pairwise comparisons were performed using Tukey test. Statistical significance was set at p ≤ 0.05 (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). All figures were prepared using Adobe Photoshop CS2 (Adobe Systems Inc, San Jose, CA, USA).
3. Results
3.1. Identification of 2 novel PFN1 variants
A total of 100 FALS, 27 SALS/FTD, and 358 SALS cases (n = 485) were screened for nucleotide variants within the 3 coding exons of PFN1 by direct Sanger sequencing. Two novel variants were identified (Table 1). The first was a novel missense variant, c.58G > A located in exon 1 of a FALS patient of Middle Eastern ethnicity that codes for an alanine to threonine amino acid change at residue 20 (A20T) and is fully conserved in mammals (Fig. 1). The change was absent in 909 in-house controls and in 4299 European-American and 2202 African-American exomes from the NHLBI Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/). No controls from Middle Eastern populations were available to screen for the A20T variant. Unfortunately, no DNA was available from family members to test for segregation of this mutation with disease status.
Table 1.
PFN1 variants identified in this study
| Exon | Patient | Phenotype | Δ nucleotide | Δ amino acid | Local controls | EVS (EA) | Sex | Age of onset | Duration (mo) | Site of onset |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 1 | 1 | FALS | c.58G > A | p.A20T | 0/909 | Absent | F | 65 | Alive | Limb |
| Exon 3 | 2 | FALS | c.350_351AA > TG | p.E117G | 1/909 | 3/4297 | M | 62 | 30 | Limb |
| Exon 3 | 3 | SALS | c.350_351AA > TG | p.E117G | 1/909 | 3/4297 | F | 54 | 24 | Limb |
| Exon 3 | 4 | SALS | c.350_351AA > TG | p.E117G | 1/909 | 3/4297 | M | 74 | 38 | Limb |
| Exon 3 | 5 | SALS | c.350_351AA > TG | p.E117G | 1/909 | 3/4297 | M | 67 | 14 | Limb |
| Exon 3 | 6 | SALS | c.416A > T | p.Q139L | 0/909 | Absent | M | 52 | 72 | NA |
Key: EVS (EA), exome variant server, European Americans; FALS, familial amyotrophic lateral sclerosis; NA, not available; SALS, sporadic amyotrophic lateral sclerosis.
Fig. 1.
Residue conservation of mammals and viper showing mutations identified in this study. The residues, indicated with a red star, for A20, E117, and Q139 are fully conserved in all mammalian species and viper. Non-mammalian species were not included because of the lack of a clear one to one orthologous relationship in the profilin family.
The second novel variant, c.416A > T coding for a glutamine to lysine change at position 139 in the C-terminus of the protein (Q139L) was identified in exon 3 of a Caucasian sporadic ALS patient, was absent from the ESP database and 909 in-house controls, and the residue position is also fully conserved within mammals.
The previously described dinucleotide variant (c.350–351AA > GT), coding for an E117G residue change was observed in 4 Caucasian ALS cases from our cohort (1 FALS and 3 SALS) (4/485 total cases), present once in 909 in-house controls and 3 times in the EVS (Table 1). The residue is conserved in mammals as previously described (Fig. 1).
3.2. The E117G variant is an ALS risk factor
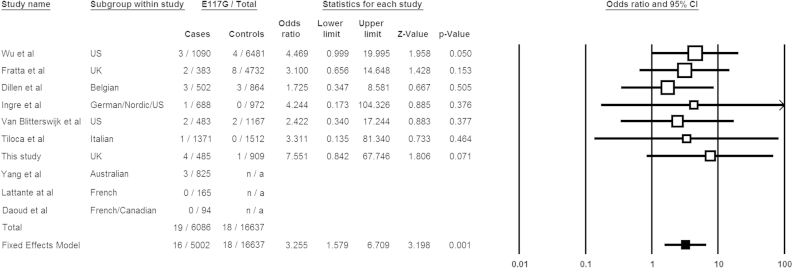
To assess whether the E117G variant is an ALS risk factor, we reviewed all published and public domain data available from European cohorts. Numbers for E117G positive ALS ± FTD cases and frequencies in local controls were examined. The frequency of E117G from the ESP data set (4300 European-American samples) was included and current as of April 2014 (http://evs.gs.washington.edu/EVS/). We excluded all East Asian studies and African-American controls from the ESP as E117G has only been identified in European ALS ± FTD patients and available control datasets. All data combined yielded a total of 19 E117G positive cases (n = 6086) and 18 in controls (n = 16,637); odds ratio of 2.90 and p = 0.002 (Fisher exact test). We then conducted a meta-analysis of those studies that assayed both cases and local controls (Dillen et al., 2013; Fratta et al., 2013; Tiloca et al., 2012; van Blitterswijk et al., 2013; Yang et al., 2013). The heterogeneity between studies was calculated using the Cochran χ2-based Q-test (p = 0.962). As the heterogeneity was not significant the meta-analysis was run under a fixed effects model (Mantel-Haenszel test) which showed a statistically significant association OR = 3.26 (95% CI 1.6-6.8) p = 0.001 (Fig. 2). A random effects model gave the same result. This confirms and supports a recent study suggesting E117G to be a moderate risk factor (Fratta et al., 2013).
