Abstract
Accumulation of reactive oxygen and chlorine species (RO/CS) is generally regarded to be a toxic and highly undesirable event, which serves as contributing factor in aging and many age-related diseases. However, it is also put to excellent use during host defense, when high levels of RO/CS are produced to kill invading microorganisms and regulate bacterial colonization. Biochemical and cell biological studies of how bacteria and other microorganisms deal with RO/CS have now provided important new insights into the physiological consequences of oxidative stress, the major targets that need protection, and the cellular strategies employed by organisms to mitigate the damage. This review examines the redox-regulated mechanisms by which cells maintain a functional proteome during oxidative stress. We will discuss the well-characterized redox-regulated chaperone Hsp33, and review recent discoveries demonstrating that oxidative stress-specific activation of chaperone function is a much more widespread phenomenon than previously anticipated. New members of this group include the cytosolic ATPase Get3 in yeast, the E. coli protein RidA, and the mammalian protein α2-macroglobin. We will conclude our review with recent evidence showing that inorganic polyphosphate (polyP), whose accumulation significantly increases bacterial oxidative stress resistance, works by a protein-like chaperone mechanism. Understanding the relationship between oxidative and proteotoxic stresses will improve our understanding of both host-microbe interactions and of how mammalian cells combat the damaging side effects of uncontrolled RO/CS production, a hallmark of inflammation.
Keywords: Molecular Chaperone, Oxidative Stress, Protein Aggregation, Disulfide Bond Formation, Hsp33, Polyphosphate, Posttranslational Modifications, N-chlorination
The Origin of Oxidative Stress
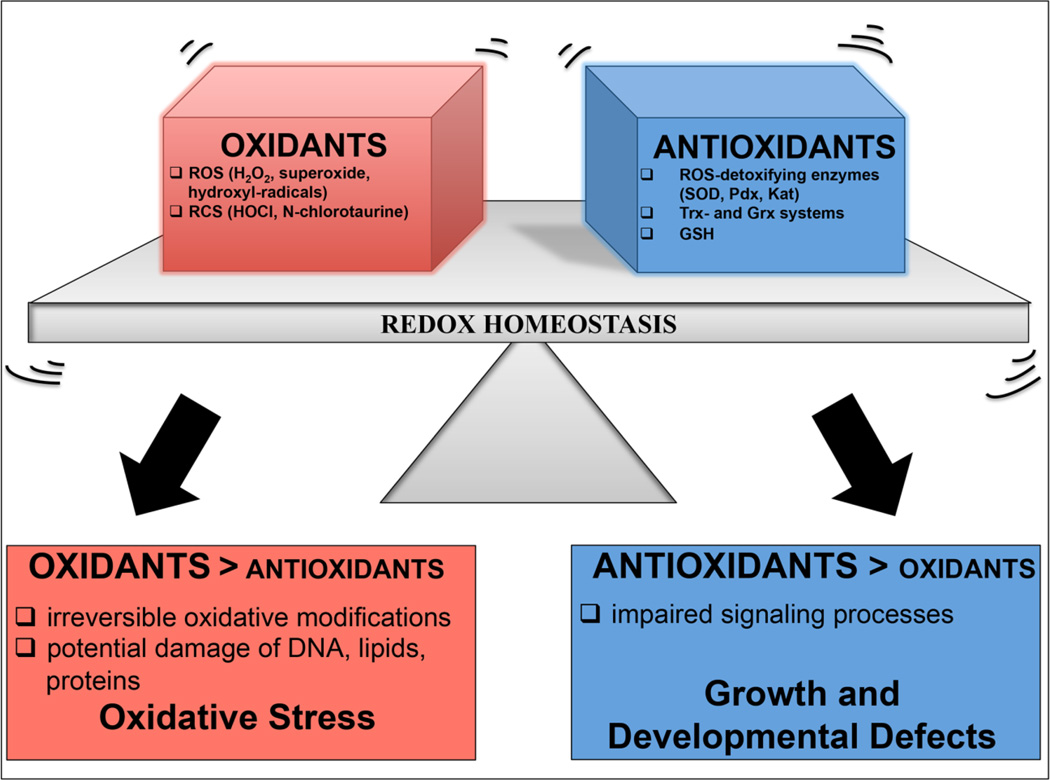
Reactive oxygen species (ROS) such as superoxide (O2•−), hydrogen peroxide (H2O2), and hydroxyl radicals (•OH) are metabolic byproducts, which occur naturally in all organisms that live an aerobic lifestyle. They are constantly produced during electron transfer in the respiratory chain (1), by enzymes such as NADPH oxidases (2, 3), and in organelles like the peroxisomes (4). Non-physiologically low levels of ROS negatively affect cell growth, development and differentiation (5–7), illustrating the importance of ROS as second messengers that control metabolic processes and signaling pathways (Figure 1) (8). Non-physiologically high levels of ROS, on the other hand, can cause irreversible oxidative modifications of virtually all cellular macromolecules, including lipids, DNA, and proteins (Figure 1) (9). Maintaining redox homeostasis requires a concerted cellular effort, involving expression of a variety of different ROS-detoxifying enzymes (e.g., superoxide dismutase, peroxiredoxin, and catalase), oxidoreductases of the thioredoxin (Trx) and glutaredoxin (Grx) systems, NADPH-(re)generating systems, such as the nicotinamide nucleotide transhydrogenase (TH), and the production of the small redox-buffering tripeptide glutathione (GSH) (10). Together, these systems keep cytosolic protein thiols reduced, and protect cells against the accumulation of toxic oxidants (Figure 1) (5, 11).
Figure 1. Redox homeostasis - A balance between oxidants and antioxidants.
Reactive oxygen species (ROS) and reactive chlorine species (RCS) are constantly produced as by-products of cellular processes. They are involved as second messenger in signaling pathways, influencing a variety of different cellular processes. Antioxidant systems including ROS-detoxifying enzymes, such as superoxide dismutase (SOD), peroxiredoxin (Prx), and catalase (Kat), oxidoreductases including the thioredoxin (Trx) and glutaredoxin (Grx) system, as well as the non-protein thiol glutathione (GSH), work together to maintain a reducing environment and prevent accumulation of oxidants beyond physiological levels. However, defects in antioxidant systems or exposure to increased concentrations of RO/CS can shift this ratio. While accumulation of RO/CS causes widespread oxidative damage and is thought to be involved in aging and many age-related diseases, diminished levels of RO/CS affect growth, development and differentiation.
However, even the best systems sometimes fail. When they do, such as during aging, age-related neurodegenerative diseases (e.g. Parkinson’s disease), or metabolic diseases (e.g. diabetes) (12), ROS begin to accumulate in the cell, causing cells to experience a stress condition commonly known as oxidative stress. Oxidative stress can be caused by a variety of different mechanisms, including defects in specific antioxidant or redox-maintaining systems, ROS production during host defense, UV-light, gamma and X-rays, pollutants and smoke, or due to metal-catalyzed Fenton reactions (13). All of these processes produce free radicals, capable of oxidizing cellular structures (14–16). Apart from lipid peroxidation and DNA damage, ROS are especially known for their high reactivity towards sulfur-containing amino acids and metal-containing cofactor sites in proteins, causing reversible and irreversible inactivation of many different proteins and representing a major threat towards the cellular proteome (17). It is thought that oxidative damage to a cell’s proteome contributes to the etiology of a variety of different human protein folding diseases, including Alzheimer’s, Parkinson’s, and prion disease (18).
The Benefit of Oxidative Stress – ROS and RCS as Physiological Antimicrobials
Oxidative stress is not always a bad thing; in fact, production of high levels of ROS and the related reactive chlorine species (RCS) (17) plays an important physiological role in the innate immune response, where it provides a powerful strategy for killing invading pathogens (19). When bacteria are taken up by neutrophils, NADPH-oxidases (NOXs) localized in the phagosomal membrane catalyze the reduction of molecular oxygen to superoxide (O2•−). After being rapidly dismuted to H2O2 by superoxide dismutases, myeloperoxidases convert H2O2 and chloride ions into the RCS hypochlorous acid (HOCl). HOCl is extremely reactive and bactericidal even at low micromolar concentrations (20), making it one of the most powerful physiological oxidants known (for a comprehensive overview see (17, 19)). Not surprisingly, HOCl is the active ingredient of household bleach, and one of the most commonly used disinfectants in medical, industrial and domestic settings (21). Once released into the phagosome, HOCl evokes a very rapid toxic effect on bacteria, contributing to the effective neutrophil-mediated killing of invading microbes (22–24).
In addition to the role of HOCl production in the antimicrobial action of neutrophils, HOCl also appears to be involved in controlling bacterial colonization of mucosal barrier epithelia, such as the airways and the intestine (25, 26). Epithelial cells express the enzyme dual oxidase (DUOX), a member of the nicotinamide adenine dinucleotide phosphate oxidase family. DUOX, like myeloperoxidase, converts peroxide into HOCl (27). Knockdown of the duox gene in the fly gut leads to increased bacterial colonization and a significantly increased rate of death caused by infections (27, 28). Duox−/− mice show a significant decrease of neutrophil invasion during the development of allergic asthma in a murine model, and increased levels of pathogens that colonize the intestinal epithelial cells (29). These results emphasize the physiological importance of oxidative stress in general and HOCl production in particular in combatting microbial pathogens and controlling the bacterial population in the host (25, 27, 30). On the downside, however, uncontrolled production of HOCl by neutrophils can cause a variety of diseases, and is thought to be involved in the tissue damage at sites of chronic inflammation as well as in arteriosclerosis (31).
