Abstract
Fibrosis is an end-stage response to tissue injury that is associated with loss of organ function as a result of excess extracellular matrix (ECM) production by fibroblasts. In skin, pathologic fibrosis is evident during keloid scar formation, systemic sclerosis (SSc), and morphea. Dermal fibroblasts in these fibrotic diseases exhibit increased Wnt/β-catenin signaling, a pathway that is sufficient to cause fibrosis in mice. However, in the context of this complex pathology, the precise pro-fibrotic consequences of Wnt/β-catenin signaling are not known. We found that expression of stabilized β-catenin in mouse dermal fibroblasts resulted in spontaneous, progressive skin fibrosis with thickened collagen fibres and altered collagen fibril morphology. The fibrotic phenotype was predominated by resident dermal fibroblasts. Genome-wide profiling of the fibrotic mouse dermis revealed elevated expression of matrix-encoding genes, and the promoter regions of these genes were enriched for Tcf/Lef family transcription factor binding sites. Additionally, we identified 32 β-catenin-responsive genes in our mouse model that are also over-expressed in human fibrotic tissues and poised for regulation by Tcf/Lef family transcription factors. Therefore, we have uncovered a matrix-regulatory role for stabilized β-catenin in fibroblasts in vivo and have defined a set of β-catenin-responsive genes with relevance to fibrotic disease.
Keywords: dermis, fibroblast, fibrosis, β-catenin, extracellular matrix
INTRODUCTION
Fibrosis is the formation of scar tissue in response to tissue injury by wounding or a disease process. It is characterized by excessive extracellular matrix (ECM) deposition by abnormal fibroblasts, which leads to tissue sclerosis and loss of organ function. This end-stage pathology is irreversible and is associated with substantial morbidity and mortality [1]. Fibrosis is a consequence of diseases of visceral organs and connective tissues, including idiopathic pulmonary fibrosis, chronic kidney disease, chronic liver disease, and systemic sclerosis (SSc). Fibroblasts are the key effector cell of fibrosis and can be derived from several potential sources: resident fibroblasts, pericytes, endothelial cells, epithelial cells, and circulating fibrocytes [2,3]. However, the relative contributions of these fibrogenic populations are not well understood and likely vary between diseases and affected tissues.
The skin is an advantageous organ in which to study fibrosis. The dermis of the skin is predominated by numerous resident fibroblasts, providing a population of healthy cells that can be directly compared to their abnormal counterparts in fibrotic skin. Resident dermal fibroblasts are a heterogeneous population of cells that are important for normal wound healing (a physiologic fibrotic process) [4]. Pathologic dermal fibrosis is evident in several diseases, including systemic sclerosis, nephrogenic systemic fibrosis, chronic graft versus host disease, and abnormal wound repair resulting in keloid or hypertrophic scarring. For some of these fibrotic conditions, there are publicly available microarray datasets that allow comparative analysis of altered gene expression in patient skin biopsies and skin-derived fibroblasts. Such studies have used fibrotic gene expression signatures to subtype patients’ responses to therapy and to identify infiltrating cell populations and dysregulated signaling pathways [5–10]. However, little is known about the gene expression changes that result from manipulating specific cell types and signaling pathways in animal models of fibrosis.
Several pro-fibrotic signaling cascades are required and sufficient for fibrosis in animal models, notably the canonical TGF-β pathway [11], but few have successfully translated to clinical use. Consequently, there is ongoing interest in novel pro-fibrotic molecules and interactions between signaling pathways [12,13]. We and others have shown that expression of a non-degradable form of β-catenin (stabilized β-catenin) in the mouse dermis causes spontaneous skin fibrosis with elevated fibroblast proliferation [14–16]. Moreover, inhibition of the Wnt/β-catenin pathway protects against fibrosis in kidney and skin [17,18]. Specifically in fibroblasts, Wnt/β-catenin signaling drives proliferation and motility and induces broad changes in expression of genes that encode transcription factors and signaling proteins [19–21]. Wnt/β-catenin signaling is active in the presence of extracellular Wnt ligands, which induce a cascade of intracellular responses that stabilize cytoplasmic β-catenin. Stabilized β-catenin, the central transducer of the pathway, translocates to the nucleus and binds to Tcf/Lef family transcriptional cofactors to regulate target gene expression in a context-specific manner [22]. We are interested in the mechanism underlying the pro-fibrotic effects of canonical Wnt/β-catenin signaling in the context of dermal fibroblasts.
We hypothesized that expression of stabilized β-catenin in dermal fibroblasts is sufficient to cause fibrotic changes in the ECM via altered expression of ECM-encoding genes. Using our mouse model in which a restricted, traceable population of skin fibroblasts expresses a stabilized form of β-catenin resulting in spontaneous dermal fibrosis, we have examined matrix composition, cellular involvement, and signaling pathway activity for changes consistent with pathological fibrosis. With genomic analysis of β-catenin-induced fibrotic tissue, we have identified a set of ECM-encoding genes whose expression levels are responsive to expression of stabilized β-catenin. Finally, we related our findings to clinical fibrosis by determining which of the experimentally-determined β-catenin-responsive genes are over-expressed in human fibrotic tissues.
MATERIALS AND METHODS
Mice
HoxB6CreERT/+; R26-YFP/+; CatnbΔex3/+ (“stabilized β-catenin”) mice were generated and genotyped as previously described [14]. Triple-transgenic stabilized β-catenin pups and double-transgenic HoxB6CreERT/+; R26-YFP/+ littermate controls were administered tamoxifen (Sigma-Aldrich, St. Louis, MO, USA; T5648) dissolved in corn oil via oral gavage to the pregnant dam at embryonic day E16.5 (3 mg tamoxifen/40 g maternal weight), born and aged to 3 weeks except where otherwise noted. All analyses were performed on four or more samples except where otherwise noted.
Ethics
All animal experiments were approved by Case Western Reserve University Institutional Animal Care and Use Committee; all procedures were in accordance with AVMA guidelines (Protocol 2013-0156 approved 11/21/2014, Animal Welfare Assurance #A3145-01).
Histology and immunohistochemistry
Tissue histology and immunohistochemistry was performed on formalin-fixed, paraffin-embedded or paraformaldehyde-fixed, cryoembedded sections. Please refer to Supporting Information for additional details. For human tissue immunohistochemistry, access to archived keloid tissue was in compliance with Case Western Reserve University Institutional Review Board for Human Studies; access to archived SSc and healthy control tissue was in compliance with Boston University Medical Center Institutional Review Board. Transmission electron microscopic (TEM) imaging was performed at the Cleveland Clinic Foundation Lerner Research Institute imaging core facility.
Dermal dissociation, fluorescence-activated cell sorting, and flow cytometry
Whole ventral skin was enzymatically digested to release cells into single cell suspension. Fluorescence-activated cell sorting (FACS) was carried out using iCyt Reflection (Sony Biotechnology, Champaign, IL, USA) and BD FACSAria (BD Biosciences, San Jose, CA, USA) cell sorters. Flow cytometry was carried out using a BD LSR II flow cytometer (BD Biosciences) with post-analysis using WinList 7 (Verity Software House, Topsham, ME, USA). Equipment, software, and technical assistance were provided by CWRU Cancer Center Cytometry Core.
Quantitative PCR
Total RNA was extracted from FACS-purified cells or whole dermis using standard Trizol/isopropanol protocol followed by RNeasy MinElute cleanup kit with DNase treatment (Qiagen, Venlo, NL; 74204, 79254), then converted to cDNA using the High Capacity RNA-to-cDNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA, USA; 4387406). Relative mRNA quantities were determined using a StepOnePlus Real-Time PCR System (Life Technologies) and ΔΔCt method. Axin2, Col1a1, Serpine1, Thbs4, Ccn3/Nov, and Fbln1 mRNA quantities were measured relative to Actb using Taqman primers (Life Technologies). Fbn1 mRNA quantity was measured relative to Gapdh using SYBR Green reagents (Life Technologies).
