Key Points
Enterovirus D68 can infect adult patients with hematologic malignancy and HCT recipients and lead to severe respiratory disease.
Current diagnostic assays for enterovirus D68 are limited, and further studies on improved rapid diagnostic tools are needed.
Abstract
The United States Centers for Disease Control and Prevention reported over 1000 cases of severe respiratory disease in pediatric patients associated with enterovirus D68 (EV-D68) in the fall of 2014. We sought to identify and define the clinical burden of disease due to EV-D68 in adult patients with hematologic malignancy or undergoing hematopoietic cell transplant (HCT). Real-time reverse-transcriptase polymerase chain reaction (PCR) for EV-D68 was performed on all respiratory samples positive for human rhinovirus (HRV) or negative for all respiratory viruses by a laboratory-developed respiratory viral PCR panel from August 11, 2014, to November 7, 2014. Presumptive cases were defined as those with an EV-D68 PCR cycle threshold (CT) at least 4 cycles lower than the HRV CT for HRV-positive samples or any EV-D68 CT value for HRV-negative samples. Sequencing of a 150-bp fragment of the 5′ noncoding region confirmed EV-D68 in 16 of 506 respiratory samples. Eight patients had a history of hematologic malignancy, and 6 of these had undergone HCT. Presentation ranged from mild upper respiratory symptoms to respiratory failure. EV-D68 can infect adult patients with hematologic malignancy and HCT recipients and may be associated with severe respiratory disease. Current commercial diagnostic assays cannot differentiate EV-D68 from other enteroviruses or HRV, and improved rapid diagnostic tools are needed.
Introduction
Enterovirus D68 (EV-D68) was first identified in California in 19621 and has since been associated with several small outbreaks, both in the United States and internationally, from 2009 to 2013.2-7 In the summer of 2014, hospitals in Missouri and Illinois began to report increases in severe respiratory illnesses in children, with clinical polymerase chain reaction (PCR) on multiplex platforms testing positive for rhinovirus/enterovirus. Further investigation by the US Centers for Disease Control and Prevention (CDC) later identified several of these cases as EV-D68 infection, and to date, over 1000 cases of EV-D68 have been confirmed.8 Currently, there is no commercially available clinical assay that specifically detects EV-D68 infection, and all US cases have been confirmed by state or CDC laboratories.
Most confirmed cases of EV-D68 infection have been in children, occurring primarily in patients with underlying lung disease such as asthma or a history of wheezing. Several patients were severely ill, requiring hospitalization and mechanical ventilation. Currently, the CDC is investigating the role of EV-D68 in a number of deaths and in children with acute flaccid paralysis. There has only been 1 recent report of adults with confirmed EV-D68 infection that included solid organ transplant recipients; no cancer patients were described in this study.9 Further investigation in immunocompromised hosts has been recommended.10
Immunocompromised hosts, including hematopoietic cell transplant (HCT) recipients and patients receiving chemotherapy for cancer, are uniquely susceptible to and can have serious complications from respiratory viral infections, including pneumonia, respiratory failure, and even death. In order to better understand the clinical manifestations of EV-D68 in these high-risk patients, we used molecular techniques to assess clinical respiratory samples from a tertiary university hospital with a large freestanding ambulatory care center. We describe the clinical presentation and outcomes of 8 presumptive cases of EV-D68 infection in adult immunocompromised patients, 6 of which were confirmed by sequencing.
Methods
The Fred Hutchinson Research Center Institutional Review Board reviewed and approved this research activity. Respiratory tract samples collected as part of routine clinical care from symptomatic adult patients were tested for 12 respiratory viruses by real-time reverse-transcriptase polymerase chain reaction (RT-PCR) assays, including respiratory syncytial virus, human metapneumovirus, influenza viruses A and B, parainfluenza viruses 1 to 4, adenovirus, human coronaviruses, human rhinovirus (HRV), and human bocavirus.11-13 Samples positive for HRV during the time period of interest (August 11, 2014, to November 7, 2014) then underwent real-time RT-PCR testing for EV-D68 with forward primer GCGTTGGCGGCCTACTC and a previously published reverse primer and 5′ FAM-labeled probe.14 Additionally, samples negative for all respiratory viruses in our panel were tested using the same EV-D68 RT-PCR assay. Samples with EV-D68 PCR cycle threshold (CT) values at least 4 cycles lower than the HRV CT from HRV-positive samples, or any EV-D68 CT value from HRV-negative samples, were considered presumptive EV-D68 cases. All presumptive cases were then sequenced if adequate virus was present (CT < 32).
