Abstract
In order to investigate associations of SCNN1A, SCNN1G and SCNN1B genes with blood pressure (BP) in Han Chinese population, we included 2 880 participants did not use antihypertensive medication in the month prior to the baseline survey in the current analysis. Forty-four tag-SNPs in epithelial sodium channel (ENaC) genes were selected and genotyped and nine BP measurements were obtained during 3-day examination. In single-marker analyses, we identified significant associations of SCNN1A marker rs13306613 with diastolic BP (DBP) and SCNN1B marker rs12447134 with systolic BP (SBP) under codominant model after Bonferroni correction (P= 2.82×10−5 and 4.63×10−4, respectively). In addition, 5 SNPs in SCNN1G and 4 SNPs in SCNN1B achieved nominal significance for SBP, DBP or mean arterial pressure (MAP) under the additive model. For example, the minor C allele of rs5735 in SCNN1G gene was associated with decreased SBP, DBP and MAP (P=0.016, 5.41×10−3, and 4.36×10−3, respectively). Gene-based results showed significant associations of SCNN1G and SCNN1Bwith BP levels. This study suggested that ENaC genes play important roles in BP regulation in the Han Chinese population. Future studies are warranted to replicate these findings and functional studies are needed to identify true causal variants in ENaC genes.
Keywords: epithelial sodium channel, genetic variants, blood pressure, hypertension, GenSalt
INTRODUCTION
Hypertension is a challenging public health problem worldwide due to its high prevalence and associated increases in risks of cardiovascular disease (CVD). 1-3As the leading risk factor for mortality globally, approximately 13.5% of premature deaths were attributable to high blood pressure (BP) in 2001. 3 High BP is influenced by multiple environmental and genetic factors, and heritability studies indicate that about 30-60% of the BP variation in the population is genetically determined. 4 Although some loci were reported to be associated with the increased BP, the majority of the genetic contribution to variation in BP is still unclear.
Kidney, as a critical organ, is very important in BP regulation by its effect on sodium balance. The epithelial sodium channel (ENaC), which locates at the distal nephron, plays an essential role in controlling sodium balance and extracellular fluid volume through the renal sodium reabsorption and excretion. 5, 6 ENaC is composed of α, β and γ homologous subunits which are encoded by SCNN1A, SCNN1B and SCNN1G genes, respectively. It is mostly expressed in the luminal membrane of connecting tubule cells and principle cells of collecting duct. 7
Some studies have reported that rare mutations in the SCNN1B and SCNN1G genes lead to Liddle's syndrome, a severe and hereditary form of early onset hypertension. 8, 9 In addition, Kzkizoe et al has found that inappropriate ENaC expression and activation might be the underlying mechanisms for salt-sensitive hypertension and organ damages developed in Dahl salt-sensitive rats. 10 Although results are inconsistent, SCNN1B and SCNN1G genes have been implicated in the physiological variation of systolic BP (SBP). 11-13 Thus, the association between common variants of ENaC genes and essential hypertension in humans should be given more attention. 14, 15
The present study aimed to investigate associations of SCNN1A,SCNN1G and SCNN1B genes with SBP, diastolic BP (DBP) and mean arterial pressure (MAP) using both single-marker and gene-based analyses in 2 880 Han Chinese participants of the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) study and provide insights into the molecular pathogenesis of hypertension or elevated BP.
METHODS
Study population
The GenSalt study was conducted in six rural villages in Northern China from 2003 to 2005. Detailed information on the study design and methods of GenSalt study has been published elsewhere. 16 Briefly, a community based BP screening was carried out among persons aged 18-60 years to identify potential probands. Those with mean SBP of 130-160 mmHg and/or DBP of 85-100 mmHg and no use of antihypertensive medication were recruited, as well as with their parents, siblings, spouses, and offspring. Individuals, who had stage-2 hypertension, secondary hypertension, a history of CVD or diabetes, or were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study. Finally, 3 142 individuals in 633 families participated in the GenSalt study and all participants were of Han Chinese ethnicity. Two hundred and sixty-two participants were excluded from the current analysis due to antihypertensive treatment in the month prior to baseline survey.
Written informed consents were obtained from all GenSalt participants after detailed explanation of the study. The study has been approved by the Institutional Review Board at all the participant institutions.
