Abstract
L-ascorbate, commonly known as vitamin C, serves as an antioxidant and cofactor essential for many biological processes. Distinct ascorbate biosynthetic pathways have been established for animals and plants, but little is known about the presence or synthesis of this molecule in invertebrate species. We have investigated ascorbate metabolism in the nematode Caenorhabditis elegans, where this molecule would be expected to play roles in oxidative stress resistance and as cofactor in collagen and neurotransmitter synthesis. Using high-performance liquid chromatography and gas-chromatography mass spectrometry, we determined that ascorbate is present at low amounts in the egg stage, L1 larvae, and mixed animal populations, with the egg stage containing the highest concentrations. Incubating C. elegans with precursor molecules necessary for ascorbate synthesis in plants and animals did not significantly alter ascorbate levels. Furthermore, bioinformatic analyses did not support the presence in C. elegans of either the plant or the animal biosynthetic pathway. However, we observed the complete 13C-labeling of ascorbate when C. elegans was grown with 13C-labeled Escherichia coli as a food source. These results support the hypothesis that ascorbate biosynthesis in invertebrates may proceed by a novel pathway and lay the foundation for a broader understanding of its biological role.
Keywords: Ascorbic acid, vitamin C, Caenorhabditis elegans, biosynthetic pathway, invertebrates
3. INTRODUCTION
L-ascorbate (vitamin C) is a widely used metabolite in plants and animals that functions as a cofactor and as an antioxidant by undergoing two successive one-electron oxidation reactions to yield monodehydroascorbate radical and dehydroascorbate (1,2). These reactions can participate in biosynthetic reactions, neutralize reactive oxygen species that lead to DNA, lipid, and protein damage, and regulate processes involved in development and stress resistance (3–10). Evidence has been presented that administering increased amounts of ascorbate to humans can boost the immune system (11), reduce the risk of chronic diseases (12,13), and improve the recovery of critically ill patients (14,15). In humans and other vertebrates, ascorbate prevents the disease scurvy by ensuring that the iron cofactors in the prolyl 3-, prolyl 4-, and lysyl hydroxylases, enzymes responsible for the formation and stability of the collagen triple helix structure, are in the enzymatically active, reduced ferrous state (16,17). Other iron- and copper-dependent monooxygenases also rely on ascorbate as a reductant, including the enzymes involved in the formation of neurotransmitters derived from tyrosine (17) and in carnitine biosynthesis (18,19), as well as the hypoxia-inducible factor 1 (HIF-1) that regulates metabolism in response to molecular oxygen levels (20).
The presence of ascorbate has been well documented in plants and vertebrates. All photosynthetic organisms studied to date synthesize ascorbate (21), including higher plants, which can contain up to 19% of their total soluble leaf carbon content as ascorbate (22,23), bryophytes (24), green algae (25), photosynthetic protists (22), and cyanobacteria (26,27). In these organisms, the amount of ascorbate varies depending on the tissue analyzed, age of the organism, time of day, altitude, and light intensity (22,28–30). Although ascorbate is present in cartilaginous, actinopterigian, and non-teleost fish (31,32), amphibians, and reptiles (33,34), it is not synthesized in all bat and bird species due to the inactivation of ascorbate biosynthesis genes (35). Furthermore, humans, anthropoid primates, guinea pigs, and teleost fish cannot synthesize ascorbate and require it in the diet (35).
In contrast to plants and animals, little is known about the presence or the possible biosynthesis of ascorbate in invertebrates. Ascorbate was reported to be present in various marine invertebrates (36–38) and in Drosophila melanogaster (39). In Drosophila, ascorbate was detected in flies raised on an apparent ascorbate-free diet, but no evidence for endogenous biosynthesis of the molecule was shown (39). Because no studies have recently explored the presence or possible biosynthesis of ascorbate in invertebrates, several authors have concluded that invertebrates are incapable of synthesizing this molecule (33,40,41). Based on the important roles of ascorbate in metabolism and in resistance to reactive oxygen species, we felt that it was important to establish in an invertebrate species both the levels of ascorbate and how it may be biosynthesized.
Three distinct ascorbate biosynthetic pathways have now been described for plants, vertebrates, and protist species. In plants, the Smirnoff-Wheeler pathway involves ten enzymes that participate in the conversion of D-glucose to L-ascorbate via GDP-D-mannose and L-galactose (3). VTC2, which catalyzes the conversion of GDP-L-galactose to L-galactose-1-phosphate, regulates the first committed step in this pathway (42). In animals, ascorbate biosynthesis can be initiated with the conversion of UDP-glucuronate to D-glucuronate (43). After a reduction reaction that results in an inversion of configuration to give L-gulonate and a lactonization reaction, ascorbate is formed by the L-gulono-1,4-lactone oxidase (43). The absence of the gene encoding this oxidase in teleost fish, or its mutation in humans, anthropoid primates, guinea pigs, and other species, results in the loss of the ability to make ascorbate in these animals (35,44). Interestingly, although some bat and bird species are unable to synthesize ascorbate as previously mentioned due to the inactivation of this oxidase gene by mutation, other species have incurred further mutations that have reactivated the oxidase gene and restored ascorbate synthesis (35). A third biosynthetic pathway using D-galacturonate has been reported in some protist and green plant species (21,45–47). Finally, it has been determined that fungi do not synthesize ascorbate, but rather produce D-erythroascorbate, a molecule with similar antioxidant properties (48–51). The pathway by which this molecule is made has not been fully elucidated, but it is believed that the formation of D-erythroascorbate proceeds through D-arabinose (43,48).
Although there has been abundant research on the functions of ascorbate, it was not until recently that the plant, animal, and protist biosynthesis pathways were clarified (3,21,43). Nevertheless, modifications to these pathways are still being made to accommodate new intermediates and interactions between the pathways (28,52), opening the possibility that other ascorbate biosynthesis pathways may exist in other organisms.
In the present work, we focused on ascorbate in the invertebrate nematode worm Caenorhabditis elegans. It was previously discovered by serendipity that the C10F3.4 (mcp-1) gene is a homolog to the plant VTC2 gene that encodes the enzyme catalyzing the rate-limiting step in the plant ascorbate biosynthetic pathway (42,53). This observation prompted the question of whether or not ascorbate is present in C. elegans. Although the mcp-1 gene has a function in C. elegans distinct from ascorbate biosynthesis (53), ascorbate was suggested to be present in worm extracts (42,53,54). In the present work, we expand upon these findings using high-performance liquid chromatography and gas chromatography-mass spectrometry approaches to quantitate ascorbate in C. elegans. We now show that ascorbate is present and synthesized by C. elegans, thereby providing evidence that invertebrate organisms are capable of synthesizing this important molecule.
4. MATERIALS AND METHODS
4.1. C. elegans husbandry
Bristol N2 C. elegans were used in this study and obtained from the Caenorhabditis Genetics Center (University of Minnesota, Saint Paul, MN). C10F3.4 (mcp-1) mutant strain tm2679 animals were obtained from the C. elegans Core Facility at Tokyo Women’s Medical University. The strains were maintained at 20 or 25 °C on 10 cm × 1.5 cm petri dish plates containing nematode growth medium (NGM, 20 g/l bacto agar (BD Biosciences, catalog # 214030), 2.5 g/l bacto peptone (BD Biosciences, catalog # 211820), 3 g/l NaCl, 1 ml of 1 M CaCl2 per liter, 1 ml of 1 M MgSO4 per liter, 25 ml of 1 M potassium phosphate, pH 6, per liter, and 1 ml of 5 mg/ml cholesterol (prepared in ethanol) per liter). Each plate contained a spot of about 150 µl of streptomycin-resistant E. coli OP50-1 (approximately 5 × 1010 cells/ml) grown in Luria Broth (LB, BD Biosciences, catalog # 244610) supplemented with 50 µg/ml streptomycin (Sigma, catalog # S6501). The worms on these plates were fed every 3–4 days with OP50-1 and maintained for 2–3 weeks.
4.2. Egg harvesting by bleach treatment
Mixed populations of N2 animals were transferred from maintenance plates to approximately 100 fresh NGM plates containing OP50 E. coli and were incubated at 25 °C for 3 days to obtain gravid adults. The plates were washed with ice-cold M9 minimal medium (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl, and 1 mM MgSO4) and the animals were transferred to 15 ml polypropylene conical tubes. The worms were centrifuged at 600 × g for 2 min at 4 °C (Beckman Coulter, Allegra X-15R) and the supernatant was aspirated. The animals were resuspended in 10 ml of ice-cold 30% sucrose and centrifuged at 2,095 × g for 5 min at 4 °C. The floated animals were transferred to a 15 ml conical tube with ice-cold M9 medium, pelleted at 2,000 × g for 2 min, and the supernatant was aspirated. The worms were subsequently washed approximately 5 times with ice-cold M9 medium and by centrifuging at 600 × g for 2 min. After the final wash, the worm pellet was resuspended in 5 ml of fresh bleaching solution (12.5 ml Clorox bleach (6.2 percent sodium hypochlorite), 6 ml 5 M NaOH, 31.5 ml water) and pelleted in a clinical centrifuge at setting 5 (~796 × g) for 30 s (International Clinical Centrifuge, model CL). The supernatant was aspirated and the pellet resuspended in 10 ml of bleaching solution. The worms were fragmented by alternating between 30 s of vortexing at setting 10 and 15 s on ice, and the release of eggs from gravid adults was monitored by microscopy. After no more than 6 min, the samples were centrifuged in a clinical centrifuge at setting 5 for 30 s and the eggs in the pellet were washed with ice-cold M9 medium 5 times to remove residual bleaching solution. C. elegans eggs were analyzed directly for ascorbate content or used to generate L1 larvae as described below.
