Abstract
Pancreatic ductal adenocarcinoma (PDAC) is nearly uniformly lethal, with a median overall survival in 2014 of only 6 months. The genetic progression of sporadic PDAC (SPC) is well established, with common somatic alterations in KRAS, p16/CDKN2A, TP53, and SMAD4/DPC4. Up to 10 % of all PDAC cases occur in families with two or more affected first-degree relatives (familial pancreatic cancer, FPC), but these cases do not appear to present at an obviously earlier age of onset. This is unusual because most familial cancer syndrome patients present at a substantially younger age than that of corresponding sporadic cases. Here we collated the reported age of onset for FPC and SPC from the literature. We then used an integrated approach including whole exomic sequencing, whole genome sequencing, RNA sequencing, and high density SNP microarrays to study a cohort of FPC cell lines and corresponding germline samples. We show that the four major SPC driver genes are also consistently altered in FPC and that each of the four detection strategies was able to detect the mutations in these genes, with one exception. We conclude that FPC undergoes a similar somatic molecular pathogenesis as SPC, and that the same gene targets can be used for early detection and minimal residual disease testing in FPC patients.
Keywords: Pancreatic ductal adenocarcinoma (PDAC), Familial pancreatic cancer (FPC), Germline predisposition, Cancer driver genes, Next generation sequencing (NGS)
Introduction
While patients with several different forms of cancer survive longer after diagnosis than in the past, the 5-year survival rate of patients with pancreatic ductal adenocarcinoma cancer (PDAC) has remained relatively unchanged over the past 5 decades [1]. As many as 10 % of PDACs have a hereditary component (familial PDAC, FPC), defined as a family with at least two first-degree relatives with PDAC [2, 3]. Known susceptibility genes include BRCA1, BRCA2, PALB2, ATM, STK11, PRSS1, SPINK1, and DNA mismatch repair genes, but all together these explain less than 20 % of familial pancreatic cancer cases [4–7]. PDAC is notoriously lethal because patients present late in the disease process and the cancers are chemorefractory. Importantly, the 9 % of cases that present with the tumor confined to the pancreas have a 5-year survival rate of 24 %, supporting the notion that lesions detected early enough can be cured [1]. To focus early detection resources, it is important to identify patients at particularly high risk, such as those with familial predispositions.
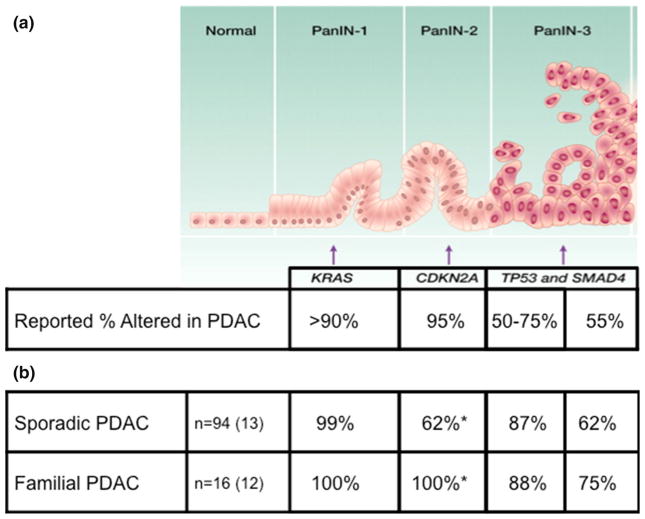
The molecular progression of SPC is well-established both histologically and molecularly [8]. The high-prevalence SPC driver genes are KRAS (>90 % of PDAC), CDKN2A/p16 (95 %), TP53 (50–75 %), and SMAD4/DPC4 (55 %) [9–12]. PDACs commonly arise from pancreatic intraepithelial neoplasia (PanIN) or Intraductal Papillary Mucinous Neoplasm (IPMN) precursor lesions. While the pathology of FPC has been shown not to differ from that of apparently sporadic disease, FPC patients have been shown to have significantly more precursor lesions as well as higher grade precursor lesions when compared to patients with sporadic disease [13–15]. Knowing the genes involved in FPC molecular progression is essential to designing effective early detection strategies [16].
Early onset is a hallmark of most familial cancer syndromes, including hereditary breast and ovarian cancer (BRCA1, BRCA2), familial adenomatous polyposis (APC), hereditary non-polyposis colorectal cancer (MLH1, MSH2, MSH6, PMS2), and familial atypical multiple mole melanoma (FAMMM) syndrome (p16) [17–20]. In contrast, an earlier age of onset is not an obvious hallmark of FPC [21, 22]. How can one possibly inherit a predisposition to a cancer without an obvious acceleration of the phenotype? This question challenges our current understanding of familial cancer syndromes and the canonical two-hit hypothesis [23, 24].
In the study, we first collated the age of onset in FPC and SPC reported in the literature to validate the general notion that the age of onset of familial and sporadic PDAC cases was similar. We then determined the status of known SPC driver genes in our own FPC cohort, a unique resource of eighteen FPC cell lines that we have generated over the past decade. We used an integrated approach including high density SNP microarrays, exomic sequencing, whole genome sequencing, and RNA-sequencing to investigate those genes involved in FPC progression. Finally, having established a consensus for each gene in each sample, we examined the ability of each tool to detect the mutations.