Data collection
Information on family structure, demographic characteristics, personal and family medical history was collected by a trained staff using a standardized questionnaire on the first day of baseline observation. Body weight and height were measured twice with the participant in light indoor clothing without shoes. Body mass index (BMI) was calculated as kilograms per square meter (kg/m2). BP was measured three times on each day of the 3-day baseline period when the participants were in the sitting position after 5 minutes of rest by the same trained and certified technician using a Hawksley random-zero sphygmomanometer (Hawksley& Sons Ltd, Lancing, UK; zero range 0–20 mmHg) according to the protocols recommended by the American Heart Association. 17 The appropriate cuff size was selected based on each participant's arm circumference. 17 Additionally, participants were advised to avoid drinking, cigarette smoking, having coffee/tea or doing exercise for at least 30 minutes before their BP measurements. BP levels were calculated as the average of the nine measurements. MAP was calculated as 1/3 SBP plus 2/3 DBP.
Genotype data and quality control
SCNN1A, SCNN1G and SCNN1B genes were identified based on their potential biological effect on BP regulation. Seventeen SNPs were genotyped using ABI SNPlex platform (Applied Biosystems, Foster City, CA) and additional 42 SNPs were genotyped on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, CA) in a subsample of GenSalt participants (N=1,881). SNPs with minor allele frequency (MAF) less than 1%, genotyping call rate less than 90% and deviation from Hardy-Weinberg equilibrium (HWE) after Bonferroni correction for multiple testing were excluded. After quality control, we selected tag-SNPs from these genes with pairwise r2 thresholds of less than 0.9. A total of 44 tag-SNPs were included in the current analysis. Haploview software (version 4.2, http://www.broad.mit.edu/mpg/haploview) was used to conduct quality control and tag-SNPs selection. 18 Detailed information on all tag-SNPs, including the gene symbol, chromosome, physical position, major/minor alleles, call rate, MAF, HWE P value and genotyping platform, has been shown in Supplementary Table 1.
Statistical analysis
The Mendelian consistency of the SNP genotype data was assessed by PLINK software (http://pngu.mgh.harvard.edu/purcell/plink/) and PedCheck. 19, 20 If Mendelian inconsistencies were found, genotypes for related SNPs in those families were set to be missing. Baseline characteristics and BP were presented as mean±SD for continuous variables and as percentages for categorical variables. Mixed-effect linear models were used to assess associations of SNPs with BP to account for the non-independence of family members under additive, codominant, dominant, and recessive genetic models. Age, gender, BMI, room temperature of BP measurement, and field center were adjusted in the mixed models. All single-marker analyses were conducted using SAS statistical software (version 9.2, SAS Institute, Cary, NC).
The truncated product method (TPM), which combines P values from single-marker association analyses, was used to evaluate the overall association of a candidate gene with BP levels. 21 Truncation point was set as τ=0.10, and the P value for TPM was estimated by 1,000,000 simulations. Sensitivity analyses were conducted using the TPM after excluding the most significant SNP within a gene. Gene-based gene-gene interaction was evaluated by GG_tProd method by combining P values from single-marker based interaction term under additive model.22 Gene-based analyses and Gene-based gene-gene interaction were performed using R software (Version 3.0.1; http://www.r-project.org).
Bonferroni correction was used to adjust for multiple testing. The α-thresholds were 1.14×10−3 (0.05/44) and 0.017 (0.05/3) for single-marker analyses and gene-based analyses, respectively.
RESULTS
Table 1 presents the characteristics of 2 880 GenSalt participants. On average, participants were 48.5 years of age, had a mean BMI of 23.0 kg/m2, and mean baseline SBP, DBP and MAP of 122.4 mmHg, 73.7 mmHg and 89.9mmHg, respectively. The average room temperature of BP measurement was 22.1°C. A total of 1 501 (51.2%) participants were male.
Table 1.