For sequencing, RNA was transcribed into complementary DNA (cDNA) using random hexamers and a Moloney murine leukemia virus reverse transcriptase kit (Invitrogen, Carlsbad, CA). The cDNA was amplified in a seminested PCR with Expand High Fidelity polymerase (Roche Molecular Systems) using outer primers forward GCACTTCTGTCTCCCC and reverse (OL27) CGGACACCCAAAGTAG and inner primers forward GCACTTCTGTCTCCCC and reverse GCCGCCAACGCAGCCT, which amplify an approximately 150-bp fragment of the 5′ noncoding region.15 The amplified product bands were purified from an agarose gel using the QIAquick Gel Extraction Kit (Qiagen, Toronto, ON) and sequenced using the inner forward and reverse primers (GeneWiz Sequencing Corporation, South Plainfield, NJ). Sequences were analyzed using Sequencher 4.1 and compared with reference sequences of EV-D68 species acquired from GenBank. Sequences with >98% match to GenBank sequences were identified as EV-D68 genotypes.
Presumptive cases were defined as those with differential EV-D68 and HRV CT values as described above; proven cases were defined as those with EV-D68 confirmed by sequencing.
Results
Virologic testing
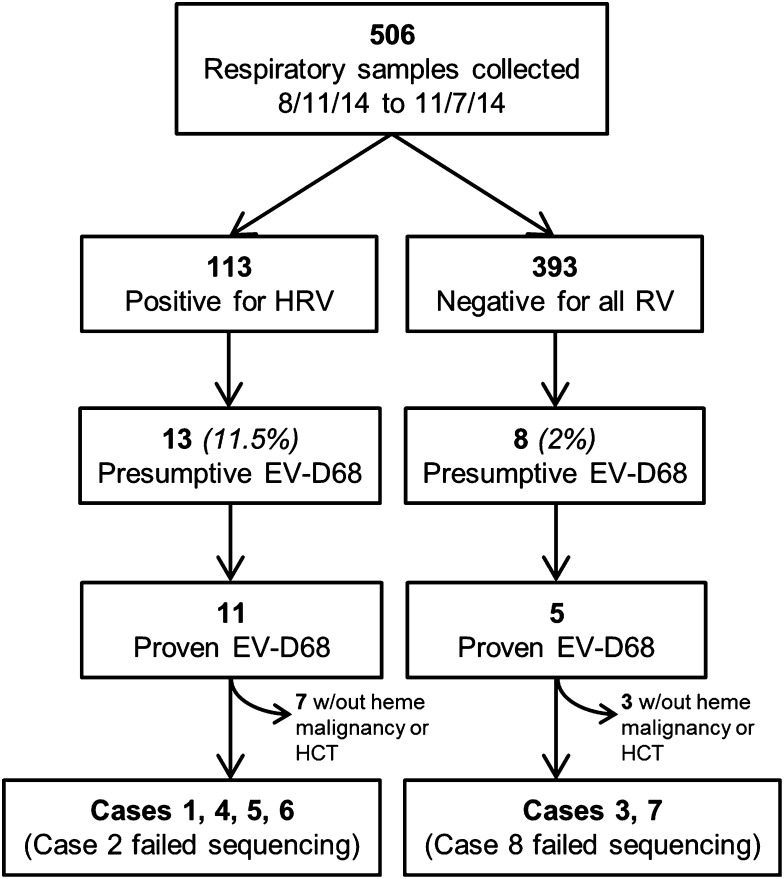
From August 11, 2014, to November 7, 2014, 506 respiratory samples were either negative for all respiratory viruses (n = 393) or positive for HRV (n = 113) by our clinical respiratory virus PCR panel (Figure 1). Twenty-one samples (4.2%) were presumptively positive for EV-D68 based on criteria described above (13 initially HRV positive, 8 initially HRV negative). Sequencing was attempted for 18 samples with sufficient viral load, and EV-D68 was confirmed in 16 samples. Two samples produced no sequencing data, possibly due to low viral loads.