Dermal RNA sequencing and differential expression analysis
RNA was extracted from isolated ventral dermis as for qPCR. Libraries were prepared in the CWRU Genomics Sequencing Core using TruSeq Stranded Total RNA kit (Illumina, San Diego, CA, USA; RS-122-2301). Paired-end sequencing was carried out on the Illumina HiSeq 2500 platform. Resulting 101-base paired-end reads were mapped to mouse genome release mm10. Mapped raw reads were counted, normalized to total mapped reads, and used for analysis of differential gene expression (fold change>1.5, p<0.05 with Benjamini and Hochberg adjustment for false discovery rate) [24]. Normalized mapped reads are publicly accessible via Gene Expression Omnibus using accession GSE60569 [31,32].
Human fibrotic tissue microarray analysis
Microarrays publicly available via Gene Expression Omnibus (GEO) were analyzed using the built-in GEO2R tool using sample groups indicated in Table 2 [31,32].
Table 2.
Publicly available microarray studies of fibrotic tissues and cells.
| GEO accession [publication] | Experimental setup | Tissues used |
|---|---|---|
| GSE26910 [46] |
|
Stroma isolated from tissue sections by laser capture microdissection |
| GSE45001 [47] | Tumor v. non-tumor stroma from intrahepatic cholangiocarcinoma patients | Stroma isolated from tissue sections by laser capture microdissection |
| GSE9285 [8] |
|
Forearm skin biopsies |
| GSE32413 [6] | Healthy controls v. DSSc (untreated) | Skin biopsies |
| GSE2052 [48,49] | Normal histology lung v. IPF | Lung biopsy or explant |
| GSE24206 [50] |
|
Lung biopsy |
| GSE3886 [5] |
|
Cultured skin fibroblasts |
| GSE7890 [10] | Normal scar v. keloid scar (untreated) | Cultured scar fibroblasts |
| GSE1724 [51] |
|
Cultured lung fibroblasts |
| GSE31356 | Non-lesional v. lesional tissue from Dupuytren contracture patients | Cultured fibroblasts |
IPF, idiopathic pulmonary fibrosis; SSc, systemic sclerosis (d, diffuse; l, limited)
Additional Materials and Methods are provided in Supporting Information.
RESULTS
Expression of stabilized β-catenin in skin fibroblasts results in fibrosis
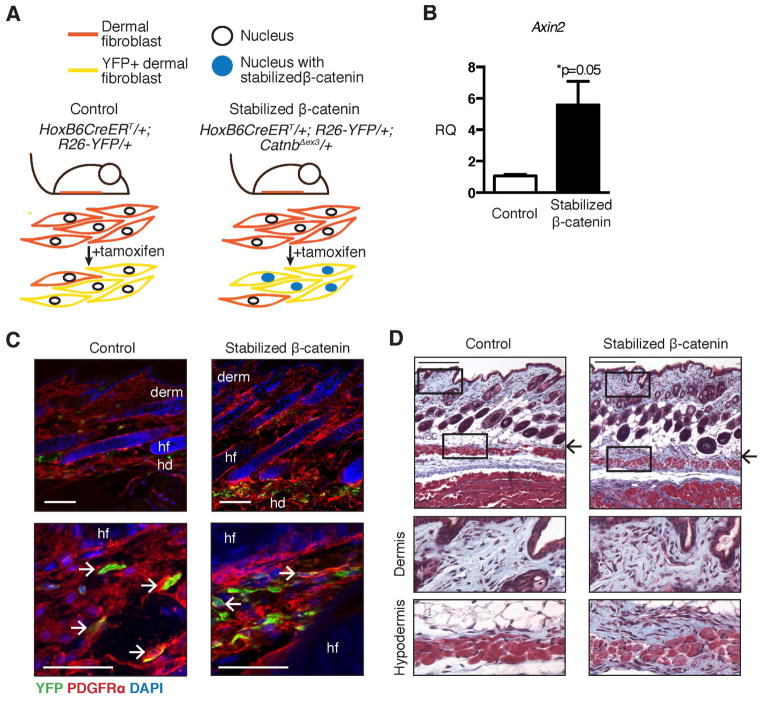
To test the consequences of increased canonical Wnt signaling in fibroblasts in vivo, we induced expression of a non-degradable form of β-catenin (“stabilized β-catenin”) in a restricted, traceable population of mouse ventral skin fibroblasts (Figure 1A). We measured a >5-fold increase in relative expression of Axin2, a β-catenin-responsive gene, in mutant YFP-labeled fibroblasts purified by FACS (Figure 1B), consistent with increased β-catenin nuclear localization [40]. There was no difference in the relative number of YFP-labeled cells between control and mutant skin (Supporting Table 1). In control and mutant mice, the YFP-labeled cells were restricted to the ventral skin and contributed to the dermis and hypodermis. All of the YFP+ cells expressed PDGFRα, a pan-fibroblast marker in mouse skin (Figure 1C, Supporting Figure 1) [4]. We did not detect any YFP-labeled adipocytes, endothelial cells, or pericytes (Supporting Figure 1B and 1C). Therefore, we define the YFP-labeled population of cells as fibroblasts that reside in the dermis and hypodermis within the skin.
Figure 1. Expression of stabilized β-catenin in mouse ventral dermal fibroblasts causes skin fibrosis.
(A) Schematic of the experimental regimen showing tamoxifen administration at E16.5 to induce Cre-mediated expression of yellow fluorescent protein (YFP) and stabilized β-catenin in the ventral dermal fibroblasts of mutant animals. (B) Relative expression levels of Axin2 in FACS-purified YFP+ cells from stabilized β-catenin and control skin analyzed by real-time PCR (n=2). (C) Fluorescent immunohistochemistry showing co-expression of PDGFRα and YFP in control and stabilized β-catenin skin (view of Z-stack). (D) Transverse sections through ventral skin at 3 weeks of age stained with Masson’s trichrome to show collagen in blue. Arrows indicate expanded collagenous tissue in mutant skin compared to corresponding region in control skin. Rectangles indicate areas selected for higher-magnification images of dermis and hypodermis, below. Abbreviations: derm, dermis; hf, hair follicle; hd, hypodermis. All scale bars = 100 μm.
In this transgenic mouse model, expression of stabilized β-catenin in ventral skin fibroblasts resulted in spontaneous, progressive dermal fibrosis [14]. Stabilized β-catenin mice at 3 weeks of age had a thickened hypodermis that stained blue for collagen with Masson’s trichrome stain (Figure 1D). The hypodermis and dermis of stabilized β-catenin mutant animals was hypercellular compared to control dermis, consistent with our previous observation of elevated fibroproliferation in these tissues (Figure 1D) [14]. Therefore, expression of stabilized β-catenin in ventral dermal fibroblasts causes a skin fibrosis phenotype in the absence of any injury or other stimulus.