Materials and methods
Case selection
This study was reviewed and approved by the Institutional Review Board at Johns Hopkins Medical Institutions, and informed consent was obtained from all study participants. Familial pancreatic cancer was defined as a pancreatic cancer that arose in a proband with at least one first-degree relative with pancreatic cancer (i.e. a family with two or more affected first-degree relatives). Cancer cell lines were established from familial pancreatic cancers and matched normal DNA from the patients was obtained from Epstein–Barr virus (EBV) transformed lymphoblasts or frozen tissue [25]. Tumor-normal pair matching was confirmed by STR analysis of nine loci and Amelogenin using ABI Profiler kit (Life Technologies, Carlsbad, CA) and size-separated on an ABI CE3130xl instrument (Life Technologies). The data from 94 SPC and 7 FPC (four from discovery, three from prevalence) were previously reported [11]. An unpaired, two-tailed t test of our cohorts was used to determine if the mean age of onset difference between our familial and sporadic cases was statistically significant.
Collation of reported age of onset
Literature reporting age of onset in FPC (excluding hereditary pancreatitis) and SPC were collected from PubMed. Only the most recent study was used when multiple studies employed the same patient registry, on the assumption that previous reported families would be included in subsequent reports and therefore exclude redundant cases. Studies were stratified based on study type (population or referral) and statistic reported (mean or median).
Preparation of genomic DNA and RNA
Genomic DNA was extracted from early passage cell lines and matched normal EBV-transformed lymphoblasts or frozen normal tissue using QIAamp DNA mini kit (Qiagen, Valencia, CA), per manufacturer’s instruction. RNA was extracted from cell lines using RNeasy mini kit (Qiagen), per manufacturer’s instruction. A HPDE (human pancreatic ductal epithelium) cell line was used as a normal control for RNA-Sequencing [26].
High density SNP microarray
The Omni2.5 array (Illumina, San Diego, CA) was used to analyze cancer cell lines and matched normal samples at 2,379,855 (2.5 M) SNP loci. Analysis was carried out with Genome Studio with the following criteria: an average LogR Ratio (LRR) ≤ −2.0 for homozygous deletions (HDs); LRR of 0–0.53 and B Allele Frequency of 0 or 1 for loss of heterozygosity (LOH); and an average LRR ≥ 1.4, with at least one SNP LRR ≥ 2.0, for amplifications. At least four SNPs must fit criteria for the region to be called an alteration and boundaries were the first and last SNPs that meet criteria. Adjacent deleted or amplified regions (within 100 kb) were considered to be one alteration. Given that half or more of the p16 and SMAD4 inactivations are HDs, we excluded the 4 FPC and 81 SPC cases without SNP microarray data, in the analysis of p16 and SMAD4 genes.
Genomic DNA libraries and exomic sequencing
Genomic DNA libraries were prepared using 1 μg of genomic DNA and human exome capture was performed following a modified protocol from Agilent’s SureSelect Paired-End Version 2.0 Human Exome Kit (Agilent, Santa Clara, CA) as previously described [27]. Briefly, captured DNA libraries were sequenced with a GAIIx Genome Analyzer, yielding 150 bp (2 × 75 bp) from the final library fragments, to 200X coverage. Sequencing reads were analyzed and aligned to human genome hg18 with the Eland algorithm in CASAVA 1.7 software (Illumina). The Database of Single Nucleotide Polymorphisms was used in the analysis of whole-exome sequencing data (dbSNP). Mutations were visually confirmed in the aligned files.
Whole genome sequencing
Sequencing on an Illumina HiSeq 2000 (Illumina) was carried out at 60X coverage for cancers and 30X coverage for matched normal by Personal Genome Diagnostics (Baltimore, MD) using 3 μg of genomic DNA and generating 200 bp (2 × 100 bp paired reads) per fragment. Reads were aligned to human genome (hg19) with Eland v.2 algorithm in CASAVA 1.7 software (Illumina).
cDNA libraries and RNA sequencing
A total of 5 μg of total RNA was depleted of ribosomal RNA using ribominus and cDNA libraries were prepared using TruSeq Stranded Total RNA Sample Preparation (Illumina), as per the manufacturer’s instructions. Paired-end sequencing, resulting in 100 bp reads was carried out on an Illumina HiSeq to a level of 50 M reads. RSEM was used to align the sequences to human genome hg19 [28]. Alterations were visually confirmed using Integrated Genomics Viewer [29].