Fig. 2.
Meta-analysis of published E117G case-control frequencies and those identified in this study. Data are presented as a forest plot. Squares and lines correspond to the study specific odds ratios (ORs) and 95% confidence intervals (CIs), respectively. The bottom row (filled square) shows the meta-analysis OR and CIs under fixed and random effects models. The fixed effect model is preferred as the studies showed minimal heterogeneity (p = 0.962, χ2-based Q-test), although there was no difference between the two methods. The area of the squares reflects the study specific weights (inverse of the variance). Variant counts from 4300 ESP European Americans and the 1000 Genomes project have been incorporated into the Wu et al. (2012) control data. Studies by Yang et al. (2013), Daoud et al., 2013 and Lattante et al. (2012) did not screen local control cohorts and so did not contribute to the final meta-analysis calculation.
3.3. Clinical presentation of patients harboring PFN1 mutations
All patients harboring the A20T, Q139L, or E117G variants had classical ALS symptoms with no evidence of FTD. The average age of onset for these patients was 66.2 years with limb onset. The patient carrying the A20T mutation (patient 1) is female, of Middle Eastern ethnicity with an age of onset of 63 years, and is still alive at the present age of 68 years (Fig. 3A). She presented with a predominant lower motor neuron phenotype with clear spinal onset, in the presence of upper motor neuron signs, and was wheelchair bound after 2 years. Her father presented with symptoms of ALS at 68 years of age, and her great grandmother was reported to have a paralysis problem occurring in her 60s. Patient 2, who carries the p.E117G variant, was a Caucasian male with an age of onset of 62 years and 30 months duration of disease (Fig. 3B). His mother was asymptomatic and died at the age of 91 years. However, his maternal first cousin had ALS and died at the age of 40 years. Clinical information on the sporadic cases (patients 3–6) is limited with available information listed in Table 1.
Fig. 3.
Familial gene mutations in PFN1. Pedigrees of PFN1 mutation positive cases identified in this study are shown for (A) A20T - Family 1 and (B) E117G - Family 2. Gender information has been withheld and denoted by diamonds. The arrow denotes the proband of each kindred. DNA was unavailable from other affected and unaffected members to test segregation of PFN1 mutations.
3.4. Modeling of PFN1 mutations
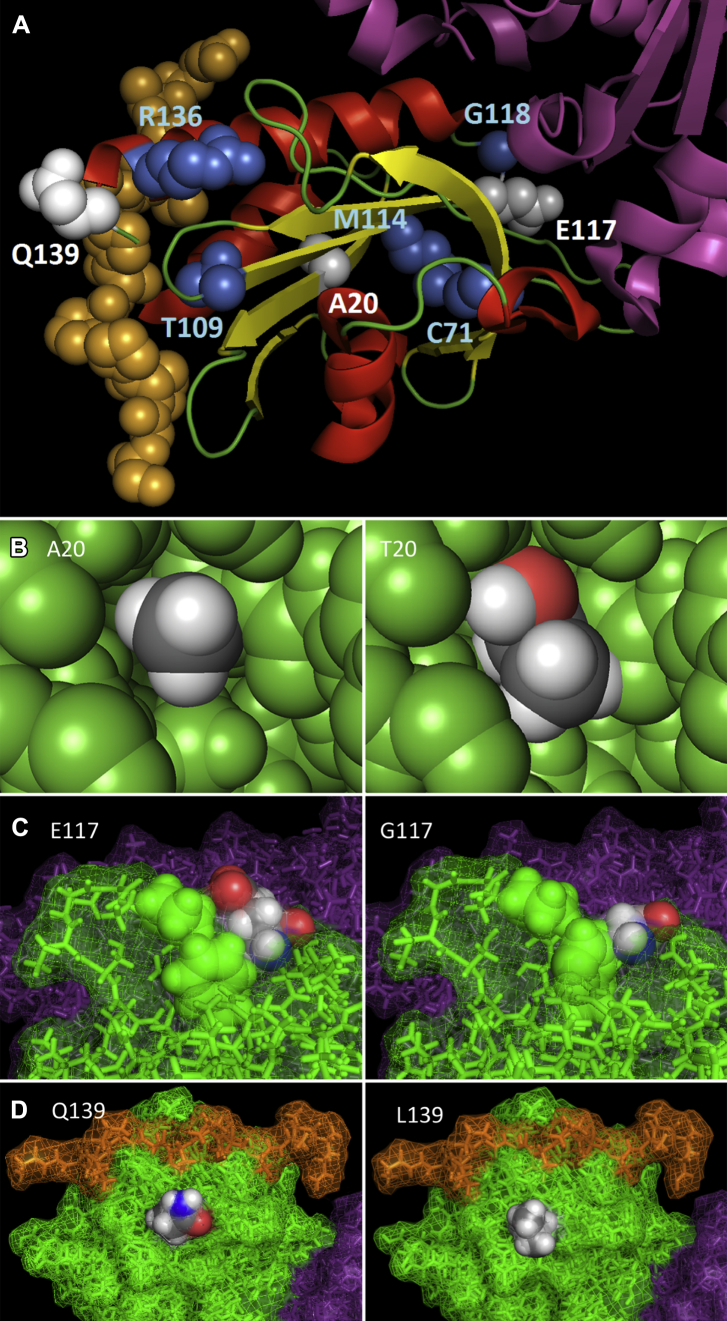
Fig. 4A shows the Pymol rendered alpha-beta structure of PFN1 (PDB:2PAV) that co-crystallizes with actin (ACTA1) and the proline-rich central portion of vasodilator-stimulated phosphoprotein (VASP). Modeling of all published mutations and those identified in this study demonstrates that the A20, C71, and M114 residues are internal to the protein, and of all PFN1 mutations identified to date these are consistently predicted to be the most damaging across the various prediction tools (Supplementary Table 2). The A20 change to a larger threonine creates internal conflicts at several residues, chiefly the backbone carboxyl of the neighboring asparagine residue at position 19 (Fig. 4B, T20) and would theoretically change the conformation of the beta sheet that runs through the core of the protein. For E117 the wild-type glutamic acid (Fig. 4C, E117) potentially has the role of stabilizing the conformation of the adjacent loop at residues 89–100, which the much smaller glycine mutation (Fig. 4C, G117) would be unlikely to fulfill. The glutamine at position 139 is the penultimate residue in the PFN1 protein chain and lies on the external surface of the structure close to the binding groove for the proline-rich region of VASP (Fig. 4D, Q139). However, neither the wild type (Fig. 4D, Q139) nor the mutant leucine residue (Fig. 4D, L139) directly interact with VASP and from currently available data it is not clear what the consequence, if any, of the Q139 mutation might be.