Non-dermal fibroblast cell types do not have measurable involvement in stabilized β-catenin-induced skin fibrosis
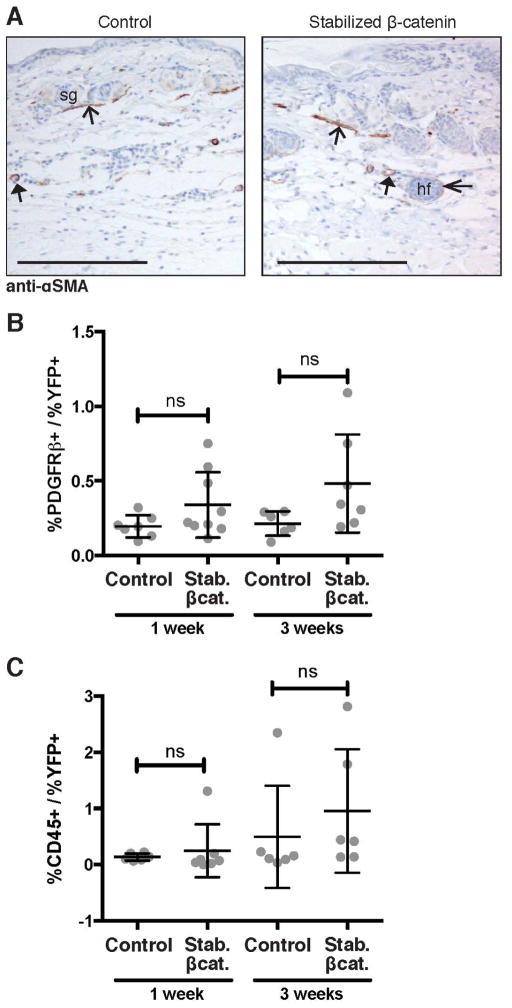
Next, to determine whether the stabilized β-catenin-induced skin fibrosis phenotype involved other cell types, we examined for the presence of myofibroblasts, which are often observed in fibrotic tissues [57]; pericytes, which have been hypothesized to contribute to fibrosis of skin and other organs [3]; and monocytes, which include fibrocytes, a potential fibrogenic cell population, as well as macrophages and other hematopoietic cells [3]. First, we performed immunohistochemical staining for α-smooth muscle actin (α-SMA), a marker for myofibroblasts. We found comparable expression of α-SMA in control and stabilized β-catenin mutant blood vessels, arrector pili, and hair follicles, and did not detect α-SMA+ fibroblasts in skin at age 3 weeks or 4 months (Figure 2A, Supporting Figure 2). Next, we performed flow cytometry on suspensions of single cells extracted from whole skin for PDGFRβ, a marker for pericytes. We found no statistically significant difference in the relative number of PDGFRβ+ cells between control and mutant skin at one week of age (before the appearance of the fibrotic phenotype) or 3 weeks of age (after fibrosis had developed) (Figure 2B). Similarly, we tested for infiltration of bone marrow-derived monocyte-lineage cells in the fibrotic tissue. Flow cytometric detection of CD45, a surface marker for monocytes, showed no statistically significant difference in the relative number of marker-positive cells between stabilized β-catenin mutant skin and control skin at one week or 3 weeks of age (Figure 2C). These data suggest that the stabilized β-catenin-induced fibrosis phenotype is predominated by resident dermal fibroblasts, without infiltration or increased numbers of other classically pro-fibrotic cell types.
Figure 2. Stabilized β-catenin dermis and hypodermis do not have increased numbers of known pro-fibrotic cell populations.
(A) Transverse sections through ventral skin at age 3 weeks with immunohistochemical staining showing comparable expression of α-smooth muscle actin (SMA) in control and stabilized β-catenin skin. Positive control expression in arrector pili muscle (open arrowhead, visible in cross-section in dermis), blood vessel walls (closed arrowhead, visible in subcutaneous fat region), and faintly in the dermal sheath of hair follicles (thin open arrowhead). Scale bar = 100 μm. (B, C) Comparable percentage of total cells express (B) PDGFRβ and (C) CD45 in control and stabilized β-catenin skin at both 1 week and 3 weeks of age (n≥6). Abbreviations: sg, sebaceous gland; hf, hair follicle.
Stabilized β-catenin-induced skin fibrosis occurs without altered canonical TGF-β signaling
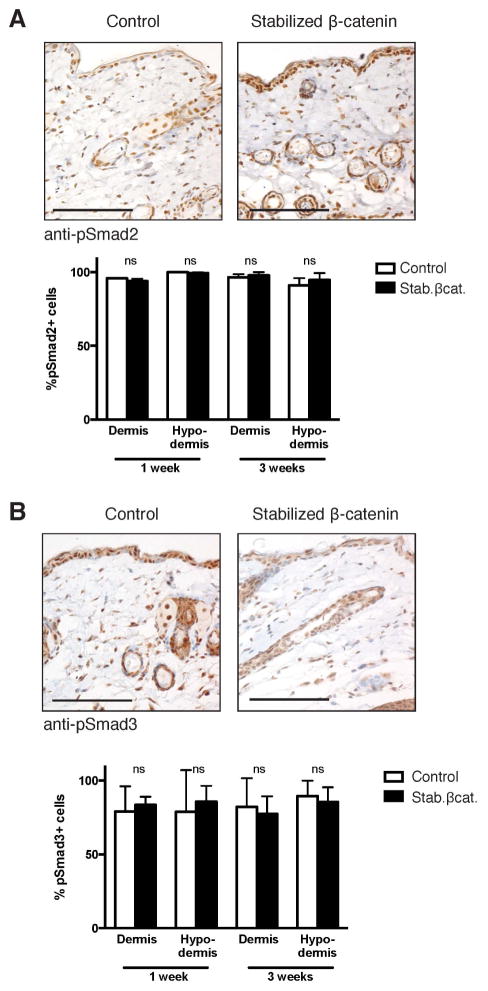
We next investigated potential crosstalk between stabilized β-catenin and canonical TGF-β signaling, which is the prototypic pro-fibrotic signaling pathway [41]. We found ~1.5-fold increased expression of PAI-1 mRNA, a gene responsive to TGF-β and Wnt/β-catenin signaling [42,43], in stabilized β-catenin dermis compared to control dermis by qPCR (Supporting Figure 3A). To directly test for activation of TGF-β signaling, we performed immunohistochemistical staining for phosphorylated (active) Smad2 (pSmad2) and Smad3 (pSmad3), the intracellular mediators of canonical TGF-β signaling. Comparable percentages of dermal and hypodermal cells expressed nuclear pSmad2 and pSmad3 in both control and stabilized β-catenin mutant skin at 1 week and 3 weeks of age (Figure 3, Supporting Figure 3B,C). These data suggest that β-catenin-induced fibrosis occurs independently of altered canonical TGF-β signaling.
Figure 3. Canonical TGFβ-signaling activity is not affected by expression of stabilized β-catenin in skin fibroblasts.
(A, B) Representative images of transverse sections through ventral skin of control and stabilized β-catenin mutants at age 3 weeks. Scale bars = 100 μm. Immunohistochemical staining for (A) phosphorylated Smad2 (pSmad2) or (B) phosphorylated Smad3 (pSmad3) followed by quantification of nuclear expression in fibroblast morphology cells shows comparable pSmad2 and pSmad3 activity in control and stabilized β-catenin skin at 1 week and 3 weeks of age.
