Abstract
The archetypical fluorescent nucleoside analog, 2-aminopurine (2Ap) has been used in countless assays, though it suffers from very low quantum yield, especially when included in double strands, and from the fact that its residual emission frequently does not represent biologically relevant conformations. To conquer 2Ap’s deficiencies, deoxythienoguanosine (dthG) was recently developed. Here, steady-state and time-resolved fluorescence spectroscopy was used to compare the ability of 2Ap and dthG, to substitute and provide relevant structural and dynamical information on a key G residue in the (−) DNA copy of the HIV-1 primer binding site, (−)PBS, both in its stem loop conformation and in the corresponding (+)/(−)PBS duplex. In contrast to 2Ap, this fluorescent nucleoside when included in (−)PBS or (−)/(+)PBS duplex fully preserves their stability and exhibits a respectable quantum yield and a simple fluorescence decay, with marginal amounts of dark species. In further contrast to 2Ap, the fluorescently detected dthG species reflect the predominantly populated G conformers, which allows exploring their relevant dynamics. Being able to perfectly substitute G residues, dthG will transform nucleic acid biophysics by allowing, for the first time, to selectively and faithfully monitor the conformations and dynamics of a given G residue in a DNA sequence.
For almost five decades, 2-aminopurine (2Ap, 1) has been the fluorescent nucleoside of choice for the community interested in nucleic acid structure, dynamics and recognition.1 Despite its isomerized base-pairing face, numerous fluorescence-based assays have used this isomorphic nucleoside analog as an emissive replacement for adenosine and guanosine (Figure 1), due to its small footprint, high emission quantum yield (QY = 0.68), and availability.1a,2 Challenges have, however, been recognized, including 2-Ap’s propensity to mispair with C and its tendency to perturb the dynamics and structure of DNA.3 Additionally, 2-Ap’s strong emission quenching upon incorporation into single-stranded and particularly double-stranded oligodeoxynucleotides (ODNs) has been commonly observed.2b,4 What appears to have been largely neglected is that the residual emission observed for such DNA and RNA constructs, although sufficient for numerous biophysical applications, frequently does not represent biologically relevant conformations of the native nucleoside replaced. The structural and dynamics information thus gathered might not actually reflect the behavior of the native system of interest. Here we demonstrate that this is indeed the case for the primer binding site (PBS) of the human immunodeficiency virus type 1 (HIV-1), and present 2-aminothieno[3,4-d]pyrimidin-4(3H)-one-7-β-D-2′-deoxyribofuranoside5 (deoxythienoguanosine, dthG) (2) as a truly faithful emissive and responsive surrogate for G in single- and double-stranded ODNs, which actually reproduces the structural context and dynamics of the parent native nucleoside.
Figure 1.
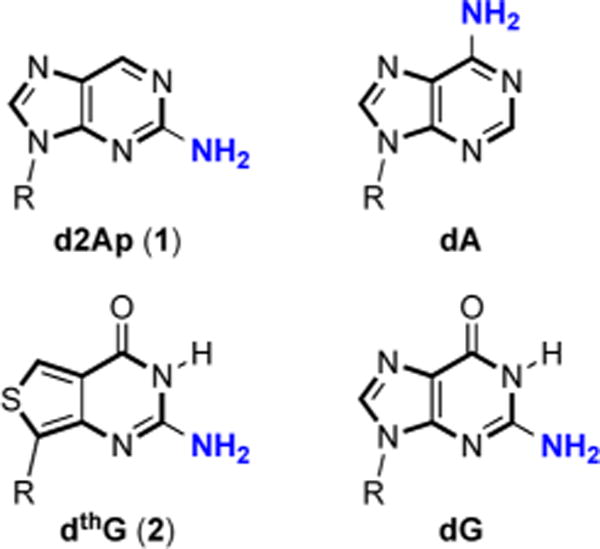
Structure of d2Ap (1), dthG (2), and the naturally occurring purines (R = 2′-deoxy-D-ribose).