4.3. Sample preparation for high-performance liquid chromatography (HPLC) ascorbate analysis
Mixed animal populations were washed from 2–80 NGM plates with ice-cold M9 medium into a 15 ml conical tube and centrifuged at 600 × g for 2 min. The supernatant was aspirated and the animals were washed an additional time with M9 medium. The pellet was transferred to a microcentrifuge tube, weighed, and resuspended in 150 µl of ascorbate extraction buffer consisting of 5% meta-phosphoric acid (Sigma, catalog # M6288), 2 mM disodium EDTA, and 2 mM tris(2-carboxyethyl) phosphine hydrochloride (Sigma, catalog # C4706). This reagent reduces dehydroascorbate to ascorbate; our assays then reflect the amount of both of these molecules. Approximately 50–100 µl of washed sea sand (Fisher Scientific, catalog # S25) was added and the samples were ground on ice with a disposable pestle (Fisher Scientific, catalog # 12-141-364) for 100 revolutions by hand or using a lab stirrer for 1 min set to 100 rpm (Fisher Scientific, LR400A). The sand was removed and the samples were centrifuged at 20,000 × g for 10 min at 4 °C (Eppendorf, Model 5417R). The supernatant was transferred to a new microcentrifuge tube and stored for no more than 24 h at −80 °C until analysis to minimize sample degradation. Ascorbate levels in eggs were determined from approximately 80 NGM plates by performing the bleach treatment and lysis procedure described above. Eggs were allowed to hatch in 70 ml of M9 medium at 20 °C and 160 rpm for at least 24 h to determine the presence of ascorbate in L1 larvae.
In general, 60 µl of extract or L-ascorbate standard (Fluka, catalog # 95209) dissolved in ascorbate extraction buffer were adjusted to a final pH of ~5 by adding 9 µl of 1 M sodium citrate, ~pH 8 (final concentration of 130 mM), and separated on a HP 1090 II liquid chromatograph with a Phenomenex C18 reversed-phase column (Catalog # 00G-4252-E0, 5 µm, 4.6-mm inner diameter, 250-mm length). Analyses were performed at room temperature with an injection volume of 30–55 µl and a flow rate of 1 ml/min. Buffer A consisted of 20 mM triethylammonium acetate in water, pH 6, and buffer B was 20 mM triethylammonium acetate in 40% acetonitrile, pH 6. The HPLC gradient was as follows: isocratic for 7 min at 100% A, a 1 min linear gradient from 100% A to 0% A, 5 min at 0% A, a 1 min linear gradient from 0% A to 100% A, and isocratic for 15 min at 100% A. Ascorbate was detected at a wavelength of 265 nm and eluted between 3.6–4 min depending on the column condition. The presence of ascorbate was verified by noting the loss of the peak after 60 µl of sample was incubated with 2 units of ascorbate oxidase (AAO, Fisher Scientific, catalog # 50–230–3657, dissolved in 100 mM sodium phosphate and 0.5 mM EDTA, pH 5.6) and 9 µl of 1 M sodium citrate, pH 8, for 30–120 min and chromatographed. The amount of ascorbate was calculated based on the area of the ascorbate peak at 265 nm using an extinction coefficient of 14,500 M−1 cm−1. Ascorbate levels in samples were normalized either to the approximate number of worms analyzed as determined by microscopy, or by assuming 1 g of wet weight worm pellet was equivalent to 1 ml of worm volume.
4.4. Addition of ascorbate precursors to C. elegans extracts and intact animals
For incubations with gulono-1,4-lactone with fractions of worms, mixed populations of animals were washed with ice-cold M9 medium from 20 NGM plates and pelleted in a 15 ml conical tube at 600 × g and 4 °C for 2 min. After the supernatant was aspirated, the pellet was washed two times with ice-cold M9 medium. The pellet was weighed (~410 mg) and 820 µl of protein extraction buffer (10 mM Tris-HCl, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 50 mM sucrose, 1 mM DTT) was added. The animals were freeze thawed three times with liquid nitrogen before lysis (Branson Sonic Power Co., W-350 Sonifier) on ice with 10 5-s sonicator pulses (50% duty cycle, intensity 4) in a total time of 5 min. The samples were placed in microcentrifuge tubes and centrifuged at 600 × g for 1 min to obtain the “intact worm” pellet of unbroken and fragmented animals. The supernatant was transferred to new microcentrifuge tubes and centrifuged at 20,000 × g for 15 min to obtain the “worm membrane” pellet. The supernatant was transferred to a new microcentrifuge tube and labeled “worm cytosol.” An additional 100 µl of protein extraction buffer was added to the “intact worm” and “worm membrane” pellets. In a total volume of 75 µl, 25 µl of “intact worms,” “worm membrane,” or “worm cytosol” were incubated in 20 µl of water, 5 µl of 100 mM L-gulono-1,4-lactone (Sigma, catalog # 310301, final concentration of 6.7 mM), and 25 µl of 100 mM sodium phosphate and 0.5 mM EDTA, pH 5.6, for 2 h at room temperature. The reactions were terminated by adding 75 µl of 10% trifluoroacetic acid and 140 µl of sample was injected on the HPLC and ascorbate was characterized as described above. To confirm the position of ascorbate, 2 units of AAO was incubated with the above reactions for 1 h at 4 °C and separated by HPLC.
For incubations of live worms with gulono-1,4-lactone and other possible ascorbate precursors, eggs were obtained from 80 NGM plates containing a mixed population of animals as described above and incubated in 5 ml of M9 medium at 20 °C and 160 rpm for approximately 24 h. The L1 larvae were centrifuged at 3,000 × g for 5 min and the supernatant was aspirated. The worms were washed once with ice-cold M9 medium and a pellet with L1 larvae and residual M9 medium (~900 µl) was obtained. 100 µl of L1 larvae in suspension were subsequently incubated with 20 µl of either protein extraction buffer, 10 mM L-gulono-1,4-lactone, 10 mM D-(+)-galacturonic acid (Fluka, catalog # 48280), 10 mM D-glucuronic acid (Sigma, catalog # G5269), 10 mM D-galactose (Fisher Scientific, catalog # BP656), or 10 mM D-glucose (Fisher Scientific, catalog # D16). After a 2 h incubation at 20 °C and 160 rpm, 50 µl of washed sea sand was added to each reaction to lyse the animals and the ascorbate content was analyzed by HPLC as described above.
4.5. Gas chromatography-mass spectrometry (GC-MS) analysis
After obtaining the animals as described above, pellets of worms were transferred to a microcentrifuge tubes and 150–300 µl of GC-MS extraction buffer (0.5 mM butylated hydroxytoluene (BHT, Sigma, catalog # B1378) in methanol) was added. Approximately 50–100 µl of washed sea sand was added to each sample and the animals were lysed with 100 rotations of a plastic pestle. After the sand was removed, the samples were centrifuged at 20,000 × g for 10 min. The supernatant was removed and transferred to amber microcentrifuge tubes until use.
Acid washed, amber micro-v glass vials (Thermo Scientific, catalog # C4000-V2) were used for the GC-MS analysis. Approximately 300–800 µl of sample was dried in GC vials in an unheated vacuum centrifuge (Savant Instruments Inc., model # SVC100H and RH 20–12 rotor). After the samples were dried, 100 µl of benzene was added to the vials and the samples were dried under nitrogen gas for approximately 15 min to remove residual water. The samples were derivatized in 50 µl of a 99:1 mixture of N, O-bis (trimethylsilyl) trifluoracetamide: trimethylchlorosilane (BSTFA/TMCS) (Supelco, catalog # 33148) at 60 °C for 90 min. Ascorbate standards were prepared in 0.5 mM BHT in methanol and derivatized using the same protocol described above.
Samples were analyzed using an Agilent 6890 GC-MS with 5975 mass selective detector and helium carrier gas. The autosampler syringe was washed 3 times with chloroform before and after each injection. 1 µl of derivatized sample was injected and the GC-MS was set to have a 50:1 split injection, helium flow rate of 1.3 ml/min, and a 250 °C injection port temperature. Samples were separated on an Agilent JW HP-5-MS column (30 m length, 250 µm inner diameter, and 0.25 µm film thickness) with the following 17.7 min method: 1 min at 140 °C, increase temperature 15 °C/min until 300 °C, 6 min at 300 °C. A 3 min solvent delay was applied to prolong detector life. GC-MS chromatograms were analyzed and molecules identified using the Agilent Enhanced Chemstation, NIST Mass Spectral Search Program 2.0f, and the NIST Mass Spectral Library 08. Ascorbate eluted at approximately 8.6–8.7 min and levels in C. elegans samples were normalized by comparing the ratio between the extracted ion current (EIC) areas of ascorbate (332 m/z) to serine (204 m/z).
To determine if ascorbate is present in the buffers and E. coli used to grow C. elegans, 100 µl of LB, 400 µl of E. coli (~8 × 109 cells) lysed by sonication in 0.5 mM BHT in methanol, 100 µl of a 5% sucrose solution in water, 200 µl of M9 medium, 200 µl of S medium (to every 100 ml of S Basal (5.84 g/l NaCl, 50 ml of 1M KH2PO4, pH 6, per liter, 1 ml of 5 mg/ml cholesterol (prepared in ethanol) per liter) the following components were added: 1 ml of 1 M potassium citrate, pH 6, 1 ml of trace metals solution, 0.3 ml of 1 M CaCl2, and 0.3 ml of 1 M MgS04), 100 µl of 0.5 mM BHT in methanol, and a mixed population of C. elegans extract washed from 10 NGM plates and prepared as previously described, were dried by vacuum centrifugation and derivatized as described above. The total ion current (TIC) and EIC of these samples were analyzed by GC-MS.
4.6. Paraquat and ethanol stress on C. elegans ascorbate levels
A mixed population of animals were collected from approximately 6 NGM plates, washed without a sucrose float, and incubated in flasks with 100 mL of S medium and OP50 E. coli at 20 °C and 160 rpm. After approximately 24 h, paraquat (Sigma, catalog # 856177) was added and the flasks were allowed to incubate for another 7 days. The samples were subsequently centrifuged at 600 × g for 2 min, the supernatant was aspirated, and the pellet was washed once with M9 medium. 0.5 mM BHT in methanol was added to the pellet in a 1:1 ratio (g pellet: ml extraction buffer) and lysed by sonication as described above. GC-MS analysis was performed as described above using 400 µl of extract. For ethanol stress, a mixed population of animals was obtained from 80 NGM plates as described above. Approximately 250 µl of worms were transferred to microcentrifuge tubes with ethanol (Acros, 200 proof, catalog # 61509-0010) diluted in M9 medium. After a 39 h incubation, the animals were pelleted, washed, lysed, and analyzed for ascorbate by reversed-phase HPLC as described above.