Sustained expression of stabilized β-catenin in dermal fibroblasts is sufficient for abnormal ECM in vivo
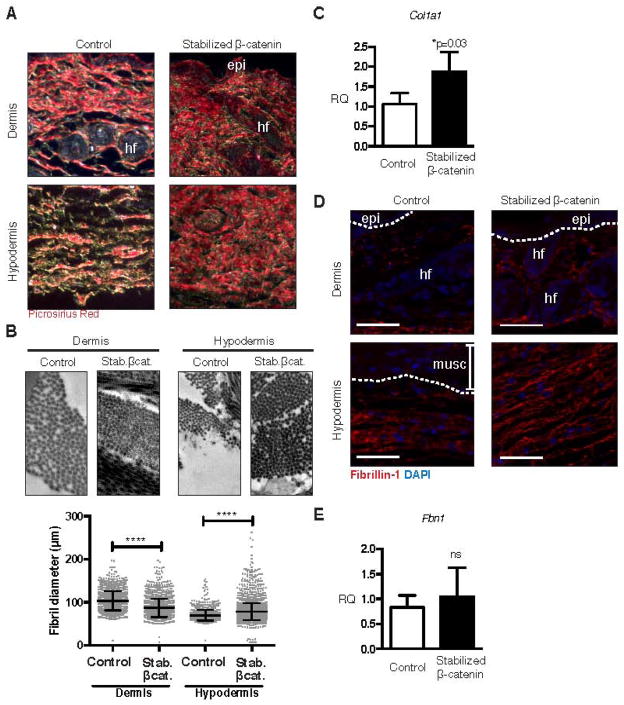
The key pathological feature of fibrosis in any organ, including skin, is the accumulation of excess ECM with aberrant morphology [44,45]. We investigated for altered ECM morphology in the stabilized β-catenin skin at 3 weeks of age, when excess collagen is first evident in the mutant hypodermis. We found that thickened collagen fibres, visualized by picrosirius red stain viewed under crossed polarized filters, predominated in the mutant skin, with very few visible thin green birefringent fibres (Figure 4A). We measured cross-sectional fibril thickness in the dermis and hypodermis in images obtained by TEM (Figure 4B, representative images from n = 2 control and mutant animals). In the dermis, average collagen fibril diameter was reduced by 16% in the stabilized β-catenin mutant compared to control tissue (mutant diameter 86.91 +/− 0.49 nm from n = 1896 fibrils versus control diameter 103.6 +/− 0.48 nm from n = 2128 fibrils, p < 0.0001). In contrast, in the stabilized β-catenin mutant hypodermis, average collagen fibril diameter was increased by 12% compared to control tissue (mutant diameter 78.38 +/− 0.33 nm from n = 3735 fibrils versus control diameter 70.10 +/− 0.33 nm from n = 1374 fibrils, p < 0.0001). In FACS-purified stabilized β-catenin mutant cells, we measured a 1.8-fold increase in relative expression of Col1a1 mRNA, which encodes the α1 chain of type I collagen, the predominant type of collagen in skin (Figure 4C, n = 4 control and mutant animals). In addition, we found altered distribution of fibrillin-1 protein by immunofluorescent staining; rather than being limited to the lower border of hypodermal connective tissue as in the control, fibrillin-1 immunoreactivity was observed throughout the thickened mutant hypodermis (Figure 4D). However, relative levels of Fbn1 mRNA were not significantly different between control and mutant cells (Figure 4E) [37]. Collectively, these data suggest that the stabilized β-catenin-induced fibrotic response is not only via altered expression of genes that encode ECM proteins such as Col1a1, but also by altered activity of regulators of matrix protein post-translational assembly or turnover.
Figure 4. Expression of stabilized β-catenin in skin fibroblasts is sufficient for abnormal extracellular matrix.
(A) Transverse sections through ventral skin stained with picrosirus red stain and viewed under crossed polarized filters showing increased thick collagen fibres in stabilized β-catenin skin compared to control skin. (B) Electron microscopy of collagen fibrils and measurement of cross-sectional fibril diameter shows thicker hypodermal and thinner dermal fibrils in stabilized β-catenin skin compared to control skin (p<0.0001 for both comparisons). Scale bars = 1 μm. (C) Relative expression of Col1a1 mRNA is elevated in stabilized β-catenin mutant versus control FACS-purified YFP+ fibroblasts by real-time PCR (n=4). (D) Immunofluorescent staining for fibrillin-1 showing expanded expression in the thickened mutant hypodermis. Scale bars = 50 μm. (E) Relative expression of Fbn1 mRNA, measured by real-time PCR in whole skin, is not significantly different between control (n=2) and stabilized β-catenin mutant skin (n=4). epi, epidermis; hf, hair follicle; musc, muscle.
Fibrosis resulting from expression of stabilized β-catenin is associated with increased expression of matrisome-encoding genes
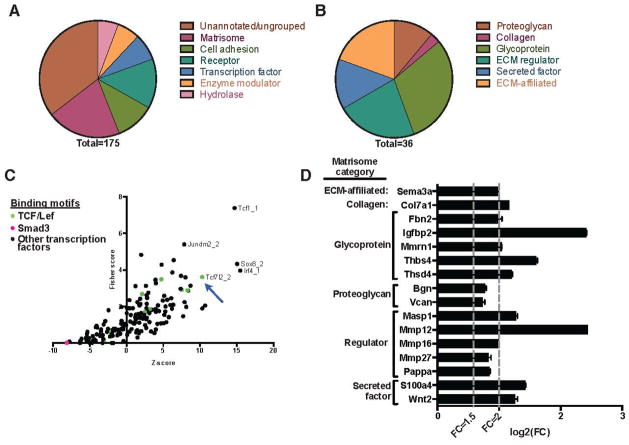
To determine which matrisome-encoding genes are responsive to dermal fibroblast β-catenin levels in vivo, we performed whole transcriptome sequencing of RNA extracted from the dermis of control and stabilized β-catenin mice at 3 weeks of age (n = 3) (Supporting File 1). There were 176 annotated genes that showed significantly different expression in the stabilized β-catenin dermis versus control. We confirmed increased expression of 3 of these genes, Thbs4, Ccn3/Nov, and Fbln1, in 2 additional biological replicates by qPCR (Supporting Figure 4A), and corresponding increased protein expression of CCN3/NOV by immunohistochemistry (Supporting Figure 4B). All differentially expressed genes had increased expression in the mutant dermis (fold change > 1.5, adjusted p-value < 0.05) (Supporting File 1). This set of β-catenin-responsive genes was strongly enriched for gene ontology terms related to ECM and cell adhesion (Table 1). We categorized 175 of these genes and found that 36 (20.6%) encode matrisome proteins as defined by Naba et al. [30] (Figure 5A, Figure 5B, Supporting File 2). These matrisome-encoding genes include the three whose increase expression we verified by qPCR: Thbs4, Ccn3/Nov, and Fbln1, which encode glycoproteins thrombospondin-4, CCN3/NOV, and fibulin-1. We performed over-representation analysis of the promoter regions of the 36 matrisome-encoding genes and found relative enrichment of predicted binding sites for Tcf7l2, Tcf1 (HNF1-α), Sox8, Jundm2, and Irf4 transcription factors. Two predicted Smad3 binding sites were also queried in the over-representation analysis and were depleted in our matrisome-encoding gene set relative to binding sites for other transcription factors (Figure 5C). Of the 36 β-catenin-responsive matrisome-encoding genes, 16 have predicted Tcf/Lef family transcription factor binding sites (TFBSs) in their promoter regions (Figure 5D). These data show that expression of stabilized β-catenin in skin fibroblasts is sufficient for elevated dermal expression of several genes related to the ECM, and a subset of these β-catenin-responsive genes is poised for regulation via Tcf/Lef family transcription factors.
Table 1.
Enriched DAVID functional annotation clusters in stabilized β-catenin dermis.
| Annotation cluster | Representative annotation term(s) | Enrichment score |
|---|---|---|
| 1 | Glycoprotein/disulfide bond | 18.04 |
| 2 | Secreted/extracellular region | 4.58 |
| 3 | Extracellular matrix | 4.67 |
| 4 | Immunoglobulin | 4.39 |
| 5 | Cell adhesion | 4.15 |
Figure 5. Stabilized β-catenin mouse dermis has elevated expression of matrix-encoding genes.
(A) Over-expressed genes in stabilized β-catenin dermis compared to control, categorized using PANTHER categories and the proteomically defined matrisome. (B) The differentially expressed genes include 36 that encode matrisome proteins. (C) Transcription factor binding site over-representation analysis using oPOSSUM showing the promoter regions of the 36 over-expressed matrisome-encoding genes are enriched for predicted binding sites for transcription factors including Tcf7l2 (Tcf4). (D) Calculated log2(fold change) of the 16 β-catenin-responsive matrisome genes that have predicted Tcf/Lef transcription factor binding sites in their promoter regions.
