Abstract
The C≡N stretching frequency and fluorescence quantum yield of p-cyanophenylalanine are sensitive to environment. As such, this unnatural amino acid has found broad applications, ranging from studying how proteins fold to determining the local electric field of membranes. Herein, we demonstrate that the fluorescence of p-cyanophenylalanine can be quenched by selenomethionine through an electron transfer process occurring at short distances, thus further expanding its spectroscopic utility. Using this fluorophore-quencher pair, we are able to show that short polyproline peptides (1–4 prolines) are not rigid; instead, they sample a bimodal conformational distribution.
Keywords: Fluorescence quenching, FRET, Cyanophenylalanine, Polyproline, Protein folding
1. INTRODUCTION
The unnatural amino acid p-cyanophenylalanine (PheCN) is a versatile spectroscopic probe in proteins because (1) its C≡N stretching vibration is sensitive to the local environment and thus can serve as a site-specific infrared (IR) probe,1–4 (2) its fluorescence quantum yield and lifetime in protic solvents, especially water, are significantly different than those in aprotic solvents,5 (3) its fluorescence can be quenched by various metal ions,6 (4) it can be used as an efficient fluorescence resonance energy transfer (FRET) donor to tryptophan (Trp) and other amino acid fluorophores,7–12 (5) its fluorescence can be selectively excited by using an excitation wavelength of 240 nm, even in the presence of other fluorescent amino acids, i.e., phenylalanine (Phe), tyrosine (Tyr) and Trp,13 (6) it can be easily incorporated into biological molecules using various methods including amber codon suppression,14 and (7) it has a similar size to Phe and Tyr, making substitution of these amino acids with PheCN minimally perturbing to the target protein’s native structure. As such, PheCN has been utilized in many biological studies to investigate, for example, the thermodynamics and kinetics of protein folding,15 protein conformation and conformational distribution,16,17 protein-peptide interactions,13,18 the kinetics and mechanism of amyloid formation,19 and protein and membrane electrostatics.20,21 To further extend the spectroscopic utility of PheCN, herein, we show that selenomethionine (SeMet), a naturally-occurring and non-fluorescent amino acid, is an efficient quencher of PheCN fluorescence and, thus, this fluorophore-quencher pair is ideally suited to probe protein conformational changes occurring over very short distances (<20 Å).
Quenching has been widely used to study the structure and dynamics of proteins. For example, the quenching of Trp fluorescence in proteins by amino acids, such as cysteine (Cys), methionine (Met), or by small molecules, such as acrylamide, has been used to determine the rate of contact formations in disordered peptides,22 peptide binding to proteins,23 and the degree of Trp residue solvent exposure.24 Additionally, Trp has been used as a quencher of oxazine and rhodamine fluorophores through photoinduced electron transfer (PET).25 FRET, depending on the donor-acceptor pair, typically occurs over a distance range of 20–100 Å.26 Fluorescence quenching, on the other hand, requires overlap of the electron orbitals of the fluorophore and quencher, and thus occurs at a shorter length scale.26 Therefore, for certain applications, fluorescence quenching is more desirable than FRET. Additionally, fluorescence quenching experiments are often simpler in practice, in comparison to FRET measurements, since the quencher does not directly contribute to the fluorescence signal.
We hypothesize that PheCN-SeMet constitutes an efficient fluorophore-quencher pair based on a previous study which suggested that SeMet is capable of quenching the fluorescence of Trp in a protein environment.23 To verify this hypothesis, we conducted both steady-state and time-resolved fluorescence measurements on a SeMet-PheCN dipeptide. Indeed, our results show that SeMet can efficiently quench PheCN fluorescence. Using the Stern-Volmer analysis and numerical simulations, we are able to further determine the underlying electron-transfer parameters that dictate the distance-dependent fluorescence quenching rate. Furthermore, we employed this fluorophore-quencher pair to investigate the conformational distribution of a series of short polyproline (Pro) peptides, i.e., SeMet-(Pro)n-PheCN (n = 1–4), and found that two conformations are populated in each case. Taken together, these results lead us to believe that PheCN-SeMet is a useful fluorophore-quencher pair for protein structural and conformational studies. Additionally, using SeMet as a quencher in this regard affords the following advantages: (1) it can be synthetically incorporated into a peptide through solid phase peptide synthesis, or into a protein, through expression vectors27 or cell-free expression,28 and (2) it is a natural derivative of Met and, thus, a Met to SeMet substitution is expected to cause only minimum structural perturbation to the protein of interest.