Figure 1.
Flowchart of the EV-D68 testing algorithm. Presumptive EV-D68: EV-D68 PCR cycle threshold (CT) values at least 4 cycles lower than the HRV CT (for HRV-positive samples) or any EV-D68 CT value (for HRV negative samples). Proven EV-D68: confirmed by sequencing. RV, respiratory viruses.
Patients
Of the 21 presumptive cases of EV-D68, 8 (38%) were patients with a hematologic malignancy. The remaining patients had underlying conditions including asthma (n = 2), HIV (n = 3), heart disease (n = 2), granulomatosis with polyangiitis (n = 1), and alcohol abuse (n = 1). Two were admitted for respiratory symptoms, including 1 patient who required brief intensive care unit care and mechanical ventilation due alcohol overdose and altered mental status; this patient did not have significant respiratory symptoms after extubation. Four additional samples originated from patients at outside facilities, and their clinical circumstances were not known.
Of the 8 presumptive EV-D68 cases in patients with hematologic malignancies (Table 1), 6 were proven to be EV-D68 by sequencing. Samples from the nonconfirmed cases (cases 2 and 8) were still presumed to be EV-D68 given the differential between the EV-D68 and HRV CT values. These 8 patients ranged in age from 22 to 69 years; 6 had a history of HCT (5 allogeneic and 1 autologous). Infection with EV-D68 occurred at a median of 485 days following HCT (range, 51-1833 days). Four patients had a history of acute and/or chronic graft-versus-host disease and were receiving treatment with prednisone plus other immunosuppressive therapy at the onset of their respiratory infection (Table 1). In the 4 patients without graft-versus-host disease, only 1 patient was not on any immunosuppressive therapy at the time of infection, and the 3 others received chemotherapy at a median duration of 7 days prior to onset of infection (range, 5-13 days; cases 1, 3, and 7).
Table 1.
Presumptive EV-D68 cases in patients with hematologic malignancy and HCT recipients
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | |
|---|---|---|---|---|---|---|---|---|
| Patient info | 62-y-old F with AML | 22-yo M with ALL | 61-y-old M with MDS, RAEB | 29-y-old F with ALL | 37-y-old F with AML | 54-y-old M with ALL | 69-y-old M with diffuse large B-cell lymphoma | 34-y-old M with cutaneous T-cell lymphoma |
| Therapy | Myeloablative MURD PBSCT with relapse (6 mo prior); decitabine (day −5); MEC (day +7) | Myeloablative MURD BMT with persistent minimal residual disease (1 y prior); imatinib (start 8 mo prior) | Autologous HCT for glioblastoma (5 y prior); G-CLAM (days −48 and −13) | Hyper-CVAD × 5 cycles IT, chemotherapy × 4 (5 mo prior); cylcophosphamide (day +12); CAR T-cell infusion (day +15) | Myeloablative MURD PBSCT (2 y prior), in remission | Nonmyeloablative MURD PBSCT (day −51); dasatinib daily (start day −10); IT methotrexate (days −10, +11) | R-CHOP cycle 5 (day −7) | Myeloablative MURD PBSCT (1.5 y prior); corticosteroids, sirolimus, imatinib, rituxan (5 mo prior); prednisone and IL-2 (start 2 mo prior) |
| Setting | Outpatient/inpatient | Inpatient/ICU | Inpatient/ICU | Outpatient | Outpatient | Outpatient/inpatient | Inpatient/ICU | Inpatient |
| Symptoms | Malaise, productive cough, nasal congestion, clear rhinorrhea, dyspnea, cough | Parasternal chest pain, dyspnea, cough, fever | Productive cough, sputum, fever, altered mental status | Sore throat, cough, headache, malaise, mild rhinorrhea | Sore throat, malaise, cough | Sore throat, rhinorrhea, nonproductive cough, fever (day +23) | Fever, dry cough, fatigue, malaise, chest tightness, syncope, altered mental status | Fever, productive cough, fatigue, diarrhea |