Results
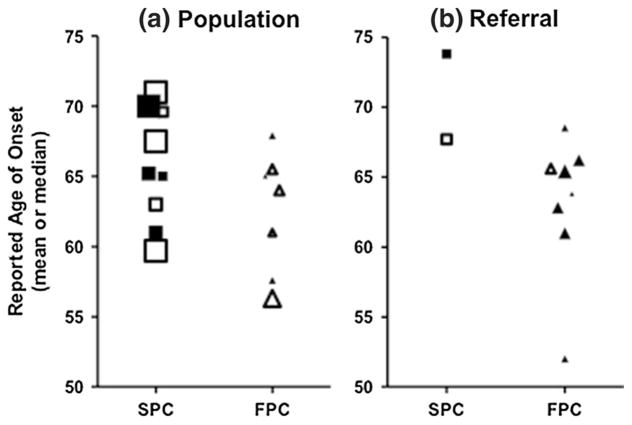
Previous investigations have noted a similar age of onset of SPC and FPC. To comprehensively examine this, we culled studies reporting FPC and SPC age of onset and published from 1991 to 2013 (n = 15). To avoid overweighting the same families, we used only the most recent study when multiple studies were reported through time from the same institution or consortium. The collated studies have reported mean or median ages of 60–74 for SPC patients and 52–69 for FPC patients (Fig. 1, Supplemental Table 1). Due to potential ascertainment bias, we separated the studies that reported age of onset in a population unselected based upon family history (Fig. 1a) versus those from family registries (Fig. 1b). The mean age of PDAC diagnosis from 1973 to 2000 SEER data is 70 years [30]. In our small cohorts of FPC and SPC, there was no obvious difference in age (FPC cohort: mean 64 years (range: 42–81); SPC cohort: mean 66 years (range: 36–85)) (Table 1, Supplemental Table 2). The lower age of both of the cohorts we analyzed (FPC and SPC) compared to SEER may be attributable to ascertainment or referral bias. There appears to be a greater difference in the referral-based studies, likely because the vast majority of samples in our study underwent surgical resection. We have intentionally omitted statistical comparison of the groups because of the invalidity in comparing means and medians, and the small sample size that would exist without pooling these two statistics.
Fig. 1.
Reported age of onset for SPC and FPC, collated from the literature. Literature was separated based on a population-based or b referral cohorts, reported as means (filled symbols) or medians (empty symbols). Symbol sizes are adjusted according to the number of individuals in the study [2*log(n)]. There are no obvious differences in the age of onset for FPC (triangles) compared to SPC (squares)
Table 1.
FPC cohort demographics
| Case | Age at diagnosis | Sex | Additional family history of pancreatic cancer |
|---|---|---|---|
| Pa007C* | 80s | F | Father, mother |
| Pa009C* | 60s | F | Brother, paternal uncle |
| Pa011X* | 70s | F | Father |
| Pa018X* | 70s | F | Brother |
| Pa101C | 40s | M | Father |
| Pa102C | 60s | F | Sister |
| Pa147X* | 70s | M | Brother, mother |
| Pa170X* | 40s | M | Brother |
| Pa212X* | 60s | M | Mother |
| Pa222C | 70s | M | Father, paternal grandfather, paternal uncle |
| Pa223C | 60s | F | Brother, cousin |
| Pa227C | 60s | F | Father |
| Pa228C | 60s | F | Brother |
| Pa229C | 40s | M | Mother |
| Pa230C | 60s | F | Father |
| Pa231C | 60s | M | Father, mother, paternal grandfather |
Samples previously reported in Jones et al. [11] Science
The molecular progression of SPC is well-documented, with common somatic alterations in the four driver genes KRAS, CDKN2A/p16, TP53, and SMAD4/DPC4, in addition to many other low prevalence genes [11, 12]. In an attempt to identify new FPC predisposition genes, we performed a comprehensive genomic analysis of our 16 FPC cell lines. No strong candidates for predisposition genes were identified in these samples. We also determined the mutational status of the four SPC driver genes in these 16 FPC samples as assessed by each of the four methods.
Overall, the prevalence of alterations in the four SPC driver genes was similar in the 16 FPC PDACs and the 94 SPC PDACs (Fig. 2a, b). Activating KRAS mutations were identified in 16/16 (100 %) of FPC PDACs, predominately at codon 12 (94 %:63 % G12D, 19 % G12V, and 13 % G12R) but with one case at codon 61 (6 %, Q61H) (Fig. 2b, Supplemental Table 3). Of the 94 sporadic PDACs, all but one had an activating KRAS mutation (99 %). The majority of KRAS mutations in the SPC PDACs were also at codon 12 (95 %:50 % G12D, 31 % G12V, and 12 % G12R). Four SPC PDACs had codon 61 mutations (3 Q61H, 1 Q61R), and one SPC PDAC had two different activating KRAS mutations (G12V and G13C).
Fig. 2.
Summarized alterations in PDAC molecular progression genes, for SPC and FPC. a PDAC molecular progression model with reported percent alterations of the four driver genes in PanIN lesions, figure modified from Iacobuzio-Donahue, et al. Clin Cancer Res 2012 [56]. b Percent alterations of molecular progression genes in PDAC cancers from SPC and FPC cohorts. As expected, the mutation prevalence in PDACs in panel b are higher than the early PanIN lesions in panel a. *CDKN2A, p = 0.04
CDKN2A/p16 was inactivated in 100 % (12/12, the four cases without SNP microarray data were excluded) by homozygous deletion (9/12, 75 %) or single base substitution with LOH (3/12, 25 %), of the FPC PDACs, compared to only 62 % (8/13, the cases without SNP microarray data were excluded) of the SPC PDACs (p = 0.04, Fig. 2b, Supplemental Table 4). Alterations of the CDKN2A gene are reported to occur in 95 % of SPC PDACs, with epigenetic silencing accounting for about 15 % of this inactivation [31]. As we did not assess epigenetic changes, the actual fraction of cases with somatically altered CDKN2A in Supplemental Table 4 is likely an underestimate.
