Abstract
Glutamine is an abundant amino acid that plays pivotal roles in cell growth, cell metabolism and neurotransmission. Dysregulation of glutamine-utilizing pathways has been associated with pathological conditions such as cancer and neurodegenerative diseases. 6-Diazo-5-Oxo-L-Norleucine (DON) is a reactive glutamine analog that inhibits enzymes affecting glutamine metabolism such as glutaminase, 2-N-amidotransferase, L-asparaginase and several enzymes involved in pyrimidine and purine de novo synthesis. As a result, DON is actively used in preclinical models of cancer and neurodegenerative disease. Moreover, there have been several clinical trials using DON to treat a variety of cancers. Considerations of dose and exposure are especially important with DON treatment due to its narrow therapeutic window and significant side effects. Consequently, a robust quantification bioassay is of interest. DON is a polar unstable molecule which has made quantification challenging. Here we report on the characterization of a bioanalytical method to quantify DON in tissue samples involving DON derivatization with 3N HCl in butanol. The derivatized product is lipophilic and stable. Detection of this analyte by mass spectrometry is fast, specific and can be used to quantify DON in plasma and brain tissue with a limit of detection in the low nanomolar level.
Keywords: 6-Diazo-5-Oxo-L-Norleucine, glutamine, DON Bioanalysis, DON derivatization
Glutamine is one of the most abundant amino acids in the human body playing a critical role in cell growth, cell metabolism and neurotransmission. Dysregulation of glutamine utilizing pathways has been associated with a variety of pathologies including cancer and neurodegenerative disease.
Glutamine serves as a nitrogen donor for purine and pyrimidine production which is required for de novo nucleotide synthesis [1]. Since de novo synthesis of nucleotides is upregulated to support DNA replication and RNA expression for rapid growth and division of cancer cells [2], inhibition of amidotransferases, the enzymes involved in the transfer of the amide group of glutamine to other molecules to initiate nucleotide synthesis, has been suggested as potential cancer therapy. Glutamine is also a major source of energy for neoplastic cells via glutaminolysis where glutaminase converts glutamine to glutamate which is further converted to α-ketoglutarate to enter the citric acid cycle [3]. In support of this hypothesis, glutaminase inhibition has been shown to be efficacious in models of cancer [4, 5]
In addition, glutaminase-catalyzed hydrolysis of glutamine to glutamate is a major source of glutamate in the brain [6]. Normal synaptic transmission in the central nervous system (CNS) involves the use of glutamate as the major excitatory amino acid neurotransmitter. Under certain pathological conditions excessive glutamatergic signaling, termed excitotoxicity [7], is postulated to cause CNS damage in several neurodegenerative diseases like stroke [8], amyotrophic lateral sclerosis [9], Huntington’s disease [10], Alzheimer’s disease [11] and HIV associated dementia [12]. Consequently, inhibition of glutaminase has been suggested as a possible way to ameliorate high levels of glutamate in neurodegenerative diseases. In support of this hypothesis, glutaminase inhibition has been efficacious in models of CNS neurodegeneration [12–17]
6-Diazo-5-oxo-L-norleucine (DON) is an amino acid analog of glutamine that is an inhibitor of glutamine utilizing enzymes. DON inhibits 2-N-amidotransferase to block purine synthesis [4]. DON was one of the earliest inhibitors to be identified for glutaminase [18]. It binds to the active site of glutaminase in an irreversible manner [18–20]. As an inhibitor of glutamine metabolizing pathways DON has been used both as a tool compound in preclinical in vivo models and also as a clinical candidate. There have been several clinical trials using DON [21–26]; unfortunately it was not well tolerated at efficacious doses. Recently, phase II clinical trials were reported for DON in combination with pegylated glutaminase with the goal of improving efficacy by co-administration with the glutamine depleting enzyme [27]. DON is still commonly used as a tool compound in glutamine related research due to its solubility and efficacy in various in vivo models [28, 29]. However, DON, with its polar structure and reactive moiety, would be expected to have difficulty reaching its target. Thus a quantification assay for DON is of interest when using DON in animal models. DON quantification has been carried out in the past by several methods that include HPLC of derivatized DON followed by absorbance and fluorescence detection [30], ionpaired HPLC followed by absorbance detection [31], radioisotope labeled DON [32] and a microbiological assay [33]. HPLC analysis of DON suffers from interference from other materials in the sample; analysis may require boiling samples to confirm results [26] and often assays are not sensitive. Using radiolabeled DON has the issues of working with radioactivity but more importantly, the assay does not differentiate intact DON from degraded DON or from metabolized or covalently bound DON that retains the radiolabel. Microbiological assays are time consuming, labor intensive, could suffer from nonspecific effects and may not differentiate between DON and its metabolites [33].
Here we report a novel robust bioanalytical method to quantify DON in tissue samples. Derivatization of DON forms a stable analyte that is detected unambiguously by mass spectrometry. In this report we used this bioanalytical assay to quantify DON exposure following systemic administration in rodent plasma and, for the first time, also in brain tissue.
METHODS
DON Derivatization
DON was derivatized in the presence of 3N HCl ± n-butanol. DON (Sigma Aldrich) was first dissolved in water at a concentration of 10 mM. An aliquot (10 μL) of this stock solution was added to 3N HCl ± n-butanol (250 μL) in a low retention micro-centrifuge tube. The solution was then heated at 60°C for 30 minutes in a shaking water bath. After heating, the sample was dried at 45°C under a nitrogen stream, resuspended in 50 μL of water/acetonitrile (70:30), vortexed and centrifuged at 16,000 × g. Supernatants were transferred to LC vials and an aliquot (2 μL) was used for liquid chromatography mass spectrometry (LC-MS) or liquid chromatography tandem mass spectrometry (LC MS/MS) analysis.