β-catenin-responsive genes in mouse dermis have elevated expression in human fibrotic tissues
Next, we used publicly available microarray datasets comparing fibrotic and healthy human tissues to determine which β-catenin-responsive genes were over-expressed in human disease. We generated lists of differentially expressed genes (fold change > 1.5, adjusted p-value < 0.05) in breast and prostate cancer stroma, systemic sclerosis (SSc) skin biopsies, idiopathic pulmonary fibrosis (IPF) lung biopsies, and cultured fibroblasts from SSc skin and lung, morphea, keloid scar, IPF lung, and Dupuytren contracture (superficial fibromatosis) (Table 2). Separately, we converted our list of 176 β-catenin-responsive mouse genes to their human homologs; 5 of the mouse genes did not have corresponding human homologous gene in the two databases that we queried and were dropped from further analysis (Supporting File 3). Comparison of the list of 171 remaining genes to the list of differentially expressed genes from each microarray experiment yielded an overlap of 41 genes (Supporting File 4). All the overlapping genes were over-expressed in microarrays from biopsied tissue, not from cultured cells. Therefore, we have identified a list of genes whose expression responds to expression of stabilized β-catenin in the dermis and which are over-expressed in at least one type of human fibrotic tissue. Of these 41 genes, 32 have predicted Tcf/Lef family TFBSs in their promoter regions (Table 3). These disease-associated genes are both β-catenin-responsive in vivo and poised for regulation via Tcf/Lef family transcription factors. These findings suggest that increased Wnt/β-catenin signaling in fibrotic conditions such as IPF, diffuse SSc, and tumor stroma may indirectly influence broader gene expression changes by regulating expression of transcription factors Sp5 and Osr2. In addition, β-catenin may directly regulate expression of matrisome genes that encode proteins with diverse roles in the extracellular matrix, including glycoproteins such as fibulin-1, thrombospondin-4, and CCN3/NOV; proteoglycans such as biglycan; and regulatory enzymes such as matrix metalloproteinase-16.
Table 3.
β-catenin-responsive genes that are over-expressed in human fibrotic tissue and contain Tcf/Lef predicted binding sites within 5kb of the transcription start site.
| Gene symbol | Associated fibrotic tissue | # predicted Tcf/Lef sites | ||
|---|---|---|---|---|
| SSc skin | Tumor stroma | IPF lung | ||
| ADAMTS8 | X | 3 | ||
| ASPN | X | X | 1 | |
| BGN | X | 8 | ||
| C1QTNF3 | X | 7 | ||
| CADPS | X | 162 | ||
| COL7A1 | X | 12 | ||
| CYP26B1 | X | 15 | ||
| ENTPD1 | X | 15 | ||
| EPHA3 | X | 4 | ||
| EPHB2 | X | 27 | ||
| FBLN1 | X | 5 | ||
| HLA-DMA | X | 18 | ||
| HLA-DMB | X | 1 | ||
| IGFBP2 | X | 10 | ||
| IL13RA2 | X | 2 | ||
| ITGA4 | X | 8 | ||
| LHFPL2 | X | 19 | ||
| MASP1 | X | 12 | ||
| MMP12 | X | 3 | ||
| MMP16 | X | 13 | ||
| OSR2 | X | 22 | ||
| PTPRO | X | X | 12 | |
| QPCT | X | 8 | ||
| SEMA3A | X | 30 | ||
| SP5 | X | 26 | ||
| THSD4 | X | X | 13 | |
| THY1 | X | 4 | ||
| TMEM150C | X | 1 | ||
| TWIST1 | X | X | 12 | |
| VCAN | X | 33 | ||
| WISP2 | X | 2 | ||
| WNT2 | X | 22 | ||
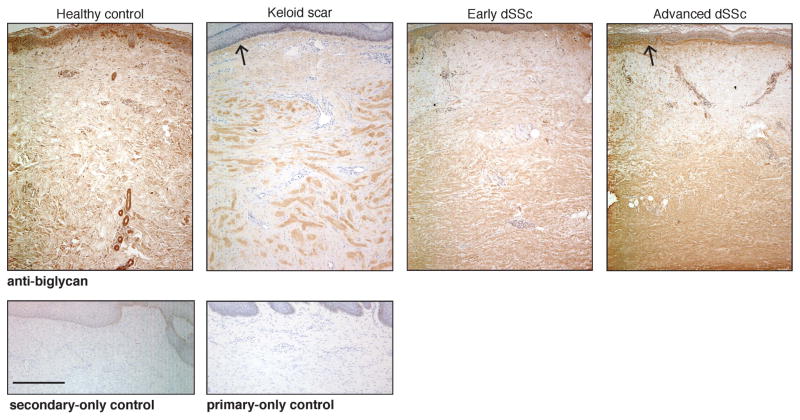
Of the disease-associated, β-catenin-responsive, matrisome-encoding genes that we identified, we next focused on Bgn, which encodes biglycan, a small leucine-rich proteoglycan. Bgn is over-expressed in tumor stroma (Table 3, Supporting File 4) and systemic sclerosis skin [52]. Biglycan binds to type I collagen and is associated with thick collagen fibrils [53,54]. We found altered distribution of biglycan protein expression in the reticular dermis of diffuse SSc and in the nodules of keloid scars (Figure 6) by immunohistochemistry using antiserum that detects biglycan and its precursor, pro-biglycan. Biglycan protein expression is variably increased in the hypodermis of stabilized β-catenin mice (Supporting Figure 5). Since both SSc dermis and keloid scar tissue have elevated fibroblast nuclear β-catenin (Supporting Figure 6) [55,56], this finding supports a model in which nuclear β-catenin in fibroblasts up-regulates the expression of matrix-encoding genes such as Bgn to contribute to aberrant, fibrotic ECM.
Figure 6. Fibrotic human skin has altered distribution of biglycan.
Immunohistochemical detection of biglycan showing elevated biglycan immunoreactivity in the keloid nodule compared to overyling papillary dermis and in the reticular dermis of early (< 2 years) and advanced (> 2 years) diffuse systemic sclerosis (dSSc). This is in contrast to homogenous staining throughout the papillary and reticular dermis in healthy control skin. Open arrowheads indicate elevated biglycan immunoreactivity just beneath the basal layer of the epidermis in advanced dSSc and keloid tissue. Representative images from n = 5 healthy controls, n = 6 each early dSSc, dvanced SSc, and keloids. Scale bar (in secondary-only control image) = 500 μm; all images photographed at same magnification.
DISCUSSION
Numerous studies in human tissues and animal models have yielded many insights into the pathobiology of fibrosis and revealed both common and distinct features of fibrosis between various tissues and diseases. By developing a tractable and cell-type restricted model to study the mechanism underlying the pro-fibrotic effects of β-catenin, we have uncovered a specific role for β-catenin in the regulation of ECM-encoding gene expression. We found that expression of stabilized β-catenin in dermal fibroblasts is sufficient for abnormal ECM morphology in the skin with concomitant increases in expression of matrisome-encoding genes. Our study elucidates an underlying mechanism for the pro-fibrotic role of dermal fibroblast nuclear β-catenin. Future studies are required to confirm the pro-fibrotic role of stabilized β-catenin in mature dermal fibroblasts, since our manipulation of fetal dermal fibroblasts may have altered their development.
Studies using genetically modified mice and cell lineage tracing are able to delineate the roles of specific cell types and the relationships between these cell types in fibrotic tissue. By manipulating the stability of a single signal transduction molecule specifically in dermal fibroblasts, we have dissected out a specific role for β-catenin: to promote the expression of matrix-encoding genes that contribute to the fibrotic ECM. This broad regulatory role for β-catenin in fibrosis is consistent with the finding that inhibition of β-catenin is sufficient for reversal of bleomycin-induced skin fibrosis [18]. We have demonstrated that stabilized β-catenin-induced dermal fibrosis does not involve expansion of pericyte or monocyte populations, or the presence of detectable α-SMA-expressing myofibroblasts. Our result contrasts with the increase of α-SMA+ cells reported in the skin of mice with post-natal induction of stabilized β-catenin expression in fibroblasts, suggesting that the age or identity of resident dermal fibroblasts may affect the appearance of α-SMA+ cells in skin fibrosis [16]. Myofibroblasts are well-documented in fibrosis but not universally present during pathologic fibrosis [57–59]. Our findings show that, in clinical fibrosis, β-catenin signaling in dermal fibroblasts is likely a downstream event from the involvement of these other cell types.
Several studies have addressed the question of interaction between Wnt/β-catenin signaling and canonical TGF-β signaling, showing that each pathway mediates the other pathway’s induction of collagen gene expression in whole skin [15,16,55]. By examining expression of phosphorylated Smad2 and Smad3 proteins, we were unable to identify any changes in canonical TGF-β signaling activity in stabilized β-catenin-induced fibrosis. Moreover, the matrisome-encoding gene set that we identified in our model is depleted for predicted Smad3 binding sites. Additional studies are required to rigorously distinguish whether β-catenin nuclear localization in fibroblasts is a downstream or parallel event to canonical TGF-β signaling in fibrosis.
