Abstract
DNA sequencing using reversible terminators, as one sequencing by synthesis strategy, has garnered a great deal of interest due to its popular application in the second-generation high-throughput DNA sequencing technology. In this review, we provided its history of development, classification, and working mechanism of this technology. We also outlined the screening strategies for DNA polymerases to accommodate the reversible terminators as substrates during polymerization; particularly, we introduced the “REAP” method developed by us. At the end of this review, we discussed current limitations of this approach and provided potential solutions to extend its application.
Keywords: DNA polymerases, Sequencing technology, Modified nucleotide, Primer extension, Reversible terminator, Sequencing by synthesis
Introduction
Dideoxy sequencing method, developed by Frederick Sanger et al. in 1977, contributed substantially to development of biological sciences in the past several decades [1]. As the core technology, it brought the great successful completion of the Human Genome Project (HGP) [2]. However, high cost and low throughput inherited within the method had limited its application to meet the ever-expanding appetite for large scale genome sequencing projects, including de novo sequencing of more species, whole-genome resequencing, and deep sequencing. Therefore, the second-generation sequencing technology, characterized by its high speed, high throughput, and lower cost, appeared on the scene [3–5].
Since 2005, the second-generation sequencing technologies have experienced a rapid development: different sequencing platforms came to market and data produced by these new technologies mushroomed exponentially. These achievements not only provided large amount of raw data for scientific research, but also uncovered new scientific ideas, and revolutionarily changed our ways of working in life sciences [3–7]. Now, the third-generation sequencers, represented by single molecule real-time sequencer (SMAT) from PacBio [8], are also on the horizon. But due to the relatively high single read error rate (∼15%) [9], the second-generation sequencing technologies are still the mainstream in whole-genome sequencing markets.
So far, technologies employed in the second-generation sequencing platforms are of two main types based on sequencing chemistry: “sequencing by synthesis” and “sequencing by ligation” [10,11]. Reversible termination sequencing is one of the sequencing-by-synthesis strategies popularized by Illumina/Solexa due to its wide adoption in the worldwide second-generation sequencing market (with 80–90% market share) [12]. In the following sections, we give a brief introduction on the developmental history and current status of this technology, discuss the limitations, and suggest potential solutions of its uses in detail.
The history and classification of reversible termination sequencing technology
Reversible termination sequencing technology was first reported by Dr. Jingyue Ju from Colombia University [13]. The prime difference between this approach and the traditional Sanger sequencing method is that the former uses modified nucleotide analogous to terminate primer extension reversibly, while the latter employs dideoxynucleotide to irreversibly terminate primer extension (Figure 1) [5].
Figure 1.
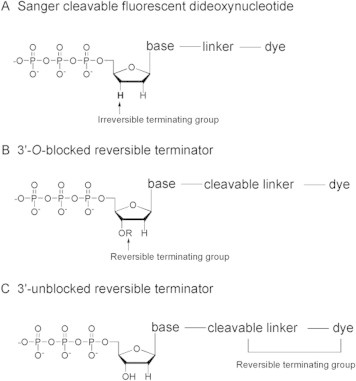
Structure schematic of irreversible and reversible terminators used in sequencing technologies
With the development over the last decade, several reversible terminators were produced. They can be classified into two types based on the difference of the reversible blocking groups [11–23]. One type is 3′-O-blocked reversible terminators. As shown in Figure 1B, the blocking group –OR [reversible terminating (capping) group] is linked to the oxygen atom of the 3′-OH of the pentose, while the florescence label is linked to the base, which acts as a reporter and can be cleaved [12–18]. The other type is 3′-unblocked reversible terminators [19–23] as shown in Figure 1C. In this case, the reversible termination group is linked to the base as well as the florescence group which not only is a reporter but also functions as part of the reversible terminating group for termination of the primer extension. These two types of reversible terminators have their advantages and disadvantages: the 3′-O-blocked reversible terminator contains a 3′ reversible blocking group, thus should render better termination effect; the 3′-unblocked reversible terminator, on the other hand, is easier to be accepted by the DNA polymerases due to the lack of a modified moiety at the 3′-OH. Because polymerases have evolved for billions of years to discriminate between ribonucleoside triphosphates and 2′-deoxyribonucleoside triphosphates, they have evolved to inspect the 2′- and 3′-positions of their substrates closely. For example, although the only difference between dideoxyribonucleoside and deoxyribonucleoside is the presence or absence of oxygen atom at the 3′ position, accepting a dideoxyribonucleoside for all known DNA polymerases will prevent further catalytic elongation of additional nucleoside afterward [17,24].