Analysis of derivatized DON by LC-MS
Derivatized DON samples (2 μL) prepared as described above were injected and separated on an Agilent 1290 LC equipped with an Agilent Eclipse Plus 2.1 × 100 mm, 1.8 micron Rapid Resolution C18 column over a 5.5 minute gradient from 30–70% acetonitrile + 0.1% formic acid. Analytes were detected with an Agilent 6520 quadrupole time-of-flight (QTOF) mass spectrometer in positive mode with drying gas at 350 °C, 11 L/min and 40 psi. The fragmenter was set at 70V and the VCAP at 4000V.
Analysis of derivatized DON by LC-MS/MS
Analysis of derivatized DON after 3N HCl ± n-butanol by LC-MS/MS was carried out in the same manner as for LC-MS except the precursor mass (m/z = 218.0942) was selected in the first quadrupole and the compound was made to collide with nitrogen gas with a collision energy of 15V in MS/MS mode to afford the daughter ions with m/z = 162.032 and 116.026.
Analysis of underivatized DON by LC-MS
Methanol (250 μL) was added to plasma samples containing DON (50 μL) and vortexed; samples were centrifuged for 5 min at 16,000 × g to precipitate proteins. An aliquot of the supernatant (200 μL) was dried and subsequently reconstituted in H2O (50 μL). An aliquot (20 μL) was then injected and separated on an Agilent 1290 LC equipped with a Thermo Hypercarb 2.1 × 100 mm column with isocratic 2.5% acetonitrile + 0.1% formic acid mobile phase. Analytes were detected with an Agilent 6520 QTOF in MS mode as when analyzing derivatized DON by LC-MS.
Bioanalysis of DON in plasma
When using plasma, DON was derivatized only using 3N HCl plus n-butanol and subsequently analyzed by LC-MS. In order to generate the standard curve to determine DON concentrations in plasma, DON (10 μL of 1 mM water solution) was added to untreated mouse plasma (90 μL) in a low retention micro-centrifuge tube. Standard solutions (100 μL) were then prepared by serial dilution to generate concentrations from 10 nM to 100 μM at half-log intervals. Prior to extraction, frozen plasma samples were thawed on ice. N-butanol (250 μL) containing 3N HCl was added directly to standards (50 μL), vortexed and centrifuged at 16,000 × g for 5 minutes in low retention micro-centrifuge tubes to precipitate proteins. An aliquot (200 μL) of the supernatant was transferred to a new tube and incubated at 60°C for 30 minutes in a shaking water bath to carry out the derivatization reaction. After derivatization, an aliquot of the reaction mixture (2 μL) was injected and analyzed by LC-MS as stated above. The area under the curve (AUC) representing the signal intensity of the extracted ion (m/z 218.0942) for each sample was used to generate the standard curve using Agilent Mass Hunter Quantitative analysis software. Plasma samples obtained from mice treated with DON were treated in exactly the same manner except exogenous DON was not added. DON concentrations in plasma samples were determined by interpolation using the standard curve.
Bioanalysis of DON in brain
When using brain, DON was derivatized only using 3N HCl plus n-butanol and subsequently analyzed by LC-MS. In order to generate the standard curve to determine DON concentrations in brain, frozen brain samples from untreated mice were thawed on ice. Tissue was weighed in low retention micro-centrifuge tubes to which 5 μL n-butanol containing 3N HCl were added per mg tissue. Tissue was then homogenized with a pestle and vortexed. Known amounts of DON from a 1 mM stock solution in water were mixed with n-butanol containing 3N HCl and spiked to brain tissue to prepare standards at concentrations from 10 nM to 100 μM at half-log intervals. Samples were centrifuged at 16,000 × g for 5 minutes in low retention micro-centrifuge tubes to precipitate proteins. An aliquot (200 μL) of the supernatant was transferred to a new tube and incubated at 60°C for 30 minutes in a shaking water bath to carry out the derivatization reaction. After derivatization, an aliquot of the reaction mixture (2 μL) was injected and analyzed by LC-MS as stated above. Brain samples obtained from mice treated with DON were treated in exactly the same manner except exogenous DON was not added. DON concentrations in brain samples were determined by interpolation using the standard curve.
Animal studies
All protocols were approved by the animal care and use committee at Johns Hopkins University. C57BL/6 male mice (4 – 5 week old) were administered DON at different doses either intravenously (i.v.) or intraperiotoneally (i.p.) as indicated. DON working solution was diluted in PBS each day from aliquots of a 100 mM stock solution in PBS stored at −80°C. At the indicated time points after DON administration mice were euthanized in a CO2 chamber and blood was collected transcardially. When collecting brains, mice were perfused with PBS before brain collection. Samples were frozen immediately at −80°C and kept frozen until bioanalysis. Plasma and brain samples were processed and analyzed as stated in the bioanalysis of DON in plasma and brain sections.
RESULTS
LC-MS analysis after DON derivatization in 3N HCl ± n-butanol shows the presence of a chlorine-containing derivative
During DON derivatization using 3N HCl plus n-butanol we found DON reacted and rearranged to form a stable and quantifiable derivative (Fig 1A). The high resolution mass and the isotopic abundances in the observed mass spectrum were used to generate the molecular formula C10H16ClNO2. The isotopic abundances closely match the predicted isotopic abundance of the proposed chemical structure. The 3:1 isotopic abundance ratio between the molecular ion (218.0942) and the M+2 (220.0912) unambiguously indicated that a chlorine atom had been incorporated into the derivatized product (Fig 1B). The corresponding chromatographic trace of the molecular ion of derivatized DON is shown in Fig 1C. When DON was incubated with 3N HCl in water in the absence of butanol, a peak with molecular mass 162.0316 m/z was observed in the mass spectrum (Fig 2). The molecular formula generated included a chlorine-containing derivative: C6H8ClNO2. The structure that fits the molecular mass and molecular formula is the same as the derivatized structure in Fig 1 without the butyl moiety (Fig 2). This is as expected since butanol was not used in this derivatization reaction.
Fig 1.