The PBS DNA sequence is an 18-mer stem-loop ODN of known 3D structure,6 which is involved in the second strand transfer of HIV-1 reverse transcription (Figure 2).7 This strand transfer, relying on the annealing of (−)PBS with its complementary (+)PBS sequence,8 is required for completing the viral DNA synthesis. To compare the ability of d2Ap and dthG to provide structural and dynamic information on the stem-loop and the corresponding perfect and mismatched duplexes, we substitute the critical G7 loop residue with the two emissive deoxynucleosides and thoroughly analyze the biophysical and photophysical features of all constructs (see Supporting Information for synthetic and additional experimental details).
Figure 2.
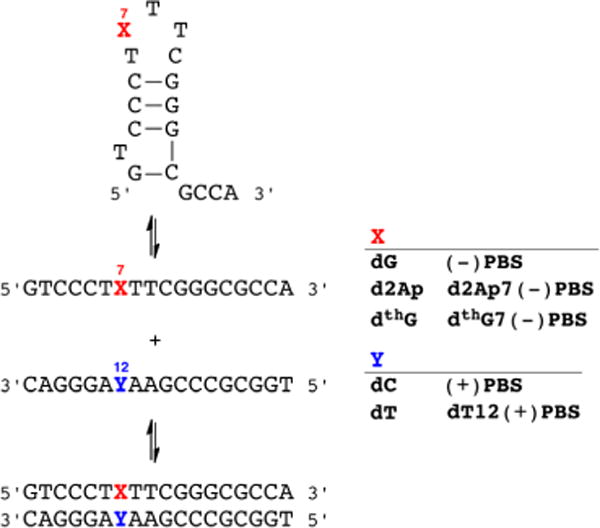
DNA sequence of the HIV-1 primer binding site (−)PBS shown as a single strand (middle), stem loop (top), and duplex with (+)PBS (bottom). Also shown are the site-specifically modified sequences containing d2Ap and dthG.
Thermal denaturation experiments reveal that replacement of G7 by dthG has a minimal impact on the stability of the (−)PBS stem-loop (50 ± 1 and 51 ± 1 °C, respectively) (Table 1). Similarly, the identical melting temperature of the native and the dthG7(−)/(+)PBS duplexes (67 ± 1 and 67 ± 2 °C, respectively) indicate that dthG also perfectly substitutes for dG in the duplex. Additionally, replacement of the pairing C12 by T in (+)PBS, forming a dthG-dT mismatch, results in a 6 °C decrease in the Tm, in excellent agreement with the ΔTm = −7 °C observed for the corresponding dG-dT mismatch.5b While substitution by d2Ap only slightly affects the stability of the (−)PBS stem-loop, it decreases the stability of the (−)/(+)PBS duplex by 7 °C, likely due to the formation of an unstable d2Ap-dC mismatch.9 Notably, the “perfect duplex” d2Ap7(−)/T12(+)PBS is still 5 °C less stable than the native or the dthG7(−)/(+)PBS duplex, indicating that in contrast to dthG, d2Ap does not faultlessly substitute for dG in this context.
Table 1.
Time-Resolved Fluorescence Parameters of d2Ap- and dthG-Labeled ODNsa
| Tm | QY | τ1 | α1 | τ2 | α2 | τ3 | α3 | τ4 | α4 | 〈τ〉 | α0 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d2Ap | 0.68b | 10.2 | 1 | 10.2 | ||||||||
| d2Ap7(−)PBS | 48 ± 1 | 0.08 | 0.15 | 0.16 | 0.66 | 0.10 | 2.6 | 0.15 | 7.4 | 0.11 | 2.4 | 0.48 |
| d2Ap7(−)/T12(+)PBS | 62 ± 1 | 0.013 | 0.18 | 0.20 | 0.44 | 0.27 | 1.4 | 0.01 | 5.2 | 0.01 | 0.4 | 0.51 |
| dthG | 0.46 | 19.6 | 1 | 19.6 | ||||||||
| dthG7(−)PBS | 51 ± 1 | 0.10 | 0.5 | 0.32 | 2.8 | 0.40 | 12.3 | 0.28 | 4.7 | <0.1 | ||
| dthG7(−)/(+)PBS | 67 ± 2 | 0.20 | 1.1 | 0.17 | 11.3 | 0.83 | 9.6 | <0.1 | ||||
| dthG7(−)/T12(+)PBS | 61 ± 1 | 0.38 | 0.8 | 0.07 | 3.9 | 0.09 | 28.2 | 0.57 | 22.3 | 0.27 |
Tm is the melting temperature (°C), QY is the fluorescence quantum yield, τi are the fluorescence lifetimes (ns), and αi are their amplitudes. The amplitude α0 of the dark species as well as the amplitudes αi were calculated as described in the Supporting Information. 〈τ〉 is the mean fluorescence lifetime (ns). Excitation and emission wavelengths were 315 and 370 nm for d2Ap and 315 and 500 nm for dthG. SDs for the lifetimes and amplitudes are <20%. SDs for QY are <10%.