TP53 was mutated in 88 % (14/16) of FPC PDACs, by single base substitution with LOH (10/14), frameshift with LOH (2/14), or biallelic mutation (2/14) (Fig. 2b, Supplemental Table 5). Of the 94 sporadic PDACs, 82 (87 %) had inactivating TP53 mutations. The mutation types included biallelic mutations (1/82), single base substitutions with LOH (64/82), frameshifts with LOH (15/82), and HDs (2/82).
SMAD4/DPC4 was inactivated in 75 % (9/12, the four cases without SNP microarray data were excluded) of FPC PDACs, by homozygous deletion (5/9), single base substitution with LOH (3/9), and frameshift with LOH (1/9) (Fig. 2b, Supplemental Table 6). Of the 13 sporadic PDACs (the cases without SNP microarray data were excluded), 62 % had inactivated SMAD4. The mutation types included single base substitutions with LOH (2/8), frameshifts with LOH (3/8), and HDs (3/8).
Having established a consensus gene mutation status, we retrospectively determined the ability of each genome-wide tool to detect the mutations. We first categorized the mutations as HDs, point mutations (including single base substitutions, frameshift deletions and insertions), and loss-of-heterozygosity (LOH) events. We then studied the ability of each tool to detect these three types of mutations in the four driver genes (Table 2).
Table 2.
Relative power of each method to detect common alterations
| Gene | Gene type* | Alteration type | # Samples altered | WES | WGS | SNP array | RNA-Seq |
|---|---|---|---|---|---|---|---|
| KRAS | OG | Mut | 7 | 7 (100 %) | 7 (100 %) | 0 (0 %) | 7 (100 %) |
| TP53 | TSG | Mut | 7 | 7 (100 %) | 7 (100 %) | 0 (0 %) | 7 (100 %) |
| HD | 0 | na | na | na | na | ||
| CDKN2A/p16 | TSG | Mut | 3 | 3 (100 %) | 3 (100 %) | 0 (0 %) | 3 (100 %) |
| HD | 4 | 4 (100 %) | 4** (100 %) | 4 (100 %) | 4 (100 %) | ||
| SMAD4/DPC4 | TSG | Mut | 2 | 2 (100 %) | 2 (100 %) | 0 (0 %) | 2 (100 %) |
| HD | 4 | 4 (100 %) | 4 (100 %) | 4 (100 %) | 4 (100 %) |
OG oncogene, TSG tumor suppressor gene, MUT mutation (includes frameshifting indels and point mutations), HD homozygous deletion, WES whole exome sequencing, WGS whole genome sequencing
1 p16 mutation was not reported as an alteration by the standard Illumina pipeline, but visualization of the bam file clearly showed a homozygous deletion
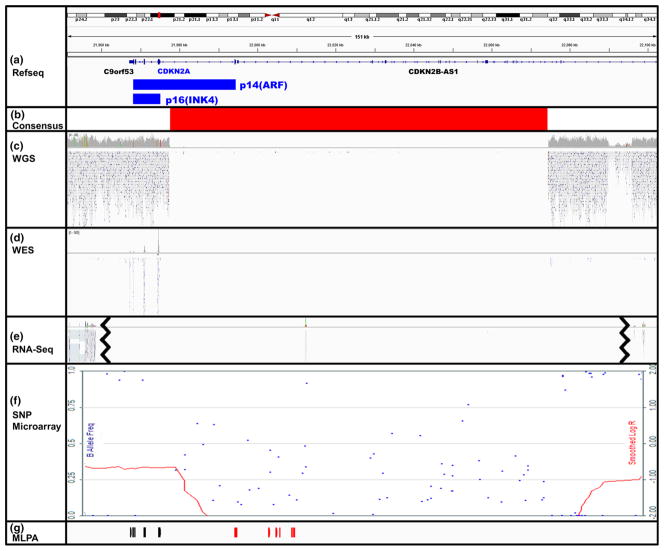
Homozygous deletions are common in the tumor suppressors CDKN2A/p16 and SMAD4/DPC4 and were detected reliably by SNP microarray, whole exomic sequencing (WES), whole genome sequencing (WGS), and RNA sequencing (RNA-Seq). For only one homozygous deletion (Fig. 3a, b), the standard WGS Illumina pipeline for calling copy number alterations missed a p16 homozygous deletion (sample PA222C), clearly deleted by visual inspection of WGS data (Fig. 3c). The homozygous deletion included 17 kb of the 5′ end of p16 transcript variant 4 (NM_058195), but did not result in the deletion of any DNA sequence corresponding to transcript variants 1,3, 5 (NM_000077, NM_058197, and NM_001195132) (Fig. 3a, b). The later transcript variants encode the p16(INK4) isoform, a CDK inhibitor, while transcript variant 4 encodes a structurally distinct p14(ARF) which stabilizes TP53 by sequestering MDM2. Both isoforms are normally expressed in the pancreas. Importantly, neither the p14ARF or p16INK4a transcripts are expressed according to the RNA-Seq data (Fig. 3e), a result of the loss of p14ARF’s first exon and p16(INK4)’s promoter sequence, respectively. This 97 kb homozygous deletion was identified by WES, RNA-Seq, and high density SNP microarray, and the deletion’s breakpoints were remarkably concordant across these methods (Fig. 3b–f). Because the WGS results were initially discordant, we used multiplex ligation-dependent probe amplification (MLPA) to confirm the homozygous deletion (Fig. 3g). That only one of the four alternative transcripts is included in the homozygous deletion explains why this was missed by the WGS using the standard Illumina pipeline. This highlights the importance of the reference transcript used in a NGS mapping algorithm and the potential utility of remapping to known deletions, such as p16 in the case of PDAC, especially at lower read depths.