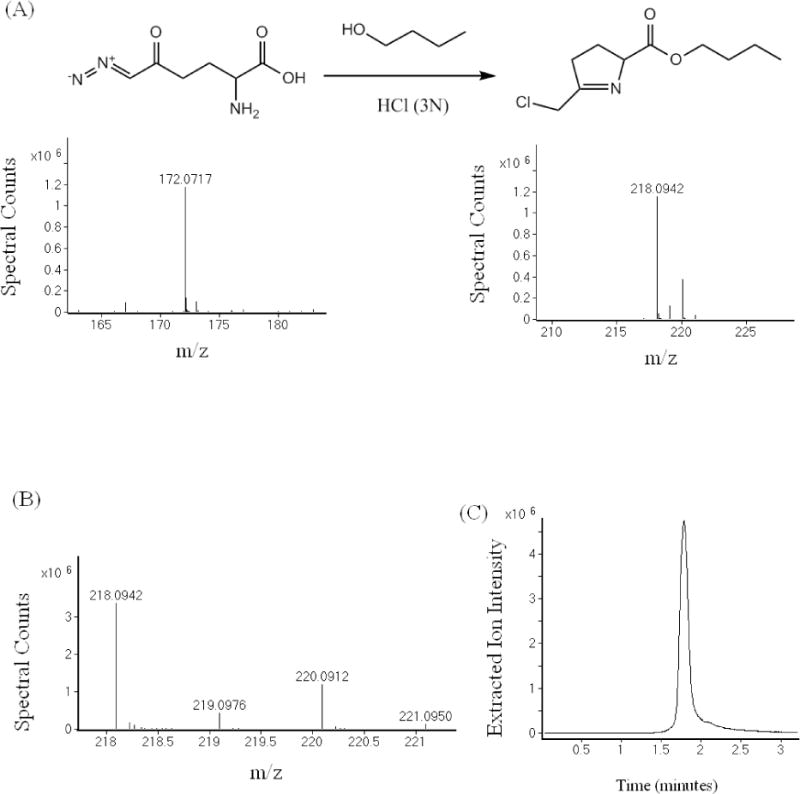
(A) Derivatization of DON in 3N HCl plus n-butanol followed by LC-MS analysis – DON was heated at 60°C for 30 minutes in n-butanol containing 3N HCl to derivatize the molecule into a stable quantifiable analyte of molecular mass [M+H]+ 218.0942. (B) The high resolution mass and isotopic abundance were used to generate the molecular formula C10H16ClNO2. The observed isotopic abundance closely matched the predicted isotopic abundance of the proposed chemical structure incorporating a chlorine atom. This is apparent from the 3:1 ratio of [M] (218.0942) and [M+2] (220.0912). (C) UPLC trace of derivatized DON. Derivatized DON was injected on an Agilent 1290 LC equipped with a C18 column and detected with an Agilent 6520 QTOF mass spectrometer.
Fig 2.
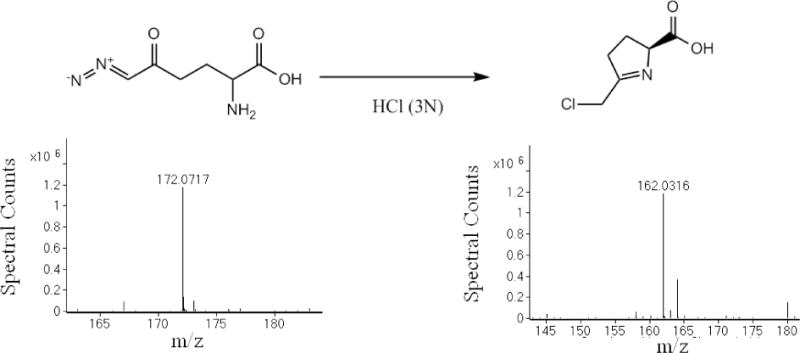
Derivatization of DON in 3N HCl without butanol followed by LC-MS analysis – Derivatized product exhibited a mass consistent with the proposed methylene chlorine substituted 1-pyrroline structure, but lacking the butyl ester.
LC-MS/MS analysis of fragmentation pattern of ester-containing derivative confirms the presence of cyclic structure and chlorine atom
In a separate experiment, DON was derivatized with 3N HCl ± n-butanol and subsequently analyzed by LC-MS/MS. The resulting product ions of 162.0318 and 116.0262 match the loss of the butyl ester and a radical formed after the loss of the entire carboxylate-ester moiety respectively (Fig 3A). These product ions are consistent with the expected DON derivative structure. When using 3N HCl during derivatization, the resulting product ion of 116.0258 is consistent with the expected DON derivative structure in the absence of butanol (Fig 3B).
Fig 3.