Data from ref 1a.
The free dthG nucleoside (2) emits in the blue with a QY of 0.46 ± 0.02 in buffer (Figure 3 and Table 1). When incorporated into position 7 in the (−)PBS loop, the QY drops to 0.10 ± 0.01, but increases 2-fold upon hybridization to its perfect complement to form dthG7(−)/(+)PBS (Table 1). In sharp contrast to dthG, the near UV emission of d2Ap (1) is severely quenched upon incorporation into ODNs, with 8-fold decrease for the stem-loop, and above 50-fold decrease upon forming the d2Ap7(−)/T12(+)PBS duplex (Table 1). Although displaying a significantly higher QY as a free nucleoside, its short emission wavelength and dramatic quenching in ODNs makes 2Ap a rather inferior emissive surrogate for G. The high QY of dthG in duplexes constitutes, therefore, an obvious asset over d2Ap for monitoring the single to double strand transition and for characterizing the dynamic properties of the substituted base, as discussed below. Moreover, while nearly no wavelength shift is observed for d2Ap in its distinct states, shifts of 5 and 12 nm were observed in the emission maxima of dthG7(−)PBS and dthG7(−)/ (+)PBS, respectively (Figure 3), as compared to the free nucleoside 2. This dthG’s responsiveness provides an additional spectroscopic handle for monitoring the biomolecular environment of this surrogate nucleoside.
Figure 3.
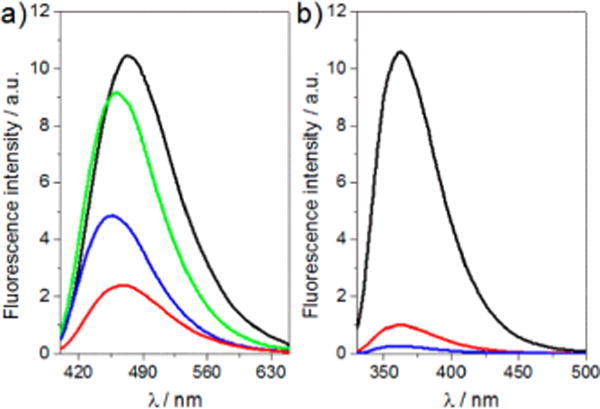
Emission spectra of (a) dthG- and (b) d2Ap-labeled (−)PBS sequences. Emission spectra of (a) dthG free nucleoside (black), dthG7(−)PBS (red), dthG7(−)/(+)PBS (blue), and dthG7(−)/T12-(+)PBS (green); (b) d2Ap free nucleoside (black), d2Ap7(−)PBS (red), and d2Ap7(−)/T12(+)PBS (blue). Excitation was at 380 nm for dthG and 315 nm for d2Ap. Nucleoside and ODN concentration was 6 μM for dthG and 4 μM for d2Ap in 25 mM TRIS-HCl buffer (pH 7.5), 30 mM NaCl, and 0.2 mM MgCl2.