MATERIALS AND METHODS
Sample Preparation
All peptides were prepared by standard 9-fluorenylmethoxy-carbonyl (Fmoc) solid phase peptide synthesis methods on a PS3 automated peptide synthesizer (Protein Technologies, MA) and purified by reverse-phase high-performance liquid chromatography (HPLC). For each peptide, the mass (see Supplementary Information) and its identity were characterized using either liquid-chromatography mass spectrometry (LC-MS) or matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). All peptide samples used in the fluorescence measurements were prepared by dissolving lyophilized peptide in Millipore water, and the pH of the solutions was around 5.3. The peptide concentration, determined optically using the absorbance of PheCN at 280 nm (ε = 850 M−1 cm−1),6 was between 10–25 µM for static measurements and 200 µM for the time-resolved experiments. For the Stern-Volmer measurements, the concentration of PheCN was kept constant at 165 µM.
Static and Time-Resolved Fluorescence Measurements
All static fluorescence measurements, except the Stern-Volmer titrations, were collected on a Jobin Yvon Horiba Fluorolog 3.10 fluorometer at 25 °C in a 1 cm quartz cuvette with spectral resolution of 1.0 nm, an integration time of 2.0 s/nm, and an excitation wavelength of 270 nm. The Stern-Volmer experiments were conducted on an Agilent Technologies Cary Eclipse fluorometer under the same conditions. Time-resolved fluorescence measurements were collected on a time-correlated single photon counting (TCSPC) system with a 1 cm cuvette at 25 °C. The details of the TCSPC system have been described elsewhere.29 Briefly, a home-built femtosecond Ti:Sapphire oscillator operating at 850 MHz was used to generate the 270 nm excitation pulse. Selective emission at 340 nm was monitored using a subtractive double monochromator with a MCP-PMT detector (Hamamatsu R2809U) and a TCSPC board (Becker and Hickl SPC-730). Fluorescence decays were deconvoluted with the experimental instrument response function (IRF) and were fit either to a single-exponential decay (Gly-PheCN-Gly) or a bi-exponential decay (SeMet-containing peptides) using FLUOFIT (Picoquant GmbH). For comparison, fluorescence decays were also fit using the maximum entropy method (MEM).30 The optical density of the samples at the excitation wavelength of 270 nm was approximately 0.01 for the static measurements and 0.1 for the Stern-Volmer and time-resolved measurements.
Molecular Dynamics (MD) Simulation
The starting geometry of Met-PheCN was produced using a trans peptide bond. MD simulations were carried out using the NAnoscale Molecular Dynamics (NAMD) program (version 2.7)31 and the CHARM22 force field.32 The force field parameters for PheCN were derived by Zoete and coworkers.33 The peptide of interest was immersed in 5857 TIP3P water molecules34 in a 40 Å × 40 Å × 40 Å cubic box with periodic boundary conditions and a cutoff of 12 Å for nonbonded interactions. After energy minimization of the entire system, a production run of 100 ns was performed with a pressure of 1 atm, maintained using the Nosé-Hoover Langevin piston method. The temperature was increased from 0 to 298 K with an increment of 20 K every 500 timesteps. The final temperature of 298 K was held constant and was controlled by the Langevin thermostat. Full electrostatics were evaluated at every time step via the Particle-Mesh Ewald (PME) method.35 A 2 fs time step was used to integrate the equations of motion and the structural coordinates were saved every 1 ps for further analysis. The SHAKE algorithm was used to restrict the motion of all bonds involving hydrogen. Convergence of the simulation was checked by monitoring the distance between the sulfur atom in Met and the center-of-mass (COM) of the PheCN sidechain in 10 ns intervals. This distance distribution was then fit to the sum of two Gaussians. The simulation was run until the centers, amplitudes, and widths of the two Gaussians were constant over a period of 50 ns. For this reason, only the frames from the last 50 ns of the full 100 ns trajectory were used for further analysis.
RESULTS AND DISCUSSION
Quenching PheCN Fluorescence by SeMet
In order to verify whether SeMet quenches PheCN fluorescence, we first compared the apparent fluorescence quantum yields of two peptides, SeMet-PheCN and Gly-PheCN-Gly. As shown (Figure 1), under the same experimental conditions the PheCN fluorescence intensity of SeMet-PheCN is ten times less than that of Gly-PheCN-Gly, indicating that SeMet, when in close proximity, can efficiently quench the fluorescence of PheCN. Additionally, while it is known that the natural amino acid Met quenches PheCN fluorescence,12 SeMet is a more effective quencher of PheCN fluorescence, which decreases the fluorescence by a factor of three when compared to a Met-PheCN peptide (Figure 1). To further quantify the efficiency of the SeMet quenching process, we carried out time-resolved fluorescence measurements. As shown (Figure 2), the fluorescence decay of Gly-PheCN-Gly fits well (i.e., χ2 < 1.2) to a single-exponential, with a lifetime of 7.5 ns (Table 1), in good agreement with previously reported fluorescence lifetime of PheCN.5 Further assessment of this decay using the MEM yielded consistent results, which show a single lifetime distribution centered at 7.1 ns (inset of Figure 2). In comparison, the fluorescence decay of SeMet-PheCN is significantly faster than that of Gly-PheCN-Gly (Figure 2), further confirming the role of SeMet as a PheCN fluorescence quencher. Surprisingly, however, the fluorescence decay of SeMet-PheCN is best described by a bi-exponential function with the following lifetime (relative amplitude): 2.0 ns (47%) and 0.2 ns (53%). This deviation from single-exponential behavior, which is further verified by the MEM analysis (inset of Figure 2), is indicative of two peptide conformations that have distinctively different separation distances between the sidechains of SeMet and PheCN.