Characteristics of 2 880 participants in GenSalt study
| Variables | Mean ± SD | Range |
|---|---|---|
| Age, years | 48.5±16.5 | (15.0, 93.0) |
| Male, n (%) | 1501 (52.1) | |
| Body Mass Index, kg/m2 | 23.0±3.2 | (14.3-37.8) |
| Baseline Systolic Blood Pressure, mmHg | 122.4±19.3 | (80.9-222.2) |
| Baseline Diastolic Blood Pressure, mmHg | 73.7±10.6 | (38.9-115.1) |
| Baseline Mean Arterial Pressure, mmHg | 89.9±12.1 | (58.7-146.5) |
| Room temperature (°C) | 22.1±3.2 | (14.3-29.7) |
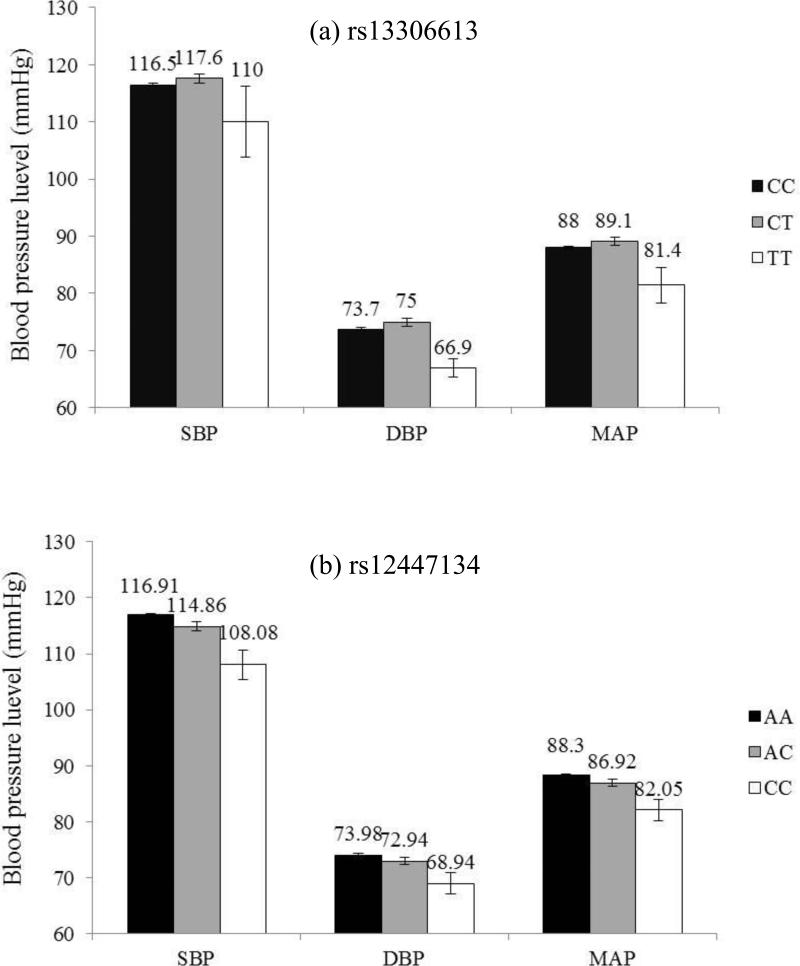
Analysis of 44 tag-SNPs in the 3 ENaC genes revealed their associations with BP levels. As shown in Table 2, significant associations of SCNN1A marker rs13306613 with DBP and SCNN1B marker rs12447134 with SBP were identified under codominant model even after Bonferroni correction (P= 2.82×10−5 and 4.63×10−4, respectively). Mean DBP (95% CIs) were 73.7 (73.2, 74.2), 75.0 (73.6, 76.4) and 66.9 (63.7, 70.1) mmHg for participants with rs13306613 genotype C/C, C/T and T/T, respectively [Figure 1(a)], and mean DBP (95% CIs) were 116.9 (116.3, 117.6), 114.9 (113.3, 116.4) and 108.1 (102.9, 113.2) mmHg for participants with rs12447134 genotype A/A, A/C and C/C, respectively [Figure 1(b)]. Although the associations were not significant after adjustment for multiple testing, 5 tag-SNPs in the SCNN1G gene (rs4073291, rs5735, rs4299163, rs4401050, rs4499238) and 4 tag-SNPs in the SCNN1B gene (rs6497659, rs7205273, rs12447134 and rs63982) achieved nominal significance for SBP, DBP or MAP under the additive genetic model (Table 3). SBP, DBP and MAP were inversely associated with the number of minor C allele of rs5735 with the regression coefficients of -1.58, -1.08, and -1.24, respectively. Other detailed information, including mean SBP, DBP and MAP according to genotypes and corresponding P values from single-marker analyses of each SNP, has been presented in Supplementary Table 2 and Supplementary Table 3.