Fig. 4.
Pymol rendering of novel mutations identified in this study and all published PFN1 reports. (A) A schematic diagram of PFN1 bound to VASP and actin. VASP is shown as orange spheres, actin as purple ribbons, and PFN1 as red helices, yellow beta sheets, and green loops. Mutations identified in this study are indicated by white spheres, whereas those described elsewhere are shown as blue spheres. B–D show the reference amino acid (left diagram) and variant amino acid (right diagram) for the A20T, E117G, and Q139L mutations, respectively. The A20T mutation in the core of the protein potentially destabilizes a beta sheet because of steric hindrance, the E117G mutation potentially destabilizes an adjacent surface loop, whereas the surface Q139L mutation has no apparent structural impact. All PFN1 atoms are colored green, actin purple, and VASP orange, except those belonging to the highlighted residue side chain which are colored according to atom (gray = carbon, white = hydrogen, red = oxygen, blue = nitrogen). All diagrams are derived from PDB: 2PAV and rendered with PyMol (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.). Abbreviations: PDB, protein data bank; PFN1, profilin 1; VASP, vasodilator-stimulated phosphoprotein.
3.5. Patients with PFN1 variants have classic TDP-43 neuropathology
Postmortem tissue was available from 2 patients who harbored the E117G variant (patients 2 and 3) and also the sporadic case carrying a Q139L change (patient 6). In all 3 cases, the right half of the brain was available for neuropathologic examination. None of the brains showed evidence of cerebral atrophy. In patient 2 (E117G), no spinal cord was available, but in the other 2 cases (patients 3 and 6), there were segments of cord that showed evidence of anterior nerve root thinning macroscopically.
The histology from patient 2 (E117G) revealed neuronal loss in the XIIth nerve nucleus and motor cortex. In the XIIth nerve nucleus, this was associated with neuronal cytoplasmic inclusions (NCIs) that were positive for p62 and TDP-43 (Fig. 5A). No NCIs were seen in the motor cortex, but both the XIIth nerve nucleus and the motor cortex revealed glial p62 positive, TDP-43 positive cytoplasmic inclusions. No p62 positive or TDP-43 positive inclusions were seen in the hippocampus, neocortex, or cerebellum. In addition, there was Lewy body pathology (identified with α-synuclein) confined to the brainstem. Staining with PFN1 antibodies did not reveal any PFN1-positive inclusions.
Fig. 5.

TDP-43 immunohistochemistry in 3 cases showing PFN1 mutations. (A) TDP-43 immunopositive skein-like neuronal cytoplasmic inclusion (NCI) seen in the XIIth nerve nucleus of the medulla (patient 2; E117G). (B) A globular TDP-43 immunopositive NCI seen in the anterior horn of the spinal cord (patient 3; E117G). (C) A TDP-43 immunopositive glial cytoplasmic inclusion (GCI) in the spinal cord (patient 6; Q139L). No PFN1 immunopositivity was seen in any of the regions examined in any of the 3 cases. Scale bar: (A) 40 μm, (B) 40 μm, and (C) 15 μm. Abbreviation: PFN1, profilin 1.