Proteins - The Primary In Vivo Targets of Oxidative Damage
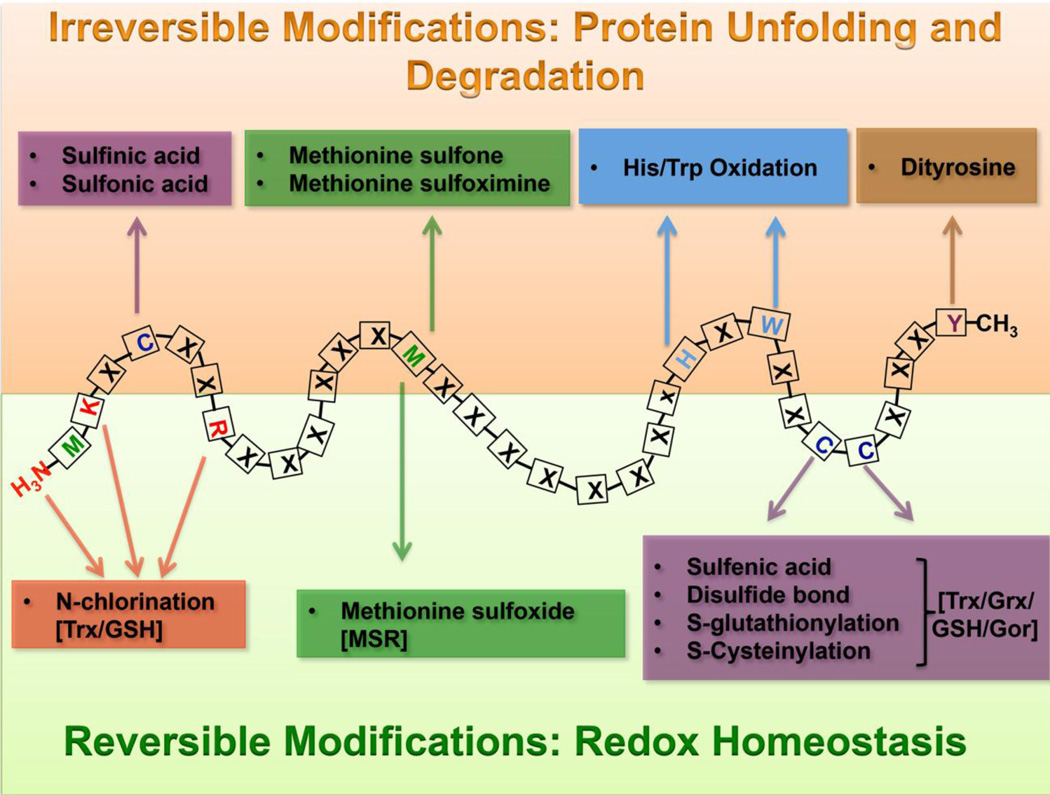
Almost 70% of all oxidized molecules in oxidatively stressed cells are of proteinaceous nature (32), indicating that proteins are the most prominent in vivo targets of oxidants. RO/CS cause numerous posttranslational protein modifications, including oxidation of sulfur-containing side chains, chlorination of side chain amines, oxidation of histidines and tryptophans, dityrosine formation, and others (Figure 2) (17, 33). These oxidative side chain modifications can lead to oligomerization, fragmentation, destabilization, aggregation and/or enhanced degradation of proteins (34–37). While some RO/CS-mediated posttranslational modifications are intentional, reversible, and part of redox-regulated processes (see below), irreversible protein modifications are typically destabilizing and capable of a triggering a major secondary stress on the proteostasis network of the cell (Figure 2).
Figure 2. Reversible and irreversible protein modifications by RO/CS.
RO/CS cause oxidative modification of a number of different residues in proteins. Oxidation of histidines and tryptophans, and the formation of dityrosines, sulfinic/sulfonic acids and methionine sulfone/sulfoximine intermediates are irreversible modifications, and lead to protein unfolding, aggregation and degradation. Disulfide bond formation, methionine sulfoxide formation and N-chlorination are reversible protein modifications, and often used to regulate protein function in response to oxidative stress. The systems responsible for reducing oxidative protein modifications in vivo are listed in brackets: Grx, glutaredoxin; Trx, thioredoxin; GSH, glutathione; MSR, methionine sulfoxide reductase.
The most vulnerable (i.e., reactive) targets in proteins are the sulfur-containing side chains of methionine and cysteine residues (Figure 2) (17, 38). With reaction rates in the 106 – 107 M −1s−1 range, HOCl rapidly chlorinates cysteine thiols (R-SH) (39). This chlorination is followed by an exchange with H2O and the formation of sulfenic acid (R-SOH). The reaction of peroxide with cysteines, which also yields sulfenic acid intermediates, is up to six orders of magnitude slower except for proteins like peroxiredoxin that contain unusually peroxide-reactive cysteines in their active site (40, 41). Due to its highly unstable nature, any sulfenic acid intermediate typically reacts very quickly with other protein thiols to form either intra- or intermolecular disulfide bonds (R1-S-S-R2) (Figure 2). Reactions with non-protein thiol antioxidants, such as GSH or free cysteines, result in the formation of mixed disulfides known as S-glutathionylation (R-S-S-GSH) and S-cysteinylation (R-S-S-RCys), respectively. Sulfenic acids can also react with vicinal primary or secondary amino-groups, thereby forming reversible sulfonamides (17). Alternatively, sulfenic acids can be further oxidized by RO/CS to sulfinic (R-SO2H) or sulfonic (R-SO3H) acid (Figure 2); two typically irreversible thiol modifications that often lead to increased rates of protein degradation. The only known example of reversible sulfinic acid formation was found in select eukaryotic peroxiredoxins, which are reduced by the ATP-dependent sulfinic acid reductase sulfiredoxin (42, 43).
Disulfide bonds, whether intramolecular, intermolecular or mixed, are fully reversible thiol modifications, which can be reduced by members of the thioredoxin or glutaredoxin family (Figure 2) (44). Both types of oxidoreductases reduce oxidized protein thiols via direct thiol-disulfide exchange (45). However, whereas Trx reduces predominantly disulfide bonds and sulfenic acid intermediates, Grx preferentially reduces S-glutathionylated proteins (46). Both proteins enter the reaction in their reduced form, and leave the reaction in their oxidized form. Trx is subsequently reduced by thioredoxin reductase, whereas reduction of oxidized Grx is achieved by GSH. Its oxidation product GSSG is subsequently reduced by glutathione oxidoreductase (Gor) (46, 47). Each of these reductases receive their reducing equivalents from NADPH, closely tying cellular redox homeostasis to the levels of NADPH (46, 48).
Methionine residues are also very oxidation-sensitive. Methionine can be oxidized by RO/CS to methionine sulfoxide at rates of up to 3.4×107 M−1 s−1 (for HOCl) (49, 50). Since the role of methionine oxidation and the methionine sulfoxide reductase (MSR) enzymes involved in repairing oxidized methionines in protein structure, function, and resistance to oxidative damage has been recently reviewed (51–53), we will touch only briefly on this topic here. It has become clear, however, that in many types of cells the repair of oxidized methionine is intimately connected to chaperone function and maintenance of proteostasis under redox stress conditions. In yeast, for example, MSR enzymes preferentially reduce oxidized methionines in unfolded protein substrates (54). In humans, MSR-catalyzed methionine sulfoxide reduction restores chaperone function to oxidatively inactivated a-crystallin (55). In the bacterial pathogen Helicobacter pylori, resistance to oxidative stress depends on MSR not only for repairing oxidized methionines in the ROS-degrading enzymes catalase and alkyl hydroperoxide reductase, but also for restoring activity to oxidatively-inactivated GroEL chaperones (56–58). Like cysteine, under severe oxidizing conditions methionine can also be further oxidized to methionine sulfone or methionine sulfoximine. These are irreversible and toxic endproducts, which likely lead to protein degradation (59)
Apart from sulfur-containing residues, HOCl and to a lesser extent H2O2, also react with histidine, tryptophan, lysine, tyrosine, and arginine side chains, as well as with the amino-terminus of proteins (Figure 2) (60). While the reaction rates are significantly lower than with cysteines (39), most of the reaction products are irreversible, increasing the tendency of proteins to aggregate or to be degraded. In addition, HOCl reacts with a protein’s primary and secondary amines, resulting in chloramine formation. Chloramines themselves are effective oxidants, able to directly chlorinate and oxidize other biomolecules. However, in comparison to HOCl, chloramines are four to five orders of magnitude less reactive (19, 60).
The Danger of Oxidative Stress: Protein Unfolding and Aggregation
As outlined above, oxidative stress-derived protein modifications can lead to the loss of a protein’s secondary or tertiary structure, which in turn impacts its activity, stability and solubility. Moreover, the integrity of the cellular proteome is also highly dependent on regulated posttranslational protein modifications, such as phosphorylation, acetylation, ubiquitination, or methylation. These processes are catalyzed by enzymes, which can also fall victim to oxidative modifications (61, 62). In vivo studies agreed with this notion, and revealed extensive oxidative protein aggregation in HOCl-treated bacteria (63). This result was independently confirmed by the fact that HOCl-treatment of bacteria triggers the heat shock response, a highly conserved transcriptional response which is known to be induced by the accumulation of protein folding intermediates (64, 65). While H2O2 treatment in E. coli yields in little to no protein aggregation and no significant heat shock response, exposure to disulfide stress (either caused by diamide or genetic depletion of the Trx/GSH systems) revealed a considerable overlap between heat shock and oxidative stress response pathways in both Gram-positive and Gram-negative bacteria (66–68). Eukaryotes appear to experience oxidative protein unfolding during peroxide treatment (69). The reason for this discrepancy is unclear. One possibility is increased production of highly reactive hydroxyl radicals in eukaryotes, which could be fostered by the presence of a more significant pool of Fenton metals. Since hydroxyl radicals are extremely reactive, and known to cause protein unfolding and aggregation, increased production of hydroxyl radicals would certainly lead to a massive stress on the proteome (70).
The Challenge of Oxidative Stress: Maintaining Proteostasis
Cells employ a large proteostasis network to maintain proteome stability and functionality during non-stress and stress conditions. This network consists of numerous different chaperones, folding catalysts and proteolytic components (37, 71). Under stress conditions, the most important task is to sequester unfolding proteins, reducing the amount of aggregation-sensitive folding intermediates, and preventing the accumulation of protein aggregates. This is the job of molecular chaperones, which harbor binding sites for unfolding proteins, and prevent aggregate formation. Several different families of chaperones, including members of the HtpG (Hsp90), GroEL (Hsp60), DnaK (Hsp70), DnaJ (Hsp40) and IbpA/B (sHsps) family contribute to maintaining proteostasis during stress conditions (72–74). The majority of these proteins are under heat shock control, and are among the first responders when protein unfolding intermediates accumulate. The Hsp90, Hsp70 and Hsp60 chaperones are ―foldases‖: typically energy-dependent proteins that use ATP binding and hydrolysis to support refolding of damaged proteins once stress conditions subside (75). The sHsp “holdases” are energy-independent chaperones that prevent aggregation without supporting refolding (76, 77). The second arm of the proteostasis network during stress consists of ATP-dependent proteases, which degrade proteins that are irreversibly damaged and/or unable to refold. The mechanism(s) by which proteins are triaged between these two fates is still subject of very active investigations (78, 79).
Transcriptional and translational processes take time, involve numerous oxidation-sensitive proteins and require ATP. Yet sudden exposure to highly proteotoxic oxidants like HOCl or hydroxyl radicals require an immediate response, transiently inactivate transcription and translation processes, and substantially reduce cellular ATP levels (80, 81). That oxidative stress reduces cellular ATP levels has been observed in every organisms studied. This phenomenon was originally thought to be due to the oxidative inactivation of glyceraldehyde dehydrogenase (GAPDH), a central enzyme of glycolysis.
GAPDH has long been known for its exquisite sensitivity to RO/CS, which cause rapid modification of its active site cysteine (reviewed in (82)). This block in glycolysis together with the oxidative modification of other ATP-generating systems was thought to cause the very rapid decline in ATP levels (83, 84). While this might still be the case in eukaryotic cells, recent studies in E. coli revealed that the observed ATP-decline in HOCl-stressed bacteria is due to the active re-routing of ATP into long chains of inorganic polyphosphates (see below) (85).