Table 2.
P values for the association of SNPs in ENaC genes and BP levels under codominant, dominant and recessive modelsa
| Gene | SNP | SBP | DBP | MAP |
|---|---|---|---|---|
| Codominant | ||||
| SCNN1A | rs13306613 | 0.347 | 2.82×10−5 b | 0.029 |
| rs11064153 | 0.055 | 0.054 | 0.038 | |
| SCNN1G | rs4421986 | 0.571 | 0.292 | 0.020 |
| rs5735 | 0.047 | 0.015 | 0.016 | |
| rs4401050 | 0.067 | 0.048 | 0.020 | |
| rs4499238 | 0.037 | 0.322 | 0.122 | |
| SCNN1B | rs6497659 | 0.132 | 0.031 | 0.038 |
| rs7205273 | 0.061 | 0.057 | 0.033 | |
| rs12447134 | 4.63×10−4 b | 0.010 | 1.61×10−3 | |
| rs63982 | 0.011 | 0.052 | 0.020 | |
| rs250567 | 0.176 | 8.65×10−3 | 0.024 | |
| Dominant | ||||
| SCNN1A | rs11064153 | 0.030 | 0.061 | 0.032 |
| SCNN1G | rs4073291 | 0.029 | 0.086 | 0.039 |
| rs5735 | 0.014 | 0.018 | 8.45×10−3 | |
| SCNN1B | rs6497659 | 0.049 | 8.85×10−3 | 0.011 |
| rs12447134 | 4.78×10−3 | 0.047 | 0.014 | |
| Recessive | ||||
| SCNN1A | rs13306613 | 0.292 | 2.51×10−3 | 0.028 |
| SCNN1G | rs5735 | 0.404 | 0.030 | 0.077 |
| rs4299163 | 0.293 | 0.028 | 0.072 | |
| rs4401050 | 0.099 | 0.054 | 0.029 | |
| rs4499238 | 0.041 | 0.191 | 0.089 | |
| SCNN1B | rs7205273 | 0.028 | 0.018 | 0.011 |
| rs239345 | 0.048 | 0.043 | 0.028 | |
| rs12447134 | 1.43×10−3 | 9.12×10−3 | 2.67×10−3 | |
| rs63982 | 3.51×10−3 | 0.015 | 5.18×10−3 | |
| rs250567 | 0.080 | 0.041 | 0.038 |
Adjustment for age, gender, body mass index, room temperature of BP measurement, and field center;
Statistically significant after Bonferroni correction.
Figure 1.
Systolic blood pressure (SBP), diastolic blood pressure (DBP) and mean arterial pressure (MAP) levels according to genotype of SNP rs13306613 in the SCNN1A gene (a) and rs12447134 in the SCNN1B gene (b) after adjustment for age, gender, BMI, room temperature of BP measurement, and field center. Values are means with standard error bars.
Table 3.
Association of SNPs in ENaC genes with BP levels under additive model a
| Gene | SNP | SBP |
DBP |
MAP |
|||
|---|---|---|---|---|---|---|---|
| β(SE) | P | β(SE) | P | β(SE) | P | ||
| SCNN1G | rs4073291 | −1.23 (0.60) | 0.040 | −0.77 (0.44) | 0.079 | −0.93 (0.46) | 0.043 |
| rs5735 | −1.58 (0.66) | 0.016 | −1.08 (−0.39) | 5.41×10−3 | −1.24 (0.43) | 4.36×10−3 | |
| rs4299163 | −1.32 (0.77) | 0.089 | −0.96 (0.48) | 0.047 | −1.07 (0.53) | 0.045 | |
| rs4401050 | −1.57 (0.75) | 0.035 | −0.94 (0.44) | 0.032 | −1.13 (0.48) | 0.020 | |
| rs4499238 | −1.51 (0.68) | 0.027 | −0.58 (−0.49) | 0.232 | −0.89 (0.51) | 0.085 | |
| SCNN1B | rs6497659 | −0.62 (0.44) | 0.154 | −0.69 (0.33) | 0.041 | −0.65 (0.34) | 0.055 |
| rs7205273 | −0.99 (0.49) | 0.045 | −0.52 (0.31) | 0.098 | −0.68 (0.34) | 0.047 | |
| rs12447134 | −2.50 (0.78) | 1.28×10−3 | −1.33 (0.57) | 0.019 | −1.72 (0.60) | 4.23×10−3 | |
| rs63982 | −1.00 (0.39) | 0.010 | −0.47 (0.29) | 0.108 | −0.64 (0.30) | 0.031 | |
Adjustment for age, gender, body mass index, room temperature of BP measurement, and field center.