4.7. Labeling of C. elegans using 13C-labeled OP50 E. coli
5 ml of LB medium supplemented with 50 µg/ml of streptomycin was inoculated with OP50 E. coli. After an incubation at 37 °C and 250 rpm for approximately 8 h, 100 µl of the culture was transferred to a flask containing 12.5 ml of a E. coli M9 minimal medium with 10.9 mM D-glucose-13C6 (Cambridge Isotope Laboratories, catalog # CLM-1396-0), 93.7 mM KH2PO4, 56.3 mM K2HPO4, 62.2 mM Na2HPO4, 13.5 mM K2SO4, 20.6 mM NH4Cl, 17.5 mM uracil, 4.9 mM MgCl2, 0.2 mM CaCl2, 0.1 mM FeSO4, 28.5 µM MnCl2, 16.5 µM CoCl2, 11.9 µM ZnSO4, 8.6 µM CuCl2, 1.6 µM H3BO3, 4.6 µM (NH4)6Mo7O24, and 65.8 µM EDTA. After this culture was incubated at 37 °C and 250 rpm for approximately 14 h, all 12.5 ml were transferred to a flask containing 497.5 ml of the 13C-glucose containing medium described above and cultured for 9 h. The cell pellet (~1.5 g wet weight) was obtained by centrifugation at 6,000 × g for 5 min at 4 °C and washed once with C. elegans M9 medium before it was resuspended in 1.5 ml C. elegans M9 medium. 150 µl of this concentrated E. coli was spotted onto NGM agarose plates and allowed to dry. For animals incubated in liquid culture, 1.5 ml of concentrated E. coli was added to 70 ml of M9 medium in a 250 ml beveled flask. Unlabeled E. coli cells were prepared using the same protocol with the exception that D-glucose (Fisher Scientific, catalog # D16) was substituted for the 13C-labeled glucose.
Mixed populations of C. elegans were used in the labeling experiments and prepared as described above from 46 NGM plates. 100 µl of animals (~1200 mixed stage worms and ~2970 eggs) were pipetted onto each of 10 NGM agarose plates and allowed to incubate in a 20 °C incubator for 85 h. For liquid culture, 1 ml of animals (~12,000 mixed stage worms and ~29,700 eggs) were added to the 70 ml flasks and incubated at 20 °C and 160 rpm for 85 h. The animals were harvested, washed, and analyzed by GC-MS as described above.
5. RESULTS
5.1. Ascorbate is detected in N2 C. elegans extracts and is not present in growth media, E. coli cells, or the extraction reagent
HPLC and LC-MS/MS had previously been used to provide evidence for the existence of ascorbate in C. elegans (42,53,54). In this work, we optimized a reversed-phase HPLC method to detect ascorbate, which eluted on our system between 3.6 and 4 min (Fig. 1A). To verify the presence of ascorbate, ascorbate oxidase (AAO) was used to convert ascorbate into dehydroascorbate, which has a decreased absorbance at 265 nm (55,56) (Fig. 1A). When an extract from a mixed population of N2 animals was analyzed, a species was detected at an elution time similar to ascorbate (Fig. 1B). The addition of AAO to this sample resulted in a decreased absorbance at 265 nm, suggesting that this peak was ascorbate (Fig. 1B). To verify the presence of ascorbate, we analyzed extracts of C. elegans by GC-MS. Ascorbate derivatized in BSTFA/TMCS displays several major GC-MS fragment ions from data in the NIST mass spectral library, including a relatively abundant species with a peak at 332 m/z (Fig. 2A). We therefore analyzed a mixed population of C. elegans by GC-MS and found a strong 332 m/z EIC signal at 8.7 min (Fig. 2B). An analysis of the fragmentation pattern of the material eluting at 8.7 min (Fig. 2C) revealed a pattern matched to the ascorbate standard found in the NIST mass spectral library with a probability score of 93.9%, a Match score of 799 (comparison of the sample to the library spectrum; 700–800, fair match; >800, good match), and an R. Match score of 848 (comparison that ignores peaks in the sample not present in the library spectrum) (Figure 2C). To verify that ascorbate elutes at this position, we analyzed a derivatized ascorbate standard by GC-MS and found a species eluting at 8.7 min identified in the NIST library as ascorbate with high probability (94.9%, Match score of 740, R. Match score of 747) (Fig. 2D). We found no evidence for the presence in C. elegans of D-isoascorbate, the 5-epimer of L-ascorbate, which can be resolved from ascorbate on the GC system (data not shown). Combined, these results demonstrate the presence of ascorbate in C. elegans extracts.
Figure 1. HPLC detection of ascorbate in extracts of mixed populations of C. elegans.
(A) Ascorbate standard separated by reversed-phase HPLC. The black line represents 3.9 µg of ascorbic acid dissolved in a final concentration of 1.7% metaphosphoric acid, 1.7 mM EDTA, 1.7 mM tris(2-carboxyethyl) phosphine hydrochloride, and 130 mM sodium citrate, pH 8, and separated on a reversed-phase HPLC column as described in the “Materials and Methods” section. 2 units of ascorbate oxidase (AAO) were added to the standard to confirm the elution of ascorbate as described in “Materials and Methods” (red line). (B) Reversed-phase HPLC analysis of a C. elegans extract prepared from a mixed population of animals obtained by washing two NGM plates (~142,000 worms) as described in the “Materials and Methods.” 55 µl of sample was injected onto the HPLC (black line). 2 units of AAO was added to confirm the elution of ascorbate (red line).
Figure 2. GC-MS detection of ascorbate in mixed populations of C. elegans.
(A) GC-MS fragmentation pattern of ascorbate derivatized with BSTFA/TMCS from the NIST Mass Spectral Library 08. An enlarged view of the m/z for the specific fragments labeled a–g are shown in the panels at the top. In the structures represented below the panel, the fragment of ascorbate is shown in red; when multiple fragments are possible for a particular m/z, the alternative fragment is shown in blue. Among the peaks, the 332 m/z fragment represents one of the most intense signals. (B). GC-MS analysis of a BSTFA/TMCS derivatized extract of mixed population of C. elegans obtained from ten NGM plates was performed as described in the “Materials and Methods.” The total ion current (TIC) and 332 m/z extracted ion current (97) are represented by the black and red traces, respectively. The asterisk (*) denotes the ascorbate peak. (C) The MS fragmentation pattern of the species denoted by the asterisk identified in panel B. (D) GC-MS analysis of a standard of 150 ng of ascorbate derivatized in BSTFA/TMCS to confirm the elution time of ascorbate as described in the “Materials and Methods.” The TIC and 332 m/z EIC are represented by the black and red traces, respectively. The asterisk (*) denotes the ascorbate peak. The peak located at 7.4 min is an artifact identified as derivatized triethylene glycol.
It is possible that some or all of the observed ascorbate was derived from the extraction solution or the C. elegans growth media components. We thus investigated if the OP50 bacterial food source of C. elegans or the LB medium used to grow the bacteria contains ascorbate. In addition, we also analyzed C. elegans growth media (M9 and S media), the sucrose involved in separating animals from E. coli, and the extraction buffer (0.5 mM BHT in methanol) to determine if ascorbate was present. However, GC-MS analysis did not indicate the presence of ascorbate in any of the solutions or E. coli extracts tested (Fig. 3A and B). Although sucrose and E. coli extracts did have a small EIC signal at the 332 m/z, the fragmentation pattern at this elution time did not match ascorbate (Fig. 3C and D). Taken together, these HPLC and GC-MS analyses have confirmed that C. elegans contain ascorbate and that it is not a derived from the media or the bacterial food source.
Figure 3. Lack of detectable ascorbate in C. elegans growth media and extraction reagent.
(A) TIC and (B) 332 m/z EIC were analyzed by GC-MS of an extract derived from a mixed population of C. elegans, 0.5 mM BHT in methanol extraction buffer, S medium, M9 medium, sucrose, E. coli extract, and LB medium. Samples were derivatized with BSTFA/TMCS as described in the “Materials and Methods.” The dashed red line denotes the elution position of ascorbate. Although small peaks were found in the TIC and 332 m/z EIC of sucrose and E. coli, further analysis of the mass spectrum at these elution times for the sucrose (C) and E. coli (D) samples reveal that the fragmentation patterns do not match ascorbate, but rather weakly match derivatized ribofuranose and phosphate, respectively.
5.2. mcp-1 (tm2679) animals have similar ascorbate levels to wild type N2 animals
MCP-1 (C10F3.4) is a C. elegans protein with homology to VTC2, an enzyme in the ascorbate biosynthesis pathway in plants (3,42,53,57,58). Because plants deficient in VTC2 have decreased ascorbate levels (58), we hypothesized that if MCP-1 was important for ascorbate biosynthesis in C. elegans, mutants should also result in decreased levels. We therefore tested the ascorbate levels in mcp-1 (tm2679) deletion animals and compared them to those found in wild type. The analysis of mcp-1 (tm2679) extracts by HPLC confirmed the presence of ascorbate in animals (Fig. 4A) and the levels were quantified to be 15±7 fmol of ascorbate/worm, a concentration not significantly different to N2 animals (36±16 fmol/worm) (Fig. 4B). To further confirm our findings, we used GC-MS to determine the ratio of ascorbate (332 m/z) to serine (204 m/z) in samples. Our analysis did not find a statistically significant difference between the ratio in N2 (0.024±0.008) and mcp-1 (tm2679) (0.045±0.04) animals. These results suggest MCP-1 is not essential for C. elegans ascorbate biosynthesis. A similar conclusion was previously reached with other methods (53,54).
Figure 4. C. elegans deficient in mcp-1 have ascorbate levels equivalent to wild type.