There is little doubt that decreasing cellular ATP levels is a clever strategy to protect the most oxidation sensitive targets, such as newly synthesized proteins (75), against oxidative damage. However, loss of ATP-equivalents also constitutes a major problem, particularly for maintaining proteostasis, since most chaperones and proteases rely on ATP for their function. Cells appear to deal with this dilemma by employing stress-specific, ATP-independent holdase chaperones. These chaperones, which are mostly inactive under non-stress conditions, are able to directly sense protein-unfolding oxidants, and rapidly respond with an increase in their chaperone function.
In this review, we will focus exclusively on the chaperone arm of the proteostasis network, since regulation of the proteasomal system and its role during oxidative stress has been expertly reviewed in the very recent past (78, 79, 86). We will discuss several groups of redox-regulated chaperones; first the prototypical prokaryotic 33 kDa heat shock protein (Hsp33), then a number of multifunctional cellular components that gain chaperone activity under oxidative stress conditions: i) the eukaryotic ATPase Get3/TRC40; ii) the bacteria enamine/imine deaminase RidA; iii) the mammalian protein α2-macroglobulin; and iv) the prebiotic inorganic polymer polyphosphate (polyP).
Whereas both Hsp33 and Get3 sense oxidants via highly sensitive cysteine residues, whose oxidation status directly affects their ability to interact with unfolded client proteins, RidA and α2-macroglobulin appear to be activated by non-cysteine side chain modifications. We will conclude with polyP, whose production seems to be largely responsible for the drop in cellular ATP levels upon HOCl-stress, and which acts like a holdase chaperone in bacteria, preventing protein aggregation.
Hsp33 - The Inaugural Member of the Redox-Regulated Chaperone Family
Hsp33 (gene name hslO) was first identified in 1993, when Fred Blattner revisited the σ32-controlled heat-shock response in E. coli (87). Transcriptional analyses confirmed that Hsp33 is expressed under non-stress conditions but that its expression is dramatically increased when bacteria experience stress conditions that induce protein unfolding conditions (87, 88). What made the protein sequence of Hsp33 unique among known heat shock-proteins at that time was the presence of four absolutely conserved cysteine residues, located in the C-terminus of the protein. These four cysteine residues were suggestive of metal binding, and instigated our investigations into the function of this highly conserved protein (89).
Extensive studies over the past 16 years revealed that Hsp33 indeed uses these four cysteines as high affinity zinc ligands. However, instead of serving a purely structural role, we found that the four cysteines function as posttranslational redox switch, effectively translating changes in ROS levels into structural and functional changes in Hsp33. While inactive as chaperone when reduced and zinc coordinated, Hsp33 converts into a highly effective chaperone holdase when disulfide bonded and zinc-free (63, 75). Activators of Hsp33’s chaperone function are reactive chlorine species (e.g. HOCl) (63) or combinations of peroxide stress and unfolding conditions, induced by either elevated temperatures (89–91) or as recently noted by bile salts (92). Bacteria might encounter any of one of these stress conditions during the innate immune response (see earlier sections), on their way through the gastrointestinal tract (92), or during inflammation and fever-episodes (93). All of these conditions cause oxidative protein unfolding, and lead to the aggregation of many essential proteins in the absence of Hsp33 in vivo (63, 92, 94). Not surprisingly, bacteria lacking Hsp33 are significantly more sensitive towards these stress conditions (92, 94). Since Hsp33 is highly conserved among prokaryotes and unicellular parasites such as Leishmania but absent from higher eukaryotes, it is a potentially promising drug target to diminish the virulence of pathogenic bacteria such as E. coli or Vibrio cholerae. However, virulence studies have yet to be conducted to test this idea.
The Activation and Inactivation Mechanism of Hsp33
Probably one of the most intriguing questions regarding Hsp33 concerned its mechanism of activation; how can a protein be activated under protein unfolding conditions, which are detrimental for a host of different proteins, including several chaperones (63, 75)? To our surprise, we found that upon exposure to oxidative protein unfolding conditions, Hsp33 also oxidatively unfolds. Yet instead of losing activity and aggregating, Hsp33 uses these massive structural rearrangements to specifically activate its chaperone function (63, 89, 92). This was a truly unexpected discovery, which immediately raised the next question as to how this would work from a mechanistic point of view. Reduced, chaperone-inactive Hsp33 consists of a compactly folded N-terminal domain, a highly flexible ~40 aa-long linker region, and a C-terminal redox-sensing domain, containing the Cys232- X- Cys234 – X27–32 - Cys265 – XX - Cys268 redox switch motif (95). The four highly conserved cysteines coordinate one zinc ion with high affinity in a tetrahedral arrangement (96). The Zn2+ ion stabilizes the cysteine thiols in their highly reactive thiolate anion form (96–98), bringing the cysteines into close proximity for disulfide bond formation to occur, and contributing significantly to the thermodynamic stability of the zinc binding domain. Once exposed to the appropriate oxidative stress conditions, the four cysteines engage in two disulfide bonds, zinc is released and the zinc-binding domain unfolds. What is truly crucial for the activation of Hsp33, however, is the unfolding of the central linker region (99), which connects Hsp33’s N-terminus with the C-terminal redox switch domain. Not particularly conserved but highly charged, this linker region is stably folded under reducing, non-stress conditions yet rapidly unfolds upon the formation of both disulfide bonds. Destabilization of the linker region by site-specific mutagenesis successfully uncouples linker unfolding from disulfide bond formation and zinc release, and yields a constitutively active yet fully reduced and zinc-coordinated Hsp33 (99). These results provided strong evidence that the redox switch domain serves primarily as a rheostat, whose function is to control the stability of the linker region via its own thiol/disulfide status.
As previously mentioned, Hsp33 is only activated by slow-acting oxidants, such as H2O2, when combined with protein unfolding conditions. This mechanism seems to assure that Hsp33 is only activated when proteins unfold under oxidative stress conditions. Peroxide treatment alone is capable of oxidizing the two distal cysteine residues (Cys232 & 234) in Hsp33, but not the two proximal cysteines (Cys265 & 268). This oxidation causes a loss in zinc binding, which in turn destabilizes the linker region. However, the destabilization is not sufficient to trigger linker unfolding under otherwise non-stress conditions (90, 95). Upon exposure to unfolding conditions, such as elevated temperatures or bile salts, however, the linker region unfolds, causing activation of chaperone function. In addition, formation of the second disulfide bond (Cys232-Cys234) is now facilitated, which locks the linker region in the unfolded state and maintains the chaperone activity of Hsp33 until reducing non-stress conditions are restored. Fast-acting oxidants like HOCl gain access to all four cysteine residues and thus cause very rapid oxidative unfolding and the activation of Hsp33’s chaperone function (63, 92). Subsequent dimerization and potentially even higher oligomer formation of oxidized Hsp33 monomers follows (91), potentially increasing the client binding interface (99, 100).
In reverse of the activation of Hsp33, which requires both oxidizing and unfolding conditions, Hsp33’s inactivation requires both reducing and refolding conditions (77). Reducing conditions convert oxidized Hsp33 dimers into reduced Hsp33 dimers, which remain bound to their client proteins. This mechanism likely assures that Hsp33 does not release its client proteins until ATP levels are restored and fully functional ATP-dependent foldase systems such as the DnaK/DnaJ/GrpE-chaperone system are available (77, 100). Once reducing and non-stress conditions are restored, Hsp33 transfers its client proteins to the DnaK/DnaJ/GrpE-chaperone system, which supports their refolding to the native state. The precise mechanism of client release and the role of the DnaK//DnaJ/GrpE-chaperone system during this release process still remain enigmatic.
Hsp33 – A Conditionally Disordered Chaperone
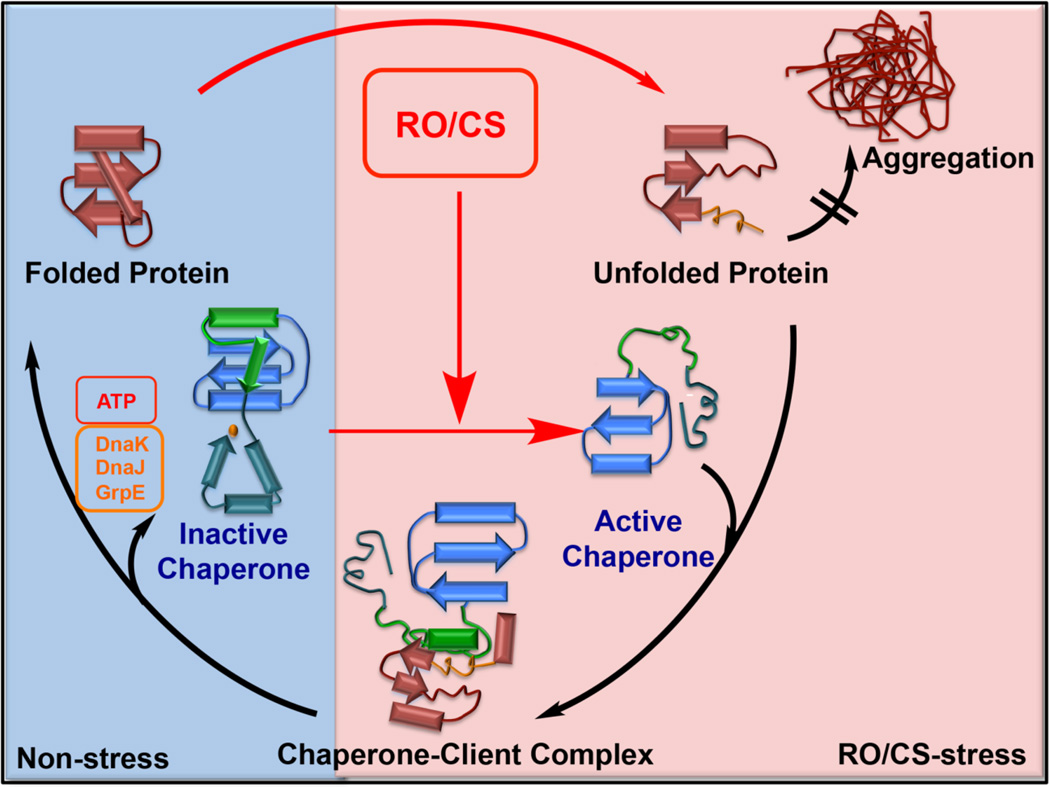
Hsp33 undergoes substantial unfolding during its activation process. This mechanism made Hsp33 the founding member of a new category of so-called conditionally disordered chaperones (100–102). Over the past few years, several other chaperones joined this category, protecting organisms against high heat (e.g., Hsp26) (103–105) or very low pH (e.g., HdeA) (106–108). What these proteins have in common is that they are fully folded and chaperone-inactive under non-stress conditions. During exposure to these rapidly proteotoxic stress conditions, they adopt a partially disordered conformation, which appears to be essential for their ability to bind protein folding intermediates and protect them against aggregation (101) (Figure 3). These results of course raised the intriguing question as to the role that the intrinsically disordered regions play in chaperone function. It appears that the disordered structure contributes to the increased plasticity of client binding (100), and promotes promiscuity (101, 104, 107). Moreover, H/D exchange studies combined with mass spectrometry provided the first evidence that Hsp33’s linker region might be in fact directly involved in client-chaperone interactions. The results of this study revealed that Hsp33 linker region regains stability upon client binding, suggesting that Hsp33 uses partially unfolding client proteins as scaffold to refold its linker region and thereby increase complex stability (100, 101) (Figure 3). Future structural studies are needed to uncover further details about the Hsp33-client interactions and define the extent to which intrinsic disorder is directly involved in the interaction with client proteins.