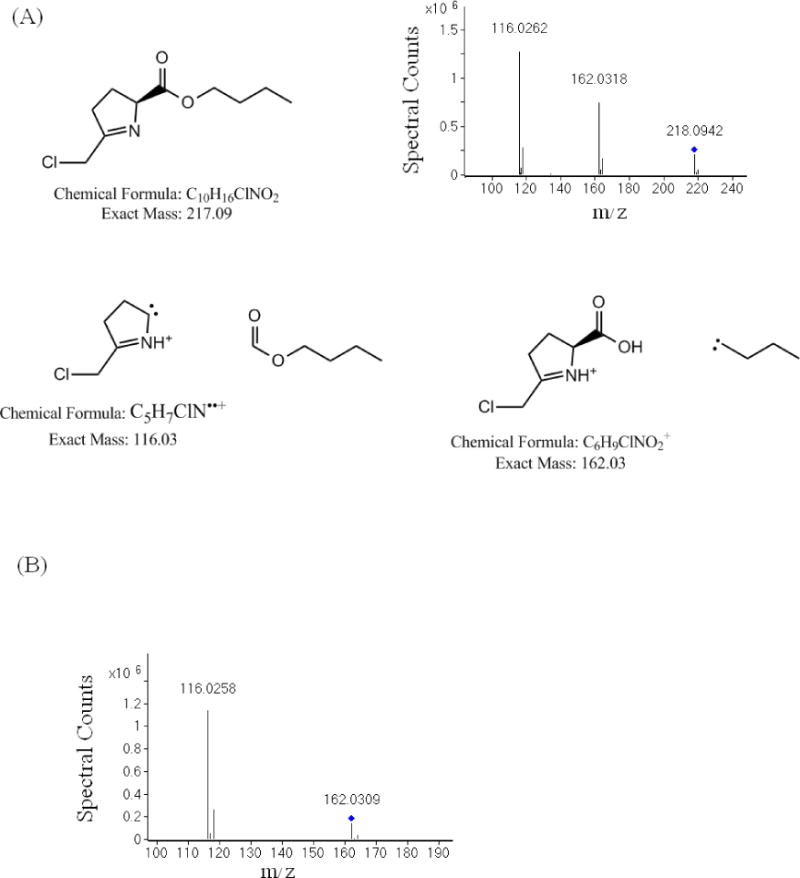
Tandem mass spectrometry (MS/MS) of derivatized DON – (A) Collision induced dissociation of DON after derivatization with acidified butanol – DON was derivatized with n-butanol containing 3N HCl and analyzed by LC-MS/MS. The resulting product ions of 162.03 and 116.03 match the loss of the butyl ester and the radical formed after the loss of the entire carboxyl-ester moiety. These product ions are consistent with the expected DON derivative structure. (B) Collision induced dissociation of DON after derivatization with 3N HCl – DON was derivatized with 3N HCl without butanol and analyzed by LC-MS/MS. The resulting product ion of 116.03 is consistent with the expected DON derivative structure in the absence of butanol.
DON derivative was quantified from plasma and brain tissue using LC-MS
In order to verify that the DON derivatization protocol was adequate to use to determine DON concentrations in biological matrices, known concentrations of DON were added to mouse plasma and brain followed by derivatization using 3N HCl in n-butanol. Derivatized samples were then analyzed by LC-MS and a standard curve for each matrix was generated. In each case there was a linear correlation between signal response and the concentration of derivatized material. Standard curves were linear in the 30 nM – 100 μM range for both plasma (Fig 4A) and brain (Fig 4B).
Fig 4.
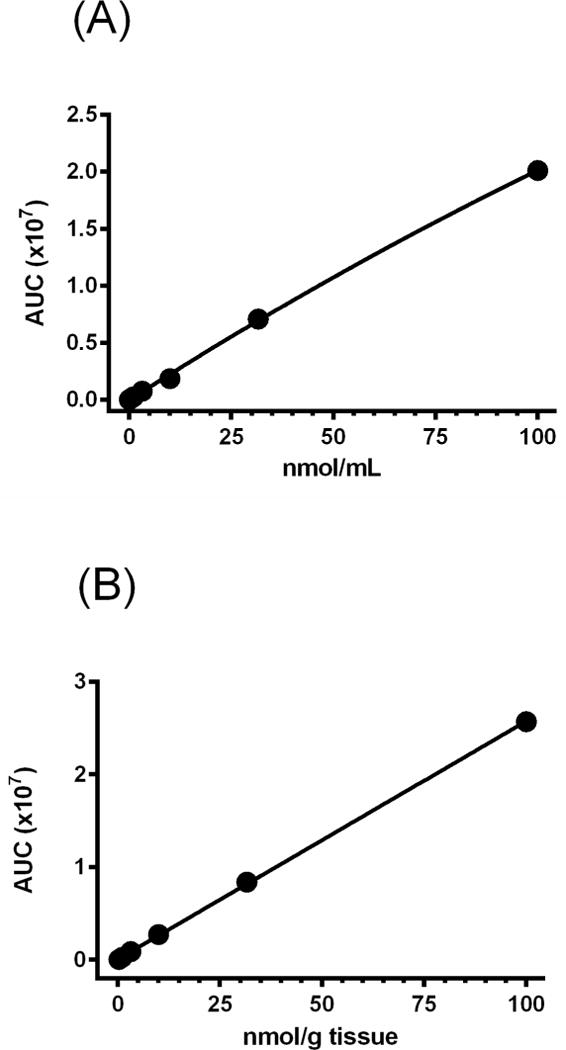
(A) Standard curve of DON in plasma after derivatization with acidified butanol and LC-MS analysis – DON was spiked into untreated mouse plasma to generate standards at various concentrations. DON derivatization was carried out with n-butanol containing 3N HCl. After centrifugation to separate denatured proteins, supernatant was incubated at 60°C for 30 minutes. Derivatized DON was detected by LC-MS. Standards in the 30 nM to 100 μM range were used to generate a standard curve. (B) Standard curve of DON in brain after derivatization with acidified butanol and LC-MS analysis – DON was spiked into untreated mouse brain to generate standards at various concentrations. DON derivatization was carried out with n-butanol containing 3N HCl. After centrifugation to separate denatured proteins, supernatant was incubated at 60°C for 30 minutes. Derivatized DON was detected by LC-MS. Standards in the 30 nM to 100 μM range were used to generate a standard curve.
DON was measured in plasma and brain using the new bioanalysis procedure
The new derivatizing procedure was used to determine DON concentrations in plasma and brain following i.v. and i.p. administration. In the first study mice were given DON (1.6 mg/kg, i.v.) and blood was collected at 0.25, 0.5, 1, 2, 4 and 6 h. The exposure of DON estimated from the area under the curve (AUC) was 8 nmol h/mL with a plasma half-life of 1.2 h (Fig 5A). In the second study mice were given DON (0.6 mg/kg, i.p.) and both plasma and brain were harvested 1 h after DON administration. DON concentrations in plasma and brain were 1.7 ± 0.5 nmol/mL and 0.22 ± 0.18 nmol/g tissue respectively (Fig 5B), suggesting a brain to plasma ratio of approximately 0.1.