The other E117G case (patient 3) showed marked loss of the anterior horn neurons of the spinal cord and the motor neurons of the motor cortex. There were p62 positive and TDP-43 positive NCIs noted in the cord (Fig. 5B) and motor cortex, but they were relatively infrequent and outnumbered by p62 positive TDP-43 positive glial cytoplasmic inclusions (GCIs) in the cord and motor cortex. Occasional p62 and TDP-43 positive GCIs were seen in the XIIth nerve nucleus, but no NCIs. No Lewy Bodies were seen, and no p62 or TDP-43 positive inclusions were identified in the neocortex, cerebellum, or hippocampus. The lateral corticospinal tracts showed only mild loss of myelin. No PFN1 positivity was seen in inclusion bodies. Staining with PFN1 antibodies did not reveal any PFN1 positivity in the inclusions.
The Q139L case (patient 6) showed marked loss of the anterior horn neurons of the spinal cord and mild neuronal loss in the motor cortex. In the cord, there were no p62 positive or TDP-43 positive NCIs, but there were p62 positive, TDP-43 positive GCIs (Fig. 5C). Numerous p62 positive, TDP-43 positive GCIs were also seen in the motor cortex, again without associated NCIs. Very occasional p62 positive NCIs were seen in the XIIth nerve nucleus, but there was no TDP-43 neuronal positivity, although GCIs positive for both TDP-43 and more commonly p62 were seen in this region. There was evidence of moderate loss of myelin in the lateral corticospinal tracts of the cord. No PFN1 positivity was seen in any inclusions, and no Lewy bodies were identified.
3.6. Mutant PFN1 forms cytoplasmic granules in vitro
To assess whether the variants identified in this study showed a pathologic cellular phenotype, V5-tagged wild type PFN1 (V5-PFN1WT) and constructs with the A20T (V5-PFN1A20T), E117G (V5-PFN1E117G), and Q139L (V5-PFN1Q139L) mutations identified in our cohort were transiently transfected into HEK293T cells (Fig. 6A). In addition, the previously identified C71G (V5-PFN1C71G) mutation (Wu et al., 2012) was used as a positive control. As expected, the V5-PFNWT showed predominantly diffuse cytoplasmic localization with only a small percentage of cells (0.6%) showing any granular or punctate staining. In contrast, the V5-PFN1C71G mutant showed a significant increase in the percentage of transfected cells with distinct granules in the cytoplasm (41%, p < 0.05, Kruskal-Wallis 1-way ANOVA on ranks with Dunnett posttest). The V5-PFN1A20T mutant also had an increased percentage of cells with granular staining (12%), although this did not reach significance (Fig. 6B). This suggests that this mutation is proaggregant, but to a lesser extent than V5-PFN1C71G. In comparison, the V5-PFN1E117G and V5-PFN1Q139L mutations showed mainly diffuse cytoplasmic staining with similar percentages of cells with granules to that seen for V5-PFN1WT (both 0.7%).
Fig. 6.
Immunocytochemistry of transfected cells shows granule accumulation of mutant PFN1 in the cytoplasm. (A) Immunofluorescent images of HEK293T cells expressing V5-PFN1WT, V5-PFN1E117G, and V5-PFN1Q139L show diffuse anti-V5 staining (green). The cells expressing V5-PFN1C71G and V5-PFN1A20T show distinct punctate granular structures in the cytoplasm. Nuclei are counterstained with DAPI (blue) in the merged image. Scale bar, 10 μm. (B) Quantification of cells containing granules shows that the V5-PFN1C71G and V5-PFN1A20T constructs have an increased percentage of cells with granules compared with V5-PFN1WT, whereas V5-PFN1E117G and V5-PFN1Q139L are similar to the wild type (*p < 0.05). Bars represent means ± SEM. Abbreviations: PFN1, profilin 1; SEM, standard error of the mean.
3.7. Mutant PFN1 forms detergent-insoluble oligomers capable of recruiting wild-type PFN1
The aggregated appearance of the C71G variant, as well as the A20T variant, led us to next investigate the detergent solubility of PFN1 mutants. Aggregated proteins often show reduced solubility in denaturing detergent-based buffers. The V5-PFN1C71G and V5-PFN1A20T, but not V5-PFN1E117G or V5-PFN1Q139L, variants showed a shift in the proportions of soluble and insoluble protein compared with V5-PFN1WT (Fig. 7A and B). The proportion of V5-PFN1C71G and V5-PFN1A20T variant proteins which were soluble was significantly reduced compared with V5-PFN1WT (p < 0.001, 1-way ANOVA with Dunnett posttest). This loss of soluble protein correlated with significantly more insoluble protein for V5-PFN1C71G and V5-PFN1A20T than V5-PFN1WT (p < 0.05, Kruskal-Wallis 1-way ANOVA on ranks with Dunnett posttest, Fig. 7A and C).