(A) mcp-1 (tm2679) extracts were prepared from two NGM plates containing a mixed population of worms (approximately 208,000 animals) and analyzed by reversed-phase HPLC as described in the “Materials and Methods” (black line). The extracts were treated with 2 units of AAO to confirm the existence of ascorbate (red line). (B) Ascorbate levels were quantified by HPLC and compared between the N2 wild type and mcp-1 (tm2679) extracts prepared from two NGM plates containing a mixed population of worms (N2, 43,700–142,400 animals; mcp-1 (tm2679), 106,000–318,500) as described in the “Materials and Methods.” The points represent three independent samples, the horizontal line the average, and the error bars represent standard deviations. Statistical significance was determined using an unpaired, two-tailed Student’s t-test. (C) N2 and mcp-1 (tm2679) extracts were prepared in 0.5 mM BHT in methanol and analyzed by GC-MS as described in the “Materials and Methods.” The area of the ascorbate 332 m/z EIC peak in each sample was determined and compared against the area of the serine 204 m/z EIC peak. The points represent four independent samples, the horizontal line is the average, and the error bars represent standard deviations. Statistical significance was determined using an unpaired, two-tailed Student’s t-test.
5.3. Ascorbate levels vary between growth stages
Previous research has indicated that ascorbate levels decline with age in organisms, including humans (59), rats (60), plants (29), and potentially Drosophila (39). To determine if levels also decline in C. elegans, we analyzed ascorbate concentrations at different growth stages. The HPLC analysis of C. elegans extracts derived from eggs, L1 larvae, and mixed populations found ascorbate levels to be the highest in the egg stage (0.144 ± 0.023 mM), compared to L1 larvae (0.055 ± 0.028 mM) and mixed population (0.021 ± 0.014 mM) (Figure 5). These levels of ascorbate are low compared to higher plants and vertebrates, but are only somewhat lower than those found in the algae Chlamydomonas reinhardtii (Table 1). Ascorbate levels in C. elegans are also comparable to those of erythroascorbate in the yeast Saccharomyces cerevisiae (Table 1).
Figure 5. Ascorbate levels are highest in the egg stage of C. elegans.
Extracts of eggs, L1 larvae, and mixed populations derived from 9–83, 5–69, and 38–455 mg wet weight of animals, respectively, were prepared as described in the “Materials and Methods” section. Ascorbate was separated by reversed-phase HPLC and quantified. Five, sixteen, and twelve independent samples were analyzed for the eggs, L1 larvae, and mixed population of worms, respectively. The data points represent independent samples, the horizontal line is the average, and the error bars represent standard deviations. Statistical significance was determined using an unpaired, two-tailed Student’s t-test.
Table 1.
Comparison of ascorbate levels in C. elegans to other organisms.
| Organism | Ascorbate (mM)a | References |
|---|---|---|
| Myrciaria dubia (camu camu) | 136–170 | Justi et al. (98) Gest et al. (22) |
| Arabidopsis thaliana (flowering plant) | 4–9 | Smirnoff et al. (21) |
| Terrapene carolina (box turtle) | 1.6–5.5 | Rice et al. (34) |
| Thamnophis sirtalis (common garter snake) | 1.1–3.7 | Rice et al. (34) |
| Fragaria ananassa (strawberry) | 1.5–3.3 | Tulipani et al. (99) Wang et al. (100) Gest et al. (22) |
| Mus musculus (mouse) | 0.4–3 | Tsao et al. (101) |
| Xenopus laevis (African clawed frog) | 1–2.3 | Rice et al. (34) |
| Himantura signifer (freshwater stingray) | 0.04–1.2 | Wong et al. (41) |
| Chlamydomonas reinhardtii (green algae) | 0.1–0.8 | Urzica et al. (25) |
| Saccharomyces cerevisiaeb (budding yeast) | <0.1–0.4 | Spickett et al. (102) |
| Caenorhabditis elegans (nematode) | 0.144 ± 0.023 (eggs) 0.055 ± 0.028 (L1 larvae) 0.021 ± 0.014 (Mixed population) |
This study |
Ascorbate concentrations from the literature were converted to mmol/l by assuming 1 gram of fresh/wet weight material is equivalent to 1 ml.
Concentration shown is that of the ascorbic acid analog erythroascorbic acid.
5.4. Sugar precursors from known ascorbate biosynthetic pathways and oxidative stressors do not increase ascorbate levels
Plants and animals utilize one of two major alternative ascorbate biosynthetic pathways. To determine if there are proteins in C. elegans that are homologous to those associated with ascorbate synthesis in plants or animals, we compared C. elegans by protein BLAST search to M. musculus and A. thaliana. In the comparison to M. musculus, three proteins, UDP-glucose pyrophosphorylase, UDP-glucose dehydrogenase, and β-glucuronidase, represented apparent orthologs with mutual best hits (Table 2). Additionally, the mouse UDP-glucuronosyltransferase and glucuronate reductase appear to have similar species in worms although they are not mutual best hits. Most importantly, however, the final two enzymes in the animal biosynthetic pathway (the lactonase and oxidase) do not appear to be found in worms (Table 2). A comparison to the plant biosynthetic pathway likewise identified probable C. elegans enzymes for eight steps of the pathway. However, there was no apparent ortholog in worms of the plant epimerase and the final L-galactono-1,4-lactone dehydrogenase. These results suggest that a complete set of genes encoding all of the enzymes of either the animal or plant pathway is not present in C. elegans. Finally, D-galacturonate has been determined to be an intermediate in a third pathway of ascorbate biosynthesis in some protists and green plants in a reaction forming L-galactonate (21, 45–47). Although C. elegans has an apparent ortholog of the enzyme catalyzing this step (Y39G8B.1), this organism still appears to lack the final two enzymes that are shared with plant pathway to form ascorbate.
Table 2.
Comparison of the animal and plant ascorbate pathways to C. elegans.
| Animal (Mus musculus) | ||||||
|---|---|---|---|---|---|---|
| Protein | Uniprot ID | Reaction | C. elegans Protein | % AA identity |
E-value |
Mutual best hit |
| UDP-glucose pyrophosphorylase** | Q91ZJ5 | D-glucose-1-phosphate→UDP-glucose | K08E3.5**** | 67% | 0 | Yes |
| UDP-glucose dehydrogenase* | O70475 | UDP-glucose→UDP-glucuronate | F29F11.1* | 66% | 0 | Yes |
| UDP-glucuronosyltransferase* | Q62452 | UDP-glucuronate→acceptor-beta-D-glucuronoside | C08F11.8*** | 27% | 7×10−56 | 2nd |
| β-glucuronidase** | P12265 | a beta-D-glucuronoside→D-glucuronate | Y105E8B.9**** | 40% | 93×10−154 | Yes |
| Glucuronate reductase** | Q540D7 | D-glucuronate→L-gulonate | Y39G8B.1**** | 47% | 4×10−97 | 2nd |
| Gulonolactonase* | Q64374 | L-gulonate→L-gulono-1,4-lactone | EO3H4.3**** | 28% | 1.2 | 51st |
| L-gulonolactone oxidase* | P58710 | L-gulono-1,4-lactone→L-ascorbate | F45D5.12**** | 27% | 73×10−9 | 25th |
| Plant (Arabidopsis thaliana) | ||||||
| Protein | Uniprot ID | Reaction |
C. elegans Protein |
% AA identity |
E-value |
Mutual best hit |
| Hexokinase* | Q42525 | D-glucose→D-glucose-6-P | F14B4.2* | 37% | 13×10−79 | Yes |
| Phosphoglucose isomerase* | P34795 | D-glucose-6-P→D-fructose-6-P | Y87G2A.8** | 47% | 13×10−151 | Yes |
| Phosphomannose isomerase* | Q9M884 | D-fructose-6-P→D-mannose-6-P | ZK632.4*** | 34% | 63×10−71 | Yes |
| Phosphomannomutase* | O80840 | D-mannose-6-P→D-mannose-1-P | F52B11.2*** | 56% | 43×10−91 | Yes |
| GDP-D-mannose pyrophosphorylase* | O22287 | D-mannose-1-P→GDP-D-mannose | C42C1.5*** | 55% | 33×10−143 | Yes |
| GDP-D-mannose 3’,5’-epimerase* | Q93VR3 | GDP-D-mannose→GDP-L-galactose | D2096.4** | 27% | 23×10−23 | 21st |
| GDP-L-galactose phosphorylase* | Q8RWE8 | GDP-L-galactose→L-galactose-1-P | C10F3.4/MCP-1* | 25% | 33×10−20 | Yes |
| L-galactose-1-P phosphatase* | Q9M8S8 | L-galactose-1-P→L-galactose | F13G3.5* | 40% | 73×10−55 | Yes |
| L-galactose dehydrogenase* | O81884 | L-galactose→L-galactono-1,4-lactone | F37C12.12**** | 37% | 3×10−65 | Yes |
| L-galactono-1,4-lactone dehydrogenase* | Q9SU56 | L-galactono-1,4-lactone→L-ascorbate | Y50D7A.7** | 30% | 43×10−5 | 7th |
Protein existence according to Uniprot
Evidence at protein level
evidence at transcript level
inferred from homology
predicted
We wanted to explore the possibility that C. elegans may still have the missing enzyme activities of either the animal or plant pathway expressed by more distant or even unrelated genes. To directly address this question, mixed populations of animals were first prepared either as intact animals, or as membrane and cytosolic protein fractions from cell lysates. These materials were subsequently incubated with the last precursor of the animal pathway, L-gulono-1,4-lactone, and analyzed by HPLC. In comparison to the membrane fraction alone, membranes incubated in the presence of the sugar did not have significantly altered ascorbate levels under our assay conditions (Fig. 6A). A similar result was observed for the cytosolic protein fraction and intact animals (Fig. 6B, C). We next tested if live animals could use sugars from both the plant and animal pathways to synthesize ascorbate. In order to analyze a uniform population and eliminate the possibility that the bacteria present as a food source would use the ascorbate precursors, starved L1 larvae that were prepared in media lacking bacteria were incubated with D-galactose, L-gulono-1,4-lactone, D-galacturonic acid, D-glucose, or D-glucuronic acid. Once again, no difference in the level of ascorbate was observed by HPLC with the addition of the sugar precursors in comparison to the control (Fig. 6D). Although it is possible that our in vitro incubation conditions and/or the gene expression status of the live worms were not conducive to detect ascorbate formed from the added precursors, these results open the possibility that ascorbate synthesis in C. elegans may occur through a novel pathway.