Figure 1.
Normalized fluorescence spectra of SeMet-PheCN, Met-PheCN, and Gly-PheCN-Gly, as indicated.
Figure 2.
Fluorescence decay kinetics of Gly-PheCN-Gly (red) and SeMet-PheCN (blue). The smooth lines are fits of these data to a single-exponential function (Gly-PheCN-Gly) and a bi-exponential function (SeMet-PheCN). The residuals of the fits are shown in the top panel and the resulting fitting parameters are listed in Table 1. Shown in the inset are the lifetime distributions obtained from MEM analysis for both peptides.
Table 1.
Fluorescence lifetime (τ) and relative amplitude (A) determined from fitting the fluorescence decay of each peptide to a bi-exponential function. For the polyproline peptides, the corresponding fluorophore-quencher separation distance (r) was calculated for each component using k0 = 42.6 ns−1, β = 1.6 Å−1, and a0 = 7.0 Å.
| τ1 (ns)a | A1(%) | r1 (Å) | τ2 (ns)a | A2(%) | r2 (Å) | |
|---|---|---|---|---|---|---|
| Gly-PheCN-Gly | 7.5 | 100 | ------ | ------ | ------ | ------ |
| SeMet-(Pro)4-PheCN | 6.0 | 71 | 11.6 | 0.6 | 29 | 9.1 |
| SeMet-(Pro)3-PheCN | 5.3 | 53 | 11.3 | 0.7 | 47 | 9.2 |
| SeMet-(Pro)2-PheCN | 3.8 | 58 | 10.7 | 0.9 | 42 | 9.4 |
| SeMet-Pro-PheCN | 2.6 | 44 | 10.3 | 0.8 | 56 | 9.4 |
| SeMet-PheCN | 2.0 | 47 | 10.1 | 0.2 | 53 | 8.5 |
The error in these values was estimated to be ±5%
Stern-Volmer Experiments
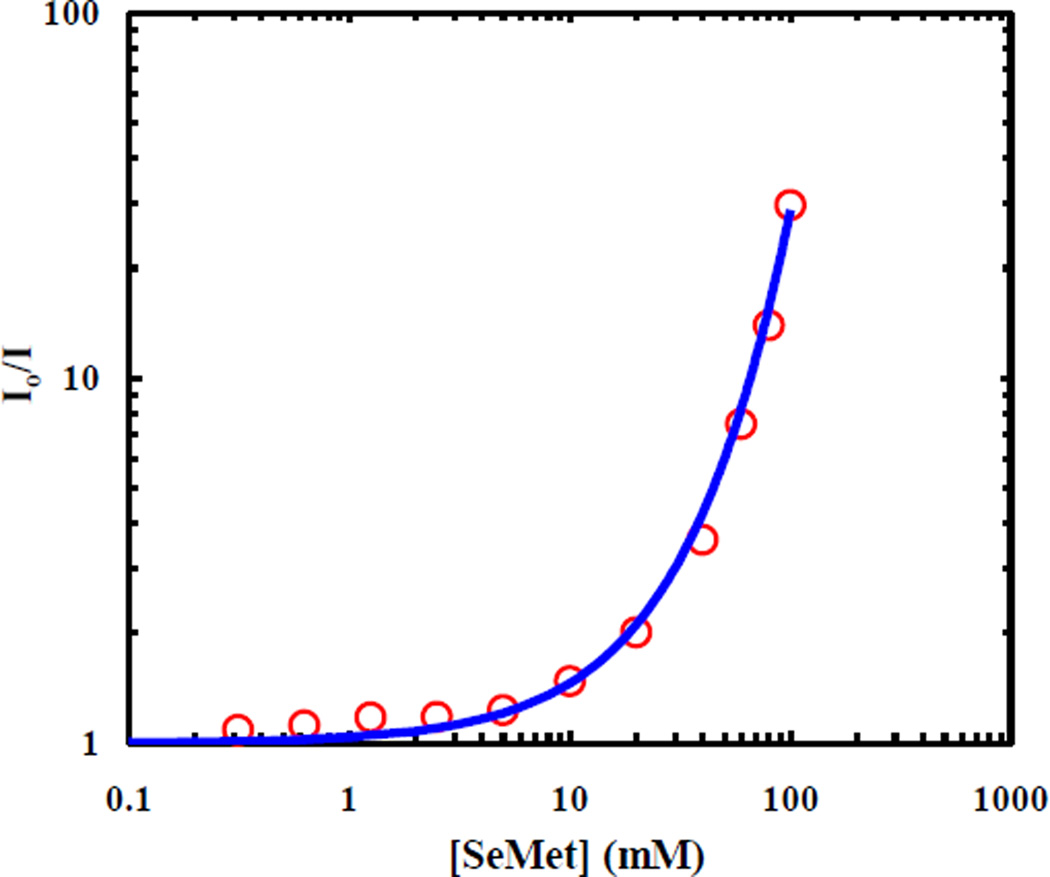
To determine the underlying quenching mechanism, we carried out further static fluorescence quenching experiments using free SeMet and PheCN. As shown, (Figure 3), the resultant Stern-Volmer plot, determined after correction of the inner filter effect of SeMet, exhibits an upward curvature, indicative of a distance-dependent quenching rate,26
| (1) |