Fig. 3.
CDKN2A/p16 homozygous deletion initially missed by WGS in one case (PA222C). Visualization of WGS reads for p16 gene region (hg19, chr9:21,951,176–22,102,475) in PA222C sample using IGV (Broad Institute, version 2.3.31) and Karyostudio (Illumina, version 1.4). The 2 protein isoforms of p16 (p14(ARF) and p16(INK4), blue) are shown as well as the adjacent genes, C9orf53 and CDKN2B-AS1 (a). There is clear agreement across the methods for the consensus 97 kb homozygous deletion boundaries (b). The homozygous deletion of p16 was not called by the standard Illumina pipeline for WGS data, despite a clear confirmation of the 5′ deletion by visual inspection, due to reference transcript choice (c). The homozygous deletion was detected by WES, as evidenced by the lack of reads (d). RNA-Seq produced no high quality reads that mapped to this deleted region, but there were reads upstream (MTAP) and downstream (DMRTA1) of the homozygous deletion (e line break indicates upstream or downstream reads shows). High density SNP microarray detected the homozygous deletion (red LogR line drops to −2.00 and scattered B allele frequencies) and the flanking LOH regions (red LogR line at −1.00 and B allele frequencies at 0 or 1) (f). MLPA probes were used to confirm the upstream LOH (black) and homozygous deletion (red) regions (g)
Point mutations and LOH in KRAS, p16, TP53, and SMAD4 were all detected by WES, WGS, and RNA-Seq. High density SNP microarray could of course not detect any of the point mutations in the four driver genes. However, it is likely that a custom SNP microarray could be designed to detect mutations in hotspots in KRAS. Where LOH in p16, TP53, and SMAD4 genes was detected, it was detected equally by all of the methods. In the two cases with biallelic TP53 mutations, there was no evidence of LOH, as expected.
We also investigated the mutation status of genes implicated at a lower frequency in PDAC, but reported to be mutated in cystic precursors, pancreatic neuroendocrine tumors (PanNETs), or implicated as FPC predisposition genes. MLL3 has been reported to be mutated in 9 % of PDACs [11, 32]. Here, MLL3 was mutated in 17 % (2/12) FPC PDACs. Both cases were single base substitutions (nonsense mutation with LOH in PA11X and bi-allelic missense mutations in PA18C). The genes implicated in pancreatic cystic lesions (GNAS, RNF43, CTNNB1, and VHL) were not mutated in any FPC case [33, 34]. ATRX, DAXX, and MEN1 are reported to be mutated in PanNETs, and ATRX was homozygously deleted in 1 (8 %) FPC case (PA102C) [27]. DAXX and MEN1 however, were not mutated in any FPC PDACs, and no clearly deleterious mutations were identified in ATM, STK11, PRSS1, PALB2, BRCA2, or SPINK1.
Discussion
We confirm, through our qualitative analysis of the literature, that most studies do not indicate a large difference in the age of onset between SPC and FPC. While some studies do show a slight difference in the age of onset, part of this difference could be due to ascertainment biases. Some studies have shown a slightly lower age of onset in FPC compared to SPC in their cohorts [35–41]. One study even showed a slightly later onset in their FPC group [42], however most studies to date have shown a similar age of onset (Supplemental Table 1) [43–52]. The field would benefit from a rigorous meta-analysis of FPC versus SPC age of onset.
Our study also showed that FPC PDACs harbor the same high prevalence genetic alterations that have been identified in SPC PDACs (Fig. 2b). One purpose of analyzing driver data prevalence is to identify “holes”, genes with lower than expected mutation prevalence, under the hypothesis that a homologue or pathway-related gene could be defective in the germline. A similar approach led to the elegant discovery of germline MYH mutations in familial colorectal cancers that were phenotypically similar to attenuated familial adenomatous polyposis, but lacked germline APC mutations [53]. From our data, there are no such “holes.” This finding confirms and builds upon a previous study that found that familial and sporadic pancreatic cancers had similar prevalence of mutations in the three SPC driver genes they assessed [31].