Figure 6. The addition of ascorbate precursors from known ascorbate biosynthetic pathways does not increase ascorbate levels in C. elegans.
(A–C) C. elegans membrane, cytosolic, and intact worm fractions were incubated with a final concentration of 6.7 mM L-gulono-1,4-lactone for 2 h at room temperature and analyzed by HPLC as described in the “Materials and Methods.” Asterisks (*) denote the elution position of ascorbate. (D) L1 larvae obtained from 80 NGM plates were incubated in a final concentration of 1.7 mM D-galactose, L-gulono-1,4-lactone, D-galacturonic acid, D-glucose, or D-glucuronic acid for 2 h at 20 °C and 160 rpm, before the samples were lysed and analyzed by reversed-phase HPLC according to the “Materials and Methods” section. The variability in ascorbate elution between panels A–C and D is due to a change in HPLC column.
Previous research in Chlamydomonas found that oxidative stress can increase ascorbate levels (25). We therefore hypothesized that oxidative stress may increase ascorbate accumulation in C. elegans and incubated mixed populations of N2 animals with OP50 E. coli and 50, 100, or 200 µM paraquat, a dipyridyl oxidative stress agent. After a 7 day incubation, we observed no significant difference by GC-MS analysis in the ascorbate:serine ratio of treated animals in comparison to nontreated animals (Fig. 7A). We also tested animals incubated in the presence of 0.5% and 1% ethanol and found no increase in ascorbate concentration (Fig. 7B). Unsurprisingly, ascorbate levels declined sharply when 10% ethanol was added, a concentration associated with poor survival of C. elegans (61), further suggesting that C. elegans may synthesize ascorbate (Figure 7B).
Figure 7. Paraquat and ethanol stress do not increase ascorbate levels in C. elegans.
(A) A mixed population of animals was obtained from approximately six NGM plates and incubated in S-medium supplemented with OP50 E. coli and 0, 50, 100, and 200 µM paraquat for 7 d as described in the “Materials and Methods.” GC-MS analysis was performed as described in the “Materials and Methods” and the amount of ascorbate in samples was analyzed by comparing the EIC area of ascorbate (332 m/z) to that of serine (204 m/z). The points represent four independent samples, the horizontal line is the average, and the error bars denote standard deviations. Statistical significance was determined using an unpaired, two-tailed Student’s t-test. (B) Mixed populations of animals from 80 NGM plates were incubated in a microcentrifuge tube at 20 °C and 160 rpm in M9 medium supplemented with 0, 1%, 5%, or 10% ethanol. After incubation for 39 h, the animals were pelleted, washed, lysed, and analyzed by reversed-phase HPLC according to the “Materials and Methods” section. The data points represent two independent samples and the horizontal line denotes the average. Statistical significance was determined using an unpaired, two-tailed Student’s t-test.
5.5. C. elegans synthesizes ascorbate
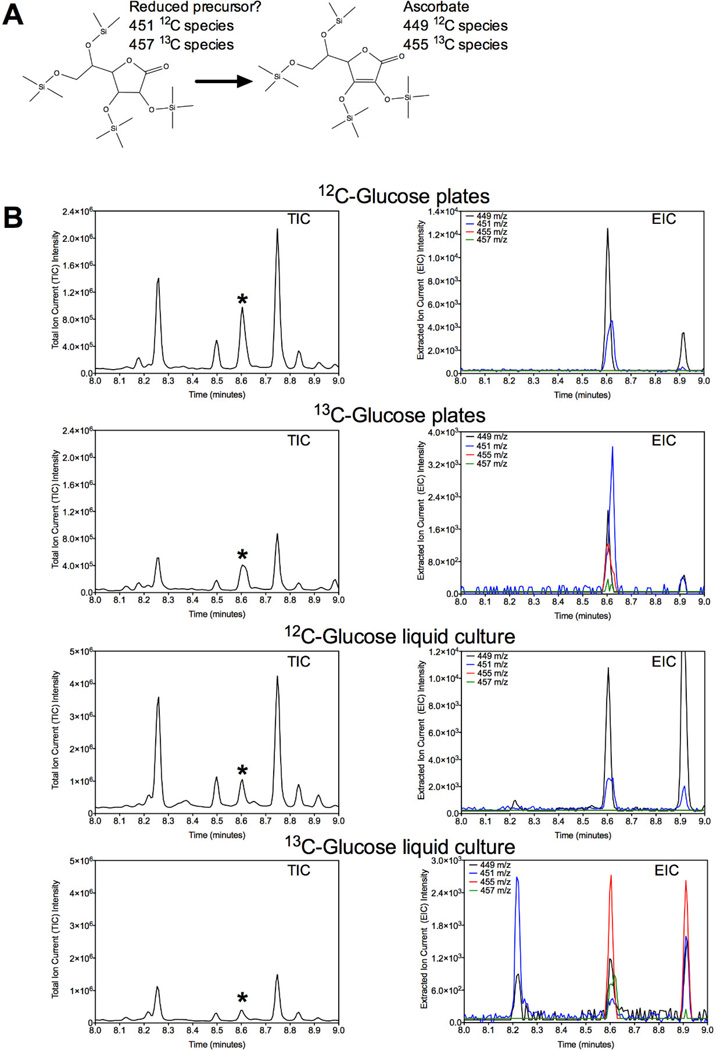
To directly demonstrate that C. elegans synthesizes ascorbate and to elucidate a possible pathway, we labeled a mixed population of animals with either [12C6]- or [13C6]-glucose-labeled E. coli on plates or in liquid media. We then analyzed the fragmentation pattern of ascorbate purified by GC-MS from extracts of these worms. In the 12C-labeled animals from both plates and liquid cultures, we detected the 449 m/z fragment corresponding to the all 12C-ascorbate species, as well as a 451 m/z species that could represent the hydrogenated lactone species that proceeds ascorbate in the plant and animal pathways (Fig. 8, compare to the L-ascorbate standard in Fig. 2A). For the corresponding material from 13C-labeled animals, we detected not only the 449 and 451 ions, but also the 455 and 457 m/z ions, which would represent the fully 13C-labeled species (Fig. 8). In the control animals grown on 12C-E. coli, there were no 455 or 457 m/z ions detected (Fig. 8). Because the 451 and 457 m/z ions that possibly correspond to the hydrogenated lactone precursor of ascorbate were detected at the same GC-MS elution time as ascorbate, it was not possible to definitely assign these ions to this possible precursor. Finally, we searched for molecules at other elution times in the GC-MS that could be precursors in the biosynthesis pathway by comparing the fragmentation patterns across GC fractions of the 12C- and 13C-labeled samples. Although we found several known species that incorporated the 13C-label (such as amino acids), we did not identify any possible precursors to ascorbate.
Figure 8. Incorporation of 13C into C. elegans ascorbate from 13C-labeled E. coli food.
(A) Expected mass change in ascorbate and the possible reduced precursor species due to uniform13C labeling. (B) A mixed population of animals were incubated with 12C6-D-glucose or 13C6-D-glucose-labeled E. coli on plates or in liquid culture at 20 °C for 85 h as described in the “Materials and Methods” section and analyzed by GC-MS. The asterisk denotes the position of ascorbate.
We then analyzed each of the ascorbate GC-MS fragments to confirm that ascorbate was 13C-labeled. C. elegans incubated with 12C-E. coli had ascorbate fragment species whose mass-to-charge ratios closely matched that of the NIST mass spectral library database (Fig. 2A and Fig. 9). Because the 117, 147, 205, 259, 304, 332, and 449 m/z species correspond to 2, 2, 2, 4, 3, 4, and 6 carbon species of ascorbate, respectively, the incorporation of heavy carbon in all positions would result in fragments of 119, 149, 207, 263, 307, 336, and 455 m/z. With the exception of the 147 m/z fragment, full labeling was observed for the remaining species in animals incubated on plates and in liquid culture with 13C-labeled bacteria (Fig. 9). Combined, these data support the conclusion that C. elegans is capable of synthesizing ascorbate. However, as described above, we found no evidence that this biosynthesis utilized either the known plant or animal pathways.
Figure 9. Ascorbate fragmentation pattern from C. elegans fed unlabeled or 13C-labeled E. coli.
Mixed populations of animals were incubated with unlabeled OP50 E. coli or OP50 E. coli labeled with 13C6-D-glucose on (A) NGM plates or (B) in liquid culture for 85 h at 20 °C and analyzed by GC-MS as described in Figure 8 and the “Materials and Methods.” Ascorbate fragmentation data for animals incubated with unlabeled or 13C-labeled OP50 E. coli is shown in blue and red, respectively, and overlaid for direct comparison. The peaks labeled a, b, c, d, e, f, and g correspond to major ascorbate fragmentation peaks shown in Figure 2 and include the 117, 147, 205, 259, 304, 332, and 449 m/z ions. A magnified view of these fragmentation peaks is shown in the inset figures in panels A and B, with the y-axis representing relatively intensity and the x-axis denoting the m/z.
6. CONCLUSIONS
Ascorbate and erythroascorbate are important reducing agents found in many vertebrates, plants, and fungi. To our knowledge, no other study has confirmed the biosynthesis of ascorbate in an invertebrate organism. However, work on crustaceans (62) and insects, such as the corn borer Diatraea grandiosella (63), the corn earworm Helicoverpa zea (64,65), the moth Plusia signata (66), and fruit fly Drosophila melanogaster (39) suggest that ascorbate is present and has biological roles potentially associated with metamorphosis, aging, and preventing oxidative stress. Furthermore, a study involving the parasitic nematode Brugia malayi found ascorbate to be important for the L3 to L4 molt (67). In C. elegans, ascorbate is an important molecule associated with handling oxidative stress, although its role in influencing longevity in wild type animals is unclear (68–72). The addition of ascorbate not only alleviates defective phenotypes in animals exposed to pro-oxidants (73), but can also increase survival and restore normal longevity to short lived mutant animals with elevated reactive oxygen species (ROS) (74–76). Interestingly, ascorbate abolishes life span extension in long lived mutant animals with elevated superoxide and normal ROS levels (77). In the present study, ascorbate was confirmed to exist in C. elegans by two independent methods, and the biosynthesis of ascorbate in this organism is supported by in vivo labeling experiments. Therefore, this study supports the hypothesis that invertebrate organisms are also able to synthesize ascorbate.