| Chest exam at diagnosis | Respirations regular and clear, productive cough | Clear lungs | Clear lungs | Diminished breath sounds, normal effort | Lungs clear | Respirations regular, cough, rhinorrhea | Decreased breath sounds L base, cough | Lungs clear |
| Maximum oxygen requirement | 3 L via nasal cannula | Mechanical ventilation | None | None | None | None | Mechanical ventilation | 2 L via nasal cannula |
| Radiologic findings | CXR (day +8): new consolidation L base, R pleural effusion | CXR (day +3): diffuse L lung disease and patchy R lung disease; possible bilateral pleural effusion; CT chest (day +18): worsening ground-glass L upper lobe, patchy ground-glass R right upper and middle lobes (see Figure 2) | CXR (day −3): bibasilar consolidation; CT chest (day −1): patchy consolidation R lower lobe, nodules in both lower lobes; mild interlobular septal thickening (see Figure 2) | CXR (day +8): lungs clear | NA | CXR (day +12): bronchial wall thickening in upper lungs | CXR (day +1): large L pleural effusion, L lower lobe atelectasis; CT chest (day +4): interval increase R pleural effusion; large L pleural effusion; new patchy consolidation R middle lobe; bilateral lower lobe consolidation (see Figure 2) | CXR (day 0): right basilar consolidation |
| Day posttransplant EV-D68 positive | 192 | 374 | 1833 | NA | 666 | 51 | NA | 596 |
| Day postchemotherapy EV-D68 positive | 5 | NA | 13 | 150 | NA | NA | 7 | NA |
| Coinfections | Coagulase-negative Staphylococcus bacteremia (second admission) | Mycobacterium fortuitum on prior BAL; Candida albicans, Aspergillus niger, and Enterobacter cloacae from BAL (day +21) | Streptococcus mitis septicemia | None | None | CMV viremia (day −4); admitted for fever and neutropenia day +24-26, no etiology identified | Clostridium difficile colitis | None |
| ANC at diagnosis (109 cells/L) | 0.58 | 11.3 | 0 | 3.68 | 7.42 | 6.33 | 0.01 | 5.19 |
| ALC at diagnosis (109 cells/L) | 0.42 | 0.41 | 0 | 0.43 | 0.84 | 1.54 | 0.12 | 0.67 |
| Duration of symptoms | 55 d | >52 d | 6 d | 11 d | 3 d | 33 d | 15 d | 7 d |
| Duration of initial hospitalization | 4 d | >54 d, ICU 13 d | 27 d, ICU 2 d | NA | NA | 3 d | 14 d, ICU 3 d | 3 d |
| Outcome | Death at day +55 due to respiratory failure in setting of CHF, fluid overload and paraplegia due to compression fracture | Remains admitted on nasal cannula, persistent respiratory symptoms in setting of pneumonia and bronchiolitis obliterans | In remission, being evaluated for HCT; respiratory symptoms resolved | Successful CAR T-cell therapy with planned non-myeloablative matched related HCT; respiratory symptoms resolved | In remission, remains on therapy for chronic GVHD; respiratory symptoms resolved | In remission, developed mild gut GVHD; respiratory symptoms resolved | Discharged to skilled nursing facility; respiratory symptoms resolved | Respiratory symptoms resolved |
| Select relevant medications at time of diagnosis | Sirolimus, prednisone, budesonide, triamcinolone cream | Budesonide, cyclosporine, prednisone, imatinib, albuterol, fluticasone-salmeterol, montelukast | Prednisone, budesonide | Cyclosporine, dasatinib, mycophenolate mofetil, prednisone | Prednisone, IL-2 (subcutaneous), fluticasone, montelukast | |||