where a0 is the donor-to-quencher separation distance when they are in van der Waals contact, k0 is the corresponding quenching rate (i.e., when r = a0), and β is a constant characteristic of the fluorophore-quencher pair.36 The minimum value of the parameter a0 was determined to be 6.23 Å from the sum of the van der Waals radii of the quencher, Se (1.90 Å), and the fluorophore, PheCN (4.33 Å). However, due to the solvent-cage and steric effects, the value of a0 is likely larger. Thus, following previous practice,37 we set the value of a0 to be 7.0 Å. To solve for k0 and β, we numerically fit the Stern-Volmer curve based on the methods described in the literature.26,37–41 Briefly, the fluorescence decay signal, I(t), is described as:
| (2) |
where τ0 is the intrinsic fluorescence lifetime of the fluorophore, is the bulk concentration of the quencher, and time-dependent quenching rate, k(t), is calculated based on the following equation:
| (3) |
where CQ(r, t) is the quencher concentration at a distance r and time t, which is governed by the rate of diffusion and the rate of quenching, given by:
| (4) |
A normalized concentration of quencher, y(r,t) was defined to simplify the calculation:
| (5) |
Using appropriate initial conditions and boundary conditions,
| (6) |
| (7) |
| (8) |
we numerically solved for CQ(r, t) and then k(t) and I(t) for each , allowing us to fit the Stern-Volmer curve with k0 = 42.6 ns−1 and β = 1.6 Å−1, as shown (Figure 3). These parameters are comparable with values obtained for other distance-dependent quenching processes.36,41–42 For example, for a fixed value of a0 = 8.0 Å, k0 and β were determined to be 60 ns−1 and 1.3 Å−1, respectively, for the fluorescence quenching of p-Bis[2-(5-phenyloxazolyl)]benzene (POPOP) by 1,2,4-Trimethoxybenzene.41
Figure 3.
Relative fluorescence intensity of free PheCN versus concentration of free SeMet. The smooth line is a fit to the data using the approach discussed in the text and the following fitting parameters: k0 = 42.6 ns−1, β = 1.6 Å−1 and a0 = 7.0 Å.