Fig. 7.
Fractionation of cell lysates shows that mutant PFN1 is more insoluble than the wild type. (A–C) Solubility fractionation of HEK293T cell transfected with V5-PFN1 constructs showing lysate (L), NP-40 soluble (S), and urea soluble/NP-40 insoluble (I) fractions. The WT protein was predominantly soluble as were the E117G and Q139L proteins. Both the C71G and A20T constructs showed significant decreases in the soluble fraction compared with the WT (***p < 0.001) alongside significant increases in the insoluble fraction (*p < 0.05). Bars represent means ± SEM. There was also an increase in high molecular weight detergent resistant oligomers in the C71G and A20T insoluble fractions. (D–F) Solubility fractionation of fibroblasts from a patient with the A20T mutation and a control individual showing lysate (L), NP-40 soluble (S), and urea soluble/NP-40 insoluble (I) fractions. PFN1 was predominantly found in the soluble fraction in both cell lines. Treatment with MG132 resulted in a significant shift of PFN1 from the soluble to the insoluble fraction in the A20T cell line (***p < 0.001). There was no change to the solubility of PFN1 in the control line (ns = nonsignificant). Bars represent means ± SEM. Abbreviations: PFN1, profilin 1; SEM, standard error of the mean; WT, wild type.
V5-PFN1C71G and V5-PFN1A20T also showed increased levels of insoluble high molecular weight species (approximately 37 kDa), which corresponded to the size of dimer between transfected V5-PFN1 (approximately 24 kDa) and recruited endogenous PFN1 (approximately 12–15 kDa). High molecular weight species of approximately 50 kDa likely represent transfected V5-PFN1 homodimer. Smears of insoluble V5-PFN1C71G and V5-PFN1A20T species of >50 kDa may indicate oligomers. These dimers and oligomers represent hyperstable interactions, resistant to NP40 and urea solubilization, as well as to boiling in SDS and reducing agent (dithiothreitol).
3.8. Fibroblasts from patients with the A20T variant show decreased PFN1 solubility under stress
To determine whether mutant PFN1 was insoluble when expressed at physiological levels, we examined endogenous PFN1 in fibroblasts grown from the patient with the A20T mutation. These cells contain 1 wild type and 1 mutant copy of the gene. With vehicle treatment (0.01% DMSO), there was no difference in the amount of soluble PFN1 between the A20T mutant patient fibroblasts and those from a control individual (1-way ANOVA with Holm-Sidak posttest, Fig. 7D and E). Surprisingly, there was a lower level of insoluble PFN1 in the mutant cell line in these conditions although this did not reach significance (Kruskal-Wallis 1 way ANOVA on ranks with Tukey posttest, Fig. 7D and F).
However, when the fibroblasts were cultured in the presence of the proteasome inhibitor MG132, there was a significant increase in the amount of insoluble PFN1 in the A20T cell line compared with vehicle-treated conditions (p < 0.001, Kruskal-Wallis 1-way ANOVA on ranks with Tukey posttest, Fig. 7D and F). Impaired proteostasis in ALS is implicated by genetic causes of disease which affect protein degradation pathways and by the accumulation of proteins labeled for degradation but which fail to be cleared. The addition of MG132 also resulted in an increase in high molecular weight oligomeric species in the A20T cell line, although not in the WT line. This was similar to the profile seen when the A20T variant of PFN1 was overexpressed in HEK293T cells.
4. Discussion
Genetic characterization of a local cohort of predominantly UK ALS cases has identified 2 novel PFN1 mutations; p.A20T (in a familial patient) and p.Q139L as well as an additional 4 E117G variants in 1 familial and 3 sporadic patients (n = 6/485, 1.2%). PFN1 mutations therefore account for a minor proportion of ALS cases and occur at similar mutation frequencies to VCP, OPTN, UBQLN2, and SQSTM1 mutations in Caucasian ALS populations (Abramzon et al., 2012; Deng et al., 2011; Kwok et al., 2013; van Blitterswijk et al., 2012b). We have identified 2 additional novel PFN1 variants and demonstrated that the A20T mutation forms detergent resistant granules in transfected cells and displays an increased tendency to aggregate in patient fibroblasts.