The presence of ascorbate in C. elegans is perhaps unsurprising because many of its proteins have mammalian homologs that require ascorbate for proper function. In particular, the ascorbate-dependent prolyl 4-hydroxylase is found in C. elegans and is important for the hydroxylation of collagen in other animals (78,79). Approximately 1% of the genes in C. elegans are devoted to the synthesis of collagen, which composes 80% of the cuticle exoskeleton and is replaced in each molting cycle (80–82). Various mutations in the dpy-18 and phy-2 genes that encode the prolyl 4-hydrolase result in abnormal morphology and lethality (78,79). Additionally, the gene let-268 encodes the only lysyl hydroxylase in C. elegans and is important for type IV collagen processing (83). It is therefore reasonable to expect C. elegans to have ascorbate in order to maintain these Fe2+/α-ketoglutarate-dependent dioxygenases and ensure the proper formation of the eggshell and cuticle. Finally, the mammalian enzymes associated with carnitine, tyrosine, and peptide hormone metabolism that use ascorbate as a cofactor, including ε-N-trimethyl-L-lysine/γ-butyrobetaine hydroxylases, dopamine beta hydroxylase, peptidylglycine alpha-amidating monooxygenase, and the 4-hydroxyphenylpyruvate dioxygenase (17), are highly homologous to the GBH-1/GBH-2, TBH-1, PGAL-1, and HPD-1 proteins, respectively, in C. elegans.
Other than serving as an antioxidant and protein cofactor, ascorbate may also have a symbiotic role in supporting the microbial environment of C. elegans. It has been suggested that C. elegans, like other organisms, can not only benefit from their microbiota in terms of the nutrients, vitamins, and energy they provide, but the worm can also support bacteria with shelter and nutrients (84). Although it is thought that bacteria are unable to synthesize ascorbate, E. coli has an ascorbate transporter that may enable the fermentation of ascorbate and its use as a carbon source under anaerobic conditions (85–88). Furthermore, C. elegans has homologs to the nucleobase ascorbate transporter (NAT) family of proteins found in other organisms (89). In a preliminary analysis of used liquid M9 medium after a C. elegans culture, we were able to find a concentration of approximately 1 µM ascorbate (data not shown). Although this may be due to the release of ascorbate from the lysed cells of dead animals, it is also possible that C. elegans may actively export ascorbate, either to neutralize oxidative stresses in the environment or provide a carbon source to bacteria, an organisms in which it is integrally entwined with in nature.
In our study, we did not observe ascorbate in the unused culture media alone or in the E. coli used to grow the animals. Only C. elegans extracts were found to contain ascorbate at average concentrations ranging between 21 µM in mixed populations to 144 µM in egg preparations. Interestingly, preliminary experiments indicate ascorbate levels in mixed animal populations are not significantly different when they are fed heat killed OP50 bacteria (data not shown), indicating ascorbate synthesis may not depend on bacterial viability. However, it is possible that feeding C. elegans a bacterial strain other than OP50 may result in an alteration in ascorbate levels. Previous studies have found that the bacterial diet of C. elegans can influence gene expression, fat storage, metabolism, and longevity (75,90–94), and future studies could use diet to dissect the ascorbate biosynthesis pathway. It is also interesting to note that the concentration of ascorbate drops from the highest in eggs to the lowest level in the L1 larva and mixed populations. This result suggests ascorbate is required at different levels during development for either added protection against oxidative stress or for specific enzymatic processes. The hypothesis that ascorbate levels are linked with enzymatic activities is possibly supported by the observation that the expression of one prolyl 4-hydroxylase catalytic subunit, the PHY-3 isoform, is limited to the early embryo, late larval, and adult nematode stages (95). Additionally, a recent study has determined that the expression of several genes associated with collagen formation in C. elegans is regulated (96). More research is needed to determine how ascorbate levels are regulated in C. elegans.
Through the use of 13C-labeled E. coli, the present work has provided evidence for the first time of ascorbate biosynthesis in an invertebrate organism. Although 13C-labeled ascorbate was observed when C. elegans were grown on plates with 13C-labeled E. coli, unlabeled species were also present. Interestingly, 13C-incorporation appeared to be greater for animals grown in liquid culture in comparison to those grown on plates. Unpublished work by Reinke et al.1 found only 1% of genes differentially expressed when comparing animals grown under these two different conditions. Interestingly, the genes upregulated in liquid culture were associated with oxidative stress protection and included thioredoxin, glutathione-S-transferase, and the mitochondrial NADP transhydrogenase. It is therefore possible that genes linked to ascorbate synthesis may also be altered in liquid culture.
Finally, this study did not observe any significant alteration in the ascorbate levels of C. elegans exposed to oxidative stresses. Although these oxidative agents may be increasing ascorbate levels, we may not be able to observe any concentration change due to the use of ascorbate to neutralize these molecules. We also found that C. elegans incubated with precursors from the plant and animal pathways also do not synthesize increased levels of ascorbate. Because our study only incubated L1 larvae with these precursors, an increased level of ascorbate may be observed using different larval stages or specific growth conditions. Nevertheless, a similar negative result was observed when D. melanogaster, an organism that has ascorbate in its extracts, was incubated with L-gulono-1,4-lactone (39). It is therefore possible that invertebrates share a common ascorbate biosynthetic pathway that is significantly different from that found in animals, plants, and fungi.
Highlights.
Little is known about the presence and biosynthesis of ascorbate in invertebrates.
HPLC and GC-MS analysis found ascorbate in Caenorhabditis elegans extracts.
C. elegans lacks orthologs to key enzymes in known ascorbate biosynthesis pathways.
Worms fed 13C-labeled bacteria synthesize 13C-ascorbate.
C. elegans is the first invertebrate organism confirmed to biosynthesize ascorbate.
ACKNOWLEDGEMENTS
This work was supported by a Ruth L. Kirschstein National Research Service Award GM007185 to ANP, and by NIH grant GM026020, a Senior Scholar in Aging Award from the Ellison Medical Foundation, and funds from the Elizabeth and Thomas Plott Chair in Gerontology of the UCLA Longevity Center to SGC. We would like to acknowledge Gregory Khitrov from the UCLA Molecular Instrumentation Center, Kym Faull, and Christopher Ryan for their help and advice on GC-MS analysis. We would also like to thank Brian Young and Carole Linster for their contributions to the early stages of this work, and Alison Frand for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Personal communication from Valerie Reinke, Frederick G. Mann, Christina M. Whittle, and Jason D. Lieb. Data is available at the NCBI Gene Expression Omnibus (GEO) database under GSE21526.
REFERENCES
- 1.Du J, Cullen JJ, Buettner GR. Ascorbic acid: chemistry, biology and the treatment of cancer. Biochim. Biophys. Acta. 2012;1826:443–457. doi: 10.1016/j.bbcan.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buettner GR, Jurkiewicz BA. Catalytic metals, ascorbate and free radicals: combinations to avoid. Radiat. Res. 1996;145:532–541. [PubMed] [Google Scholar]
- 3.Linster CL, Clarke SG. L-ascorbate biosynthesis in higher plants: the role of VTC2. Trends Plant Sci. 2008;13:567–573. doi: 10.1016/j.tplants.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Q, Zhang L. Ultraviolet-B-induced oxidative stress and antioxidant defense system responses in ascorbate-deficient vtc1 mutants of Arabidopsis thaliana . J. Plant Physiol. 2008;165:138–148. doi: 10.1016/j.jplph.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Barth C, De Tullio M, Conklin PL. The role of ascorbic acid in the control of flowering time and the onset of senescence. J. Exp. Bot. 2006;57:1657–1665. doi: 10.1093/jxb/erj198. [DOI] [PubMed] [Google Scholar]
- 6.Muller-Moule P, Golan T, Niyogi KK. Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol. 2004;134:1163–1172. doi: 10.1104/pp.103.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pignocchi C, Foyer CH. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr. Opin. Plant Biol. 2003;6:379–389. doi: 10.1016/s1369-5266(03)00069-4. [DOI] [PubMed] [Google Scholar]
- 8.Barth C, Moeder W, Klessig DF, Conklin PL. The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1 . Plant Physiol. 2004;134:1784–1792. doi: 10.1104/pp.103.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraga CG, Motchnik PA, Shigenaga MK, Helbock HJ, Jacob RA, Ames BN. Ascorbic acid protects against endogenous oxidative DNA damage in human sperm. Proc. Natl. Acad. Sci. U. S. A. 1991;88:11003–11006. doi: 10.1073/pnas.88.24.11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duthie SJ, Ma A, Ross MA, Collins AR. Antioxidant supplementation decreases oxidative DNA damage in human lymphocytes. Cancer Res. 1996;56:1291–1295. [PubMed] [Google Scholar]