Figure 3. Redox-mediated activation of conditionally disordered chaperones.
RO/CS can cause substantial structural changes in proteins, leading to protein unfolding and aggregation. To sequester unfolding intermediates and prevent accumulation of toxic protein aggregates, bacteria employ the stress-specific molecular chaperone Hsp33. Under non-stress conditions, Hsp33 is well-folded and chaperone-inactive. Its four conserved cysteine residues are reduced and involved in high affinity binding of zinc. Upon exposure to oxidative protein unfolding conditions, Hsp33 undergoes oxidative disulfide bond formation, zinc release and massive structural rearrangements, including significant protein unfolding. In this partially intrinsically disordered conformation, Hsp33 is chaperone-active and able to interact with partially unfolded protein intermediates. Upon return to non-stress conditions, the disulfide bonds are reduced and the client proteins are transferred to the DnaK/DnaJ/GrpE system, which uses ATP to refold the client proteins. Hsp33 is specific for prokaryotes and unicellular eukaryotes. Very recently, Get3 has been identified to serve as the likely functional analogue of Hsp33 in yeast.
Get3 – A Redox-Regulated Dual-Function Protein in Eukaryotes
Hsp33, while highly conserved in prokaryotes, is absent from higher eukaryotes. This raised the obvious question as to how eukaryotic cells defend themselves against oxidative protein unfolding. One group of proteins that might be involved in this process are 2-Cys peroxiredoxins, which have been shown to gain chaperone activity upon peroxide-mediated overoxidation of their active site cysteine (for a more detailed overview see (109–111)). Recent studies however suggest that overoxidation is not essential and that other triggers, including high temperature and low pH (112, 113) might activate the chaperone function of peroxiredoxin as well. This makes peroxiredoxin less of a specialized and more of a general ATP-independent chaperone, which protects cells against a variety of different stress conditions, including oxidative stress. The other candidate, Get3, appears more Hsp33-like in its properties as a redox-regulated chaperone, and has been recently shown to protect eukaryotic cells against oxidative protein unfolding (114). On the surface, Get3 and Hsp33 have little in common. Reduced Hsp33 is a monomeric, non-ATP binding protein, which coordinates zinc using four highly conserved cysteines. Reduced Get3 is a dimeric ATPase, involved in binding and targeting tail-anchored (TA) proteins to the ER membrane. It too binds zinc, but zinc binding involves a Cys-XX-Cys pair from both monomers, stabilizing the reduced Get3 dimer (114). Upon closer inspection, however, the two proteins do have some intriguing similarities; like Hsp33, the yeast protein Get3 i) contains a Cys-X-Cys- X40–50-Cys-X-X- Cys motif; ii) protects unfolding proteins against aggregation in vitro when purified under aerobic conditions (115); iii) shows oxidation sensitivity in vivo (114, 115), and most importantly, iv) leads to an oxidative stress sensitive phenotype when deleted in yeast (114). Growth studies of a get3-deficient yeast strain under various stress conditions identified two of the four conserved cysteine residues (Cys285/Cys288) as essential for complementation of the growth defects (116).
We therefore decided to investigate the potential chaperone activity of Get3 in more detail. Indeed, we found that upon exposure to hydroxyl radicals, whose in vitro production can be elicited via the Fenton reaction using a combination of Cu2+ and peroxide (114), Get3 forms two disulfide bonds and releases Zn2+ (114). These redox changes trigger massive structural rearrangements, causing the inactivation of Get3’s ATPase activity and the formation of tetrameric and higher oligomeric species with high, ATP-independent chaperone holdase activity (114). These results are fully consistent with previously reported co-localization studies of Get3, which showed that Get3 associates with both unfolding proteins and other chaperones under ATP-depleted conditions in vivo (115). Importantly, all of the conformational and functional changes in Get3 are fully reversible, requiring both reducing conditions and the presence of ATP (114).
What makes the Get3 case particularly fascinating is the fact that Get3 has an apparently completely different function in its reduced state. In fact, reduced Get3 was originally identified as the central member of the GET (Guided Entry of Tail-Anchored proteins) pathway. In this role, Get3 shuttles tail-anchored (TA-proteins from the ribosome to the ER membrane using ATP binding and hydrolysis to modulate client binding and release (117, 118). Site directed mutagenesis studies revealed that the chaperone function of oxidized Get3 is independent of, and potentially mutually exclusive with the membrane targeting function of reduced Get3 (114). Phenotypic studies using a mutant Get3 variant, which was no longer capable of sorting TA proteins but had wild-type-like redox-regulated chaperone activity, revealed that the chaperone function and not the sorting function of Get3 is responsible for the growth deficit that is observed in get3-deficient cells during oxidative stress (114). These findings are consistent with previous reports about a potential dual function in the Get3 homologue TRC40 in Caenorhabditis elegans. Deletion of TRC40 was found to cause severe growth deficits and increased sensitivity towards cisplatin (119), an anticancer drug thought to cause oxidative stress in vivo (120). Importantly, complementation studies using either wild-type TRC40 or a mutant variant lacking two of the four cysteines revealed that both proteins rescued the growth defect of a TRC40 deletion strain. However, only wild-type TRC40 was able to complement for the observed cisplatin sensitivity. These results strongly suggest that Get3’s redox-regulated chaperone functions plays a crucial role in oxidative stress protection in higher eukaryotes as well (119). These are exciting findings but many questions remain open: How does Get3 recognize its client proteins? What fates do client proteins have after the stress conditions subside? What is the role of the other GET pathway components, and might they possibly be involved in client release, refolding, and/or restoration of proteostasis after oxidative stress? Time will tell how Get3 (TRC40) balances its two important functions in the cell.
Chaperone Activation by Non-Cysteine Oxidation
HOCl causes a number of other oxidative modifications in proteins, including N-chlorination, methionine oxidation and dityrosine formation (Figure 1). These mechanisms have now also been found to play a regulatory role in the activation of chaperone proteins during oxidative stress.
E. coli RidA - Activation by reversible N-chlorination
N-chlorination of proteins occurs during severe HOCl stress. It most likely affects the ε-amino groups of lysine residues, the guanidinium groups of arginines, and the terminal amino groups of polypeptides. Chloramine formation is known to have detrimental effects on molecular level, including protein inactivation, unfolding, and aggregation. In addition, protein fragmentation due to halogen transfer as well as cross-linking events have been reported in response to treatment with chloramines (35, 121–123). Intriguingly, the E. coli protein RidA was recently identified to specifically sense RCS via reversible N-chlorination, using these modification to turn into a chaperone-active state (124).
RidA is member of the functionally diverse YjgF/ER057c/DUK114 family of proteins, which is highly conserved in all domains of life (124). In Enterobacteriaceae, such as E. coli and Salmonella enterica, RidA is an enamine/imine deaminase, which speeds up the IlvA-catalyzed deamination of threonine into 2-ketobutyrate (124, 125). Redox proteomic studies under nitrosative stress revealed that the only cysteine residue of RidA is oxidatively modified during peroxynitrite treatment, resulting in the loss of the stimulatory effect of RidA on the activity of IlvA (126).
Very surprisingly, however, in the presence of HOCl or other RCS such as monochloramine, RidA not only failed to stimulate but in fact strongly inhibited the activity of IlvA (124), indicating that peroxynitrite and HOCl treatment cause a markedly different outcome for RidA’s activity. The results furthermore suggested that HOCl-treated RidA might form a tight complex with IlvA, thereby inhibiting its catalytic activity. This led to the hypothesis that HOCl-treated RidA might function as a protein-binding holdase chaperone. In vitro aggregation studies using a variety of different client proteins, including unfolded IlvA, agreed with this conclusion and demonstrated that HOCl-treated RidA but not untreated RidA prevents protein aggregation under stress conditions (124). Notably, RidA appeared to specifically sense RCS, since no other tested oxidants, including peroxide or diamide had any effect on its chaperone function. Even more astounding was the finding that HOCl-treatment of the cysteine-free RidA variant had precisely the same chaperone-activating effect, excluding the possibility that modification of RidA’s sole cysteine is involved in the activation process. Instead, HOCl-treated chaperone-active RidA trimers were found to associate into higher oligomers with substantially decreased levels of free amino groups, and, based on MS analyses, up to seven different N-chlorination sites (124). These results led to the conclusion that chlorination of several lysine and/or arginine residues might contribute to the observed increase in hydrophobicity, a hallmark of binding sites in chaperones (127). Deletion of RidA increased E. coli’s sensitivity to HOCl and caused the accumulation of aggregated proteins, suggesting that RidA is a true member of the proteostasis network during oxidative stress. N-chlorination of RidA is fully reversible, and treatment with DTT, ascorbic acid or with the physiological redox systems Trx or GSH abolished RidA’s chaperone activity in vitro. RidA’s chaperone function appears to not be specific for E. coli since HOCl treatment of the Drosophila melanogaster homologue DUK114 also activates the chaperone function in vitro (albeit irreversibly) (124). One challenge remains to identify the residue(s) whose chlorination contributes to the chaperone function of RidA, and to define the precise place that RidA holds in the cellular proteostasis network.
α2-Macroglobulin – Activation by Methionine Oxidation/Dityrosine Formation
α2-macroglobulin is a very abundant glycoprotein in mammals, present within the highly oxidizing milieu of the blood plasma and extracellular space. The functions of α2-macroglobulin are quite diverse but it is probably best known for its ability to trap and inhibit a variety of different extracellular proteinases, irrespective of their mechanism or specificity (128). In addition, α2-macroglobulin interacts with a number of other biomolecules, including different hormones and cytokines. α2-macroglobulin has also long been known for its sensitivity towards HOCl (129). Earlier studies showed that HOCl-treatment causes the homo-tetrameric protein to dissociate into stable dimers, which no longer interact with proteinases but show increased affinity for other binding partners, including the LDL-receptor protein LRP (130). These results suggested that HOCl-mediated dissociation into dimers is not simply a random, non-specific inactivation process but might be part of a regulatory mechanism. Indeed, very recently Dobson and coworkers demonstrated that HOCl treated α2-macroglobulin dimers show substantially increased surface hydrophobicity and are highly active in preventing the aggregation of a range of different model clients in vitro (131). More specifically, HOCl-treated α2-macroglobulin was able to prevent the HOCl-mediated aggregation of several disease-associated proteins, including fibrinogen and LDL and showed increased activity towards preventing Aβ 1–42 fibril formation (131). This interaction was even more effective when Aβ 1–42 was treated with HOCl as well. Neurotoxicity assays suggested that these interactions are indeed physiologically relevant; compared to non-treated α2-macroglobulin, the researchers found that the efficacy of HOCl-treated α2-macroglobulin to protect neuroblastoma cells against Aβ1–42 toxicity was significantly improved. These results strongly suggest that α2-macroglobulin switches into a physiologically relevant chaperone upon exposure to HOCl stress.