In addition, gene-based analyses revealed that SCNN1G was significantly associated with SBP, DBP and MAP when using P values from additive model, while SCNN1B were significantly associated with SBP, DBP and MAP when using P values from recessive model (Table 4). In sensitivity analyses removing the most significant single-marker, the overall gene-based association of SCNN1G with SBP and MAP remained significant with P values of 1.00×10−3and 1.60×10−3, respectively. In addition, marginally significant SCNN1A×SCNN1G gene-based gene-gene interaction for SBP and MAP were identified (Supplementary Table 4).
Table 4.
Gene-based associations of ENaC genes with BP levels
| Gene | SBP | DBP | MAP |
|---|---|---|---|
| Additive model | |||
| SCNN1A | 0.6270 | 0.5392 | 0.4280 |
| SCNN1G | 2.00×10−4a | 6.00×10−4 | 1.00×10−5b |
| SCNN1B | 0.0820 | 0.0324 | 0.0012 |
| Codominant model | |||
| SCNN1A | 0.3612 | 0.0074 | 0.3410 |
| SCNN1G | 0.0066 | 0.0986 | 0.0046 |
| SCNN1B | 0.0798 | 0.0014 | 0.0802 |
| Dominant model | |||
| SCNN1A | 0.2854 | 0.4400 | 0.3168 |
| SCNN1G | 0.0020 | 0.0580 | 0.0018 |
| SCNN1B | 0.4098 | 0.2332 | 0.4040 |
| Recessive model | |||
| SCNN1A | 0.4500 | 0.0916 | 0.3210 |
| SCNN1G | 0.0890 | 0.0882 | 0.0714 |
| SCNN1B | 0.0016 | 6.00×10−4 | 0.0010 |
P value for sensitivity analysis is 1.00×10−3;
P value for sensitivity analysis is 1.60×10−3.
DISCUSSION
In the present study, we identified 2 common variants, rs13306613 in SCNN1A gene and rs12447134 in SCNN1B gene, were significantly associated BP levels. Although associations of variants in the SCNN1G gene with BP levels were not significant after adjustment for multiple testing, gene-based associations of SCNN1G with BP levels achieved statistical significance, as well as the association of SCNN1B gene. These findings suggest that ENaC genes play important roles in BP regulation.
SCNN1A marker rs13306613 was associated with DBP level in the current analysis, which is consistent with the previous study conducted in Korean population. 23 This marker is located in intron region of SCNN1A, with no known functional relevance. Several other SCNN1A variants have been associated with BP traits. 24-26 For example, our recent study has identified significant association of rs11064153 with longitudinal BP change, 26 Iwai and colleagues reported the association of rs3759324 with cross-sectional BP and hypertension in a Japanese population. 24 In addition, gene-based analysis also found a significant association of SCNN1A with DBP. However, the result of sensitivity analysis when removing significant single-marker rs13306613 suggested the gene-based association was driven by this marker.