The predominant effect of sustained expression of stabilized β-catenin in dermal fibroblasts in our system is altered ECM morphology and gene expression. This is in contrast to other mouse models of fibrosis which demonstrate cellular infiltrates, myofibroblasts, and TGF-β signaling activity in addition to increased type I collagen [15,16,60,61]. The discrepant changes in collagen fibril thickness between dermis and hypodermis suggest that β-catenin may have distinct regulatory consequences between dermal and hypodermal fibroblasts. Our results support a model of fibrosis in which the primary role for nuclear β-catenin, in addition to its known role in dermal fibroblast proliferation [19,20], is to regulate expression of specific matrix-encoding genes, resulting in altered morphology, distribution, and amounts of ECM proteins. Several of the β-catenin-responsive matrisome genes that we identified in this model encode proteoglycans, glycoproteins, and matrix metalloproteinases, each of which may have distinct contributions to the fibrotic phenotype. These contributions likely include structural contribution to the bulk extracellular matrix, regulation of matrix protein turnover, regulation of collagen fibrillogenesis, sequestration or potentiation of secreted signaling factors, and altered crosslinking of extracellular proteins, which collectively result in a progressive fibrotic phenotype. Some of the genes we identified, such as those encoding matrix metalloproteinases that enzymatically degrade matrix proteins, and CCN3/NOV, which is anti-fibrotic when over-expressed in mouse embryonic fibroblasts [62], may comprise a negative feedback loop in response to pro-fibrotic β-catenin.
We found that the promoter regions of the β-catenin-responsive matrix-encoding genes are enriched for predicted Tcf/Lef binding sites. Chromatin-immunoprecipitation studies and functional analysis of candidate targets are required to demonstrate which of these genes are directly regulated by β-catenin, and how their gene products contribute to fibrosis. We measured expression levels of genes in the whole dermis, not just in fibroblasts; therefore, we only detected the most robustly over-expressed transcripts. Future efforts to define gene expression changes in stabilized β-catenin mutant dermal fibroblasts may reveal novel mediators of fibrosis whose expression levels are diluted out when evaluated in whole tissue [63].
Increased Wnt/β-catenin signaling has been implicated in various fibrosing tissues including fibrous tumors [10,20,55,64–66]. In an attempt to understand if the β-catenin-responsive genes that we identified in our mouse model might be relevant to fibrotic disease, we compared the mouse dermal over-expressed genes to the genes that are over-expressed in fibrotic human tissues and cells and tumor stroma. We found 32 β-catenin-responsive genes that are over-expressed in fibrotic human tissues, including genes that encode matrisome proteins and transcription factors. As a proof-of-principle, we verified that biglycan, a proteoglycan encoded by one of these 32 genes, has altered distribution of protein expression in the reticular dermis of SSc skin and in keloid nodules. Expression of biglycan protein positively correlates with skin thickness in cultured dSSc fibroblasts [67], and biglycan protein and mRNA levels are elevated in keloids [68]. Since biglycan binds to type I collagen and is associated with thicker collagen fibrils [53,54], the over-expression of Bgn in stabilized β-catenin dermis, SSc, and keloids may be linked to the altered fibril morphology in these tissues [44,45,52]. Future studies to test whether β-catenin is required for cell-autonomous expression of Bgn and other ECM protein-coding genes will help establish a role for β-catenin control of the aberrant matrix in fibrotic disease.
Together, our findings suggest that the increased nuclear β-catenin that has been observed in fibrotic human tissue may both directly regulate the production of fibrotic ECM, and also exert broader effects via up-regulation of specific transcription factors. Since ECM accumulation is the feature of fibrosis that is explicitly involved in loss of organ function, our findings strengthen the rationale for testing inhibitors of Wnt/β-catenin signaling in amelioration of fibrosis [69,70]. In addition, the β-catenin-responsive, disease-associated genes that we identified encode proteins that may have utility as new targets for therapeutic intervention and as biomarkers for tracking the response to Wnt/β-catenin inhibition in vivo.
Supplementary Material
Supporting Figure 1. Manipulated cells express fibroblast markers. (A) Merged and single-channel images showing YFP+ PDGFRα+ cells. Scale bars = 100 μm. (B) Lack of double immunofluorescent staining for YFP and endothelial marker PECAM or adipose marker perilipin. Scale bars = 100 μm. (C) Flow cytometric scatter plot showing lack of YFP+ PDGFRβ+ cells; values indicate percent of detected cells. hf, hair follicle.
Supporting Figure 2. α-SMA-expressing fibroblasts do not appear in advanced β-catenin-induced fibrosis. Immunohistochemistical staining for α-SMA in 4-month-old control and stabilized β-catenin mice demonstrates α-SMA+ structures (labeled), but not α-SMA+ fibroblasts. hf, hair follicle; musc, muscle; adip, adipose tissue. Scale bars = 100 μm.
Supporting Figure 3. Canonical TGF-β signaling activity is not affected in stabilized β-catenin-induced fibrosis. Stabilized β-catenin mutant skin has (A) 1.5-fold increase in relative expression of Serpine1 (PAI1) but no change in percentage of fibroblasts that express nuclear (B) pSmad2 or (C) pSmad3 at 1 week or 3 weeks of age. Scale bar (in secondary-only control image) = 200 μm; all images photographed at same magnification.
Supporting Figure 4. Validation of up-regulated genes in additional biological replicates. (A) Stabilized β-catenin mutant dermis has increased relative expression, measured by qPCR (n=2), of Thbs4, Ccn3, and Fbln1, genes which were also up-regulated upon differential expression analysis of whole transcriptome profiles. (B) Stabilized β-catenin mutant skin has increased CCN3 protein expression in dermis and hypodermis compared to control skin. Higher-magnification images correspond with dotted and solid outlined areas in corresponding lower-magnification image. Scale bars (in control images) = 200 μm; all images photographed at same magnification.
Supporting Figure 5. Protein expression of biglycan in stabilized β-catenin mouse skin. Stabilized β-catenin mutant skin has variably increased biglycan protein expression in the hypodermis compared to control skin. Higher-magnification images correspond with dotted and solid outlined areas in corresponding lower-magnification image. Scale bars (in control images) = 200 μm; all images photographed at same magnification.
Supporting Figure 6. Keloid scar nodule has elevated cytoplasmic and nuclear β-catenin. Immunohistochemical detection of β-catenin in healthy control skin and three keloid scars demonstrating elevated cytoplasmic and nuclear β-catenin immunoreactivity in the keloid nodule.
List of differentially expressed (DE) genes with corresponding log2(fold change) and adjusted p-values.
Categorization of 175 DE genes; number of genes per category; categorization of 36 matrisome-encoding genes; number of genes per matrisome category.
List of homologous human genes to 171 of the differentially expressed genes from our mouse model.
List of the human homologous genes that are also over-expressed in fibrotic tissues, with corresponding log(fold change) and adjusted p-values from GEO2R analysis.
Acknowledgments
We are grateful to Paresh M. Atit (1942–2014) for inspiring this work. We thank the patients who provided tissues for this study. Thanks to Sheldon Bai, Scott Howell, Michael Sramkoski, and Miarasa Steele for their indispensable technical assistance and Suneel Apte, Dirk Hubmacher, David Carrino, and previous and current members of the Atit laboratory for excellent discussion and advice. Funding provided by NIH NIDCR R01 DE01870 (RA), T32 GM07250 (EH), TL1 TR000441 (EH), NIA F30 AG045009 (EH), NIAMS P30AR061271 (RL); Scleroderma Research Foundation (RA), and Global Fibrosis Foundation (RA).