Quenching Mechanism
There are several mechanisms by which SeMet could quench PheCN fluorescence, including FRET, electron transfer, and the heavy atom effect. As there is no overlap between the fluorescence emission spectrum of PheCN (Figure 1) and the absorption spectrum of SeMet (Figure S1), a prerequisite for FRET to occur,26 we do not believe that the quenching of the PheCN fluorescence by SeMet is through the mechanism of FRET. Furthermore, we tentatively rule out the possibility of the heavy atom effect, which is known to increase the intersystem crossing rate of the fluorophore,41 as no detectable triplet state formation was observed for PheCN when a high concentration of SeMet (8 mM) was present (Figure S2). Thus, most likely SeMet quenches the PheCN fluorescence through an electron transfer mechanism. To further verify this possibility, we carried out cyclic voltammetry experiment on PheCN. As shown (Figure S3), the oxidation potential of PheCN was measured to be 0.921 V vs. NHE at pH 7.0, a value similar to that of other aromatic amino acids.43 Because the reduction potential of SeMet was previously determined to be 1.21 V vs NHE at pH 7.0,44 we can use the following Rehm and Weller equation41 to estimate the free energy change (ΔGET) of the corresponding electron transfer reaction:
| (9) |
where E1/2ox is the half-wave oxidation potential of the electron donor PheCN, E1/2red is the half-wave reduction potential of the electron acceptor SeMet, Es is the singlet excitation energy of the fluorophore PheCN (280 nm),5 and C is the Coulomb energy change described by:
| (10) |
where ε is the dielectric constant of water and a is the closest distance separation (7 Å). As expected, ΔGET was estimated to be −4.7 eV, a value that is consistent with the notion that the PheCN fluorescence is quenched by SeMet via an electron transfer mechanism.41
MD Simulations
To investigate the origin of the non-single-exponential fluorescence decay of SeMet-PheCN, we carried out a 100 ns MD simulation on Met-PheCN in water at 298 K, as the force field parameters for SeMet were not available to us. While this will limit the extent to which direct and quantitative comparison can be made between simulation and experimental results, it will provide a qualitative description of the conformation distribution of SeMet-PheCN. To assess the distribution of the separation distance between the quencher and fluorophore, which is defined as the distance between the S atom in Met and the COM of the PheCN sidechain, a total of 200,000 MD frames, with adjacent frames separated by 0.5 ps, were saved. As shown (Figure 4), the resultant distance distribution from the last 50 ns of the trajectory consists of two peaks and can be fit by the sum of two Gaussians, centered at 9.0 Å (σ2 = 1.9, relative amplitude = 45%) and 10.6 Å (σ2 = 0.7, relative amplitude = 55%). Using these distances and the quenching parameters determined above from the Stern-Volmer analysis, we calculated the fluorescence decay lifetimes of these two components to be 3.4 and 0.5 ns. Comparing these values to those (i.e., 2.0 and 0.2 ns) determined experimentally for SeMet-PheCN, a qualitative agreement is observed with deviations likely due to using the Met force field instead of that for SeMet. Thus, these MD simulation results support the notion that the bi-exponential fluorescence decay of SeMet-PheCN arises from two distinguishable conformational ensembles.
Figure 4.
Probability distribution of the distance between S and the COM of PheCN sidechain obtained from the MD simulation. The red line represents the best fit of this distribution to the sum of two Gaussians with centers at 9.0 Å (σ2 = 1.9, relative amplitude = 45%) and 10.6 Å (σ2 = 0.7, relative amplitude = 55%), respectively. A representative snapshot for each center distance is also shown.
Conformational Distribution of Short Polyproline Peptides
It is often assumed that polyproline adopts a rigid PPII, all-trans structure and, thus, can be used to determine the Förster distance of FRET pairs.45 Though the all-trans conformation of polyproline is dominant in aqueous solution, there is evidence that other conformations can also populate.46–52 For example, using single-molecule measurements, Best et al.50 showed that the end-to-end distance of long polyproline chains (>8 Pro) can deviate significantly from the value predicted based on the corresponding PPII structure and demonstrated that as much as 30% of the internal peptide bonds could adopt the cis configuration in water. For short polyproline sequences (<6 Pro), computer simulations53 have shown that the energy difference between the all-cis and all-trans conformations in water is small (~1 kcal/mol), allowing both states to be populated. Moreover, nuclear magnetic resonance (NMR) measurements46–48 have indicated that there could be as much as 65% cis-favored conformations for polyproline chains with 1–3 Pro residues. To further verify the notion that short polyproline peptides can adopt non-PPII structures in aqueous solution and also test the utility of the PheCN-SeMet fluorophore-quencher pair, we measured the PheCN fluorescence decay kinetics of the following peptides, SeMet-(Pro)n-PheCN (n = 1–4).