Several lines of evidence suggest that the A20T mutation is responsible for disease in patient 1. First, taking into account our own local controls and those available from the NHLBI exome sequencing project, the A20T mutation is absent in 5208 European control individuals and 2202 African-American exomes, which suggests this variant is associated with ALS. As this residue is fully conserved in mammals, an amino acid change is likely to disrupt normal protein function. Second, in silico modeling of the crystallized PFN1 molecule reveals that, similarly to the C71 and M114 residues, the A20 residue is internal to the protein and a larger threonine substitution is likely to destabilize protein structure. Third, A20T PFN1 when overexpressed in HEK293T cells forms cytoplasmic granules, visible microscopically, and detectable biochemically as detergent-insoluble species capable of recruiting wild-type PFN1. Detergent resistant aggregates are a hallmark of ALS pathology, and their formation in vitro suggests that A20T may act via a toxic gain of function involving the sequestration of PFN1 itself as well as interacting partners. Reduced solubility of A20T PFN1 was also seen in patient fibroblasts treated with the proteasome inhibitor MG132. An unveiling of decreased A20T solubility following proteosomal inhibition parallels what is seen for TDP-43 (Scotter et al., 2014), and suggests that like TDP-43 this variant may interact with environmental stress or the age-related decline in proteostatic systems (Gamerdinger et al., 2009; Tydlacka et al., 2008). Finally, the presentation of symptoms of the patient with the A20T mutation is of a predominant lower motor neuron syndrome, with a clear spinal onset which conforms with previously reported familial PFN1 cases (Ingre et al., 2013; Wu et al., 2012). This suggests that ALS patients harboring PFN1 mutations are specific for a distinct clinical presentation of symptoms.
Three sporadic patients and a familial case possess the E117G rare variant within our ALS cohort. A meta-analysis of all published E117G cases and frequencies from local controls and Exome Variant Server, including our own study as a replication cohort confirmed this association (p = 0.001). This genetic evidence suggests a functional basis to the mechanism of action; however, no visible aggregates or decreases in mutant protein solubility were seen in HEK293T cells transfected with V5-PFN1E117G. Previous studies demonstrated formation of insoluble aggregates only with MG132 treatment, and although there was a (nonsignificant) tendency for the E117G mutant to inhibit neurite outgrowth in primary motor neuron cultures, there was no affect of the mutation on the interaction with actin (Wu et al., 2012). Our modeling of this mutation predicts there should be a reduced interaction with actin, because of a loss of hydrogen bonding compared with the native glutamic acid residue. At this stage, functional and in silico modeling of the E117G variant has suggested tentative insights into a disruptive mechanism. At a genetic level however, E117G appears to be overrepresented in SALS cases compared with FALS in the literature. This suggests E117G to behave more as a low penetrant, susceptibility factor and that multiple mutations may be required to manifest a phenotype in these specific cases, which supports an oligogenic model of disease (van Blitterswijk et al., 2012a). Exome sequencing of PFN1 E117G cases might reveal additional and/or interacting disease susceptibility alleles. In contrast however, the Q139L variant is novel but does not yield either visible aggregates or decrease protein solubility, and modeling of this variant suggests no effect on the interaction with actin or VASP or destabilization of the PFN1 structure. It is plausible that Q139L is a benign private polymorphism or alternatively could be involved in as yet unidentified protein-protein interactions that our assays may not have detected.
Brain tissue was available from 2 ALS patients who carried the E117G variant (1 FALS and 1 SALS case) and also a sporadic patient with the Q139L mutation. Neuropathologic examination revealed the 3 cases to have neuronal and/or glial TDP-43 positive and p62 positive inclusions in the motor cortex or spinal cord, with no PFN1 positive inclusions detected. This confirms previous neuropathology reports of patients with the E117G variant (van Blitterswijk et al., 2013; Yang et al., 2013). As in vitro studies have failed to identify any striking cellular phenotypes because of the E117G and the Q139L mutations, it is perhaps not surprising that the neuropathology of these cases present with a classic TDP-43 proteinopathy. It remains to be seen whether the same neuropathologic signature will be seen in cases with other PFN1 mutants that form insoluble PFN1 aggregates in vitro.
The identification and characterization of novel PFN1 variants in patients with ALS adds further support to the hypothesis that these are causative of disease. These PFN1 variants span a genetic and pathobiological spectrum from rare, nonaggregant (Q139L), to common, ALS-associated (E117G), to rare and highly proaggregant (C71G, A20T). The recruitment of wild-type PFN1 to oligomers of mutant PFN1 suggests both loss and gain of function mechanisms may be at play. The cytoskeleton is central to axonal transport and morphogenesis, the stress granule response and several other cellular processes linked to ALS. As more genetic causes of ALS are identified, common targets for drug discovery will continue to emerge.
Disclosure statement
The authors declare that there are no conflicts of interest for any of them.