The mechanism(s) by which α2-macroglobulin gets activated as a chaperone remain unclear so far. Since it is present in a very oxidizing environment, the cysteines are constitutively oxidized. It has been shown that HOCl-activated α2-macroglobulin undergoes methionine oxidation, tryptophan oxidation, and dityrosine formation (129, 131). However, which combination of oxidation events is necessary or sufficient to activate the chaperone function remains to be investigated. The observed oxidative modifications are thought to be irreversible (131). However, more controlled in vitro oxidation, and careful analysis of the oxidation and activity state of α2-macroglobulin in vivo might be necessary to ultimately answer this question.
Polyphosphate: A Protein-Like Inorganic Chaperone
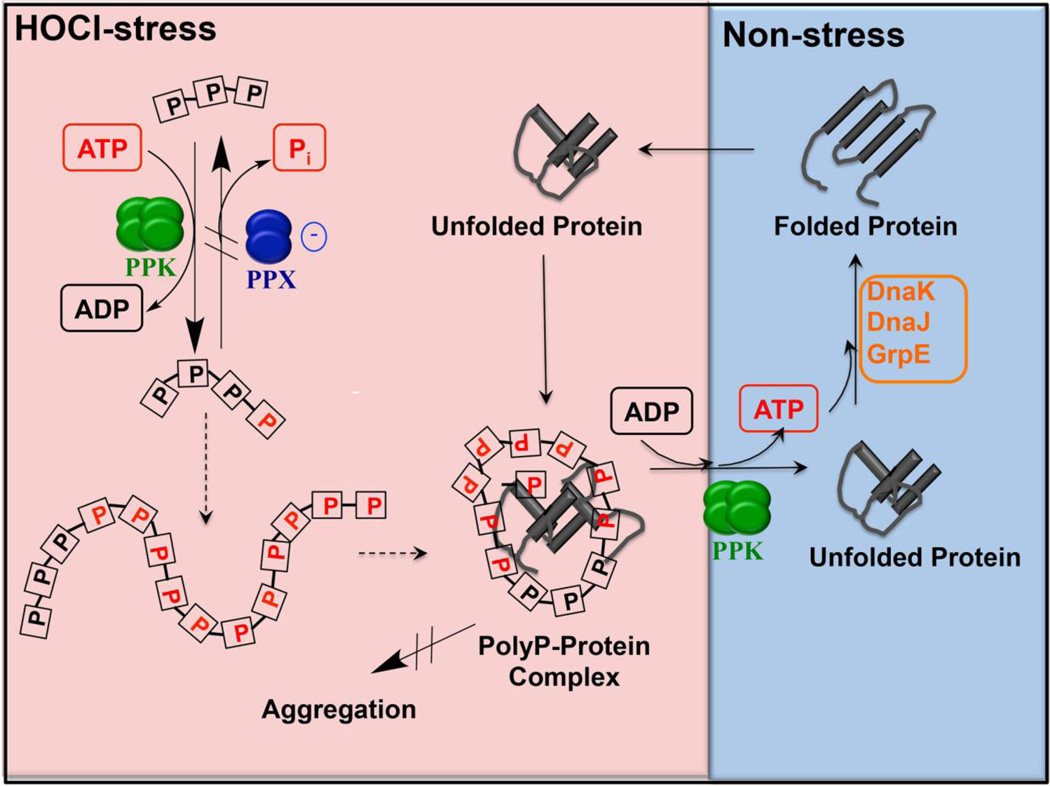
Polyphosphates (polyP) are prebiotic polymers (132): highly conserved, universal and structurally extremely simple. They exist as long, typically unbranched chains of phosphoanhydride bond-linked phosphates, which can reach lengths of up to 1000 Pi units (133). More than 20 years ago, the late Arthur Kornberg and coworkers showed that polyP-deficient bacterial cells suffer from a number of different phenotypes, including increased sensitivity towards multiple stressors such as heat shock, heavy metal exposure, peroxide, and starvation (134). The molecular reason for this effect was unknown, but speculations considered the fact that PolyP’s energy status is equivalent to ATP and thus might serve as a suitable storage compound for phosphate and energy during stress conditions (133). Alternatively, it was proposed that polyP might be involved in the regulation of the σ38-dependent general stress response system of E. coli (135, 136), which controls the transcription of various genes related to ROS resistance, including katE (encoding catalase), sodC (encoding superoxide dismutase), and the polyphosphate kinase (PPK) encoding gene ppk itself (137). In addition to increased sensitivity towards ROS, polyP-deficient prokaryotic cells display also defects in biofilm formation, virulence, and motility (133, 138). In higher eukaryotes, polyP affects blood clotting, and is involved in apoptosis, mTOR activation and neuronal signaling (139–141). It remained fascinating how a molecule that is so structurally simple can be involved in all these seemingly unrelated functions in diverse organisms. Bacterial polyphosphate kinases (PPK) reversibly catalyze the generation of polyP directly from ATP, whereas exopolyphosphatases (PPX) can degrade polyP into Pi molecules (Figure 4) (85). Numerous independent studies have reported that deletion of the ppk gene in many species of bacteria leads to increased sensitivity towards ROS (85, 133, 142, 143), and we very recently reviewed the mechanistic details of several pathways by which polyP protects bacteria from oxidative stress in a direct or indirect fashion (144). A role for polyP in protein homeostasis under severe oxidative stress conditions, however, has only very recently been deciphered (85). Measurement of ATP and polyP levels in E. coli cells lacking either PPK or PPX in comparison to wild-type cells revealed that 50% of the cellular ATP is converted into long polyP chains under severe HOCl stress (Figure 4). Investigation of ppk-deficient strains of E. coli and V. cholerae revealed that the absence of polyP results in much higher sensitivity towards HOCl, suggesting that polyP has an important function in bacterial HOCl resistance (85). However, the question arose of how polyP protects cells against HOCl. We found the answer in our observation that ppk-deficient strains accumulate large amounts of protein aggregates in vivo (85). Consistently, the expression of heat shock response genes was upregulated under HOCl stress in the absence of polyP, indicating the need for molecular chaperones to combat protein unfolding and suggesting that polyP might work as a physiologically relevant chaperone that can functionally replace proteinaceous molecular chaperones when present in sufficient amounts (85). In vitro and in vivo studies revealed that polyP prevents aggregation of a variety of denatured client proteins in a concentration- and polyP chain length-dependent manner and thus acts as a highly effective chemical chaperone (Figure 4). PolyP binds unfolded proteins, prevents their aggregation, and finally releases them to ATP-dependent foldase chaperones like the DnaK/DnaJ/GrpE system once non-stress conditions are restored (85). The demand for polyP under severe HOCl stress is directly regulated by the oxidant itself via transient oxidative inactivation of PPX (Figure 4) (85). PolyP synthesis is an elegant way for cells to prevent protein aggregation during severe oxidative stress, and an intriguing new component of the proteostasis machinery for the following reasons: polyP (i) does not require ATP for its chaperone function, (ii) is impervious to oxidative damage, (iii) works during ATP depletion, (iv) does not require time- consuming transcription/translation processes, and (v) can be reconverted into ATP by PPK, which can then be used by ATP-dependent foldases to promote protein refolding. However, several open questions remain, particularly concerning the exact mechanism by which polyP binds unfolding client proteins to prevent their aggregation. Since the chain-length of polyP molecules strongly influenced the chaperone effect of polyP (85), it is likely that polyP acts as a stabilizing scaffold. Yet how polyP distinguishes between native and unfolded proteins is still not understood.
Figure 4. Model of polyP’s chaperone function.
Treatment of bacteria with HOCl leads to the conversion of cellular ATP into long chains of polyphosphate (polyP) catalyzed by the conserved enzyme polyphosphate kinase (PPK). PolyP accumulation in the cell is a consequence of the reversible oxidative inactivation of the polyP-degrading enzyme polyP phosphatase (PPX), which contains an oxidation-sensitive cysteine in its polyP-binding site. PolyP accumulation results in a significant depletion of the cellular ATP level, affecting most ATP-dependent cellular processes, including ATP-depending chaperones, such as the DnaK/DnaJ/GrpE system. PolyP is able to compensate for the lack of ATP-dependent chaperones by serving as a scaffold that binds to unfolding proteins, preventing them from aggregation, and keeping them soluble and refolding competent. Once reducing conditions are restored, polyP is reconverted to ATP and client proteins are released. Reactivated ATP-dependent foldases then support the refolding of these client proteins.
In addition to the newly-discovered chaperone function of polyP, it has been known for some time that polyP plays a role in regulating proteolysis in bacteria. PolyP retargets the Lon protease of E. coli to degrade distinct substrate proteins, including ribosomal proteins (145) and the anti-toxin modules of toxin-antitoxin systems (146). This is thought to be important for the survival of amino acid starvation and accumulation of stress-tolerant persister cells, respectively (147, 148). What remains unknown, however, is how cells balance polyP’s chaperone activity with its role in protein degradation, particularly under protein-damaging stress conditions. This question is an interesting future direction for studies of the role of polyP in proteostasis in bacteria.
Although polyP also exists in animals, where it - among other functions - stimulates proliferation (149), influences cytokinesis (150), and affects blood coagulation by enhancing fibrin polymerization (139, 151), the gene encoding the polyP-generating polyphosphate kinase has not yet been identified. Identification of the polyP-generating enzyme(s) will help fill the gaps in our current understanding of polyP’s role in mammals. Given the fact that many neurodegenerative diseases including Alzeimer’s diseases and Parkinson’s disease have been associated with oxidative stress and are caused by the accumulation of protein aggregates, understanding the synthesis and role of polyP in mammalian cells might lead to improved therapeutic strategies for a variety of diseases.