- 11.Wintergerst ES, Maggini S, Hornig DH. Immune-enhancing role of vitamin C and zinc and effect on clinical conditions. Ann. Nutr. Metab. 2006;50:85–94. doi: 10.1159/000090495. [DOI] [PubMed] [Google Scholar]
- 12.Carr AC, Frei B. Toward a new recommended dietary allowance for vitamin C based on antioxidant and health effects in humans. Am. J. Clin. Nutr. 1999;69:1086–1107. doi: 10.1093/ajcn/69.6.1086. [DOI] [PubMed] [Google Scholar]
- 13.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGregor GP, Biesalski HK. Rationale and impact of vitamin C in clinical nutrition. Curr. Opin. Clin. Nutr. Metab. Care. 2006;9:697–703. doi: 10.1097/01.mco.0000247478.79779.8f. [DOI] [PubMed] [Google Scholar]
- 15.Goodyear-Bruch C, Pierce JD. Oxidative stress in critically ill patients. Am. J. Crit. Care. 2002;11:543–551. [PubMed] [Google Scholar]
- 16.Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 17.Englard S, Seifter S. The biochemical functions of ascorbic acid. Annu. Rev. Nutr. 1986;6:365–406. doi: 10.1146/annurev.nu.06.070186.002053. [DOI] [PubMed] [Google Scholar]
- 18.Dunn WA, Rettura G, Seifter E, Englard S. Carnitine biosynthesis from gamma-butyrobetaine and from exogenous protein-bound 6-N-trimethyl-L-lysine by the perfused guinea pig liver. Effect of ascorbate deficiency on the in situ activity of gamma-butyrobetaine hydroxylase. J. Biol. Chem. 1984;259:10764–10770. [PubMed] [Google Scholar]
- 19.Rebouche CJ. Ascorbic acid and carnitine biosynthesis. Am. J. Clin. Nutr. 1991;54:1147S–1152S. doi: 10.1093/ajcn/54.6.1147s. [DOI] [PubMed] [Google Scholar]
- 20.Hirota K, Semenza GL. Regulation of hypoxia-inducible factor 1 by prolyl and asparaginyl hydroxylases. Biochem. Biophys. Res. Commun. 2005;338:610–616. doi: 10.1016/j.bbrc.2005.08.193. [DOI] [PubMed] [Google Scholar]
- 21.Smirnoff N, Conklin PL, Loewus FA. Biosynthesis of ascorbic acid in plants: A renaissance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:437–467. doi: 10.1146/annurev.arplant.52.1.437. [DOI] [PubMed] [Google Scholar]
- 22.Gest N, Gautier H, Stevens R. Ascorbate as seen through plant evolution: the rise of a successful molecule? J. Exp. Biol. 2013;64:33–53. doi: 10.1093/jxb/ers297. [DOI] [PubMed] [Google Scholar]
- 23.Streb P, Aubert S, Gout E, Bligny R. Reversibility of cold- and light-stress tolerance and accompanying changes of metabolite and antioxidant levels in the two high mountain plant species Soldanella alpina and Ranunculus glacialis . J. Exp. Bot. 2003;54:405–418. doi: 10.1093/jxb/erg048. [DOI] [PubMed] [Google Scholar]
- 24.Paciolla C, Tommasi F. The ascorbate system in two bryophytes: Brachythecium velutinum and Marchantia polymorpha . Biol. Plant. 2003;47:387–393. [Google Scholar]
- 25.Urzica EI, Adler LN, Page MD, Linster CL, Arbing MA, Casero D, Pellegrini M, Merchant SS, Clarke SG. Impact of oxidative stress on ascorbate biosynthesis in Chlamydomonas via regulation of the VTC2 gene encoding a GDP-L-galactose phosphorylase. J. Biol. Chem. 2012;287:14234–14245. doi: 10.1074/jbc.M112.341982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tel-Or E, Huflejt ME, Packer L. Hydroperoxide metabolism in cyanobacteria. Arch. Biochem. Biophys. 1986;246:396–402. doi: 10.1016/0003-9861(86)90485-6. [DOI] [PubMed] [Google Scholar]
- 27.Obinger C, Regelsberger G, Pircher A, Strasser G, Peschek GA. Scavenging of superoxide and hydrogen peroxide in blue-green algae (cyanobacteria) Physiol. Plant. 1998;104:693–698. [Google Scholar]
- 28.Lorence A, Chevone BI, Mendes P, Nessler CL. myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004;134:1200–1205. doi: 10.1104/pp.103.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang WY, Lorence A, Gruszewski HA, Chevone BI, Nessler CL. AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/L-galactose ascorbic acid biosynthetic pathway. Plant Physiol. 2009;150:942–950. doi: 10.1104/pp.109.138453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartoli CG, Yu JP, Gomez F, Fernandez L, McIntosh L, Foyer CH. Interrelationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J. Exp. Bot. 2006;57:1621–1631. doi: 10.1093/jxb/erl005. [DOI] [PubMed] [Google Scholar]
- 31.Dabrowski K. Primitive actinopterigian fishes can synthesize ascorbic acid. Experientia. 1994;50:745–748. [Google Scholar]
- 32.Moreau R, Dabrowski K. Biosynthesis of ascorbic acid by extant actinopterygians. J. Fish Biol. 2000;57:733–745. [Google Scholar]
- 33.Chatterjee IB. Evolution and the biosynthesis of ascorbic acid. Science. 1973;182:1271–1272. doi: 10.1126/science.182.4118.1271. [DOI] [PubMed] [Google Scholar]
- 34.Rice ME, Lee EJK, Choy YY. High levels of ascorbic acid, not glutathione, in the CNS of anoxia-tolerant reptiles contrasted with levels in anoxia-intolerant species. J. Neurochem. 1995;64:1790–1799. doi: 10.1046/j.1471-4159.1995.64041790.x. [DOI] [PubMed] [Google Scholar]
- 35.Drouin G, Godin JR, Page B. The genetics of vitamin C loss in vertebrates. Curr. Genomics. 2011;12:371–378. doi: 10.2174/138920211796429736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr RS, Neff JM. Determination of ascorbic acid in tissues of marine animals by liquid-chromatography with electrochemical detection. Anal. Chem. 1980;52:2428–2430. doi: 10.1021/ac50064a046. [DOI] [PubMed] [Google Scholar]
- 37.Carr RS, Bally MB, Thomas P, Neff JM. Comparison of methods for determination of ascorbic acid in animal tissues. Anal. Chem. 1983;55:1229–1232. doi: 10.1021/ac00259a012. [DOI] [PubMed] [Google Scholar]
- 38.Dabrowski K, Hinterleitner S. Applications of a simultaneous assay of ascorbic acid, dehydroascorbic acid and ascorbic sulfate in biological materials. Analyst. 1989;114:83–87. doi: 10.1039/an9891400083. [DOI] [PubMed] [Google Scholar]
- 39.Massie HR, Shumway ME, Whitney SJ, Sternick SM, Aiello VR. Ascorbic acid in Drosophila and changes during aging. Exp. Gerontol. 1991;26:487–494. doi: 10.1016/0531-5565(91)90037-m. [DOI] [PubMed] [Google Scholar]
- 40.Gupta SD, Chaudhuri CR, Chatterjee IB. Incapability of L-ascorbic acid synthesis by insects. Arch. Biochem. Biophys. 1972;152:889–890. doi: 10.1016/0003-9861(72)90287-1. [DOI] [PubMed] [Google Scholar]
- 41.Wong SZH, Ching BY, Chng YR, Wong WP, Chew SF, Ip YK. Ascorbic acid biosynthesis and brackish water acclimation in the euryhaline freshwater white-rimmed stingray, Himantura signifer . PLoS ONE. 2013:8. doi: 10.1371/journal.pone.0066691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG. Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the Smirnoff-Wheeler pathway to ascorbic acid in plants. J. Biol. Chem. 2007:282. doi: 10.1074/jbc.M702094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linster CL, Van Schaftingen E. Vitamin C: Biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274:1–22. doi: 10.1111/j.1742-4658.2006.05607.x. [DOI] [PubMed] [Google Scholar]
- 44.Lachapelle MY, Drouin G. Inactivation dates of the human and guinea pig vitamin C genes. Genetica. 2011;139:199–207. doi: 10.1007/s10709-010-9537-x. [DOI] [PubMed] [Google Scholar]
- 45.Badejo AA, Wada K, Gao Y, Maruta T, Sawa Y, Shigeoka S, Ishikawa T. Translocation and the alternative D-galacturonate pathway contribute to increasing the ascorbate level in ripening tomato fruits together with the D-mannose/L-galactose pathway. J. Exp. Bot. 2012;63:229–239. doi: 10.1093/jxb/err275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agius F, Gonzalez-Lamothe R, Caballero JL, Munoz-Blanco J, Botella MA, Valpuesta V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003;21:177–181. doi: 10.1038/nbt777. [DOI] [PubMed] [Google Scholar]
- 47.Smirnoff N. Vitamin C booster. Nat. Biotechnol. 2003;21:134–136. doi: 10.1038/nbt0203-134. [DOI] [PubMed] [Google Scholar]
- 48.Baroja-Mazo A, del Valle P, Rua J, de Cima S, Busto F, de Arriaga D, Smirnoff N. Characterisation and biosynthesis of D-erythroascorbic acid in Phycomyces blakesleeanus . Fungal Genet. Biol. 2005;42:390–402. doi: 10.1016/j.fgb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Dumbrava VA, Pall ML. Control of nucleotide and erythroascorbic acid pools by cyclic AMP in Neurospora crassa . Biochim. Biophys. Acta. 1987;926:331–338. doi: 10.1016/0304-4165(87)90219-4. [DOI] [PubMed] [Google Scholar]
- 50.Nick JA, Leung CT, Loewus FA. Isolation and Identification of Erythroascorbic Acid in Saccharomyces-Cerevisiae and Lypomyces-Starkeyi. Plant Sci. 1986;46:181–187. [Google Scholar]
- 51.Huh WK, Lee BH, Kim ST, Kim YR, Rhie GE, Baek YW, Hwang CS, Lee JS, Kang SO. D-Erythroascorbic acid is an important antioxidant molecule in Saccharomyces cerevisiae . Mol. Microbiol. 1998;30:895–903. doi: 10.1046/j.1365-2958.1998.01133.x. [DOI] [PubMed] [Google Scholar]
- 52.Valpuesta V, Botella MA. Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. Trends Plant Sci. 2004;9:573–577. doi: 10.1016/j.tplants.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Adler LN, Gomez TA, Clarke SG, Linster CL. A novel GDP-D-glucose phosphorylase involved in quality control of the nucleoside diphosphate sugar pool in Caenorhabditis elegans and mammals. J. Biol. Chem. 2011;286:21511–21523. doi: 10.1074/jbc.M111.238774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adler LN. From vitamin C to metabolite repair: The role of novel sugar nucleotide phosphorylases. Ph.D., Biochemistry and Molecular Biology, UCLA; 2012. [Google Scholar]
- 55.Oberbacher MF, Vines HM. Spectrophotometric assay of ascorbic acid oxidase. Nature. 1963;197:1203–1204. doi: 10.1038/1971203a0. [DOI] [PubMed] [Google Scholar]