Fig 5.
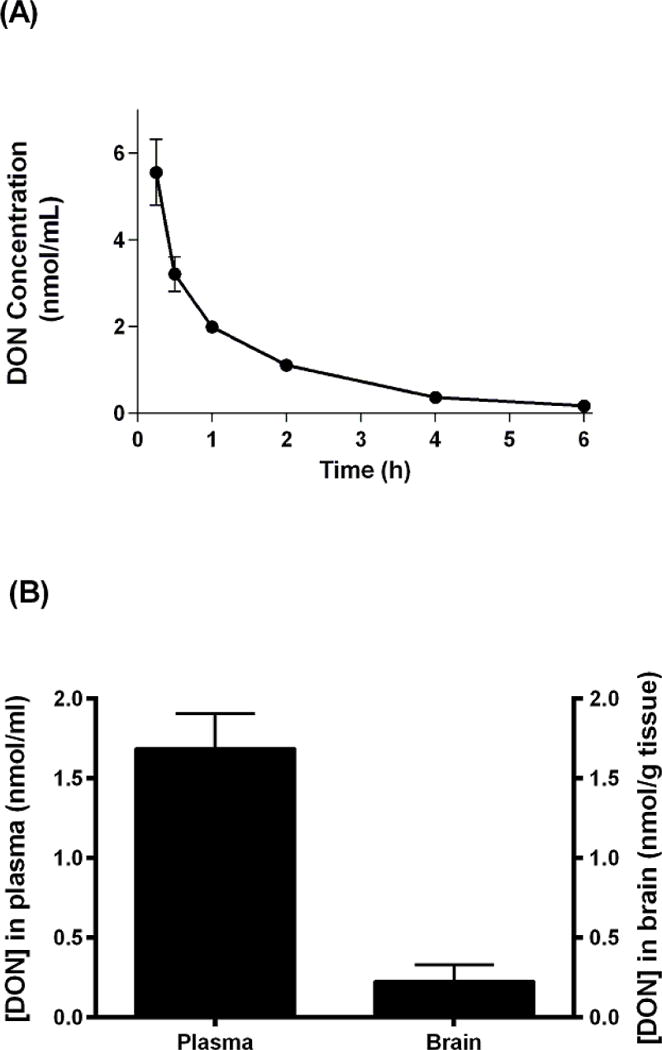
(A) DON concentrations in plasma over time – Mice were administered DON at 1.6 mg/kg (i.v.). Mice were euthanized and blood was collected transcardially 0.25, 0.5, 1, 2, 4 and 6 hours after dosing. Blood was centrifuged, plasma collected and stored at −80 °C. N-butanol containing 3N HCl (250 μL) was added directly to samples (50 μL) and centrifuged at 16,000 × g for 5 minutes to precipitate proteins. An aliquot (200 μL) of the supernatant was incubated at 60 °C for 30 min. After derivatization, the samples were analyzed by LC-MS (methods). (B) DON Plasma to Brain ratio analysis – Mice were administered DON (0.6 mg/kg, i.p.) and blood and brain were collected 1 h after DON administration. Plasma was isolated from blood samples as described in (A). Brains were collected following perfusion with PBS and frozen immediately at −80 °C. Before bioanalysis, brains were weighed and n-butanol containing 3N HCl was added to each sample (5 μL/mg tissue) and homogenized using a pestle. Resulting homogenates were centrifuged at 16,000 × g for 5 minutes to precipitate proteins and analyzed the same way as the plasma samples. All determinations in both (A) and (B) were carried out in triplicate. Error bars correspond to ± S.E.M.
Analysis of derivatized DON or intact DON gave the same results
It is conceivable that when DON is used in vivo it could form byproducts that could also form the derivatized structure. In order to determine if this was the case, DON concentration was measured both directly using a less sensitive method (LOD > 1 μM) and through acidified butanol derivatization in plasma samples collected from mice 15 min after DON administration (1.6 mg/kg i.v.). The two methods gave the same DON concentration within experimental error: 3.9 nmol/mL ± 0.3 and 4.2 nmol/mL ± 0.8 when using the underivatized and derivatization methods respectively (Fig 6).
Fig 6.
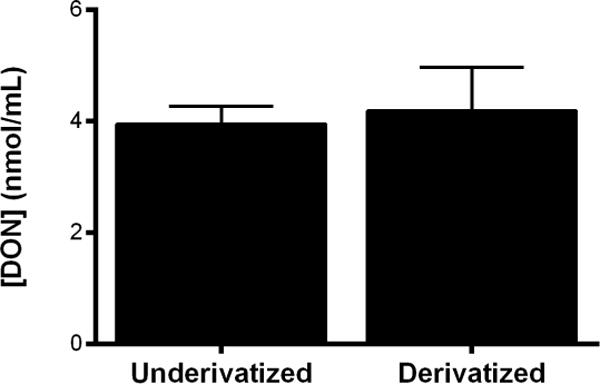
DON concentration in plasma from DON- treated mice using underivatized and derivatization protocols – Mouse plasma samples were obtained 15 min after DON administration (1.6 mg/kg, i.v.). LC-MS bioanalysis of underivatized DON from plasma: DON was extracted from plasma with methanol, dried and resuspended in H2O and separated in a Hypercarb column (methods). LC-MS bioanalysis of derivatized DON from plasma: DON was derivatized in butanol containing 3N HCl for 30 min at 60 °C, dried, reconstituted in 30% acetonitrile and separated by RPC (methods). Analytes eluting after each chromatographic separation were detected by QTOF MS. N= 3 for each treatment. Error bars correspond to ± S.D.