LIST OF ABBREVIATIONS
- SSc
systemic sclerosis
- IPF
idiopathic pulmonary fibrosis
- FACS
fluorescence-activated cell sorting
- TEM
transmission electron microscopy
- TFBS
transcription factor binding site
Footnotes
CONFLICTS OF INTEREST
No conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
EH and RA conceived and carried out experiments and analyzed data. RL provided human samples and data. GD performed genotyping and immunohistochemistry. NM collected and analyzed data. EH generated figures. EH and RA interpreted the data and wrote the manuscript. All authors had final approval of the submitted version.
References
- 1.Bitterman PB, Henke CA. Fibroproliferative disorders. Chest. 1991;99:81S–84S. doi: 10.1378/chest.99.3_supplement.81s. [DOI] [PubMed] [Google Scholar]
- 2.Krenning G, Zeisberg EM, Kalluri R. The origin of fibroblasts and mechanism of cardiac fibrosis. J Cell Physiol. 2010;225(3):631–7. doi: 10.1002/jcp.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postlethwaite AE, Shigemitsu H, Kanangat S. Cellular origins of fibroblasts: possible implications for organ fibrosis in systemic sclerosis. Curr Opin Rheumatol. 2004;16(6):733–8. doi: 10.1097/01.bor.0000139310.77347.9c. [DOI] [PubMed] [Google Scholar]
- 4.Driskell RR, Lichtenberger BM, Hoste E, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504(7479):277–81. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitfield ML, Finlay DR, Murray JI, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci USA. 2003;100(21):12319–24. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pendergrass SA, Lemaire R, Francis IP, et al. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012;132(5):1363–73. doi: 10.1038/jid.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinchcliff M, Huang C-C, Wood TA, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol. 2013;133:1979–89. doi: 10.1038/jid.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milano A, Pendergrass SA, Sargent JL, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS ONE. 2008;3(7):e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell SB, Russell JD, Trupin KM, et al. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489–96. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JC, Boone BE, Opalenik SR, et al. Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J Invest Dermatol. 2007;128(5):1298–310. doi: 10.1038/sj.jid.5701149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denton CP, Zheng B, Evans LA, et al. Fibroblast-specific expression of a kinase-deficient type II transforming growth factor beta (TGF-beta) receptor leads to paradoxical activation of TGF-beta signaling pathways with fibrosis in transgenic mice. J Biol Chem. 2003;278(27):25109–19. doi: 10.1074/jbc.M300636200. [DOI] [PubMed] [Google Scholar]
- 12.Distler A, Lang V, Del Vecchio T, et al. Combined inhibition of morphogen pathways demonstrates additive antifibrotic effects and improved tolerability. Ann Rheum Dis. 2014;73:1264–68. doi: 10.1136/annrheumdis-2013-204221. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharyya S, Wei J, Varga J. Understanding fibrosis in systemic sclerosis: shifting paradigms, emerging opportunities. Nature Reviews Rheumatology. 2011;8(1):42–54. doi: 10.1038/nrrheum.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamburg EJ, Atit RP. Sustained beta-catenin activity in dermal fibroblasts is sufficient for skin fibrosis. J Invest Dermatol. 2012;132:2469–72. doi: 10.1038/jid.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akhmetshina A, Palumbo K, Dees C, et al. Activation of canonical Wnt signalling is required for TGF-beta-mediated fibrosis. Nat Comms. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beyer C, Schramm A, Akhmetshina A, et al. Beta-catenin is a central mediator of pro- fibrotic Wnt signaling in systemic sclerosis. Ann Rheum Dis. 2012;71(5):761–7. doi: 10.1136/annrheumdis-2011-200568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surendran K, Schiavi S, Hruska KA. Wnt-dependent beta-catenin signaling is activated after unilateral ureteral obstruction, and recombinant secreted frizzled-related protein 4 alters the progression of renal fibrosis. Journal of the American Society of Nephrology. 2005;16(8):2373–84. doi: 10.1681/ASN.2004110949. [DOI] [PubMed] [Google Scholar]
- 18.Beyer C, Reichert H, Akan H, et al. Blockade of canonical Wnt signalling ameliorates experimental dermal fibrosis. Ann Rheum Dis. 2013;72:1255–58. doi: 10.1136/annrheumdis-2012-202544. [DOI] [PubMed] [Google Scholar]
- 19.Cheon SS, Nadesan P, Poon R, et al. Growth factors regulate beta-catenin-mediated TCF-dependent transcriptional activation in fibroblasts during the proliferative phase of wound healing. Experimental Cell Research. 2004;293(2):267–74. doi: 10.1016/j.yexcr.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Cheon SS, Cheah AYL, Turley S, et al. Beta-catenin stabilization dysregulates mesenchymal cell proliferation, motility, and invasiveness and causes aggressive fibromatosis and hyperplastic cutaneous wounds. Proc Natl Acad Sci USA. 2002;99(10):6973–8. doi: 10.1073/pnas.102657399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klapholz-Brown Z, Walmsley GG, Nusse YM, et al. Transcriptional program induced by Wnt protein in human fibroblasts suggests mechanisms for cell cooperativity in defining tissue microenvironments. PLoS ONE. 2007;2(9):e945. doi: 10.1371/journal.pone.0000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archbold HC, Yang YX, Chen L, et al. How do they do Wnt they do?: regulation of transcription by the Wnt/beta-catenin pathway. Acta Physiologica. 2011;204(1):74–109. doi: 10.1111/j.1748-1716.2011.02293.x. [DOI] [PubMed] [Google Scholar]
- 23.Tiedemann K, Bätge B, Müller PK, et al. Interactions of fibrillin-1 with heparin/heparan sulfate, implications for microfibrillar assembly. J Biol Chem. 2001;276(38):36035–42. doi: 10.1074/jbc.M104985200. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37 (1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Kwon AT, Arenillas DJ, Worsley Hunt R, et al. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3. 2012;2(9):987–1002. doi: 10.1534/g3.112.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi H, Muruganujan A, Casagrande JT, et al. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naba A, Clauser KR, Hoersch S, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11(4) doi: 10.1074/mcp.M111.014647. M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Research. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2012;40:D13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathelier A, Zhao X, Zhang AW, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Research. 2014;42:D142–7. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher LW, Stubbs JT, Young MF. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl. 1995;266:61–5. [PubMed] [Google Scholar]
- 37.Bayle J, Fitch J, Jacobsen K, et al. Increased expression of Wnt2 and SFRP4 in Tsk mouse skin: role of Wnt signaling in altered dermal fibrillin deposition and systemic sclerosis. J Invest Dermatol. 2007;128(4):871–81. doi: 10.1038/sj.jid.5701101. [DOI] [PubMed] [Google Scholar]
- 38.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–81. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. 2014. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jho E, Zhang T, Domon C, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22(4):1172– 83. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J. 2004;18 (7):816–27. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- 42.Verrecchia F, Mauviel A, Farge D. Transforming growth factor-beta signaling through the Smad proteins: role in systemic sclerosis. Autoimmun Rev. 2006;5(8):563–9. doi: 10.1016/j.autrev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 43.He W, Tan R, Dai C, et al. Plasminogen activator inhibitor-1 Is a transcriptional target of the canonical pathway of Wnt/beta-catenin signaling. J Biol Chem. 2010;285(32):24665–75. doi: 10.1074/jbc.M109.091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayes RL, Rodnan GP. The ultrastructure of skin in progressive systemic sclerosis (scleroderma). I. Dermal collagen fibers. American Journal of Pathology. 1971;63(3):433–42. [PMC free article] [PubMed] [Google Scholar]