While the two emissive nucleosides exhibit a single exponential decay, the corresponding modified ODNs display a more complex behavior (Table 1). Four decay components are observed for d2Ap7(−)PBS, indicating a large conformational heterogeneity of d2Ap in this loop position, as already described for other positions in the loop.10 The three short lifetimes (τ1-τ3) likely correspond to conformations where dynamic fluctuations of the loop facilitate dynamic quenching of d2Ap by its neighbors, through a charge transfer mechanism11 or relaxation into a low-lying nonemissive electronic state.12 The long-lived lifetime (τ4 = 7.4 ns), being close to that of the free nucleoside, likely corresponds to a conformation where d2Ap is extrahelical and distant from potential quenchers.2b,13 Since the difference in the mean lifetime of d2Ap7(−)PBS as compared to the free d2Ap is markedly smaller than the difference seen for the QY (4.3- vs 8.5-fold), nonemissive “dark species”, with lifetimes shorter than the detection limit of our setup (~30 ps), are present.10,11,14 This population, resulting from either static quenching or very fast dynamic quenching, represents a total of 48% (calculated from eq (1) in the Supporting Information). Only three components are needed to fit the intensity decay of dthG7(−)PBS (Table 1). The long lifetime is close to the component measured for the free nucleoside in methanol (12.3 vs 13.7 ns),5b reflecting a minimally quenched dthG in the less polar environment of the (−)PBS loop.15 The two other components are markedly shorter (0.5 and 2.8 ns), suggesting that they correspond to conformations where dthG is dynamically quenched by its neighboring nucleobases, likely through mechanisms comparable to those of d2Ap.
In contrast to d2Ap, however, comparison of the QY and mean lifetimes of dthG7(−)PBS with those of the free nucleoside reveals that the two evolve in parallel. Dark species are therefore negligible (<10%), which is a distinctive advantage over d2Ap, since all conformations of dthG in (−)PBS can therefore be monitored by the time-resolved measurements.
Differences between d2Ap and dthG become more pronounced in the (−)/(+)PBS duplex. The decay of d2Ap in d2Ap7(−)/T12(+)PBS is best fitted with four discrete lifetime components, ranging from 0.18 to 5.2 ns (Table 1). When comparing the duplex to the stem loop, a dramatic decrease in the amplitudes associated with the two long-lived lifetimes τ3 and τ4 is seen. A total of 98% of the species and thus of the d2Ap conformations in the d2Ap-labeled duplex exhibit lifetimes shorter than 0.5 ns, explaining its extremely low QY. These commonly observed features,2b,13a,16 which severely limit the use of 2Ap in duplexes, likely originate from the destabilization induced by 2Ap in its own base pair and its immediate adjacent base pairs.3 In line with the key role of conformational motions of DNA bases in charge transfer based quenching mechanisms,17 the resulting increased dynamics likely favor efficient 2Ap quenching by its neighbors, explaining the multiple and mainly short-lived fluorescence lifetimes observed for 2Ap in double-stranded DNA.
In sharp contrast to the complex decay of d2Ap7(−)/ T12(+)PBS, the decay of the corresponding dthG7(−)/(+)PBS duplex appears very simple, being characterized by only two lifetimes (1.1 and 11.3 ns) and a marginal fraction of dark species. This indicates that in contrast to d2Ap, dthG adopts better defined conformations, due to its ability to form a stable Watson–Crick base pair with C.18 Thus, we attribute the major conformation (>80%) associated with the 11.3 ns component to the paired dthG in the rather apolar environment created by the stacked base pairs within the duplex.15 This interpretation is further supported by the mismatched duplex dthG7(−)/ T12(+)PBS, where the three lifetimes (0.8, 3.9, and 28.2 ns) and the significant amount of dark species (27%) reflect a greater conformational heterogeneity of dthG, as expected from the reduced constraints imposed by the dthG-dT mismatch compared to the Watson–Crick dthG-dC base pair. Similarly, the dramatic increase in the long-lived lifetime value (28.2 vs 11.3 ns, respectively), which is comparable to the lifetime value of dthG in water, suggests higher accessibility to water, as a result of the lesser constraints imposed by the dthG-dT mismatch in the duplex.