SUMMARY AND OUTLOOK
Controlled production of RO/CS has been shown to play an important role during the mammalian host defense (26, 27, 152). Uncontrolled accumulation of RO/CS, on the other hand, has been associated with a number of patho-physiological processes and diseases (15, 153). One common effect that most RO/CS exert on cells is their damage to the proteome. Over the recent years, a number of protein and non-protein chaperones have been identified, which are specifically activated during oxidative stress conditions to reduce redox stress-induced protein aggregation. Their stress-specific, posttranslational activation together with their ATP-independent chaperone function makes them ideally suited to prevent protein aggregation under stress conditions that are known to deplete the energy status of the cell. The chaperones differ primarily in their modes of activation. Hsp33 in bacteria and Get3 in eukaryotes use the oxidation status of some of their cysteine residues to become activated. RidA/DUK114, in contrast, require N-chlorination of their lysine- and/or arginine residues to turn into active chaperone-holdases, and α2-macroglubin, an extracellular chaperone, most likely uses methionine oxidation as trigger for its functional activation. Together with polyP, which is generated from ATP and in part responsible for the ATP-depletion at least in bacteria, these chaperones are capable of binding a variety of different unfolding client proteins and prevent their irreversible aggregation. Once non-stress conditions are restored, the chaperone-activating modifications are reduced by members of the Trx/Grx?GSH systems. At the same time, polyP is reconverted into ATP, which releases the client proteins while fueling ATP-dependent chaperone systems to promote their refolding (85). Obtaining a detailed view of the precise mechanisms of molecular chaperones under oxidative stress conditions and the processes they are affecting will shed light on our understanding of how molecular chaperones and the pathways they are affecting can be manipulated to alter their oxidative stress resistance. This will help us to design new specific drugs that target these players and potentially attenuate the resistance of pathogenic bacteria towards oxidative stress or help combat the oxidative protein damage associated with human diseases. It is also particularly intriguing that so many of the cellular components with redox-activated chaperone activity are multifunctional. It remains to be seen, however, what the significance of this pattern might be.
Highlights.
Proteome is the major target of oxidative stress in vivo
Proteostasis is maintained by specialized, redox-regulated chaperones
Hsp33 and Get3 are activated by oxidant-induced disulfide bond formation
RidA and α2-macroglubin are activated by N-chlorination and methionine oxidation
Polyphosphate builds up during oxidative stress and works as protein-like chaperone
Acknowledgements
This work was supported by Grants F32GM096613 and R01GM065318 from the National Institutes of Health. J.-U.D. is supported by a postdoctoral fellowship provided from the Deutsche Forschungsgemeinschaft DFG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dröse S, Brandt U. Mitochondial Oxidative Phosphorylation. Advances in Experimental Medicine and Biology. 2012:145–169. doi: 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- 2.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nature Reviews Microbiology. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 3.Nauseef WM. Biological Roles for the NOX Family NADPH Oxidases. The Journal of Biological Chemistry. 2008;283:16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabaldón T. Peroxisome diversity and evolution. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:765–773. doi: 10.1098/rstb.2009.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Autréaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nature Reviews Molecular Cell Biology. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 6.Ray PD, Huang B-W, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cellular Signalling. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winterbourn CC. Are free radicals involved in thiol-based redox signaling? Free Radical Biology and Medicine in press. 2014 doi: 10.1016/j.freeradbiomed.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Knoefler D, Leichert LI, Thamsen M, Cremers CM, Reichmann D, Gray MJ, Wholey WY, Jakob U. About the dangers, costs and benefits of living an aerobic lifestyle. Biochem. Soc. Trans. 2014;42:917–921. doi: 10.1042/BST20140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature Reviews Microbiology. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winterbourn CC. Hydrogen Peroxide and Cell Signalling, Part C. Methods in Enzymology. 2013:3–25. doi: 10.1016/B978-0-12-405881-1.00001-X. [DOI] [PubMed] [Google Scholar]
- 11.Masip L, Veeravalli K, Georgiou G. The Many Faces of Glutathione in Bacteria. Antioxidants & Redox Signaling. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 12.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free Radical Biology and Medicine. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uttara B, Singh A, Zamboni P, Mahajan R. Reactions of Myeloperoxidase-Derived Oxidants with Biological Substrates:Gaining Chemical Insight into Human Inflammatory Diseases. Current Medicinal Chemistry. 2009;16:4419–4444. doi: 10.2174/092986706778773095. [DOI] [PubMed] [Google Scholar]
- 14.Bartosz G. Reactive oxygen species: Destroyers or messengers? Biochemical Pharmacology. 2009;77:1303–1315. doi: 10.1016/j.bcp.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chemico-Biological Interactions. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Dröge W. Free Radicals in the Physiological Control of Cell Function. Physiological Reviews. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 17.Gray MJ, Wholey WY, Jakob U. Bacterial Responses to Reactive Chlorine Species. Annual Review of Microbiology. 2013;67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nature Reviews Molecular Cell Biology. 2013;14:617–629. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- 19.Winterbourn CC, Kettle AJ. Redox Reactions and Microbial Killing in the Neutrophil Phagosome. Antioxidants & Redox Signaling. 2013;18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 20.Nagl M, Hess MW, Pfaller K, Hengster P, Gottardi W. Bactericidal Activity of Micromolar N-Chlorotaurine: Evidence for Its Antimicrobial Function in the Human Defense System. Antimicrobial Agents and Chemotherapy. 2000;44:2507–2513. doi: 10.1128/aac.44.9.2507-2513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutala WA, Weber DJ. Uses of inorganic hypochlorite (bleach) in health-care facilities. Clinical Microbiology Reviews. 1997;10:597–610. doi: 10.1128/cmr.10.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss SJ, LoBuglio AF. Phagocyte-generated oxygen metabolites and cellullar injury. Lab Invest. 1982;47:5–18. [PubMed] [Google Scholar]
- 23.Prütz WA. Hypochlorous Acid Interactions with Thiols, Nucleotides, DNA, and Other Biological Substrates. Archives of Biochemistry and Biophysics. 1996;332:110–120. doi: 10.1006/abbi.1996.0322. [DOI] [PubMed] [Google Scholar]
- 24.Green JN, Kettle AJ, Winterbourn CC. Protein chlorination in neutrophil phagosomes and correlation with bacterial killing. Free Radical Biology and Medicine. 2014;77:49–56. doi: 10.1016/j.freeradbiomed.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Bae YS, Choi MK, Lee W-J. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends in Immunology. 2010;31:278–287. doi: 10.1016/j.it.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Hassani El RA, Benfares N, Caillou B, Talbot M, Sabourin J-C, Belotte V, Morand S, Gnidehou S, Agnandji D, Ohayon R, Kaniewski J, Noël-Hudson M-S, Bidart J-M, Schlumberger M, Virion A, Dupuy C. Dual oxidase2 is expressed all along the digestive tract. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2005;288:G933–G942. doi: 10.1152/ajpgi.00198.2004. [DOI] [PubMed] [Google Scholar]
- 27.Ha EM, Oh CT, Bae YS, Lee WJ. A Direct Role for Dual Oxidase in Drosophila Gut Immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 28.Kim S-H, Lee W-J. Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Frontiers in Cellular and Infection Microbiology. 2013;3:1–12. doi: 10.3389/fcimb.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grasberger H, Zaatari El M, Dang DT, Merchant JL. Dual Oxidases Control Release of Hydrogen Peroxide by the Gastric Epithelium to Prevent Helicobacter felis Infection and Inflammation in Mice. Gastroenterology. 2013;145:1045–1054. doi: 10.1053/j.gastro.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Lee K-A, Ha E-M, Lee KM, Seo YY, Choi HK, Kim HN, Kim MJ, Cho C-S, Lee SY, Lee W-J, Yoon J. A specific and sensitive method for detection of hypochlorous acid for the imaging of microbe-induced HOCl production. Chem Communications. 2011;47:4373–4375. doi: 10.1039/c1cc10589b. [DOI] [PubMed] [Google Scholar]
- 31.Heinecke JW. Mechanisms of oxidative damage by myeloperoxidase in atherosclerosis and other inflammatory disorders. Journal of Laboratory and Clinical Medicine. 1999;133:321–325. doi: 10.1016/s0022-2143(99)90061-6. [DOI] [PubMed] [Google Scholar]
- 32.Corcoran A, Cotter TG. Redox regulation of protein kinases. FEBS J. 2013;280:1944–1965. doi: 10.1111/febs.12224. [DOI] [PubMed] [Google Scholar]
- 33.Cai Z, Yan L-J. Protein Oxidative Modifications: Beneficial Roles in Disease and Health. J Biochem Pharmacol Res. 2013;1:15–26. [PMC free article] [PubMed] [Google Scholar]
- 34.Winterbourn CC. Free-radical production and oxidative reactions of hemoglobin. Environmental Health Perspectives. 1985;64:321–330. doi: 10.1289/ehp.8564321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawkins CL, Davies MJ. Hypochlorite-induced damage to proteins: formation of nitrogen-centred radicals from lysine residues and their role in protein fragmentation. Biochem.J. 1998;332:617–625. doi: 10.1042/bj3320617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trougakos IP, Sesti F, Tsakiri E, Gorgoulis VG. Non-enzymatic post-translational protein modifications and proteostasis network deregulation in carcinogenesis. Journal of Proteomics. 2013;92:274–298. doi: 10.1016/j.jprot.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Storz G, Imlay JA. Oxidative stress. Current Opinion in Microbiology. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 39.Pattison DI, Davies MJ. Absolute Rate Constants for the Reaction of Hypochlorous Acid with Protein Side Chains and Peptide Bonds. Chem. Res. Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 40.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O’Neill JS, Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, Lunardi J. Proteomics Analysis of Cellular Response to Oxidative Stress: Evidence for in vivo overoxidation of peroxiredoxins at their active site. The Journal of Biological Chemistry. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 42.Moon JC, Kim GM, Kim E-K, Lee HN, Ha B, Lee SY, Jang HH. Reversal of 2-Cys peroxiredoxin oligomerization by sulfiredoxin. Biochemical and Biophysical Research Communications. 2013;432:291–295. doi: 10.1016/j.bbrc.2013.01.114. [DOI] [PubMed] [Google Scholar]