Several researchers have studied the role of SCNN1G gene in human essential hypertension but reported conflicting results. 11, 14, 27-30 Although most studies concluded that this subunit is unlikely to play a significant role in the pathogenesis of human essential hypertension, Wong et al found significant linkage between SBP and markers of SCNN1G gene. 11 However, results of single-marker analyses in SCNN1G gene were still inconsistent. For example, Iwai N et al observed minor allele of variant G(-173)A (also known as rs5718), located in the promoter region of SCNN1G gene, had significantly decreased SBP. 14 While a study conducted in Australian population has observed that minor allele of rs13331086, which locates in the same block as rs4401050, was associated with 1 mmHg increase in SBP and 0.52 mmHg increase with DBP. 29 Although mutations on NEDD4L, NDFIP2 or USP2 genes could influence BP or hypertension through regulating the expression of ENaC genes in either Korean or Chinese population, only nominal associations of variants in ENaC genes with BP have been observed. 23 The current study has shown similar nominal results that minor alleles of rs5735, rs4299163 and rs4401050 are associated with decreased BP. Although results were not significant after adjustment for multiple testing, the current study reported significant gene-based association of SCNN1G gene with BP even after removing the most significant single-marker. This result has confirmed the role of SCNN1G gene in BP regulation. 11 Since the single-marker effect on BP was minor and multiple variants in the same gene act jointly in leading to disease phenotype, gene-based analysis will have a higher power than single-marker analyses.
Mutations in SCNN1B gene cause Liddle's syndrome and pseudohyoaldosteronism. Investigators have made much effort to sequence the exons of SCNN1B gene to identify mutations associated with BP in general population. 8, 9, 30 However, the significances of SCNN1B SNPs appear to be more important in African ancestries than in Whites, Japanese or Korean population. 23, 31-34 While most of previous studies focused on the coding sequence of SCNN1B gene, the current study has provided some evidence on SNPs in non-coding regions. Our previous study has reported minor allele of rs7205273 is significantly associated with lower DBP in Han Chinese population with high levels of physical activity. 35 In addition, gene-based analyses have observed significant association between SCNN1B gene and MAP. The functional implications of rs7205273, rs12447134 and rs63982 are possible intronic enhancers for the regulation of gene expression. All findings contribute to the evidence suggesting a role of SCNN1B gene on regulating BP in Chinese population, but further studies are still needed to replicate the results. In order to fully interpret these associations and find the true relationship, functional studies are still needed in the future.
Our study has several important strengths. To our knowledge, this is the first study to investigate associations of ENaC genes with BP levels in Han Chinese general population. Second, participants in our study were recruited from a semi-isolated population with the purpose of minimizing genetic and environmental heterogeneity. Third, participants were excluded if they were treated with antihypertensive medications in past month prior to baseline examination to ensure accurate BP measurements. However, some potential limitations of our study should be noted. First, replication study is stilled needed to confirm these results. Second, some important variants, such as rs5718, may not be included in the current study, although our genotyping platform has good performance and considerable potential to identify candidate susceptibility or resistance genetic factors. 36
In conclusion, we provide the first evidence that genetic variants in the ENaC genes may regulate BP levels in Han Chinese population. In concordance with single-marker findings, gene-based analysis revealed effects of SCNN1G and SCNN1B on BP regulation. Replications of these results in other populations are needed and further functional studies are critically important for identifying true causal variants.
Supplementary Material
SUMMARY TABLE.
What is known about this topic
The epithelial sodium channel (ENaC) plays an important role in controlling sodium balance and extracellular fluid volume through the renal sodium reabsorption and excretion.
SCNN1A, SCNN1G and SCNN1B genes are associated with blood pressure related traits, such as Liddle's syndrome and salt-sensitivity of blood pressure.
The findings of studies of the relationship between ENaC genes and blood pressure have been inconsistent and the associations have not been well examined in Han Chinese population.
What this study adds
Significant associations of SCNN1A marker rs13306613 with diastolic blood pressure and SCNN1B marker rs12447134 with systolic blood pressure were identified under codominant model in single-marker analyses.
Gene-based analyses revealed that SCNN1G and SCNN1B genes are associated with blood pressure traits.
Our results suggest that ENaC genes play important roles in blood pressure regulation in Han Chinese population.
ACKNOWLEDGEMENT
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD.
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
Supplementary information is available at JHH's website.