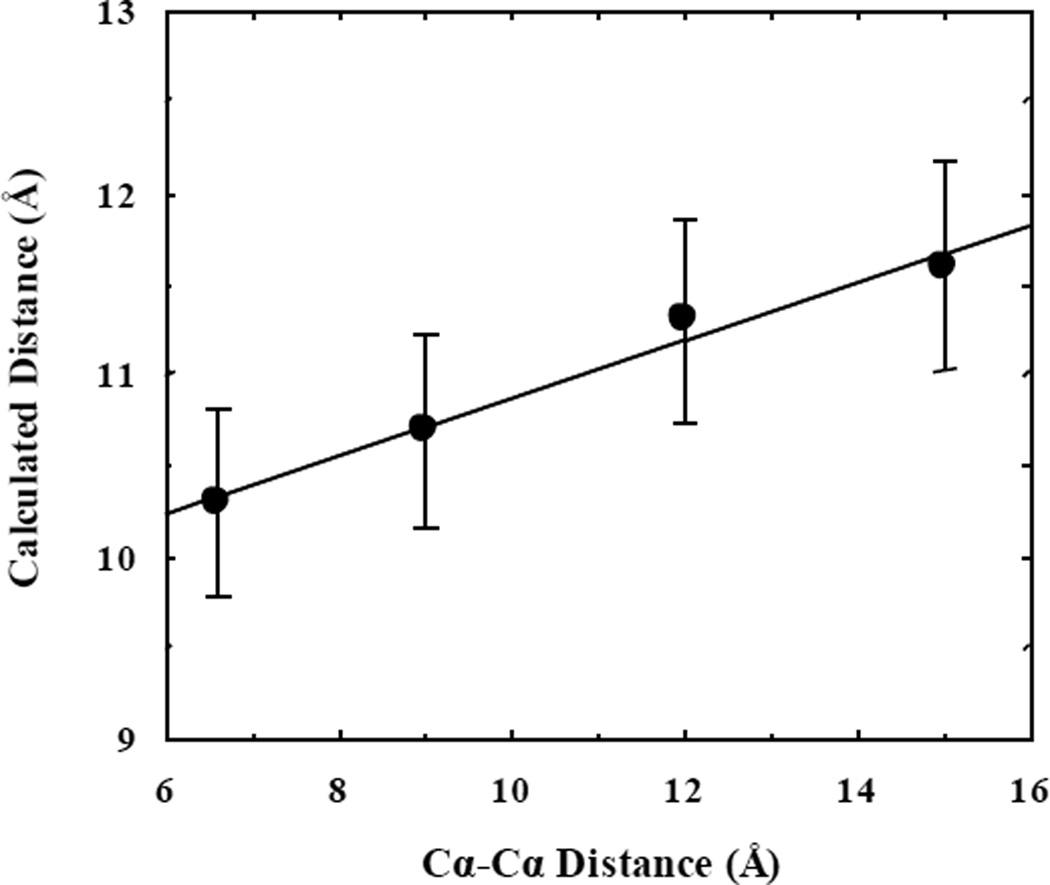
For all four peptides, as shown (Figure 5), the fluorescence decay of PheCN is reasonably well fit (i.e., χ2 < 1.2) by a bi-exponential function, suggesting that there are at least two conformations present in the lifetime of the fluorophore. Further MEM analysis yielded consistent results. Interestingly, as indicated (Table 1), the longer lifetime (i.e., τ1) in each case shows a clear increase with increasing the number of prolines, while the value of τ2 does not show such a trend. These longer lifetimes, and therefore longer fluorophore-quencher separation distances, are likely due to the all-trans conformations of the peptides, as a single cis-bond can significantly reduce the end-to-end distance of polyproline chains.50,51 More convincing evidence in support of this assignment is that the fluorophore-quencher separation distance calculated from the quenching rate (Table 1) shows a linear correlation with the Cα−Cα distance between the first and last amino acid when all the peptide bonds are trans (Figure 6). It should be noted that the increase of the calculated sidechain-to-sidechain distance with n is smaller than that of the Cα−Cα distance. This is most likely due to the fact that in these peptides the PheCN and SeMet sidechains sample a distribution of separation distances and the associated conformational transitions occur on a timescale that is faster than the PheCN fluorescence lifetime. Since a shorter fluorophore-quencher separation distance would lead to a faster quenching rate, it would contribute more toward the decrease of the fluorescence lifetime. In other words, the calculated fluorophore-quencher separation distance in this case is skewed toward the short-distance side of the true separation distance distribution, leading to a smaller increase with n. On the other hand, the shorter lifetime components should arise from non-PPII conformations. An all-cis polyproline structure, with 1.9 Å per proline residue, is shorter than an all-trans structure, with 3.1 Å per proline residue. Varying conformations of cis and trans will give rise to these shorter lifetime components. Taken together, these results validate not only the utility of the PheCN-SeMet fluorophore-quencher pair in peptide or protein conformational studies, but also the notion that polyproline is not a rigid chain and caution should be taken when use it in distance determinations.
Figure 5.
Fluorescence decay kinetics (black lines) of SeMet-Pro-PheCN (A), SeMet-(Pro)2-PheCN (B), SeMet-(Pro)3-PheCN (C), and SeMet-(Pro)4-PheCN (D). The smooth color line in each case represents the best fit of the corresponding decay to a bi-exponential function, with the corresponding residuals shown in the top panel and the fitting parameters given in Table 1. Additionally, shown in the inset is the bimodal lifetime distributions obtained from MEM analysis for each peptide.
Figure 6.
Correlation between the Cα−Cα distance between the first and last amino acid of each polyproline peptide and the corresponding fluorophore-quencher separation distance determined from the τ1 lifetime component.