| Other relevant medical history | Chronic GVHD (skin and gut) | Acute GVHD (gut), chronic GVHD (mouth), bronchiolitis obliterans | Glioblastoma multiforme s/p resection, radiotherapy and chemotherapy (5 y prior) | Asthma | Chronic GVHD (skin and gut) | Allergic rhinitis, hypertension, type 2 diabetes mellitus | Chronic left pleural effusion, coronary artery disease, history of tobacco use | Chronic GVHD (skin, eyes, mouth), restrictive pulmonary disease, osteoblastoma cervical spine, superficial melanoma, pediatric T-cell NHL of the large intestine |
AML, acute myeloid leukemia; ANC, absolute neutrophil count; ALC, absolute lymphocyte count; ALL, acute lymphoblastic leukemia; BAL, bronchoalveolar lavage; BMT, bone marrow transplant; CAR, chimeric antigen receptor; CHF, congestive heart failure; CMV, cytomegalovirus; CXR, chest radiograph; F, female; G-CLAM, granulocyte colony-stimulating factor/cladribine/cytarabine/dose-escalated mitoxantrone; GVHD, graft-versus-host disease; hyper-CVAD, cyclophosphamide/vincristine/doxorubicin/dexamethasone with methotrexate and cytarabine; IL-2, interleukin-2; IT, intrathecal; M, male; MDS, myelodysplastic syndrome; MEC, mitoxantrone/etoposide/cytarabine; MURD, matched unrelated donor; NHL, non-Hodgkins lymphoma; PBSCT, peripheral blood stem cell transplant; RAEB, refractory anemia with excessive blasts; R-CHOP, rituximab/cyclophosphamide/doxorubicin/vincristine/prednisone.
Four of the eight patients had a history of underlying lung disease (Table 1). Bronchiolitis obliterans was thought to contribute significantly to the respiratory failure seen in case 2; this patient required sustained respiratory support necessitating a prolonged intensive care unit (ICU) stay and the need for mechanical ventilation. His course was also complicated by the presence of multiple copathogens identified on bronchoalveolar lavage, but a repeat bronchoalveolar lavage on day +19 was negative for both HRV and EV-D68 by PCR. Case 7 had a preexisting pleural effusion that was larger at the time of admission; this was thought to play a role in the respiratory failure, although thoracentesis did not reveal an etiology for the effusion.
All presumptive cases were sporadic, as there were no temporal or spatial correlations identified among cases in either inpatient or ambulatory environments to suggest nosocomial transmission.
Presenting signs and symptoms
All 8 patients presented with or developed cough during their illness. Three patients reported rhinorrhea, 5 patients had malaise, and 3 patients had dyspnea; only 4 patients presented with fever. Four patients were diagnosed in the outpatient clinic. One patient was hospitalized on day +7 from infection for planned chemotherapy and became hypoxic on day +9, requiring 3 L of oxygen via nasal cannula. Another patient was hospitalized on day +23 from infection after developing fever and neutropenia. Five patients had other copathogens identified during the course of their respiratory symptoms (Table 1). No patients had simultaneous identification of another viral respiratory pathogen.
Six of 7 patients had abnormalities on chest radiograph including patchy or diffuse consolidations (Table 1). Three patients had chest computed tomography (CT) abnormalities that were consistent with lower tract disease, although 1 patient had evidence of underlying lung disease (Figure 2). Seven of these patients could be classified as lymphopenic (absolute lymphocyte count <1 × 109 cells/L) at the onset of their respiratory illness; 5 of these had an absolute lymphocyte count <0.5 × 109 cells/L. The only patient without lymphopenia at diagnosis (case 6) developed lymphopenia by day +3 after EV-D68 was diagnosed. Three patients also developed neutropenia during the course of their respiratory illness, presumably related to the recent receipt of chemotherapy.