To further cement the picture painted above, we performed time-resolved anisotropy to provide information about the local, segmental and global motions of the labeled ODNs, as well as KI quenching experiments to quantitatively assess the solvent exposure of the emissive nucleosides within the ODNs (Table 2 and Figure S7). The free nucleosides d2Ap and dthG exhibit single rotational correlation times of 80 and 120 ps, respectively. Two correlation times were observed for d2Ap7(−)PBS. The short one (θ1 = 290 ps) likely describes the local rotation of the solvent-exposed extrahelical d2Ap conformation, associated with the long-lived lifetime τ4 = 7.4 ns, which contributes to more than 60% of the labeled ODN emission (as calculated by α4τ4/〈τ〉). This conclusion is further substantiated by the very high bimolecular quenching constant, kq, observed for d2Ap7(−)PBS in iodide quenching experiments (Table 2). Indeed, this kq value being only 2-fold lower than that of the free d2Ap nucleotide, unambiguously confirms that this extrahelical conformation is highly accessible to the solvent. The long correlation time (θ2 =1.9 ns) observed for d2Ap7(−)PBS was significantly shorter than the theoretical correlation time (2.5 ns) calculated for the tumbling of a sphere representing the stem-loop structure. Therefore, this θ2= 1.9 ns component may correspond to a combination of the (−)PBS tumbling motion and a segmental motion, likely associated with the loop. In contrast, the anisotropy decay of d G7(−)PBS is adequately fitted to only one component (2.4 ns) that matches with the theoretical correlation time of the folded ODN. This indicates that the conformations of dthG, associated with the 12.3 ns lifetime, are rigidly held in the (−)PBS loop and only the tumbling of the entire ODN is perceived. This behavior is fully consistent with the NMR structure of (−)PBS, showing that the G7 residue is directed toward the loop interior and well constrained by its neighbors.6b The internal orientation of dthG with poor solvent accessibility is further supported by the low kq value observed with dthG7(−)PBS, that was more than 1 order of magnitude lower than that of the free nucleoside. Thus, time-resolved anisotropy and iodide quenching data confirm that dthG mimics the native G residue much more closely than d2Ap in the stem loop.
Table 2.
Fluorescence Anisotropy Decay Parameters and Quenching Constantsa
| θ1 | β1 | θ2 | β2 | kq | |
|---|---|---|---|---|---|
| d2Ap | 0.08b | 1.00 | 6.7 | ||
| d2Ap7(−)PBS | 0.29 | 0.52 | 1.9 | 0.48 | 3.5 |
| d2Ap7(−)/T12(+)PBS | 2.7 | 1.00 | <10−3 | ||
| dthG | 0.12 | 1.00 | 1.3 | ||
| dthG7(−)PBS | 2.4 | 1.00 | 0.09 | ||
| dthG7(−)/(+)PBS | 8.1 | 1.00 | <10−3 | ||
| dthG7(−)/T12(+)PBS | 8.4 | 1.00 | <10−3 |
θi are the rotational correlation times (in ns) and βi their amplitudes. The reported values are the means from three experiments. SDs for θi and βi are <20%. kq is the bimolecular quenching rate constant for the quenching by iodide (in 109 M−1 s−1). The kq values are the means from two experiments. SDs are <10% for this parameter.
Data from ref 10.
The anisotropy decay of d2Ap7(−)/T12(+)PBS could be fitted with a single component (2.7 ns) that is much shorter than the theoretical correlation time (9.6 ns) calculated for the tumbling motion of this duplex.19 This likely reflects the segmental motions associated with the partially stacked d2Ap conformations that dominate the emission of d2Ap7(−)/ T12(+)PBS. In contrast, the anisotropy decay of the dthG7(−)/ (+)PBS, while also displaying a single correlation time, matches well with the theoretical correlation time of the tumbling duplex. This absence of segmental motion is fully consistent with the attribution of the dominant 11.3 ns lifetime component to the dthG-dC base pair in its optimally stacked configuration. In this highly stable configuration, only the tumbling motion could be detected. Interestingly, a single correlation time (8.4 ns) describing the overall tumbling of the duplex was also observed for dthG7(−)/T12(+)PBS, indicating that the major dthG conformation associated with the 28.2 ns lifetime component is probably not extrahelical. Thus, in line with the high stability (>100 ms) of internal G-C base pairs and the absence of intrahelical dynamics (in the μs-ms range) in the central part of duplexes,3,20 our data indicate that only dthG but not 2Ap can be used to obtain relevant information on the oligonucleotide dynamics and size. Noticeably, for both d2Ap7-and dthG7-labeled duplexes, the kq values are at least 3 orders of magnitude below those of the free nucleosides, suggesting that the emissive nucleosides predominantly adopt an intrahelical conformation.