- 43.Rhee SG, Jeong W, Chang T-S, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int. 2007;72:S3–S8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 44.Berndt C, Lillig CH, Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2008;1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Holmgren A, Johansson C, Berndt C, Lönn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem. Soc. Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 46.Holmgren A, Aslund F. Methods in Enzymology. 1995:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 47.Kuntz AN, Davioud-Charvet E, Sayed AA, Califf LL, Dessolin J, Amer ESJ, Williams DL. Thioredoxin Glutathione Reductase from Schistosoma mansoni: An Essential Parasite Enzyme and a Key Drug Target. PLoS Medicine. 2007:4. doi: 10.1371/journal.pmed.0040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radical Biology and Medicine. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 49.Schöneich C. Methionine oxidation by reactive oxygen species: reaction mechanisms and relevance to Alzheimer’s disease. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2005;1703:111–119. doi: 10.1016/j.bbapap.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Storkey C, Davies MJ, Pattison DI. Reevaluation of the rate constants for the reaction of hypochlorous acid (HOCl) with cysteine, methionine, and peptide derivatives using a new competition kinetic approach. Free Radical Biology and Medicine. 2014;73:60–66. doi: 10.1016/j.freeradbiomed.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 51.Boschi-Muller S, Branlant G. Methionine sulfoxide reductase: Chemistry, substrate binding, recycling process and oxidase activity. Bioorganic Chemistry. 2014;57:222–230. doi: 10.1016/j.bioorg.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Drazic A, Winter J. The physiological role of reversible methionine oxidation. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2014;1844:1367–1382. doi: 10.1016/j.bbapap.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Kim G, Weiss SJ, Levine RL. Methionine oxidation and reduction in proteins. Biochimica et Biophysica Acta (BBA) - General Subjects. 2014;1840:901–905. doi: 10.1016/j.bbagen.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarrago L, Kaya A, Weerapana E, Marino SM, Gladyshev VN. Methionine Sulfoxide Reductases Preferentially Reduce Unfolded Oxidized Proteins and Protect Cells from Oxidative Protein Unfolding. The Journal of Biological Chemistry. 2012;287:24448–24459. doi: 10.1074/jbc.M112.374520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brennan LA, Lee W, Giblin FJ, David LL, Kantorow M. Methionine sulfoxide reductase A (MsrA) restores α-crystallin chaperone activity lost upon methionine oxidation. Biochimica et Biophysica Acta (BBA) - General Subjects. 2009;1790:1665–1672. doi: 10.1016/j.bbagen.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benoit SL, Bayyareddy K, Mahawar M, Sharp JS, Maier RJ. Alkyl Hydroperoxide Reductase Repair by Helicobacter pylori Methionine Sulfoxide Reductase. Journal of Bacteriology. 2013;195:5396–5401. doi: 10.1128/JB.01001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khor HK, Fisher MT, Schoneich C. Potential Role of Methionine Sulfoxide in the Inactivation of the Chaperone GroEL by Hypochlorous Acid (HOCl) and Peroxynitrite (ONOO-) The Journal of Biological Chemistry. 2004;279:19486–19493. doi: 10.1074/jbc.M310045200. [DOI] [PubMed] [Google Scholar]
- 58.Mahawar M, Tran V, Sharp JS, Maier RJ. Synergistic Roles of Helicobacter pylori Methionine Sulfoxide Reductase and GroEL in Repairing Oxidant-damaged Catalase. The Journal of Biological Chemistry. 2011;286:19159–19169. doi: 10.1074/jbc.M111.223677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hentchel KL, Escalante-Semerena JC. In Salmonella enterica, the Gcn5-Related Acetyltransferase MddA (Formerly YncA) Acetylates Methionine Sulfoximine and Methionine Sulfone, Blocking Their Toxic Effects. Journal of Bacteriology. 2014;197:314–325. doi: 10.1128/JB.02311-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pattison D, Davies M. Reactions of Myeloperoxidase-Derived Oxidants with Biological Substrates:Gaining Chemical Insight into Human Inflammatory Diseases. Current Medicinal Chemistry. 2006;13:3271–3290. doi: 10.2174/092986706778773095. [DOI] [PubMed] [Google Scholar]
- 61.Tanner JJ, Parsons ZD, Cummings AH, Zhou H, Gates KS. Redox Regulation of Protein Tyrosine Phosphatases: Structural and Chemical Aspects. Antioxidants & Redox Signaling. 2011;15:77–97. doi: 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- 62.Truong TH, Carroll KS. Redox regulation of protein kinases. Critical Reviews in Biochemistry and Molecular Biology. 2013;48:332–356. doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winter J, Ilbert M, Graf PCF, Özcelik D, Jakob U. Bleach Activates a Redox-Regulated Chaperone by Oxidative Protein Unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dukan S, Dadon S, Smulski DR, Belin S. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Applied and Environmental Microbiology. 1996;62:4003–4008. doi: 10.1128/aem.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Straus D, Walter W, Gross CA. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes & Development. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 66.Elsholz AKW, Hempel K, Pöther D-C, Becher D, Hecker M, Gerth U. CtsR inactivation during thiol-specific stress in low GC, Gram+ bacteria. Molecular Microbiology. 2011;79:772–785. doi: 10.1111/j.1365-2958.2010.07489.x. [DOI] [PubMed] [Google Scholar]
- 67.Runde S, Molière N, Heinz A, Maisonneuve E, Janczikowski A, Elsholz AKW, Gerth U, Hecker M, Turgay K. The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis. Molecular Microbiology. 2014;91:1036–1052. doi: 10.1111/mmi.12521. [DOI] [PubMed] [Google Scholar]
- 68.Müller A, Hoffmann JH, Meyer HE, Narberhaus F, Jakob U, Leichert LI. Nonnative Disulfide Bond Formation Activates the σ32-dependent Heat Shock Response in Escherichia coli. Journal of Bacteriology. 2013;195:2807–2816. doi: 10.1128/JB.00127-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumsta C, Thamsen M, Jakob U. Effects of Oxidative Stress on Behavior, Physiology, and the Redox Thiol Proteome of Caenorhabditis elegans. Antioxidants & Redox Signaling. 2011;14:1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu G, Chance MR. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chemical Reviews. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 71.Roth DM, Balch WE. Modeling general proteostasis: proteome balance in health and disease. Current Opinion in Cell Biology. 2011;23:126–134. doi: 10.1016/j.ceb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bukau B, Weissman J, Horwich A. Molecular Chaperones and Protein Quality Control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 73.Liberek K, Lewandowska A, Ziętkiewicz S. Chaperones in control of protein disaggregation. The EMBO Journal. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nature Reviews Molecular Cell Biology. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winter J, Linke K, Jatzek A, Jakob U. Severe Oxidative Stress Causes Inactivation of DnaK and Activation of the Redox-Regulated Chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 76.Beissinger M, Buchner J. How chaperones fold proteins. The Journal of Biological Chemistry. 1998;379:245–259. [PubMed] [Google Scholar]
- 77.Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. The EMBO Journal. 2003;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jung T, Grune T. The proteasome and the degradation of oxidized proteins: Part I—structure of proteasomes. Redox Biology. 2013;1:178–182. doi: 10.1016/j.redox.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jung T, Höhn A, Grune T. The proteasome and the degradation of oxidized proteins: Part II - protein oxidation and proteasomal degradation. Redox Biology. 2014;2:99–104. doi: 10.1016/j.redox.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colussi C, Albertini MC, Coppola S, Rovidati S, Galli F, Ghibelli L. H2O2-induced block of glycolysis as an active ADP-ribosylation reaction protecting cells from apoptosis. The FASEB Journal. 2000;14:2266–2276. doi: 10.1096/fj.00-0074com. [DOI] [PubMed] [Google Scholar]
- 81.Spragg RG, Hinshaw DB, Hyslop PA, Schraufstätter IU, Cochrane CG. Alterations in adenosine triphosphate and energy charge in cultured endothelial and P388D1 cells after oxidant injury. J. Clin. Invest. 1985;76:1471–1476. doi: 10.1172/JCI112126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brandes N, Schmitt S, Jakob U. Thiol-Based Redox Switches in Eukaryotic Proteins. Antioxidants & Redox Signaling. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cotgreave IA, Gerdes R, Schuppe-Koisstinen I, Lind C. S-glutathionylation of glyceraldehyde-3-phosphate dehydrogenase: role of thiol oxidation and catalysis by glutaredoxin. Methods in Enzymology. 2002;348:175–182. doi: 10.1016/s0076-6879(02)48636-3. [DOI] [PubMed] [Google Scholar]
- 84.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Reactive Oxygen Species Special Feature: Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JCA, Jakob U. Polyphosphate Is a Primordial Chaperone. Mol Cell. 2014;53:689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Demasi M, Simões V, Bonatto D. Cross-talk between redox regulation and the ubiquitin–proteasome system in mammalian cell differentiation. Biochimica et Biophysica Acta (BBA) - General Subjects in press. 2014 doi: 10.1016/j.bbagen.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 87.Chuang SE, Blattner FR. Characterization of twenty-six new heat shock genes of Escherichia coli. Journal of Bacteriology. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang H-J, Heo D-H, Choi S-W, Kim K-N, Shim J, Kim C-W, Sung H-C, Yun C-W. Functional characterization of Hsp33 protein from Bacillus psychrosaccharolyticus; additional function of HSP33 on resistance to solvent stress. Biochemical and Biophysical Research Communications. 2007;358:743–750. doi: 10.1016/j.bbrc.2007.04.184. [DOI] [PubMed] [Google Scholar]
- 89.Jakob U, Muse W, Eser M, Bardwell JCA. Chaperone Activity with a Redox Switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 90.Ilbert M, Horst J, Ahrens S, Winter J, Graf PCF, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Graumann J, Lilie H, Tang X, Tucker KA, Hoffmann JH, Vijayalakshmi J, Saper M, Bardwell JCA, Jakob U. Activation of the Redox-Regulated Molecular Chaperone Hsp33—A Two-Step Mechanism. Structure. 2001;9:377–387. doi: 10.1016/s0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 92.Cremers CM, Knoefler D, Vitvitsky V, Banerjee R, Jakob U. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1610–1619. doi: 10.1073/pnas.1401941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jacquier-Sarlin MR, Fuller K, Dinh-Xuan AT, Richard MJ, Polla BS. Protective effects of hsp70 in inflammation. Experientia. 1994;50:1031–1038. doi: 10.1007/BF01923458. [DOI] [PubMed] [Google Scholar]
- 94.Wholey WY, Jakob U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Molecular Microbiology. 2012;83:981–991. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vijayalakshmi J, Mukhergee MK, Graumann J, Jakob U, Saper MA. The 2.2 Å Crystal Structure of Hsp33. Structure. 2001;9:367–375. doi: 10.1016/s0969-2126(01)00597-4. [DOI] [PubMed] [Google Scholar]
- 96.Jakob U, Eser M, Bardwell JCA. Redox Switch of Hsp33 Has a Novel Zinc-binding Motif. The Journal of Biological Chemistry. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- 97.Graf PCF, Martinez-Yamout M, VanHaerents S, Lilie H, Dyson HJ, Jakob U. Activation of the Redox-regulated Chaperone Hsp33 by Domain Unfolding. The Journal of Biological Chemistry. 2004;279:20529–20538. doi: 10.1074/jbc.M401764200. [DOI] [PubMed] [Google Scholar]