Acknowledgements
This publication is dedicated to the patients and families who have contributed to this project. Postmortem tissues were provided by Medical Research Council London Neurodegenerative Diseases Brain Bank. This work was supported by grants from: the American ALS Association, the Middlemass family, Lady Edith Wolfson Trust, Heaton-Ellis Trust, Motor Neurone Disease Association, Wellcome Trust/Medical Research Council (089701/Z/09/Z), European Union (APOPIS consortium, contract LSHM-CT-2003-503330; NeuroNE Consortium; European Community's Seventh Framework Programme [FP7/2007–2013] under grant agreement number 259867, UK Medical Research Council under the aegis of the EU JPND, and Marie Curie International Incoming Fellowship), NIHR Biomedical Research Centre for Mental Health, The South London and Maudsley NHS Foundation Trust, Medical Research Council (G0900688 and MR/L021803/1), a Jack Cigman grant from King's College Hospital Charity and The Psychiatry Research Trust of the Institute of Psychiatry and Guy's and St Thomas' Charity. AriSLA co-financed with support of “5 × 1000”—Healthcare research of the Italian Ministry of Health (grants EXOMEFALS 2009 and NOVALS 2012 (N.T., V.S.). Grant support was provided by the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) (1R01NS065847 (John E. Landers) Christopher E. Shaw and Ammar Al-Chalabi receive salary support from the National Institute for Health Research (NIHR) Dementia Biomedical Research Unit at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Appendix A. Supplementary data
References
- Abramzon Y., Johnson J.O., Scholz S.W., Taylor J.P., Brunetti M., Calvo A., Mandrioli J., Benatar M., Mora G., Restagno G., Chio A., Traynor B.J. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2012;33:2231.e1–2231.e6. doi: 10.1016/j.neurobiolaging.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A., Jones A., Troakes C., King A., Al-Sarraj S., van den Berg L.H. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012;124:339–352. doi: 10.1007/s00401-012-1022-4. [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Brooks B.R. Versailles minimal dataset for diagnosis of ALS: a distillate of the 2nd Consensus Conference on accelerating the diagnosis of ALS. Versailles 2nd Consensus Conference participants. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000;1(Suppl 1):S79–S81. doi: 10.1080/14660820050515629. [DOI] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Rossi I., Casadio R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics. 2008;9(Suppl 2):S6. doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zheng Z.Z., Huang R., Chen K., Song W., Zhao B., Chen X., Yang Y., Yuan L., Shang H.F. PFN1 mutations are rare in Han Chinese populations with amyotrophic lateral sclerosis. Neurobiol. Aging. 2013;34:1922.e1–1922.e5. doi: 10.1016/j.neurobiolaging.2013.01.013. [DOI] [PubMed] [Google Scholar]
- Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud H., Dobrzeniecka S., Camu W., Meininger V., Dupre N., Dion P.A., Rouleau G.A. Mutation analysis of PFN1 in familial amyotrophic lateral sclerosis patients. Neurobiol. Aging. 2013;34:1311.e1–1311.e2. doi: 10.1016/j.neurobiolaging.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Dehouck Y., Kwasigroch J.M., Gilis D., Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.X., Chen W., Hong S.T., Boycott K.M., Gorrie G.H., Siddique N., Yang Y., Fecto F., Shi Y., Zhai H., Jiang H., Hirano M., Rampersaud E., Jansen G.H., Donkervoort S., Bigio E.H., Brooks B.R., Ajroud K., Sufit R.L., Haines J.L., Mugnaini E., Pericak-Vance M.A., Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillen L., Van Langenhove T., Engelborghs S., Vandenbulcke M., Sarafov S., Tournev I., Merlin C., Cras P., Vandenberghe R., De Deyn P.P., Jordanova A., Cruts M., Van Broeckhoven C., van der Zee J. Explorative genetic study of UBQLN2 and PFN1 in an extended Flanders-Belgian cohort of frontotemporal lobar degeneration patients. Neurobiol. Aging. 2013;34:1711.e1–1711.e5. doi: 10.1016/j.neurobiolaging.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Fratta P., Charnock J., Collins T., Devoy A., Howard R., Malaspina A., Orrell R., Sidle K., Clarke J., Shoai M., Lu C.H., Hardy J., Plagnol V., Fisher E.M. Profilin1 E117G is a moderate risk factor for amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2013;85:506–508. doi: 10.1136/jnnp-2013-306761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M., Hajieva P., Kaya A.M., Wolfrum U., Hartl F.U., Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard S.M., Korning P.G., Tolstrup N., Engelbrecht J., Rouze P., Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingre C., Landers J.E., Rizik N., Volk A.E., Akimoto C., Birve A., Hubers A., Keagle P.J., Piotrowska K., Press R., Andersen P.M., Ludolph A.C., Weishaupt J.H. A novel phosphorylation site mutation in profilin 1 revealed in a large screen of US, Nordic, and German amyotrophic lateral sclerosis/frontotemporal dementia cohorts. Neurobiol. Aging. 2013;34:1708.e1–1708.e6. doi: 10.1016/j.neurobiolaging.