Taken together, our data clearly illustrate that dthG can faithfully substitute a key G residue in this HIV-1 construct, providing reliable information on its conformations and dynamics in both the (−)PBS stem loop and (−)/(+)PBS duplex. Particularly beneficial are dthG’s reliable base pairing and its high emission QY, which is maintained in single- and double-stranded ODNs. As a result, and in sharp contrast to the corresponding d2Ap labeled ODNs, the species detected by dthG fluorescence techniques, actually reflect the predominantly populated conformers as determined by other means, such as NMR. These features make this new emissive analog a perfect tool to faithfully monitor the conformations and dynamics of G residues in oligonucleotides. This will undoubtedly open a new era with the promise of properly addressing unsolved problems in nucleic acid biophysics.
Supplementary Material
Acknowledgments
We thank Ludovic Richert for his technical help. This work was supported by a fellowship from the Ministère de la Recherche (M.S.), the European Project THINPAD “Targeting the HIV-1 Nucleocapsid Protein to fight Antiretroviral Drug Resistance” (FP7 Grant Agreement 601969), Agence Nationale de la Recherche (ANR blanc Fluometadn), Agence Nationale de Recherche sur le SIDA, and French-Ukrainian Dnipro program. Support by the U.S. National Institutes of Health (grant GM069773 to Y.T.) is gratefully acknowledged.
Footnotes
Supporting Information
Synthesis and fluorescence spectroscopy details. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.(a) Ward DC, Reich E, Stryer L. J Biol Chem. 1969;244:1228. [PubMed] [Google Scholar]; (b) Sinkeldam RW, Greco NJ, Tor Y. Chem Rev. 2010;110:2579. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hall KB. Methods Enzymol. 2009;469:269. doi: 10.1016/S0076-6879(09)69013-3. [DOI] [PubMed] [Google Scholar]
- 2.(a) Rachofsky EL, Osman R, Ross JBA. Biochemistry. 2001;40:946. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]; (b) Guest CR. Biochemistry. 1991;30:3271. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]; (c) Jean JM, Hall KB. Biochemistry. 2002;41:13152. doi: 10.1021/bi020308y. [DOI] [PubMed] [Google Scholar]
- 3.Dallmann A, Dehmel L, Peters T, Mugge C, Griesinger C, Tuma J, Ernsting NP. Angew Chem Int Ed. 2010;49:5989. doi: 10.1002/anie.201001312. [DOI] [PubMed] [Google Scholar]
- 4.(a) Nag N, Ramreddy T, Kombrabail M, Krishna Mohan PM, D’souza J, Rao BJ, Duportail G, Mely Y, Krishnamoorty G. In: Reviews in Fluorescence. Geddes C, Lakowicz JR, editors. Springer Science; New York: 2006. pp. 311–340. [Google Scholar]; (b) Stivers JT. Nucleic Acids Res. 1998;26:3837. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Law SM, Eritja R, Goodman MF, Breslauer KJ. Biochemistry. 1996;35:12329. doi: 10.1021/bi9614545. [DOI] [PubMed] [Google Scholar]; (d) Avilov SV, Godet J, Piemont E, Mély Y. Biochemistry. 2009;48:2422. doi: 10.1021/bi8022366. [DOI] [PubMed] [Google Scholar]; (e) Avilov SV, Piemont E, Shvadchak V, de Rocquigny H, Mély Y. Nucleic Acids Res. 2008;36:885. doi: 10.1093/nar/gkm1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Shin D, Sinkeldam RW, Tor Y. J Am Chem Soc. 2011;133:14912. doi: 10.1021/ja206095a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Park S, Otomo H, Zheng L, Sugiyama H. Chem Commun. 2014;50:1573. doi: 10.1039/c3cc48297a. [DOI] [PubMed] [Google Scholar]