For the first type, there are three commercially-available reversible terminators with a blocking group at 3′-OH (structures shown in Figure 2A–C): the 3′-ONH2 reversible terminator developed by Dr. Steven A. Benner and his colleagues from Foundation for Applied Molecular Evolution (FfAME) [17,18]; the 3′-O-allyl reversible terminator created by Jingyue Ju and his colleagues from Colombia University [5]; the 3′-O-azidomethy reversible terminator developed and used by Illumina Solexa [12]. All three commercial reversible terminators were reported to show good performance in reversible termination function: achieving nearly 100% of 3′-O blocking efficiency and florescent label group cleavage after primer extension termination [12–18].
Figure 2.
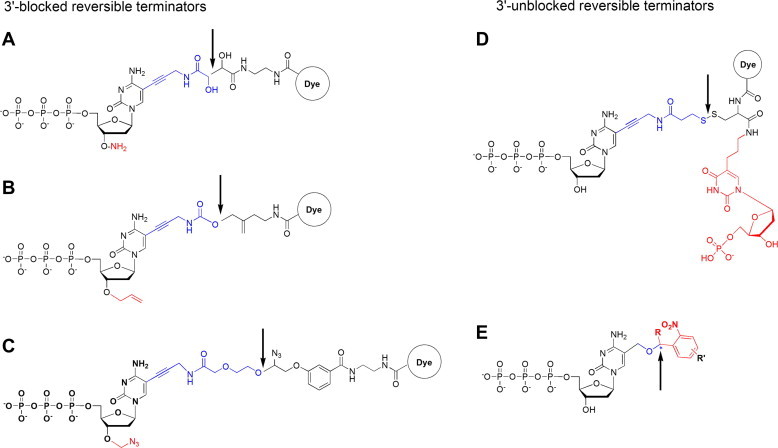
Several commercial reversible terminators Three different 3′-blocked reversible terminators were shown on the left (A–C) and two 3′-unblocked reversible terminators were shown on the right (D–E). The chemical structures in red denote the reversible terminating groups. Arrows indicate the site of cleavage separating the fluorescent groups from the nucleotide, and the chemical structures in blue denote the molecular scars that are attached to the base.
For the second type, hitherto, there is only one commercial 3′-OH unblocked reversible terminator named “virtual terminator”, which is developed by Helicos BioSciences Corporation (Figure 2D) and employed by the first “single molecule” sequencer to hit the market in the emerging third generation of DNA sequencing platform [19]. In addition, the “Lightning terminator” recently developed by Michael L. Metzker’s group also belongs to 3′-OH unblocked reversible terminator (the lower panel in Figure 2E), and is unique in using UV light to cleave the fluorescent group [20–23].
Working mechanism of reversible termination sequencing technology
Reversible termination sequencing technology is a sequencing-by-synthesis approach [3–5] that infers the sequence of a template by stepwise primer elongation. It is popularized as a second generation sequencing technology on the Illumina platform. The general reversible termination sequencing process involves (i) immobilizing the sequencing templates and primers on a solid support; (ii) primer extension by one base and termination; (iii) recognizing the color of the fluorophore carried by the extended base to identify the incorporated nucleotide after washing away the unincorporated nucleotides; (iv) removal of the fluorescent tag and the 3′-O blocking group; (v) washing again and repeating the aforementioned steps (ii–iv). The whole process can be summarized as extension–termination–cleavage–extension cycle (Figure 3).
Figure 3.
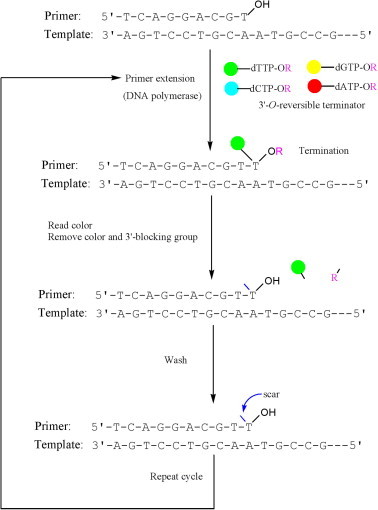
Schematic showing sequencing during synthesis using 3′-O blocking reversible terminators Each triphosphate has its 3′-OH group blocked and a fluorescent tag attached to its nucleobase via a cleavable linker. The blue arrow indicates the residual linker “scar” after cleavage of fluorescent group.