The late detection of pancreatic cancers contributes to the lethality of the disease. Much work has been done in the area of non-invasive early detection tests, using molecular signatures of pancreatic cancer—notably, the KRAS codon 12/13 mutation hotspot. Because FPC shares the SPC molecular signature mutations, these could be included in early detection tests and the gene panels currently in development could also be used in familial pancreatic kindreds for early detection and molecular relapse.
Assuming that FPC and SPC have a similar age of onset, how can one inherit a predisposition to a disease without accelerating its age of onset? Unfortunately, our study did not provide any great insights into this question and it remains unanswered. We note, however, that there is precedent in PDAC, even when the causative genes are known. For example, in patients with FAMMM syndrome, p16 germline mutations confer a significantly earlier age of onset for melanoma, but not for PDAC [20, 49, 54, 55]. These observations support the idea that it is the pancreatic tissue rather than the gene that is responsible for the curious lack of age dependence on the presence of hereditary predisposition genes. The mechanisms underlying this difference represent an important area for future study as it may shed light on PDAC pathogenesis in general.
We employed an integrative strategy to more comprehensively detect alterations in FPC than previous reports. Combining WES, WGS, and RNA-Seq allowed for a greater coverage of gene-coding regions, particularly in expressed genes (Table 2). The importance of gene transcript choice in identifying alterations, such as HDs, in next generation sequencing data was highlighted by the PA222C homozygous deletion of p16 initially missed by WGS analysis, but obvious upon visual inspection of the reads (Fig. 3). Other than this one example, the methods were remarkably concordant. High density SNP microarrays strengths are identifying LOH and large HDs, both hallmarks of tumor suppressor genes.
We conclude that FPC and SPC undergo similar pathogenesis permitting the same gene targets to be used for early detection and minimal residual disease testing.
Acknowledgments
We acknowledge Drs. Scott Kern, Ming Tseh-Lin, Gloria Petersen, Manuel Hidalgo, Elizabeth Jaffee, Steve Leach, Michael Goggins, Christine Iacobuzio-Donahue, Hiro Kamiyama, Mihoko Kamiyama, and Denise Batista for helpful discussions. We thank Alyza Skaist, Laura Morsberger, Jennifer Meyers, Diane Echavarria, Roxanne Ashworth, and Hong Liang for outstanding technical support. Grants: The Sol Goldman Pancreatic Cancer Research Center (pilot project) (JRE), R21CA164592 (JRE), PanCan/AACR Innovation Award (JRE), SPORE P50-CA62924-13 (Kern) and The Stringer Foundation (JRE).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10689-014-9755-y) contains supplementary material, which is available to authorized users.
Ethical standard This study was performed with approval by the Institutional Review Board at Johns Hopkins Medical Institution. All participants gave informed consent and details that might disclose the identity of the subjects under study have been omitted.
Contributor Information
Alexis L. Norris, Department of Pathology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Room 344, Cancer Research Building-II, 1550 Orleans Street, Baltimore, MD 21231, USA
Nicholas J. Roberts, Department of Pathology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Room 344, Cancer Research Building-II, 1550 Orleans Street, Baltimore, MD 21231, USA. Ludwig Center and Howard Hughes Medical Institute, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA
Siân Jones, Ludwig Center and Howard Hughes Medical Institute, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA.
Sarah J. Wheelan, Department of Oncology Biostatistics, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA
Nickolas Papadopoulos, Ludwig Center and Howard Hughes Medical Institute, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA. Department of Oncology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA.
Bert Vogelstein, Ludwig Center and Howard Hughes Medical Institute, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA. Department of Oncology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA.