DISCUSSION
Several quantification methods for DON have been previously described [26–29] all with limits of detection in the low micromolar level. HPLC/fluorescence, radiolabel and microbiological assays all have the potential for nonspecific signals. DON is a polar amino acid that elutes unretained in the void volume with many other polar compounds during reverse phase chromatography (RPC). Due to ion suppression and poor chromatography that result in broad irregular peak shapes, DON cannot be successfully separated and quantified from complex matrices such as brain and plasma with ordinary RPC. Direct measurement of DON quantification by LC-MS has been possible by using a porous graphitic carbonbased chromatographic column (Hypercarb) that minimizes the ion-suppression seen with the silicabased C18 column (unpublished observation); this measurement however also exhibits low sensitivity (LOD > 1 μM) so it is not an alternative for routine pharmacokinetics samples where low nanomolar levels are of interest. One alternative was to derivatize DON. Polar amino acids are often derivatized to make them more amenable to separation by RPC [34]. Esterification of the carboxylic acid on an amino acid improves RPC separation and increases the analyte mass which enhances ionization at the electrospray source of the mass spectrometer. For example, derivatization with an n-butyl ester has been used to quantify plasma methylmalonic acid [35]. However, in the case of DON, in addition to the carboxylic acid moiety there is the added complication of the diazo ketone moiety that lacks stability and is not expected to survive derivatization conditions.
In an effort to develop a reliable way to measure DON in complex biological matrices we incubated DON with butanol in 3N HCl for 30 min at 60 °C, the same procedure used to derivatize carboxylic acids to make the corresponding n-butyl ester [35]. DON derivatization under these conditions produced a chlorine containing derivative as supported by the 3:1 isotopic abundance ratio between the molecular ion (218.0942) and the M+2 (220.0912) (Fig 1). Incorporation of a chloromethyl ketone to the diazo moiety of DON has been shown previously [36]. The molecular mass (218.0942) (Fig 1A) and isotopic abundances (Fig 1B) observed in the mass spectrum are consistent with cyclization to form butyl 5-(chloromethyl)-3, 4-dihydro-2H-pyrrole-2-carboxylate (Fig 1A). When derivatization was carried out in 3N HCl in the absence of butanol, a chlorine-containing derivative that corresponds to the same cyclized product lacking the butyl group on the ester with molecular formula C6H8ClNO2 (molecular mass 162.0316 m/z) was formed (Fig 2).
In a separate effort to confirm the structure of derivatized DON, the product of derivatization was analyzed after collision induced dissociation (CID). The fragmentation pattern was consistent with the presence of an ester-containing derivative, and a 1-pyrroline ring with a methylene chlorine substitution (Fig 3A). The resulting product ions match the loss of the butyl ester and a radical formed after the loss of the carboxylate-ester (Fig 3A). Further confirmation was obtained when the 116.0258 product ion was also seen with CID of derivatized DON with 3N HCl without butanol (Fig 3B).
A possible mechanism of the derivatization reaction in the absence of butanol is illustrated in scheme A. At low pH (3N HCl), the α-carbon of the carbonyl close to the diazo moiety will abstract a proton from the solvent resulting in a diazonium ion. In the next step, the same α-carbon undergoes chlorine ion addition and concomitant N2 loss. The chloromethyl ketone undergoes cyclization and dehydration to form the 1-pyrrolinedine derivative illustrated in scheme A. When the derivatization reaction is carried out in n-butanol containing 3N HCl, the carboxylic acid moiety will also undergo standard acid-catalyzed esterification [37].
Scheme A.
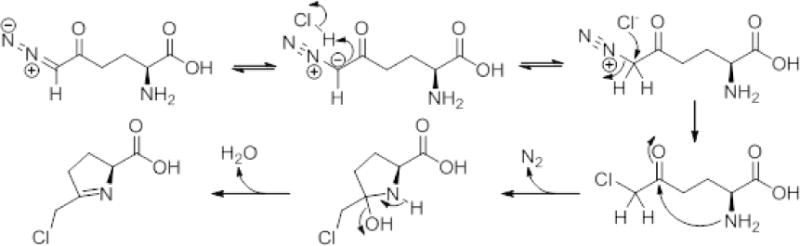
Proposed Mechanism for the derivatization reaction of DON in 3 N HCl in the absence of butanol –Reaction at the diazo moiety – At low pH the α-carbon of the carbonyl close to the diazo moiety abstracts a proton from solvent resulting in a diazonium ion. Subsequently, the same α-carbon undergoes chlorine ion addition and concomitant N2 loss. The chloromethyl ketone undergoes cyclization and dehydration to form the 5-member ring 1-pyrrolinedine with the methylene chlorine substitution. When butanol is present in the reaction mixture, the same derivatization occurs along with esterification of the carboxylate moiety with the n-butyl group.
We have used the new derivatizing procedure to determine DON concentrations in both plasma and brain from mice after DON administration. First, we generated standard curves of signal intensity vs. known concentrations of DON that were added to both plasma and brain followed by derivatization, extraction and bioanalysis. The resulting standard curves for plasma (Fig 4A) and brain (Fig 4B) were then used to determine unknown concentrations of DON in plasma and brain from mice that had been treated with DON. The results show that DON can be readily monitored using the new derivatizing procedure. We report i.v. pharmacokinetic parameters of t1/2 = 1.2 h and AUC = 8 nmol h/mL (Fig 5A) and for the first time show DON brain penetration with a brain to plasma ratio of 10% at 1 h (Fig 5B). The latter finding was surprising given DON’s polar structure. Since the animals were perfused, DON found in brain at this level is unlikely to be due to blood contamination. DON could be actively transported by an amino acid carrier into the brain; previous reports have demonstrated active DON uptake systems that can be inhibited by glutamine in leukemia cells [38] and xenopus oocytes [39].