CONCLUSIONS
Fluorescence quenching through the mechanism of electron transfer requires the fluorophore and quencher to be in van der Waals contact, thus allowing measurement of very short distances. Here, we hypothesized and experimentally verified that PheCN and SeMet constitute such a fluorophore-quencher pair. Through the use of steady-state and time-resolved fluorescence measurements, we were able to determine the quenching rate constant to be kQ = k0exp(−β(r – a0)), where k0 = 42.6 ns−1, β = 1.6 Å−1, and a0 = 7.0 Å. To further demonstrate the utility of this fluorophore-quencher pair, we investigated the end-to-end distances of a series of short polyproline chains, SeMet-(Pro)n-PheCN with n = 1–4. We found in all cases that the fluorescence decay of PheCN follows a bi-exponential function, indicating that short polyproline peptides can sample different conformations. Because of their small sizes, we believe that this amino-acid based fluorophore-quencher pair will be valuable in structural, conformational, and dynamical studies of proteins.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Christopher C. Moser in the Department of Biochemistry and Molecular Biophysics at the University of Pennsylvania for the use the cyclic voltammetry instrument and the helpful insight into the electrochemistry. We gratefully acknowledge financial support from the National Institutes of Health (GM-065978 and P41-GM104605). M.R.M. is supported by a National Science Foundation Graduate Research Fellowship (DGE-1321851).
Footnotes
SUPPORTING INFORMATION
Electronic Supplementary Information (ESI) available: Peptide Mass Data, UV-Vis Spectrum, Cyclic Voltammetry Measurements, and Nanosecond Transient Absorption Spectra.
REFERENCES
- 1.Getahun Z, Huang C-Y, Wang T, De León B, DeGrado WF, Gai F. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 2.Lindquist BA, Furse KE, Corcelli SA. Phys. Chem. Chem. Phys. 2009;11:8119–8132. doi: 10.1039/b908588b. [DOI] [PubMed] [Google Scholar]
- 3.Waegele MM, Culik RM, Gai F. J. Phys. Chem. Lett. 2011;2:2598–2609. doi: 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazewicz CG, Lipkin JS, Smith EE, Liskov MT, Brewer SH. J. Phys. Chem. B. 2012;116:10824–10831. doi: 10.1021/jp306886s. [DOI] [PubMed] [Google Scholar]
- 5.Serrano AL, Troxler T, Tucker MJ, Gai F. Chem. Phys. Lett. 2010;487:303–306. doi: 10.1016/j.cplett.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pazos IM, Roesch RM, Gai F. Chem. Phys. Lett. 2013;563:93–96. doi: 10.1016/j.cplett.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker MJ, Oyola R, Gai F. J. Phys. Chem. B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 8.Glasscock JM, Zhu Y, Chowdhury P, Tang J, Gai F. Biochemistry. 2008;47:11070–11076. doi: 10.1021/bi8012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyake-Stoner SJ, Miller AM, Hammill JT, Peeler JC, Hess KR, Mehl RA, Brewer SH. Biochemistry. 2009;48:5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- 10.Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Biochemistry. 2009;48:9040–9046. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg JM, Batjargal S, Petersson EJ. J. Am. Chem. Soc. 2010;132:14718–14720. doi: 10.1021/ja1044924. [DOI] [PubMed] [Google Scholar]
- 12.Taskent-Sezgin H, Marek P, Thomas R, Goldberg D, Chung J, Carrico I, Raleigh DP. Biochemistry. 2010;49:6290–6295. doi: 10.1021/bi100932p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tucker MJ, Oyola R, Gai F. Biopolymers. 2006;83:571–576. doi: 10.1002/bip.20587. [DOI] [PubMed] [Google Scholar]
- 14.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. J. Am. Chem. Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 15.Aprilakis KN, Taskent H, Raleigh DP. Biochemistry. 2007;46:12308–12313. doi: 10.1021/bi7010674. [DOI] [PubMed] [Google Scholar]
- 16.Urbanek DC, Vorobyev DY, Serrano AL, Gai F, Hochstrasser RM. J. Phys. Chem. Lett. 2010;1:3311–3315. doi: 10.1021/jz101367d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung JK, Thielges MC, Fayer MD. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3578–3583. doi: 10.1073/pnas.1100587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Strzalka J, Tronin A, Johansson JS, Blasie JK. Biophys. J. 2009;96:4176–4187. doi: 10.1016/j.bpj.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marek P, Gupta R, Raleigh DP. ChemBioChem. 2008;9:1372–1374. doi: 10.1002/cbic.200800052. [DOI] [PubMed] [Google Scholar]