- 98.Cremers CM, Jakob U. Oxidant Sensing by Reversible Disulfide Bond Formation. The Journal of Biological Chemistry. 2013;288:26489–26496. doi: 10.1074/jbc.R113.462929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cremers CM, Reichmann D, Hausmann J, Ilbert M, Jakob U. Unfolding of Metastable Linker Region Is at the Core of Hsp33 Activation as a Redox-regulated Chaperone. The Journal of Biological Chemistry. 2010;285:11243–11251. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. Order out of Disorder: Working Cycle of an Intrinsically Unfolded Chaperone. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bardwell JCA, Jakob U. Conditional disorder in chaperone action. Trends in Biochemical Sciences. 2012;37:517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jakob U, Kriwacki R, Uversky VN. Conditionally and Transiently Disordered Proteins: Awakening Cryptic Disorder To Regulate Protein Function. Chemical Reviews. 2014;114:6779–6805. doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 104.Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reichmann D, Jakob U. The Roles of Conditional Disorder in Redox Proteins. Current Opinion in Structural Biology. 2013;23:436–442. doi: 10.1016/j.sbi.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JCA. Protein refolding by pH-triggered chaperone binding and release. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1071–1076. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tapley TL, Korner JL, Barge MT, Hupfeld J, Schauerte JA, Gafni A, Jakob U, Bardwell JCA. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5557–5562. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Foit L, George JS, Bin W, Zhang Brooks CL, Bardwell JCA. Chaperone activation by unfolding. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1254–1262. doi: 10.1073/pnas.1222458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Groitl B, Jakob U. Thiol-based redox switches. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2014;1844:1335–1343. doi: 10.1016/j.bbapap.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kumsta C, Jakob U. Redox-Regulated Chaperones. Biochemistry. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rhee SG, Woo HA. Multiple Functions of Peroxiredoxins: Peroxidases, Sensors and Regulators of the Intracellular Messenger H2O2, and Protein Chaperones. Antioxidants & Redox Signaling. 2011;15:781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 112.Bayer SB, Maghzal G, Stocker R, Hampton MB, Winterbourn CC. Neutrophil-mediated oxidation of erythrocyte peroxiredoxin 2 as a potential marker of oxidative stress in inflammation. The FASEB Journal. 2013;27:3315–3322. doi: 10.1096/fj.13-227298. [DOI] [PubMed] [Google Scholar]
- 113.Teixeira F, Castro H, Cruz T, Tse E, Koldewey P, Southworth DR, Tomás AM, Jakob U. Mitochondrial peroxiredoxin functions as crucial chaperone reservoir in Leishmania infantum. Proceedings of the National Academy of Sciences of the United States of America in press. 2015 doi: 10.1073/pnas.1419682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Voth W, Schick M, Gates S, Li S, Vilardi F, Gostimskaya I, Southworth DR, Schwappach B, Jakob U. The Protein Targeting Factor Get3 Functions as ATP-Independent Chaperone under Oxidative Stress Conditions. Mol Cell. 2014;56:116–127. doi: 10.1016/j.molcel.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Powis K, Schrul B, Tienson H, Gostimskya I, Breker M, High S, Schuldiner M, Jakob U, Schwappach B. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. Journal of Cell Science. 2013;126:473. doi: 10.1242/jcs.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Metz J, Wachter A, Schmidt B, Bujnicki JM, Schwappach B. The Yeast Arr4p ATPase Binds the Chloride Transporter Gef1p When Copper Is Available in the Cytosol. The Journal of Biological Chemistry. 2005;281:410–417. doi: 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- 117.Chartron JW, Clemons WM, Jr, Suloway CJ. The complex process of GETting tail-anchored membrane proteins to the ER. Current Opinion in Structural Biology. 2012;22:217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Denic V, Dotsch V, Sinning I. Endoplasmic Reticulum Targeting and Insertion of Tail-Anchored Membrane Proteins by the GET Pathway. Cold Spring Harbor Perspectives in Biology. 2013;5:13334–13334. doi: 10.1101/cshperspect.a013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hemmingsson O, Kao G, Still M, Naredi P. ASNA-1 Activity Modulates Sensitivity to Cisplatin. Cancer Research. 2010;70:10321–10328. doi: 10.1158/0008-5472.CAN-10-1548. [DOI] [PubMed] [Google Scholar]
- 120.Deavall DG, Martin EA, Horner JM, Roberts R. Drug-Induced Oxidative Stress and Toxicity. Journal of Toxicology. 2012;2012:1–13. doi: 10.1155/2012/645460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hawkins CL, Davies MJ. Inactivation of Protease Inhibitors and Lysozyme by Hypochlorous Acid: Role of Side-Chain Oxidation and Protein Unfolding in Loss of Biological Function. Chem. Res. Toxicol. 2005;18:1600–1610. doi: 10.1021/tx050207b. [DOI] [PubMed] [Google Scholar]
- 122.Vissers MCM, Winterbourn CC. Oxidative damage to fibronectin. Archives of Biochemistry and Biophysics. 1991;285:53–59. doi: 10.1016/0003-9861(91)90327-f. [DOI] [PubMed] [Google Scholar]
- 123.Chapman ALP, Winterbourn CC, Brennan SO, Jordan PA, Kettle AJ. Characterization of non-covalent oligomers of proteins treated with hypochlorous acid. Biochem.J. 2003;375:33–40. doi: 10.1042/BJ20030685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Müller A, Langklotz S, Lupilova N, Kuhlmann K, Bandow JE, Leichert LIO. Activation of RidA chaperone function by N-chlorination. Nature Communications. 2014;5:5804–5818. doi: 10.1038/ncomms6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lambrecht JA, Flynn JM, Downs DM. Conserved YjgF Protein Family Deaminates Reactive Enamine/Imine Intermediates of Pyridoxal 5′-Phosphate (PLP)-dependent Enzyme Reactions. The Journal of Biological Chemistry. 2012;287:3454–3461. doi: 10.1074/jbc.M111.304477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lindemann C, Lupilova N, Muller A, Warscheid B, Meyer HE, Kuhlmann K, Eisenacher M, Leichert LI. Redox Proteomics Uncovers Peroxynitrite-sensitive Proteins That Help Escherichia coli to Overcome Nitrosative Stress. The Journal of Biological Chemistry. 2013;288:19698–19714. doi: 10.1074/jbc.M113.457556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frydman J. Folding of newly translated proteins in vivo: The role of Molecular Chaperones. Annual Review of Biochemistry. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 128.Rehman AA, Ahsan H, Khan FH. Alpha- 2- macroglobulin: A physiological guardian. Journal of Cellular Physiology. 2012;228:1665–1675. doi: 10.1002/jcp.24266. [DOI] [PubMed] [Google Scholar]
- 129.Reddy VY, Desorchers PE, Pizzo SV, Gonias SL, Sahakian JA, Levine RL, Weiss SJ. Oxidative dissociation of human alpha 2-macroglobulin tetramers into dysfunctional dimers. The Journal of Biological Chemistry. 1994;269:4683–4691. [PubMed] [Google Scholar]
- 130.Imber MJ, Pizzo SV. Clearance and binding of two electrophoretic “fast” forms of human alpha 2-macroglobulin. The Journal of Biological Chemistry. 1981;256:8134–8139. [PubMed] [Google Scholar]
- 131.Wyatt AR, Kumita JR, Mifsud RW, Gooden CA, Wilson MR, Dobson CM. Hypochlorite-induced structural modifications enhance the chaperone activity of human α2-macroglobulin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2081–2090. doi: 10.1073/pnas.1403379111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Norris V, Reusch RN, Igarashi K, Root-Bernstein R. Molecular complementarity between simple, universal molecules and ions limited phenotype space in the precursors of cells. Biol Direct. 2014;10:28–47. doi: 10.1186/s13062-014-0028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rao NN, Gómez-García MR, Kornberg A. Inorganic Polyphosphate: Essential for Growth and Survival. Annual Review of Biochemistry. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 134.Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. The Journal of Biological Chemistry. 1992;267:22556–22361. [PubMed] [Google Scholar]
- 135.Shiba T, Tsutsumi K, Yano H. Inorganic polyphosphate and the induction of rpoS expression. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schurig-Briccio LA, Farias RN, Rintoul MR, Rapisarda VA. Phosphate-Enhanced Stationary-Phase Fitness of Escherichia coli Is Related to Inorganic Polyphosphate Level. Journal of Bacteriology. 2009;191:4478–4481. doi: 10.1128/JB.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maciag A, Peano C, Pietrelli A, Egli T, De Bellis G, Landini P. In vitro transcription profiling of the S subunit of bacterial RNA polymerase: redefinition of the S regulon and identification of S-specific promoter sequence elements. Nucleic acids research. 2011;39:5338–5355. doi: 10.1093/nar/gkr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Docampo R, Ulrich P, Moreno SNJ. Evolution of acidocalcisomes and their role in polyphosphate storage and osmoregulation in eukaryotic microbes. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:775–784. doi: 10.1098/rstb.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith SA, Choi SH, Davis-Harrison R, Huyck J, Boettcher J, Rienstra CM, Morrissey JH. Polyphosphate exerts differential effects on blood clotting, depending on polymer size. Blood. 2010;116:4353–4359. doi: 10.1182/blood-2010-01-266791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moreno SNJ, Docampo R. Polyphosphate and Its Diverse Functions in Host Cells and Pathogens. PLoS Pathog. 2013;9:e1003230. doi: 10.1371/journal.ppat.1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Holmström KM, Marina N, Baev AY, Wood NW, Gourine AV, Abramov AY. Signalling properties of inorganic polyphosphate in the mammalian brain. Nature Communications. 2013;4:1362–1370. doi: 10.1038/ncomms2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jahid IK, Silva AJ, Benitez JA. Polyphosphate Stores Enhance the Ability of Vibrio cholerae To Overcome Environmental Stresses in a Low-Phosphate Environment. Applied and Environmental Microbiology. 2006;72:7043–7049. doi: 10.1128/AEM.00924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nikel PI, Chavarria M, Martinez-Garcia E, Taylor AC, de Lorenzo V. Accumulation of inorganic polyphosphate enables stress endurance and catalytic vigour in Pseudomonas putida KT2440. Microbial Cell Factories. 2013;12:50–64. doi: 10.1186/1475-2859-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gray MJ, Jakob U. Oxidative stress protection by polyphosphate—new roles for an old player. Current Opinion in Microbiology. 2015;24:1–6. doi: 10.1016/j.mib.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kuroda A, Nomura K, Ohtomo R, Kato J-I, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Role of Inorganic Polyphosphate in Promoting Ribosomal Protein Degradation by the Lon Protease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 146.Maisonneuve E, Gerdes K. Molecular Mechanisms Underlying Bacterial Persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 147.Kuroda A. A Polyphosphate-Lon Protease Complex in the Adaptation of Escherichia coli to Amino Acid Starvation. Biosci. Biotechnol. Biochem. 2014;70:325–331. doi: 10.1271/bbb.70.325. [DOI] [PubMed] [Google Scholar]
- 148.Gerdes K, Maisonneuve E. Bacterial Persistence and Toxin-Antitoxin Loci. Annual Review of Microbiology. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 149.Wang L, Fraley CD, Faridi J, Kornberg A, Roth RA. Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;100:11249–11254. doi: 10.1073/pnas.1534805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang H, Gomez-Garcia MR, Shi X, Rao NN, Kornberg A. Polyphosphate kinase 1, a conserved bacterial enzyme, in a eukaryote, Dictyostelium discoideum, with a role in cytokinesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16486–16491. doi: 10.1073/pnas.0706847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Smith SA, Morrissey JH. Polyphosphate. Current Opinion in Hematology. 2014;21:388–394. doi: 10.1097/MOH.0000000000000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Klebanoff SJ, Kettle AJ, Rosen H, Winterbourn CC, Nauseef WM. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. Journal of Leukocyte Biology. 2013;93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Aliev G, Smith MA, Seyidova D, Neal ML, Lamb BT, Nunomura A, Gasimov EK, Vinters HV, Perry G, LaManna JC, Friedland RP. The Role of Oxidative Stress in the Pathophysiology of Cerebrovascular Lesions in Alzheimer’s Disease. Brain Pathology. 2006;12:21–35. doi: 10.1111/j.1750-3639.2002.tb00419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]