REFERENCE
- 1.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Gu D, Kelly TN, Wu X, Chen J, Duan X, Huang JF, et al. Blood pressure and risk of cardiovascular disease in Chinese men and women. Am J Hypertens. 2008;21:265–272. doi: 10.1038/ajh.2007.59. [DOI] [PubMed] [Google Scholar]
- 3.Lawes CM, Vander Hoorn S, Rodgers A. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371:1513–1518. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 4.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 5.Thomas CP, Itani OA. New insights into epithelial sodium channel function in the kidney: site of action, regulation by ubiquitin ligases, serum- and glucocorticoid-inducible kinase and proteolysis. Curr Opin Nephrol Hypertens. 2004;13:541–548. doi: 10.1097/00041552-200409000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Shehata MF. Regulation of the epithelial sodium channel [ENaC] in kidneys of salt-sensitive Dahl rats: insights on alternative splicing. Int Arch Med. 2009;2:28. doi: 10.1186/1755-7682-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bankir L, Bichet DG, Bouby N. Vasopressin V2 receptors, ENaC, and sodium reabsorption: a risk factor for hypertension? Am J Physiol Renal Physiol. 2010;299:F917–F928. doi: 10.1152/ajprenal.00413.2010. [DOI] [PubMed] [Google Scholar]
- 8.Furuhashi M, Kitamura K, Adachi M, Miyoshi T, Wakida N, Ura N, et al. Liddle's syndrome caused by a novel mutation in the proline-rich PY motif of the epithelial sodium channel beta-subunit. J Clin Endocrinol Metab. 2005;90:340–344. doi: 10.1210/jc.2004-1027. [DOI] [PubMed] [Google Scholar]
- 9.Wang LP, Gao LG, Zhou XL, Wu HY, Zhang L, Wen D, et al. Genetic diagnosis of Liddle's syndrome by mutation analysis of SCNN1B and SCNN1G in a Chinese family. Chin Med J (Engl) 2012;125:1401–1404. [PubMed] [Google Scholar]
- 10.Kakizoe Y, Kitamura K, Ko T, Wakida N, Maekawa A, Miyoshi T, et al. Aberrant ENaC activation in Dahl salt-sensitive rats. J Hypertens. 2009;27:1679–1689. doi: 10.1097/HJH.0b013e32832c7d23. [DOI] [PubMed] [Google Scholar]
- 11.Wong ZY, Stebbing M, Ellis JA, Lamantia A, Harrap SB. Genetic linkage of beta and gamma subunits of epithelial sodium channel to systolic blood pressure. Lancet. 1999;353:1222–1225. doi: 10.1016/S0140-6736(98)10118-6. [DOI] [PubMed] [Google Scholar]
- 12.Niu T, Xu X, Cordell HJ, Rogus J, Zhou Y, Fang Z, et al. Linkage analysis of candidate genes and gene-gene interactions in chinese hypertensive sib pairs. Hypertension. 1999;33:1332–1337. doi: 10.1161/01.hyp.33.6.1332. [DOI] [PubMed] [Google Scholar]
- 13.Munroe PB, Strautnieks SS, Farrall M, Daniel HI, Lawson M, DeFreitas P, et al. Absence of linkage of the epithelial sodium channel to hypertension in black Caribbeans. Am J Hypertens. 1998;11:942–945. doi: 10.1016/s0895-7061(98)00092-2. [DOI] [PubMed] [Google Scholar]
- 14.Iwai N, Baba S, Mannami T, Katsuya T, Higaki J, Ogihara T, et al. Association of sodium channel gamma-subunit promoter variant with blood pressure. Hypertension. 2001;38:86–89. doi: 10.1161/01.hyp.38.1.86. [DOI] [PubMed] [Google Scholar]
- 15.Wang XF, Lu XM, Lin RY, Wang SZ, Zhang LP, Qian J, et al. Lack of association of functional variants in alpha-ENaC gene and essential hypertension in two ethnic groups in China. Kidney Blood Press Res. 2008;31:268–273. doi: 10.1159/000151286. [DOI] [PubMed] [Google Scholar]
- 16.GenSalt Collaborative Research Group GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, et al. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88:2460–2470. doi: 10.1161/01.cir.88.5.2460. [DOI] [PubMed] [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 21.Sheng X, Yang J. Truncated Product Methods for Panel Unit Root Tests. Oxf Bull Econ Stat. 2013;75:624–636. doi: 10.1111/j.1468-0084.2012.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Clark AG, Keinan A. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet. 2013;9:e1003321. doi: 10.1371/journal.pgen.1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin HS, Hong KW, Lim JE, Hwang SY, Lee SH, Shin C, et al. Genetic variations in the sodium balance-regulating genes ENaC, NEDD4L, NDFIP2 and USP2 influence blood pressure and hypertension. Kidney Blood Press Res. 2010;33:15–23. doi: 10.1159/000275706. [DOI] [PubMed] [Google Scholar]