- 20.Tang J, Yin H, Qiu J, Tucker MJ, DeGrado WF, Gai F. J. Am. Chem. Soc. 2009;131:3816–3817. doi: 10.1021/ja809007f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu W, Webb LJ. J. Phys. Chem. Lett. 2011;2:1925–1930. [Google Scholar]
- 22.Lapidus LJ, Steinbach PJ, Eaton WA, Szabo A, Hofrichter J. J. Phys. Chem. B. 2002;106:11628–11640. [Google Scholar]
- 23.Yuan T, Weljie AM, Vogel HJ. Biochemistry. 1998;37:3187–3195. doi: 10.1021/bi9716579. [DOI] [PubMed] [Google Scholar]
- 24.Eftink MR, Ghiron CA. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 25.Doose S, Neuweiler H, Sauer M. ChemPhysChem. 2009;10:1389–1398. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- 26.Lakowicz JR. Principles of Fluorescence Spectroscopy. third ed. New York: Springer; 2006. [Google Scholar]
- 27.Zhang M, Vogel H. J. Mol. Biol. 1994;239:545–554. doi: 10.1006/jmbi.1994.1393. [DOI] [PubMed] [Google Scholar]
- 28.Kigawa T, Yamaguchi-Nunokawa E, Kodama K, Matsuda T, Yabuki T, Matsuda N, Ishitani R, Nureki O, Yokoyama S. J. Struct. Funct. Genomics. 2002;2:29–35. doi: 10.1023/a:1013203532303. [DOI] [PubMed] [Google Scholar]
- 29.Hill PA, Wei Q, Troxler T, Dmochowski IJ. J. Am. Chem. Soc. 2009;131:3069–3077. doi: 10.1021/ja8100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Kondrashkina E, Kayatekin C, Matthews CR, Bilsel O. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13367–13372. doi: 10.1073/pnas.0802788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- 32.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karpus M. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 33.Gfeller D, Michielin O, Zoete V. Nucleic Acid Res. 2013;41:D327–D332. doi: 10.1093/nar/gks991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 35.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 36.Marcus R, Sutin N. Biochim. Biophys. Acta, Rev. Bioenerg. 1985;811:265–322. [Google Scholar]
- 37.Lakowicz JR, Kuśba J, Szmacinski H, Johnson ML, Gryczynski I. Chem. Phys. Lett. 1993;206:455–463. [Google Scholar]
- 38.Kuśba J, Lakowicz J. Methods Enzymol. 1994;240:216–262. doi: 10.1016/s0076-6879(94)40051-2. [DOI] [PubMed] [Google Scholar]
- 39.Lakowicz JR, Zelent B, Gryczynski I, Kuba J, Johnson ML. Photobiol. 1994;60:205–214. doi: 10.1111/j.1751-1097.1994.tb05092.x. [DOI] [PubMed] [Google Scholar]
- 40.Lakowicz JR, Zelent B, Kusba J, Gryczynski I. J. Fluoresc. 1996;6:187–194. doi: 10.1007/BF00732821. [DOI] [PubMed] [Google Scholar]
- 41.Zelent B, Gryczynski I, Johnson ML, Lakowicz JR. J. Phys. Chem. 1996;100:18592–18602. doi: 10.1021/jp9618688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moser C, Keske J, Warncke K, Farid R, Dutton P. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 43.Harriman A. J. Phys. Chem. 1987;91:6102–6104. [Google Scholar]
- 44.Mishra B, Sharma A, Naumov S, Priyadarsini KI. J. Phys. Chem. B. 2009;113:7709–7715. doi: 10.1021/jp900322z. [DOI] [PubMed] [Google Scholar]
- 45.Stryer L, Haugland RP. Proc. Natl. Acad. Sci. U. S. A. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deber C, Bovey F, Carver J, Blout E. J. Am. Chem. Soc. 1970;92:6191–6198. doi: 10.1021/ja00724a016. [DOI] [PubMed] [Google Scholar]
- 47.Tonelli A. J. Am. Chem. Soc. 1970;92:6187–6190. doi: 10.1021/ja00724a015. [DOI] [PubMed] [Google Scholar]
- 48.Chiu HC, Bersohn R. Biopolymers. 1977;16:277–288. doi: 10.1002/bip.1977.360160205. [DOI] [PubMed] [Google Scholar]
- 49.Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Best RB, Merchant KA, Gopich IV, Schuler B, Bax A, Eaton WA. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18964–18969. doi: 10.1073/pnas.0709567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doose S, Neuweiler H, Barsch H, Sauer M. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17400–17405. doi: 10.1073/pnas.0705605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radhakrishnan A, Vitalis A, Mao AH, Steffen AT, Pappu RV. J. Phys. Chem. B. 2012;116:6862–6871. doi: 10.1021/jp212637r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong H, Carlson HA. J. Chem. Theory Comput. 2006;2:342–353. doi: 10.1021/ct050182t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.