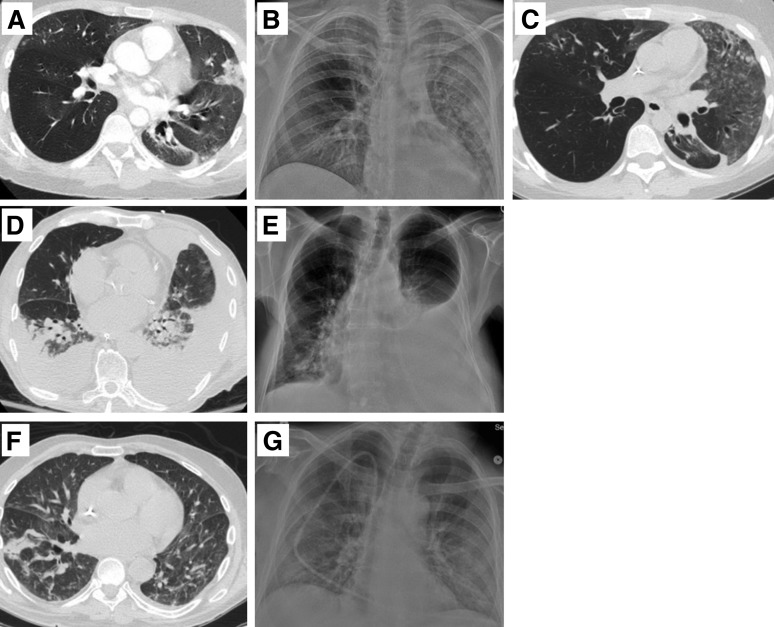
Figure 2.
Selected radiographic images from patients with presumptive EV-D68 infection. (A) Case 2, day −3 CT: ground glass opacities and nodule airspace opacities throughout, and consolidation in lingula and L lower lobe. (B) Case 2, day +3 chest radiograph (CXR): diffuse L lung disease and patchy R lung disease; possible bilateral pleural effusion. (C) Case 2, day +18 CT: worsening ground-glass L upper lobe, and patchy ground-glass R right upper and middle lobes. (D) Case 3, day −1 CT: patchy consolidation R lower lobe, and nodules in both lower lobes; mild interlobular septal thickening. (E) Case 3, day +4 CXR: bilateral basilar consolidation. (F) Case 7, day +1 CXR: increased left pleural effusion with underlying atelectasis; increased right lower lobe consolidation. (G) Case 7, day +4 CT: interval increase R pleural effusion; large L pleural effusion. New patchy consolidation R middle lobe; bilateral lower lobe consolidation.
Outcomes
The duration of symptoms ranged from 6 to 55 days, with a median of 13 days. Of hospitalized patients, the duration of hospitalization ranged from 4 to 35 days (median 9 days). All admitted patients were started on empiric antibiotics, but no antiviral agents with activity against respiratory viruses were given. Repeat respiratory viral PCR testing was done for case 2 and was negative for HRV and EV-D68. Case 4 had a subsequent sample (day +21) negative for both HRV and EV-D68, followed by a sample (day +43) positive for HRV (CT 22.4) and negative for EV-D68, suggesting a new primary infection with HRV. Two patients required ICU admission for respiratory failure, and both required mechanical ventilation. Case 2 remained on mechanical ventilation for 11 days with a max FiO2 of 60%; at the time of this report, he remained on nasal cannula. One additional patient was admitted to the ICU briefly for septic shock due to Streptococcus mitis bacteremia but did not require respiratory support.
Discussion
We describe 8 adult immunocompromised patients with proven or presumptive EV-D68 infections. To our knowledge, this report is the first to describe EV-D68 in patients with hematologic malignancies or in HCT recipients. All 6 EV-D68 cases in HCT recipients occurred relatively late after transplant; however, all transplant patients were still receiving immunosuppressive therapy at the time of infection.