- 45.Rockwell WB, Cohen IK, Ehrlich HP. Keloids and hypertrophic scars: a comprehensive review. Plastic and Reconstructive Surgery. 1989;84(5):827–37. doi: 10.1097/00006534-198911000-00021. [DOI] [PubMed] [Google Scholar]
- 46.Planche A, Bacac M, Provero P, et al. Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS ONE. 2011;6(5):e18640. doi: 10.1371/journal.pone.0018640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sulpice L, Rayar M, Desille M, et al. Molecular profiling of stroma identifies osteopontin as an independent predictor of poor prognosis in intrahepatic cholangiocarcinoma. Hepatology. 2013;58(6):1992–2000. doi: 10.1002/hep.26577. [DOI] [PubMed] [Google Scholar]
- 48.Wang XM, Zhang Y, Kim HP, et al. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med. 2006;203(13):2895–906. doi: 10.1084/jem.20061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardo A, Gibson K, Cisneros J, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. Plos Med. 2005;2(9):e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meltzer EB, Barry WT, D’Amico TA, et al. Bayesian probit regression model for the diagnosis of pulmonary fibrosis: proof-of-principle. BMC Medical Genomics. 2011;4 (1):70. doi: 10.1186/1755-8794-4-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renzoni EA, Abraham DJ, Howat S, et al. Gene expression profiling reveals novel TGF- beta targets in adult lung fibroblasts. Respir Res. 2004;5:24. doi: 10.1186/1465-9921-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardner H, Shearstone JR, Bandaru R, et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheum. 2006;54(6):1961–73. doi: 10.1002/art.21894. [DOI] [PubMed] [Google Scholar]
- 53.Schönherr E, Witsch-Prehm P, Harrach B. Interaction of biglycan with type I collagen. J Biol Chem. 1995;270:2776–2783. doi: 10.1074/jbc.270.6.2776. [DOI] [PubMed] [Google Scholar]
- 54.San Martin S, Zorn TMT. The small proteoglycan biglycan is associated with thick collagen fibrils in the mouse decidua. Cell Mol Biol. 2003;49(4):673–8. [PubMed] [Google Scholar]
- 55.Wei J, Fang F, Lam AP, et al. Wnt/beta-catenin signaling is hyperactivated in systemic sclerosis and induces Smad-dependent fibrotic responses in mesenchymal cells. Arthritis Rheum. 2012;64(8):2734–45. doi: 10.1002/art.34424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato M. Upregulation of the Wnt/beta-catenin pathway induced by transforming growth factor-beta in hypertrophic scars and keloids. Acta Derm Venereol. 2006;86(4):300–7. doi: 10.2340/00015555-0101. [DOI] [PubMed] [Google Scholar]
- 57.Hinz B, Phan SH, Thannickal VJ, et al. Recent developments in myofibroblast biology. American Journal of Pathology. 2012;180(4):1340–55. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kissin EY, Merkel PA, Lafyatis R. Myofibroblasts and hyalinized collagen as markers of skin disease in systemic sclerosis. Arthritis Rheum. 2006;54(11):3655–60. doi: 10.1002/art.22186. [DOI] [PubMed] [Google Scholar]
- 59.Lecca MR, Maag C, Berger EG, et al. Fibrotic response in fibroblasts from congenital disorders of glycosylation. J Cell Mol Med. 2011;15(8):1788–96. doi: 10.1111/j.1582-4934.2010.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith GP, Chan ESL. Molecular pathogenesis of skin fibrosis: insight from animal models. Current Rheumatology Reports. 2010;12(1):26–33. doi: 10.1007/s11926-009-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Derrett-Smith EC, Denton CP, Sonnylal S. Animal models of scleroderma: lessons from transgenic and knockout mice. Curr Opin Rheumatol. 2009;21(6):630–5. doi: 10.1097/BOR.0b013e32833130c1. [DOI] [PubMed] [Google Scholar]
- 62.Lemaire R, Farina G, Bayle J, et al. Antagonistic effect of the matricellular signaling protein CCN3 on TGF-beta- and Wnt-mediated fibrillinogenesis in systemic sclerosis and Marfan syndrome. J Invest Dermatol. 2010;130:1514–23. doi: 10.1038/jid.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu J, Krautzberger AM, Sui SH, et al. Cell-specific translational profiling in acute kidney injury. J Clin Invest. 2014;124(3):1242–54. doi: 10.1172/JCI72126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 65.Ng TL, Gown AM, Barry TS, et al. Nuclear beta-catenin in mesenchymal tumors. Modern Pathology. 2005;18:68–74. doi: 10.1038/modpathol.3800272. [DOI] [PubMed] [Google Scholar]
- 66.Chilosi M, Poletti V, Zamò A, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. American Journal of Pathology. 2003;162(5):1495–502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hesselstrand R, Westergren-Thorsson G, Scheja A, et al. The association between changes in skin echogenicity and the fibroblast production of biglycan and versican in systemic sclerosis. Clin Exp Rheumatol. 2002;20(3):301–8. [PubMed] [Google Scholar]
- 68.Hunzelmann N, Anders S, Sollberg S, et al. Co-ordinate induction of collagen type I and biglycan expression in keloids. Br J Dermatol. 1996;135(3):394–9. [PubMed] [Google Scholar]
- 69.Lafyatis R. SSc—fibrosis takes flight with Wingless inhibition. Nature Reviews Rheumatology. 2012;8(8):441–2. doi: 10.1038/nrrheum.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dees C, Distler JHW. Canonical Wnt signalling as a key regulator of fibrogenesis - implications for targeted therapies? Exp Dermatol. 2013;22(11):710–3. doi: 10.1111/exd.12255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure 1. Manipulated cells express fibroblast markers. (A) Merged and single-channel images showing YFP+ PDGFRα+ cells. Scale bars = 100 μm. (B) Lack of double immunofluorescent staining for YFP and endothelial marker PECAM or adipose marker perilipin. Scale bars = 100 μm. (C) Flow cytometric scatter plot showing lack of YFP+ PDGFRβ+ cells; values indicate percent of detected cells. hf, hair follicle.
Supporting Figure 2. α-SMA-expressing fibroblasts do not appear in advanced β-catenin-induced fibrosis. Immunohistochemistical staining for α-SMA in 4-month-old control and stabilized β-catenin mice demonstrates α-SMA+ structures (labeled), but not α-SMA+ fibroblasts. hf, hair follicle; musc, muscle; adip, adipose tissue. Scale bars = 100 μm.
Supporting Figure 3. Canonical TGF-β signaling activity is not affected in stabilized β-catenin-induced fibrosis. Stabilized β-catenin mutant skin has (A) 1.5-fold increase in relative expression of Serpine1 (PAI1) but no change in percentage of fibroblasts that express nuclear (B) pSmad2 or (C) pSmad3 at 1 week or 3 weeks of age. Scale bar (in secondary-only control image) = 200 μm; all images photographed at same magnification.
Supporting Figure 4. Validation of up-regulated genes in additional biological replicates. (A) Stabilized β-catenin mutant dermis has increased relative expression, measured by qPCR (n=2), of Thbs4, Ccn3, and Fbln1, genes which were also up-regulated upon differential expression analysis of whole transcriptome profiles. (B) Stabilized β-catenin mutant skin has increased CCN3 protein expression in dermis and hypodermis compared to control skin. Higher-magnification images correspond with dotted and solid outlined areas in corresponding lower-magnification image. Scale bars (in control images) = 200 μm; all images photographed at same magnification.
Supporting Figure 5. Protein expression of biglycan in stabilized β-catenin mouse skin. Stabilized β-catenin mutant skin has variably increased biglycan protein expression in the hypodermis compared to control skin. Higher-magnification images correspond with dotted and solid outlined areas in corresponding lower-magnification image. Scale bars (in control images) = 200 μm; all images photographed at same magnification.
Supporting Figure 6. Keloid scar nodule has elevated cytoplasmic and nuclear β-catenin. Immunohistochemical detection of β-catenin in healthy control skin and three keloid scars demonstrating elevated cytoplasmic and nuclear β-catenin immunoreactivity in the keloid nodule.
List of differentially expressed (DE) genes with corresponding log2(fold change) and adjusted p-values.
Categorization of 175 DE genes; number of genes per category; categorization of 36 matrisome-encoding genes; number of genes per matrisome category.
List of homologous human genes to 171 of the differentially expressed genes from our mouse model.
List of the human homologous genes that are also over-expressed in fibrotic tissues, with corresponding log(fold change) and adjusted p-values from GEO2R analysis.