One potential drawback of the bioanalysis procedure is that DON could cyclize in vivo to form a byproduct which in turn could convert into our analyte during derivatization. This could give artificially high DON concentrations. In order to rule out this possibility, we performed a control experiment where we compared DON concentrations obtained when using the derivatizing protocol (3N HCl + n-butanol) and when measuring DON directly. Even though measurement of underivatized DON is far less sensitive than when using the derivatizing procedure (LOD for underivatized method > 1 μM vs. LOD for derivatizing procedure = 30 nM), when measuring μM levels of DON, a side by side comparison of the two methods would unveil whether the derivatization method is measuring DON byproducts. We found that the derivatization procedure gave the same DON concentrations within error as the direct measurements of underivatized DON (Fig 6). Further, a cyclized byproduct, made by heating DON at 37 °C for 2 h then added to plasma did not convert to the derivatized product when using the derivatization protocol. The result showed the cyclized byproduct of DON was impervious to the derivatization procedure. In a separate study, the cyclized byproduct of DON was not observed by LC-MS analysis of plasma from DON-treated mice 2 h after IV treatment.
In summary, we have developed a simple robust method to quantify DON in complex biological matrices using UPLC/MS after DON derivatization with acidified butanol. DON in the sample is made to react with n-butanol containing 3 N HCl to form butyl 5-(chloromethyl)-3, 4-dihydro-2H-pyrrole-2-carboxylate. A single solvent for extraction and derivatization solution simplifies sample processing and shortens analysis time. The derivatization LC-MS method is rapid, reproducible and rigorous and has a lower limit of quantitation of 30 nM that is over 30-fold more sensitive than methods reported in the literature. Mass spectrometry is able to reduce nonspecific signal, since only one analyte with a specific molecular formula is quantified. This method was applied to monitor DON levels in plasma and brain and could readily be applied to other tissues as well.
Acknowledgments
The authors acknowledge the financial support of NIH/NIMH Grant R03 DA032470, NIMH Grant P30 MH075673-06 and of the Brain Science Institute at Johns Hopkins. We thank Dr. Takashi Tsukamoto from the Brain Science Institute for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cory JG, Cory AH. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo. 2006;20:587–589. [PubMed] [Google Scholar]
- 2.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kisner DL, Catane R, Muggia FM. The rediscovery of DON (6-diazo-5-oxo-L-norleucine) Recent Results Cancer Res. 1980;74:258–263. doi: 10.1007/978-3-642-81488-4_30. [DOI] [PubMed] [Google Scholar]
- 5.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thanki CM, Sugden D, Thomas AJ, Bradford HF. In vivo release from cerebral cortex of [14C]glutamate synthesized from [U-14C]glutamine. J Neurochem. 1983;41:611–617. doi: 10.1111/j.1471-4159.1983.tb04785.x. [DOI] [PubMed] [Google Scholar]
- 7.Marmiroli P, Cavaletti G. The glutamatergic neurotransmission in the central nervous system. Curr Med Chem. 2012;19:1269–1276. doi: 10.2174/092986712799462711. [DOI] [PubMed] [Google Scholar]
- 8.Lai TW, Zhang S, Wang YT. Excitotoxicity and stroke: identifying novel targets for neuroprotection. Prog Neurobiol. 2013;115:157–188. doi: 10.1016/j.pneurobio.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Vucic S, Kiernan MC. Utility of transcranial magnetic stimulation in delineating amyotrophic lateral sclerosis pathophysiology. Handb Clin Neurol. 2013;116:561–575. doi: 10.1016/B978-0-444-53497-2.00045-0. [DOI] [PubMed] [Google Scholar]
- 10.Sepers MD, Raymond LA. Mechanisms of synaptic dysfunction and excitotoxicity in Huntington’s disease. Drug Discov Today. 2014 doi: 10.1016/j.drudis.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Hynd MR, Scott HL, Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer’s disease. Neurochem Int. 2004;45:583–595. doi: 10.1016/j.neuint.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Potter MC, Figuera-Losada M, Rojas C, Slusher BS. Targeting the glutamatergic system for the treatment of HIV-associated neurocognitive disorders. J Neuroimmune Pharmacol. 2013;8:594–607. doi: 10.1007/s11481-013-9442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CJ, Ou YC, Chang CY, Pan HC, Liao SL, Chen SY, Raung SL, Lai CY. Glutamate released by Japanese encephalitis virus-infected microglia involves TNF-alpha signaling and contributes to neuronal death. Glia. 2012;60:487–501. doi: 10.1002/glia.22282. [DOI] [PubMed] [Google Scholar]
- 14.Jayakumar AR, Rao KV, Murthy Ch R, Norenberg MD. Glutamine in the mechanism of ammonia-induced astrocyte swelling. Neurochem Int. 2006;48:623–628. doi: 10.1016/j.neuint.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Maezawa I, Jin LW. Rett syndrome microglia damage dendrites and synapses by the elevated release of glutamate. J Neurosci. 2010;30:5346–5356. doi: 10.1523/JNEUROSCI.5966-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas AG, O’Driscoll CM, Bressler J, Kaufmann W, Rojas CJ, Slusher BS. Small molecule glutaminase inhibitors block glutamate release from stimulated microglia. Biochem Biophys Res Commun. 2014;443:32–36. doi: 10.1016/j.bbrc.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian C, Erdmann N, Zhao J, Cao Z, Peng H, Zheng J. HIV-infected macrophages mediate neuronal apoptosis through mitochondrial glutaminase. J Neurochem. 2008;105:994–1005. doi: 10.1111/j.1471-4159.2007.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman SC, McGrath TF. Glutaminase A of escherichia coli. Reactions with the substrate analogue, 6-diazo-5-oxonorleucine. J Biol Chem. 1973;248:8506–8510. [PubMed] [Google Scholar]