Overall, the immunocompromised patients infected with EV-D68 in our series presented with a wide range of potential manifestations, from relatively mild symptoms managed in the outpatient clinic to respiratory failure requiring ICU admission and mechanical ventilation. All patients developed cough with a range of additional respiratory symptoms including rhinorrhea, dyspnea, and sore throat. The universal presence of cough in this cohort may be a distinguishing feature from HRV infection,16,17 although larger, prospective comparative studies are needed to define unique clinical characteristics. HCT recipients were not noted to be more severely affected by EV-D68 when compared with patients with hematologic malignancy alone. Only 1 patient had EV-D68 isolated from the lower respiratory tract, although at least 5 other cases had presentations that were consistent with lower respiratory tract infection based on clinical and radiographic findings. Given that the lower respiratory tract was not sampled in all cases, it is difficult to determine the specific role of EV-D68 vs that of other pathogens in lower tract disease. In our cohort, half the patients had underlying lung disease, which possibly contributed to their clinical course and which is consistent with findings described in pediatric patients during the outbreak.8 Finally, most patients presented with or developed lymphopenia, a known risk factor for development of lower tract disease and/or mortality for several common respiratory viral illnesses including respiratory syncytial virus and influenza virus.18
During our study period, 11.5% of cases originally identified as HRV by our respiratory viral PCR panel were subsequently characterized as EV-D68 with the specific PCR assay, and 9.7% of these were confirmed with sequencing. The limited specificity of the HRV primers used in our assay and in many commercial assays14,19 makes an accurate discrimination between HRV and EV-D68 difficult. HRV is the most common respiratory virus in HCT recipients, accounting for between 25% and 40% of viral respiratory infections in this population.17,20,21 Thus, it is possible that EV-D68 is the true pathogen in ∼10% of HRV cases and up to 5% of overall cases during peak circulation of this enterovirus strain. In our cohort, all samples that met our criteria for presumptive EV-D68 and were successfully sequenced and confirmed as EV-D68, suggesting a PCR-based algorithm for identification of presumptive cases of EV-D68 may be useful in the clinical setting. Accurate identification may become increasingly important as new antiviral therapies become available that could have more targeted activity against either HRV or enteroviruses.
This testing methodology could also be important in the clinical setting for infection-prevention purposes. Of the 393 cases initially negative for all respiratory viruses, 8 (2%) were presumptive EV-D68. Unless persistently symptomatic, our center-based protocols would have allowed such patients with negative test results to be removed from droplet isolation. It is possible that EV-D68 could therefore lead to person-to-person transmission within these highly immunocompromised patient populations in either the clinic or hospital setting. Our current understanding of the natural history of EV-D68 and its transmission is limited, and to date no nosocomial transmissions have been documented. Our data argue that additional algorithms for confirmatory testing may be needed in clinical practice. However, these data should be interpreted with caution, and further studies with more patients are needed to make clear recommendations for infection prevention practices.
In conclusion, EV-D68 should be considered in cases of immunocompromised adults with suspected respiratory viral infection. The severity of illness described in our study suggests the need for additional studies to address clinical outcomes and risk factors for severe disease in this population. The detection of the virus in patients with negative clinical multiplex-PCR testing reminds clinicians that current respiratory viral testing systems may not be sufficient to detect and therefore prevent nosocomial transmission of EV-D68. Although there are no licensed antivirals available for the treatment of EV-D68, future studies addressing enhanced diagnostic assays for EV-D68 may also be important as novel therapies become available.
Acknowledgments
The authors thank Greg Boughton and Isabelle Palileo for laboratory testing.
This work was supported by the National Institutes of Health, National Heart Lung and Blood Institute (grant K24 HL093294-06). A.W. also received support from the Seattle Children’s Center for Clinical and Translational Research Clinical Research Scholar’s Program.
Footnotes
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.W. performed the research, collected data, analyzed data, and wrote the manuscript; S.A.P. and J.A.E. critically reviewed the manuscript; K.R.J. provided resources and critically reviewed the manuscript; M.B. designed and performed the research, analyzed data, provided resources, and wrote the manuscript; and J.K. contributed to assay development, provided resources, and wrote the manuscript.
Conflict-of-interest disclosure: S.A.P. has served as a consultant and received research support from Cubist and Merck. J.A.E. has served as a consultant for GlaxoSmithKline and received research funding from GlaxoSmithKline, Gilead, and Ansun. M.B. has received research funding from Ansun. The remaining authors declare no competing financial interests.
Correspondence: Alpana Waghmare, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave North, Seattle, WA 98109; e-mail: alpana@uw.edu.