- 56.Dawson CR, Strothkamp KG, Krul KG. Ascorbate oxidase and related copper proteins. Ann. N. Y. Acad. Sci. 1975;258:209–220. doi: 10.1111/j.1749-6632.1975.tb29281.x. [DOI] [PubMed] [Google Scholar]
- 57.Linster CL, Adler LN, Webb K, Christensen KC, Brenner C, Clarke SG. A second GDP-L-galactose phosphorylase in arabidopsis en route to vitamin C. Covalent intermediate and substrate requirements for the conserved reaction. J. Biol. Chem. 2008;283:18483–18492. doi: 10.1074/jbc.M802594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conklin PL, Saracco SA, Norris SR, Last RL. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics. 2000;154:847–856. doi: 10.1093/genetics/154.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sasaki R, Kurokawa T, Tero-Kubota S. Ascorbate radical and ascorbic acid level in human serum and age. J. Gerontol. 1983;38:26–30. doi: 10.1093/geronj/38.1.26. [DOI] [PubMed] [Google Scholar]
- 60.Lykkesfeldt J, Hagen TM, Vinarsky V, Ames BN. Age-associated decline in ascorbic acid concentration, recycling, and biosynthesis in rat hepatocytes--reversal with (R)-alpha-lipoic acid supplementation. FASEB J. 1998;12:1183–1189. doi: 10.1096/fasebj.12.12.1183. [DOI] [PubMed] [Google Scholar]
- 61.Dhawan R, Dusenbery DB, Williams PL. Comparison of lethality, reproduction, and behavior as toxicological endpoints in the nematode Caenorhabditis elegans . J. Toxicol. Environ. Health A. 1999;58:451–462. doi: 10.1080/009841099157179. [DOI] [PubMed] [Google Scholar]
- 62.Merchie G, Lavens P, Sorgeloos P. Optimization of dietary vitamin C in fish and crustacean larvae: A review. Aquaculture. 1997;155:165–181. [Google Scholar]
- 63.Chippendale GM. Ascorbic acid: An essential nutrient for a plant-feeding insect, Diatraea grandiosella . J. Nutr. 1975;105:499–507. doi: 10.1093/jn/105.4.499. [DOI] [PubMed] [Google Scholar]
- 64.Felton GW, Summers CB. Antioxidant systems in insects. Arch. Insect Biochem. Physiol. 1995;29:187–197. doi: 10.1002/arch.940290208. [DOI] [PubMed] [Google Scholar]
- 65.Summers CB, Felton GW. Antioxidant role of dehydroascorbic acid reductase in insects. Biochim. Biophys. Acta. 1993;1156:235–238. doi: 10.1016/0304-4165(93)90142-u. [DOI] [PubMed] [Google Scholar]
- 66.Saxena PN. Ascorbic acid in larval stages of Plusia Signata (Lepidoptera: Noctuidae) Nature. 1966;209:535–536. [Google Scholar]
- 67.Rajan TV, Paciorkowski N, Kalajzic I, McGuiness C. Ascorbic acid is a requirement for the morphogenesis of the human filarial parasite Brugia malayi . J. Parasitol. 2003;89:868–870. doi: 10.1645/GE-3137RN. [DOI] [PubMed] [Google Scholar]
- 68.Pallauf K, Bendall JK, Scheiermann C, Watschinger K, Hoffmann J, Roeder T, Rimbach G. Vitamin C and lifespan in model organisms. Food Chem. Toxicol. 2013;58:255–263. doi: 10.1016/j.fct.2013.04.046. [DOI] [PubMed] [Google Scholar]
- 69.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 70.Shibamura A, Ikeda T, Nishikawa Y. A method for oral administration of hydrophilic substances to Caenorhabditis elegans: Effects of oral supplementation with antioxidants on the nematode lifespan. Mech. Ageing Dev. 2009;130:652–655. doi: 10.1016/j.mad.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 71.Gruber J, Ng LF, Poovathingal SK, Halliwell B. Deceptively simple but simply deceptive - Caenorhabditis elegans lifespan studies: Considerations for aging and antioxidant effects. FEBS Lett. 2009;583:3377–3387. doi: 10.1016/j.febslet.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 72.Harrington LA, Harley CB. Effect of vitamin E on lifespan and reproduction in Caenorhabditis elegans . Mech. Ageing Dev. 1988;43:71–78. doi: 10.1016/0047-6374(88)90098-x. [DOI] [PubMed] [Google Scholar]
- 73.Khare S, Gomez T, Linster CL, Clarke SG. Defective responses to oxidative stress in protein L-isoaspartyl repair-deficient Caenorhabditis elegans . Mech. Ageing Dev. 2009;130:670–680. doi: 10.1016/j.mad.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang JZ, Lemire BD. Mutations in the C. elegans succinate dehydrogenase iron-sulfur subunit promote superoxide generation and premature aging. J. Mol. Biol. 2009;387:559–569. doi: 10.1016/j.jmb.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 75.Pang SS, Curran SP. Adaptive capacity to bacterial diet modulates aging in C. elegans . Cell Metab. 2014;19:221–231. doi: 10.1016/j.cmet.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pang SS, Lynn DA, Lo JY, Paek J, Curran SP. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 2014;5:5048. doi: 10.1038/ncomms6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans . PLoS Biol. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, Kimble J. Prolyl 4-hydroxylase is required for viability and morphogenesis in Caenorhabditis elegans . Proc. Natl. Acad. Sci. U. S. A. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winter AD, Page AP. Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans . Mol. Cell. Biol. 2000;20:4084–4093. doi: 10.1128/mcb.20.11.4084-4093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:1719–1733. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1999;283:35–35. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 82.Cox GN, Kusch M, Edgar RS. Cuticle of Caenorhabditis elegans - Its isolation and partial characterization. J. Cell Biol. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Norman KR, Moerman DG. The let-268 locus of Caenorhabditis elegans encodes a procollagen lysyl hydroxylase that is essential for type IV collagen secretion. Dev. Biol. 2000;227:690–705. doi: 10.1006/dbio.2000.9897. [DOI] [PubMed] [Google Scholar]
- 84.Cabreiro F, Gems D. Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans . EMBO Mol. Med. 2013;5:1300–1310. doi: 10.1002/emmm.201100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Esselen WB, Fuller JE. The oxidation of ascorbic acid as influenced by intestinal bacteria. J. Bacteriol. 1939;37:501–521. doi: 10.1128/jb.37.5.501-521.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Young RM, James LH. Action of intestinal microorganisms on ascorbic acid. J. Bacteriol. 1942;44:75–84. doi: 10.1128/jb.44.1.75-84.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Richter HE, Switala J, Loewen PC. Effect of ascorbate on oxygen-uptake and growth of Escherichia coli B. Can. J. Microbiol. 1988;34:822–825. doi: 10.1139/m88-140. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Z, Aboulwafa M, Smith MH, Saier MH., Jr The ascorbate transporter of Escherichia coli . J. Bacteriol. 2003;185:2243–2250. doi: 10.1128/JB.185.7.2243-2250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gournas C, Papageorgiou I, Diallinas G. The nucleobase-ascorbate transporter (NAT) family: genomics, evolution, structure-function relationships and physiological role. Mol. Biosyst. 2008;4:404–416. doi: 10.1039/b719777b. [DOI] [PubMed] [Google Scholar]
- 90.Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 91.Larsen PL, Clarke CF. Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science. 2002;295:120–123. doi: 10.1126/science.1064653. [DOI] [PubMed] [Google Scholar]
- 92.Gomez F, Monsalve GC, Tse V, Saiki R, Weng E, Lee L, Srinivasan C, Frand AR, Clarke CF. Delayed accumulation of intestinal coliform bacteria enhances life span and stress resistance in Caenorhabditis elegans fed respiratory deficient E. coli . BMC Microbiol. 2012:12. doi: 10.1186/1471-2180-12-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans . PLoS ONE. 2009:4. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yilmaz LS, Walhout AJM. Worms, bacteria, and micronutrients: an elegant model of our diet. Trends Genet. 2014;30:496–503. doi: 10.1016/j.tig.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Riihimaa P, Nissi R, Page AP, Winter AD, Keskiaho K, Kivirikko KI, Myllyharju J. Egg shell collagen formation in Caenorhabditis elegans involves a novel prolyl 4-hydroxylase expressed in spermatheca and embryos and possessing many unique properties. J. Biol. Chem. 2002;277:18238–18243. doi: 10.1074/jbc.M200895200. [DOI] [PubMed] [Google Scholar]
- 96.Hendriks GJ, Gaidatzis D, Aeschimann F, Grosshans H. Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell. 2014;53:380–392. doi: 10.1016/j.molcel.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 97.Olson SK, Greenan G, Desai A, Muller-Reichert T, Oegema K. Hierarchical assembly of the eggshell and permeability barrier in C. elegans . J. Cell Biol. 2012;198:731–748. doi: 10.1083/jcb.201206008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Justi KC, Visentainer JV, de Souza NE, Matsushita M. Nutritional composition and vitamin C stability in stored camu-camu (Myrciaria dubia) pulp. Arch. Latinoam. Nutr. 2000;50:405–408. [PubMed] [Google Scholar]
- 99.Tulipani S, Mezzetti B, Capocasa F, Bompadre S, Beekwilder J, De Vos CHR, Capanoglu E, Bovy A, Battino M. Antioxidants, phenolic compounds, and nutritional quality of different strawberry genotypes. J. Agric. Food Chem. 2008;56:696–704. doi: 10.1021/jf0719959. [DOI] [PubMed] [Google Scholar]
- 100.Wang SY, Lewers KS, Bowman L, Ding M. Antioxidant activities and anticancer cell proliferation properties of wild strawberries. J. Am. Soc. Hortic. Sci. 2007;132:647–658. [Google Scholar]
- 101.Tsao CS, Leung PY, Young M. Effect of dietary ascorbic acid intake on tissue vitamin C in mice. J. Nutr. 1987;117:291–297. doi: 10.1093/jn/117.2.291. [DOI] [PubMed] [Google Scholar]
- 102.Spickett CM, Smirnoff N, Pitt AR. The biosynthesis of erythroascorbate in Saccharomyces cerevisiae and its role as an antioxidant. Free Radic. Biol. Med. 2000;28:183–192. doi: 10.1016/s0891-5849(99)00214-2. [DOI] [PubMed] [Google Scholar]