- 19.Shapiro RA, Clark VM, Curthoys NP. Inactivation of rat renal phosphate-dependent glutaminase with 6-diazo-5-oxo-L-norleucine. Evidence for interaction at the glutamine binding site. J Biol Chem. 1979;254:2835–2838. [PubMed] [Google Scholar]
- 20.Thangavelu K, Chong QY, Low BC, Sivaraman J. Structural basis for the active site inhibition mechanism of human kidney-type glutaminase (KGA) Sci Rep. 2014;4:3827. doi: 10.1038/srep03827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earhart RH, Koeller JM, Davis HL. Phase I trial of 6-diazo-5-oxo-L-norleucine (DON) administered by 5-day courses. Cancer Treat Rep. 1982;66:1215–1217. [PubMed] [Google Scholar]
- 22.Kovach JS, Eagan RT, Powis G, Rubin J, Creagan ET, Moertel CG. Phase I and pharmacokinetic studies of DON. Cancer Treat Rep. 1981;65:1031–1036. [PubMed] [Google Scholar]
- 23.Lynch G, Kemeny N, Casper E. Phase II evaluation of DON (6-diazo-5-oxo-L-norleucine) in patients with advanced colorectal carcinoma. Am J Clin Oncol. 1982;5:541–543. [PubMed] [Google Scholar]
- 24.Rubin J, Sorensen S, Schutt AJ, van Hazel GA, O’Connell MJ, Moertel CG. A phase II study of 6-diazo-5-oxo-L-norleucine (DON, NSC-7365) in advanced large bowel carcinoma. Am J Clin Oncol. 1983;6:325–326. doi: 10.1097/00000421-198306000-00012. [DOI] [PubMed] [Google Scholar]
- 25.Sklaroff RB, Casper ES, Magill GB, Young CW. Phase I study of 6-diazo-5-oxo-L-norleucine (DON) Cancer Treat Rep. 1980;64:1247–1251. [PubMed] [Google Scholar]
- 26.Sullivan MP, Nelson JA, Feldman S, Van Nguyen B. Pharmacokinetic and phase I study of intravenous DON (6-diazo-5-oxo-L-norleucine) in children. Cancer Chemother Pharmacol. 1988;21:78–84. doi: 10.1007/BF00262746. [DOI] [PubMed] [Google Scholar]
- 27.Mueller C, Al-Batran S, Jaeger E, Schmidt B, Bausch M, Unger C, Sethuraman N. A phase IIa study of PEGylated glutaminase (PEG-PGA) plus 6-diazo-5-oxo-L-norleucine (DON) in patients with advanced refractory solid tumors. Journal of Clinical Oncology. 2008;26:2533. [Google Scholar]
- 28.Cao BB, Han XH, Huang Y, Qiu YH, Peng YP. The hypothalamus mediates the effect of cerebellar fastigial nuclear glutamatergic neurons on humoral immunity. Neuro Endocrinol Lett. 2012;33:393–400. [PubMed] [Google Scholar]
- 29.Shelton LM, Huysentruyt LC, Seyfried TN. Glutamine targeting inhibits systemic metastasis in the VM-M3 murine tumor model. Int J Cancer. 2010;127:2478–2485. doi: 10.1002/ijc.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powis G, Ames MM. Determination of 6-diazo-5-oxo-L-norleucine in plasma and urine by reversed-phase high-performance liquid chromatography of the dansyl derivative. J Chromatogr. 1980;181:95–99. doi: 10.1016/s0378-4347(00)81274-8. [DOI] [PubMed] [Google Scholar]
- 31.Nelson JA, Herbert B. Rapid Analysis of 6Diazo5-oxo-L-norleucine (DON) in Human Plasma and Urine. Journal of Liquid Chromatography & Related Technologies. 1981;4:1641–1649. [Google Scholar]
- 32.Rahman A, Smith FP, Luc PT, Woolley PV. Phase I study and clinical pharmacology of 6-diazo-5-oxo-L-norleucine (DON) Invest New Drugs. 1985;3:369–374. doi: 10.1007/BF00170760. [DOI] [PubMed] [Google Scholar]
- 33.Cooney DA, Jayaram HN, Milman HA, Homan ER, Pittillo R, Geran RI, Ryan J, Rosenbluth RJ. DON, CONV and DONV-III. Pharmacologic and toxicologic studies. Biochem Pharmacol. 1976;25:1859–1870. doi: 10.1016/0006-2952(76)90190-8. [DOI] [PubMed] [Google Scholar]
- 34.Molnár-Perl, editor. Quantitation of Amino Acids and Amines by Chromatography: Methods and Protocols. Elsevier; 2005. [Google Scholar]
- 35.Kushnir MM, Komaromy-Hiller G, Shushan B, Urry FM, Roberts WL. Analysis of dicarboxylic acids by tandem mass spectrometry. High-throughput quantitative measurement of methylmalonic acid in serum, plasma, and urine. Clin Chem. 2001;47:1993–2002. [PubMed] [Google Scholar]
- 36.Walker B, Brown MF, Lynas JF, Martin SL, McDowell A, Badet B, Hill AJ. Inhibition of Escherichia coli glucosamine synthetase by novel electrophilic analogues of glutamine–comparison with 6-diazo-5-oxo-norleucine. Bioorg Med Chem Lett. 2000;10:2795–2798. doi: 10.1016/s0960-894x(00)00565-5. [DOI] [PubMed] [Google Scholar]
- 37.Sykes P. A guidebook to mechanism in organic chemistry. Third. Longman Group Limited; 1975. [Google Scholar]
- 38.Huber KR, Rosenfeld H, Roberts J. Uptake of glutamine antimetabolites 6-diazo-5-oxo-L-norleucine (DON) and acivicin in sensitive and resistant tumor cell lines. Int J Cancer. 1988;41:752–755. doi: 10.1002/ijc.2910410519. [DOI] [PubMed] [Google Scholar]
- 39.Taylor PM, Mackenzie B, Hundal HS, Robertson E, Rennie MJ. Transport and membrane binding of the glutamine analogue 6-diazo-5-oxo-L-norleucine (DON) in Xenopus laevis oocytes. J Membr Biol. 1992;128:181–191. doi: 10.1007/BF00231811. [DOI] [PubMed] [Google Scholar]