- 6.(a) Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. Biochemistry. 2000;39:9084. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]; (b) Bourbigot S, Ramalanjaona N, Boudier C, Salgado GFJ, Roques BP, Mely Y, Bouaziz S, Morellet N. J Mol Biol. 2008;383:1112. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 7.(a) Thomas JA, Gorelick RJ. Virus Res. 2008;134:39. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Poeschla E. Virology. 2013;441:1. doi: 10.1016/j.virol.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Virus Res. 2008;134:19. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 9.Sowers LC, Fazakerley GV, Eritja R, Kaplan BE, Goodman MF. Proc Natl Acad Sci USA. 1986;83:5434. doi: 10.1073/pnas.83.15.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godet J, Ramalanjaona N, Sharma KK, Richert L, De Rocquigny H, Darlix JL, Duportail G, Mély Y. Nucleic Acids Res. 2011;39:6633. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Larsen OFA, Van Stokkum IHM, Gobets B, Van Grondelle R, Van Amerongen H. Biophys J. 2001;81:1115. doi: 10.1016/S0006-3495(01)75768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wan C, Fiebig T, Schiemann O, Barton JK, Zewail AH. Proc Natl Acad Sci USA. 2000;97:14052. doi: 10.1073/pnas.250483297. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Fiebig T, Wan C, Zewail AH. ChemPhysChem. 2002;3:781. doi: 10.1002/1439-7641(20020916)3:9<781::AID-CPHC781>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]; (d) O’Neill MA, Barton JK. J Am Chem Soc. 2002;124:13053. doi: 10.1021/ja0208198. [DOI] [PubMed] [Google Scholar]; (e) Kelley SO, Barton JK. Science. 1999;283:375. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 12.Jean JM, Hall KB. Proc Natl Acad Sci USA. 2001;98:37. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Neely RK, Daujotyte D, Grazulis S, Magennis SW, Dryden DTF, Klimašauskas S, Jones AC. Nucleic Acids Res. 2005;33:6953. doi: 10.1093/nar/gki995. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Neely RK, Magennis SW, Parsons S, Jones AC. ChemPhysChem. 2007;8:1095. doi: 10.1002/cphc.200600593. [DOI] [PubMed] [Google Scholar]
- 14.Godet J, Kenfack C, Przybilla F, Richert L, Duportail G, Mély Y. Nucleic Acids Res. 2013;41:5036. doi: 10.1093/nar/gkt164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Klymchenko AS, Shvadchak VV, Yushchenko DA, Jain N, Mely Y. J Phys Chem B. 2008;112:12050. doi: 10.1021/jp8058068. [DOI] [PubMed] [Google Scholar]; (b) Ogawa AK, Abou-Zied OK, Tsui V, Jimenez R, Case DA, Romesberg FE. J Am Chem Soc. 2000;122:9917. [Google Scholar]
- 16.(a) Gaied NB, Glasser N, Ramalanjaona N, Beltz H, Wolff P, Marquet R, Burger A, Mely Y. Nucleic Acids Res. 2005;33:1031. doi: 10.1093/nar/gki253. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kenfack CA, Piemont E, Ben Gaied N, Burger A, Mely Y. J Phys Chem B. 2008;112:9736. doi: 10.1021/jp8028243. [DOI] [PubMed] [Google Scholar]; (c) Ramreddy T, Rao BJ, Krishnamoorthy G. J Phys Chem B. 2007;111:5757. doi: 10.1021/jp068818f. [DOI] [PubMed] [Google Scholar]
- 17.O’Neill MA, Barton JK. J Am Chem Soc. 2004;126:13234. doi: 10.1021/ja0455897. [DOI] [PubMed] [Google Scholar]
- 18.Samanta PK, Pati SK. New J Chem. 2013;37:3640. [Google Scholar]
- 19.Ortega A, Garcia De La Torre J. J Chem Phys. 2003;119:9914. [Google Scholar]
- 20.Galindo-Murillo R, Roe DR, Cheatham TE., III Nat Commun. 2014;5(5152) doi: 10.1038/ncomms6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