- 24.Iwai N, Baba S, Mannami T, Ogihara T, Ogata J. Association of a sodium channel alpha subunit promoter variant with blood pressure. J Am Soc Nephrol. 2002;13:80–85. doi: 10.1681/ASN.V13180. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Li NF, Hong J, Zhang L, Zhou L, Li T, et al. [Relationship between four single nucleotide polymorphisms of epithelial sodium channel alpha subunit gene and essential hypertension of Kazakhs in Xinjiang]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2009;31:740–745. doi: 10.3881/j.issn.1000-503X.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, He J, Gu D, Hixson JE, Huang J, Rao DC, et al. Associations of Epithelial Sodium Channel Genes With Blood Pressure Changes and Hypertension Incidence: The GenSalt Study. Am J Hypertens. doi: 10.1093/ajh/hpu060. e-pub ahead of print 15 April 2014; doi: 10.1093/ajh/hpu060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persu A, Coscoy S, Houot AM, Corvol P, Barbry P, Jeunemaitre X. Polymorphisms of the gamma subunit of the epithelial Na+ channel in essential hypertension. J Hypertens. 1999;17:639–645. doi: 10.1097/00004872-199917050-00007. [DOI] [PubMed] [Google Scholar]
- 28.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, et al. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–1664. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 29.Büsst CJ, Scurrah KJ, Ellis JA, Harrap SB. Selective genotyping reveals association between the epithelial sodium channel gamma-subunit and systolic blood pressure. Hypertension. 2007;50:672–678. doi: 10.1161/HYPERTENSIONAHA.107.089128. [DOI] [PubMed] [Google Scholar]
- 30.Melander O, Orho M, Fagerudd J, Bengtsson K, Groop PH, Mattiasson I, et al. Mutations and variants of the epithelial sodium channel gene in Liddle's syndrome and primary hypertension. Hypertension. 1998;31:1118–1124. doi: 10.1161/01.hyp.31.5.1118. [DOI] [PubMed] [Google Scholar]
- 31.Dong YB, Zhu HD, Baker EH, Sagnella GA, MacGregor GA, Carter ND, et al. T594M and G442V polymorphisms of the sodium channel beta subunit and hypertension in a black population. J Hum Hypertens. 2001;15:425–430. doi: 10.1038/sj.jhh.1001182. [DOI] [PubMed] [Google Scholar]
- 32.Matsubara M, Metoki H, Suzuki M, Fujiwara T, Kikuya M, Michimata M, et al. Genotypes of the betaENaC gene have little influence on blood pressure level in the Japanese population. Am J Hypertens. 2002;15:189–192. doi: 10.1016/s0895-7061(01)02266-x. [DOI] [PubMed] [Google Scholar]
- 33.Persu A, Barbry P, Bassilana F, Houot AM, Mengual R, Lazdunski M, et al. Genetic analysis of the beta subunit of the epithelial Na+ channel in essential hypertension. Hypertension. 1998;32:129–137. doi: 10.1161/01.hyp.32.1.129. [DOI] [PubMed] [Google Scholar]
- 34.Chang H, Fujita T. Lack of mutations in epithelial sodium channel beta-subunit gene in human subjects with hypertension. J Hypertens. 1996;14:1417–1419. doi: 10.1097/00004872-199612000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Montasser ME, Gu D, Chen J, Shimmin LC, Gu C, Kelly TN, et al. Interactions of genetic variants with physical activity are associated with blood pressure in Chinese: the GenSalt study. Am J Hypertens. 2011;24:1035–1040. doi: 10.1038/ajh.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida N, Koike A, Tajima A, Ogasawara Y, Ishibashi Y, Uehara Y, et al. Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals. BMC Genomics. 2008;9:431. doi: 10.1186/1471-2164-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.