References
- 1.Schieble JH, Fox VL, Lennette EH. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol. 1967;85(2):297–310. doi: 10.1093/oxfordjournals.aje.a120693. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Clusters of acute respiratory illness associated with human enterovirus 68—Asia, Europe, and United States, 2008-2010. MMWR Morb Mortal Wkly Rep. 2011;60(38):1301–1304. [PubMed] [Google Scholar]
- 3.Imamura T, Fuji N, Suzuki A, et al. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis. 2011;17(8):1430–1435. doi: 10.3201/eid1708.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamura T, Suzuki A, Lupisan S, et al. Detection of enterovirus 68 in serum from pediatric patients with pneumonia and their clinical outcomes. Influenza Other Respi Viruses. 2014;8(1):21–24. doi: 10.1111/irv.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaida A, Kubo H, Sekiguchi J, et al. Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan. Emerg Infect Dis. 2011;17(8):1494–1497. doi: 10.3201/eid1708.110028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YC, Cheng HB, Chen HH, Liu CM, Chou CH, Sung FC. Circulating viruses associated with severe complicated enterovirus infection in Taiwan: a multi-year analysis. Pediatr Infect Dis J. 2010;29(4):334–339. doi: 10.1097/INF.0b013e3181c2a1d2. [DOI] [PubMed] [Google Scholar]
- 7.Kreuter JD, Barnes A, McCarthy JE, et al. A fatal central nervous system enterovirus 68 infection. Arch Pathol Lab Med. 2011;135(6):793–796. doi: 10.5858/2010-0174-CR.1. [DOI] [PubMed] [Google Scholar]
- 8.Midgley CM, Jackson MA, Selvarangan R, et al. Severe respiratory illness associated with enterovirus D68 - Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep. 2014;63(36):798–799. [PMC free article] [PubMed] [Google Scholar]
- 9.Poelman R, Scholvinck EH, Borger R, Niesters H, van Leer-Buter C. The emergence of enterovirus D68 in a Dutch University Medical Center and the necessity for routinely screening for respiratory viruses. J Clin Virol. 2015 doi: 10.1016/j.jcv.2014.11.011. 62:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ison MG, Danziger-Isakov L. Severe respiratory illness associated with enterovirus 68: implications for solid organ transplantation [published online ahead of print October 30, 2014]. Transplantation. doi: 10.1097/TP.0000000000000502. [DOI] [PubMed] [Google Scholar]
- 11.Kuypers J, Campbell AP, Cent A, Corey L, Boeckh M. Comparison of conventional and molecular detection of respiratory viruses in hematopoietic cell transplant recipients. Transpl Infect Dis. 2009;11(4):298-303. [DOI] [PMC free article] [PubMed]
- 12.Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119(1):e70–e76. doi: 10.1542/peds.2006-1406. [DOI] [PubMed] [Google Scholar]
- 13.Kuypers J, Wright N, Corey L, Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J Clin Virol. 2005;33(4):299–305. doi: 10.1016/j.jcv.2004.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X, Holloway B, Dare RK, et al. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46(2):533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadopoulos NG, Hunter J, Sanderson G, Meyer J, Johnston SL. Rhinovirus identification by BglI digestion of picornavirus RT-PCR amplicons. J Virol Methods. 1999;80(2):179–185. doi: 10.1016/S0166-0934(99)00045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh S, Champlin R, Couch R, et al. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clin Infect Dis. 1999;29(3):528-532. [DOI] [PubMed]
- 17.Milano F, Campbell AP, Guthrie KA, et al. Human rhinovirus and coronavirus detection among allogeneic hematopoietic stem cell transplantation recipients. Blood. 2010;115(10):2088–2094. doi: 10.1182/blood-2009-09-244152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YJ, Boeckh M, Englund JA. Community respiratory virus infections in immunocompromised patients: hematopoietic stem cell and solid organ transplant recipients, and individuals with human immunodeficiency virus infection. Semin Respir Crit Care Med. 2007;28(2):222–242. doi: 10.1055/s-2007-976494. [DOI] [PubMed] [Google Scholar]
- 19.Popowitch EB, O’Neill SS, Miller MB. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol. 2013;51(5):1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowden RA. Respiratory virus infections after marrow transplant: the Fred Hutchinson Cancer Research Center experience. Am J Med. 1997;102(3A):27-30; discussion 42-43. [DOI] [PubMed]
- 21.Hassan IA, Chopra R, Swindell R, Mutton KJ. Respiratory viral infections after bone marrow/peripheral stem-cell transplantation: the Christie hospital experience. Bone Marrow Transplant. 2003;32(1):73–77. doi: 10.1038/sj.bmt.1704048. [DOI] [PubMed] [Google Scholar]