Kenneth W. Kinzler, Ludwig Center and Howard Hughes Medical Institute, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA. Department of Oncology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA
Ralph H. Hruban, Department of Pathology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Room 344, Cancer Research Building-II, 1550 Orleans Street, Baltimore, MD 21231, USA. Department of Oncology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA
Alison P. Klein, Department of Pathology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Room 344, Cancer Research Building-II, 1550 Orleans Street, Baltimore, MD 21231, USA. Department of Oncology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA
James R. Eshleman, Email: jeshlem@jhmi.edu, Department of Pathology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Room 344, Cancer Research Building-II, 1550 Orleans Street, Baltimore, MD 21231, USA. Department of Oncology, The Sol Goldman Center for Pancreatic Cancer Research, Johns Hopkins University School of Medicine, Baltimore, MD 21231, USA
References
- 1.Howlader NNA, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2010. National Cancer Institute; 2013. SEER web site. [Google Scholar]
- 2.Petersen GM, Hruban RH. Familial pancreatic cancer: where are we in 2003? J Natl Cancer Inst. 2003;95(3):180–181. doi: 10.1093/jnci/95.3.180. [DOI] [PubMed] [Google Scholar]
- 3.Shi C, Hruban RH, Klein AP. Familial pancreatic cancer. Arch Pathol Lab Med. 2009;133(3):365–374. doi: 10.1043/1543-2165-133.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein AP, Brune KA, Petersen GM, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64(7):2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 5.Hruban RH, Canto MI, Goggins M, Schulick R, Klein AP. Update on familial pancreatic cancer. Adv Surg. 2010;44:293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haddad A, Kowdley GC, Pawlik TM, Cunningham SC. Hereditary pancreatic and hepatobiliary cancers. Int J Surg Oncol. 2011;2011:154673. doi: 10.1155/2011/154673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts NJ, Jiao Y, Yu J, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discovery. 2012;2(1):41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am J Pathol. 2000;156(6):1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldas C, Hahn SA, da Costa LT, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8(1):27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 10.Iacobuzio-Donahue CA, van der Heijden MS, Baumgartner MR, et al. Large-scale allelotype of pancreaticobiliary carcinoma provides quantitative estimates of genome-wide allelic loss. Cancer Res. 2004;64(3):871–875. doi: 10.1158/0008-5472.can-03-2756. [DOI] [PubMed] [Google Scholar]
- 11.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brune K, Abe T, Canto M, et al. Multifocal neoplastic precursor lesions associated with lobular atrophy of the pancreas in patients having a strong family history of pancreatic cancer. The American journal of surgical pathology. 2006;30(9):1067–1076. [PMC free article] [PubMed] [Google Scholar]
- 14.Shi C, Klein AP, Goggins M, et al. Increased Prevalence of Precursor Lesions in Familial Pancreatic Cancer Patients. Clin Cancer Res. 2009;15(24):7737–7743. doi: 10.1158/1078-0432.CCR-09-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhi AD, Ishida H, Ali S, et al. A histomorphologic comparison of familial and sporadic pancreatic cancers. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Trans Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250(4988):1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 18.Bunyan DJ, Shea-Simonds J, Reck AC, Finnis D, Eccles DM. Genotype-phenotype correlations of new causative APC gene mutations in patients with familial adenomatous polyposis. J Med Genet. 1995;32(9):728–731. doi: 10.1136/jmg.32.9.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myrhoj T, Bisgaard ML, Bernstein I, Svendsen LB, Sondergaard JO, Bulow S. Hereditary non-polyposis colorectal cancer: clinical features and survival. Results from the Danish HNPCC register. Scand J Gastroenterol. 1997;32(6):572–576. doi: 10.3109/00365529709025102. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein AM, Chan M, Harland M, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66(20):9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 21.Krainer M, Silva-Arrieta S, FitzGerald MG, et al. Differential contributions of BRCA1 and BRCA2 to early-onset breast cancer. N Engl J Med. 1997;336(20):1416–1421. doi: 10.1056/NEJM199705153362003. [DOI] [PubMed] [Google Scholar]
- 22.Raimondi S, Maisonneuve P, Lohr JM, Lowenfels AB. Early onset pancreatic cancer: evidence of a major role for smoking and genetic factors. Cancer Epidemiol Biomarkers Prev. 2007;16(9):1894–1897. doi: 10.1158/1055-9965.EPI-07-0341. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls EM. Somatic variation and multiple neurofibromatosis. Hum Hered. 1969;19(5):473–479. doi: 10.1159/000152255. [DOI] [PubMed] [Google Scholar]
- 24.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffin CA, Morsberger L, Hawkins AL, et al. Molecular cytogenetic characterization of pancreas cancer cell lines reveals high complexity chromosomal alterations. Cytogenet Genome Res. 2007;118(2–4):148–156. doi: 10.1159/000108295. [DOI] [PubMed] [Google Scholar]
- 26.Ouyang H, Mou L, Luk C, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157(5):1623–1631. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Internet] NCIhot (released April 2005, based on the November 2004 submission [cited 2014 Jun 24]) Surveillance Epidemiology and End Results (SEER) Program. SEER*Stat Database: Incidence—SEER 13 Regs Public-Use, Nov 2004 Sub (1973–2002 varying), National Cancer Institute DCCPS, Surveillance Research Program, Cancer Statistics Branch. http://www.seercancergov