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.U., Zoete V., Michielin O., Guex N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics. 2012;13:173. doi: 10.1186/1471-2105-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C.T., Morris A., de Belleroche J.S. Sequestosome-1 (SQSTM1) sequence variants in ALS cases in the UK: prevalence and coexistence of SQSTM1 mutations in ALS kindred with PDB. Eur. J. Hum. Genet. 2013;22:492–496. doi: 10.1038/ejhg.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lattante S., Le Ber I., Camuzat A., Brice A., Kabashi E. Mutations in the PFN1 gene are not a common cause in patients with amyotrophic lateral sclerosis and frontotemporal lobar degeneration in France. Neurobiol. Aging. 2012;34:1709.e1–1709.e2. doi: 10.1016/j.neurobiolaging.2012.10.026. [DOI] [PubMed] [Google Scholar]
- Maekawa S., Leigh P.N., King A., Jones E., Steele J.C., Bodi I., Shaw C.E., Hortobagyi T., Al-Sarraj S. TDP-43 is consistently co-localized with ubiquitinated inclusions in sporadic and Guam amyotrophic lateral sclerosis but not in familial amyotrophic lateral sclerosis with and without SOD1 mutations. Neuropathology. 2009;29:672–683. doi: 10.1111/j.1440-1789.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotter E.L., Vance C., Nishimura A.L., Lee Y.B., Chen H.J., Urwin H., Sardone V., Mitchell J.C., Rogelj B., Rubinsztein D.C., Shaw C.E. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J. Cell Sci. 2014;127(Pt 6):1263–1278. doi: 10.1242/jcs.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiloca C., Ticozzi N., Pensato V., Corrado L., Del Bo R., Bertolin C., Fenoglio C., Gagliardi S., Calini D., Lauria G., Castellotti B., Bagarotti A., Corti S., Galimberti D., Cagnin A., Gabelli C., Ranieri M., Ceroni M., Siciliano G., Mazzini L., Cereda C., Scarpini E., Soraru G., Comi G.P., D'Alfonso S., Gellera C., Ratti A., Landers J.E., Silani V. Screening of the PFN1 gene in sporadic amyotrophic lateral sclerosis and in frontotemporal dementia. Neurobiol. Aging. 2012;34:1517.e9–1517.e10. doi: 10.1016/j.neurobiolaging.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tydlacka S., Wang C.E., Wang X., Li S., Li X.J. Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J. Neurosci. 2008;28:13285–13295. doi: 10.1523/JNEUROSCI.4393-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M., Baker M.C., Bieniek K.F., Knopman D.S., Josephs K.A., Boeve B., Caselli R., Wszolek Z.K., Petersen R., Graff-Radford N.R., Boylan K.B., Dickson D.W., Rademakers R. Profilin-1 mutations are rare in patients with amyotrophic lateral sclerosis and frontotemporal dementia. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013;4:463–469. doi: 10.3109/21678421.2013.787630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M., van Es M.A., Hennekam E.A., Dooijes D., van Rheenen W., Medic J., Bourque P.R., Schelhaas H.J., van der Kooi A.J., de Visser M., de Bakker P.I., Veldink J.H., van den Berg L.H. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012;21:3776–3784. doi: 10.1093/hmg/dds199. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M., van Vught P.W., van Es M.A., Schelhaas H.J., van der Kooi A.J., de Visser M., Veldink J.H., van den Berg L.H. Novel optineurin mutations in sporadic amyotrophic lateral sclerosis patients. Neurobiol. Aging. 2012;33:1016.e1–1016.e7. doi: 10.1016/j.neurobiolaging.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Vance C., Scotter E.L., Nishimura A.L., Troakes C., Mitchell J.C., Kathe C., Urwin H., Manser C., Miller C.C., Hortobagyi T., Dragunow M., Rogelj B., Shaw C.E. ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum. Mol. Genet. 2013;22:2676–2688. doi: 10.1093/hmg/ddt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth, C.L., Burke, D.F., Blundell, T.L., 2007. Estimating the effects of single nucleotide polymorphisms on protein structure: how good are we at identifying likely disease associated mutations? Proceedings of Molecular Interactions - Bringing Chemistry to Life, 11–26.
- Wu C.H., Fallini C., Ticozzi N., Keagle P.J., Sapp P.C., Piotrowska K., Lowe P., Koppers M., McKenna-Yasek D., Baron D.M., Kost J.E., Gonzalez-Perez P., Fox A.D., Adams J., Taroni F., Tiloca C., Leclerc A.L., Chafe S.C., Mangroo D., Moore M.J., Zitzewitz J.A., Xu Z.S., van den Berg L.H., Glass J.D., Siciliano G., Cirulli E.T., Goldstein D.B., Salachas F., Meininger V., Rossoll W., Ratti A., Gellera C., Bosco D.A., Bassell G.J., Silani V., Drory V.E., Brown R.H., Jr., Landers J.E. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Fifita J.A., Williams K.L., Warraich S.T., Pamphlett R., Nicholson G.A., Blair I.P. Mutation analysis and immunopathological studies of PFN1 in familial and sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2013;34:2235.e7–2235.e10. doi: 10.1016/j.neurobiolaging.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Zou Z., Sun Q., Liu M., Li X., Cui L. Mutations in the profilin 1 gene are not common in amyotrophic lateral sclerosis of Chinese origin. Neurobiol. Aging. 2013;34:1713.e5–1713.e6. doi: 10.1016/j.neurobiolaging.2012.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.