- 31.Brune K, Hong SM, Li A, et al. Genetic and epigenetic alterations of familial pancreatic cancers. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3536–3542. doi: 10.1158/1055-9965.EPI-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao Y, Yonescu R, Offerhaus GJ, et al. Whole-exome sequencing of pancreatic neoplasms with acinar differentiation. J Pathol. 2014;232(4):428–435. doi: 10.1002/path.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Jiao Y, Dal Molin M, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA. 2011;108(52):21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Science Transl Med. 2011;3(92):92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luttges J, Stigge C, Pacena M, Kloppel G. Rare ductal adenocarcinoma of the pancreas in patients younger than age 40 years. Cancer. 2004;100(1):173–182. doi: 10.1002/cncr.11860. [DOI] [PubMed] [Google Scholar]
- 36.Lal G, Liu G, Schmocker B, et al. Inherited predisposition to pancreatic adenocarcinoma: role of family history and germ-line p16, BRCA1, and BRCA2 mutations. Cancer Res. 2000;60(2):409–416. [PubMed] [Google Scholar]
- 37.James TA, Sheldon DG, Rajput A, et al. Risk factors associated with earlier age of onset in familial pancreatic carcinoma. Cancer. 2004;101(12):2722–2726. doi: 10.1002/cncr.20700. [DOI] [PubMed] [Google Scholar]
- 38.Brandt A, Bermejo JL, Sundquist J, Hemminki K. Age of onset in familial cancer. Ann Oncol. 2008;19(12):2084–2088. doi: 10.1093/annonc/mdn527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen GM, de Andrade M, Goggins M, et al. Pancreatic cancer genetic epidemiology consortium. Cancer Epidemiol Biomarkers Prev. 2006;15(4):704–710. doi: 10.1158/1055-9965.EPI-05-0734. [DOI] [PubMed] [Google Scholar]
- 40.Eberle MA, Pfutzer R, Pogue-Geile KL, et al. A new susceptibility locus for autosomal dominant pancreatic cancer maps to chromosome 4q32-34. Am J Hum Genet. 2002;70(4):1044–1048. doi: 10.1086/339692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozcelik H, Schmocker B, Di Nicola N, et al. Germline BRCA2 6174delT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat Genet. 1997;16(1):17–18. doi: 10.1038/ng0597-17. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez E, La Vecchia C, D’Avanzo B, Negri E, Franceschi S. Family history and the risk of liver, gallbladder, and pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 1994;3(3):209–212. [PubMed] [Google Scholar]
- 43.McWilliams RR, Bamlet WR, Rabe KG, Olson JE, de Andrade M, Petersen GM. Association of family history of specific cancers with a younger age of onset of pancreatic adenocarcinoma. Clin Gastroenterol Hepatol. 2006;4(9):1143–1147. doi: 10.1016/j.cgh.2006.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartsch DK, Kress R, Sina-Frey M, et al. Prevalence of familial pancreatic cancer in Germany. Int J Cancer. 2004;110(6):902–906. doi: 10.1002/ijc.20210. [DOI] [PubMed] [Google Scholar]
- 45.Lynch HT, Fitzsimmons ML, Smyrk TC, et al. Familial pancreatic cancer: clinicopathologic study of 18 nuclear families. Am J Gastroenterol. 1990;85(1):54–60. [PubMed] [Google Scholar]
- 46.Lynch HT, Fusaro L, Lynch JF. Familial pancreatic cancer: a family study. Pancreas. 1992;7(5):511–515. doi: 10.1097/00006676-199209000-00001. [DOI] [PubMed] [Google Scholar]
- 47.Ji J, Forsti A, Sundquist J, Lenner P, Hemminki K. Survival in familial pancreatic cancer. Pancreatology. 2008;8(3):252–256. doi: 10.1159/000134272. [DOI] [PubMed] [Google Scholar]
- 48.Ghadirian P, Boyle P, Simard A, Baillargeon J, Maisonneuve P, Perret C. Reported family aggregation of pancreatic cancer within a population-based case-control study in the Francophone community in Montreal, Canada. Int J Pancreatol. 1991;10(3–4):183–196. doi: 10.1007/BF02924156. [DOI] [PubMed] [Google Scholar]
- 49.Hruban RH, Petersen GM, Ha PK, Kern SE. Genetics of pancreatic cancer. From genes to families. Surgical oncology clinics of North America. 1998;7(1):1–23. [PubMed] [Google Scholar]
- 50.Silverman DT, Schiffman M, Everhart J, et al. Diabetes mellitus, other medical conditions and familial history of cancer as risk factors for pancreatic cancer. Br J Cancer. 1999;80(11):1830–1837. doi: 10.1038/sj.bjc.6690607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Permuth-Wey J, Egan KM. Family history is a significant risk factor for pancreatic cancer: results from a systematic review and meta-analysis. Fam Cancer. 2009;8(2):109–117. doi: 10.1007/s10689-008-9214-8. [DOI] [PubMed] [Google Scholar]
- 52.Couch FJ, Johnson MR, Rabe KG, et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(2):342–346. doi: 10.1158/1055-9965.EPI-06-0783. [DOI] [PubMed] [Google Scholar]
- 53.Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C–> T: A mutations in colorectal tumors. Nat Genet. 2002;30(2):227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 54.FitzGerald MG, Harkin DP, Silva-Arrieta S, et al. Prevalence of germ-line mutations in p16, p19ARF, and CDK4 in familial melanoma: analysis of a clinic-based population. Proc Natl Acad Sci USA. 1996;93(16):8541–8545. doi: 10.1073/pnas.93.16.8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldstein AM, Struewing JP, Chidambaram A, Fraser MC, Tucker MA. Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst. 2000;92(12):1006–1010. doi: 10.1093/jnci/92.12.1006. [DOI] [PubMed] [Google Scholar]
- 56.Iacobuzio-Donahue CA, Velculescu VE, Wolfgang CL, Hruban RH. Genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing. Clin Cancer Res. 2012;18(16):4257–4265. doi: 10.1158/1078-0432.CCR-12-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]