Screening proper polymerases for their abilities to accept the reversible terminators
After design and synthesis of a reversible terminator, first we need to find a proper polymerase to accept the nucleotide analog. Due to the specificity of enzyme and the special structures of the reversible terminating nucleotides, it is difficult to get a proper polymerase to accept them with high efficiency and fidelity. Proper polymerases were usually obtained after experimenting potential candidates through primer extension screenings [17,18] (Figure 4).
Figure 4.
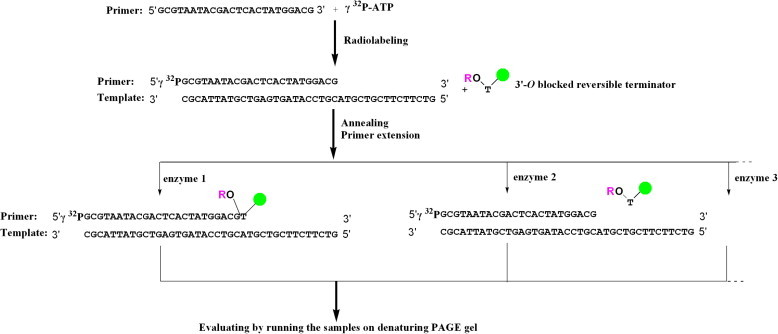
Screening the proper polymerases compatible with the reversible terminators using primer extension method γ 32P-labeled primer was first annealed with the template, followed by the addition of the reversible terminator. Primer extension was then initiated by adding the candidate polymerases, and evaluated by running the samples on denaturing PAGE gel.
Based on screening libraries, two approaches are applied to select the proper polymerases for the reversible terminators [17,18]. One simple approach is to screen the polymerases from commercially-available DNA polymerases and reverse transcriptases [18]. If no desired polymerase can be obtained in this screening, another approach can be taken to obtain the proper polymerases from mutation libraries constructed through rational design, “directed evolution” (random mutation), or semi-rational design (combination of rational design with directed evolution) [17].
Many commercially-available DNA polymerases, such as Therminator, Klenow, Bst and 9°Nm DNA polymerases, have been reported to work well with the reversible terminating nucleotides [12,15,18,20]. Jingyue Ju’s group used AmpliTaq DNA polymerase in their sequencing process (Invitrogen) due to its good compatibility to a large fluorescent group at 5′ position of pyrimidine and 7′ position of purine [15]. A mutant 9°Nm DNA polymerase was adopted by Illumina Solexa, but the mutation sites are unknown due to commercial consideration [12]. For the same reason, Helicos did not disclose the DNA polymerase used in their sequencing platform [19]. “Lightning terminator” research group screened eight commercial DNA polymerases, among which Bst DNA polymerase demonstrated the best incorporation efficiency for the “Lightning terminator” [20]. In addition, Steven A. Benner et al. found that some reverse transcriptases showed unexpected compatibility for nucleotide analogs [24].
A kind of semi-rational design strategy, reconstructed evolutionary adaptive path (REAP), developed by us, was proved to be a useful tool to genetically engineer the polymerases to accept the reversible terminator [17]. This strategy is based on the concept of “conserved but different” pattern, introduced by Steven A. Benner et al. more than two decades ago [25,26]. Figure 5 illustrates this concept. Here, sites are identified where the residue has not been absolutely conserved in all proteins in the family, but rather conserved within sub-branches of the family. The fact that the residue is conserved in sub-branches of the family means that it is under relatively strong functional constraints, implying its importance for some aspects of enzymatic function. This, in turn, infers that behavior of the polymerase is likely to change if the residue at the site is modified. The fact the residue is not absolutely conserved, however, means that the site can tolerate some degree of variations without destroying the core functionality of the protein, such as folding or catalytic activity. Therefore, such “conserved but different” sites are preferred targets for protein engineering in our REAP method. Several of the variant Taq polymerases that accept 3′-ONH2 reversible terminator have emerged through the application of this strategy [17].
Figure 5.
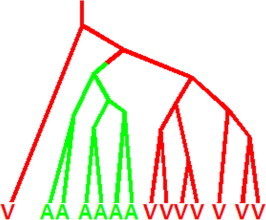
An evolutionary tree describing the amino acid at a single site in a protein family The capital letters represent different amino acid residues. Such a “conserved but different” site in a protein family is a target for protein engineering.
The opportunity and challenge of reversible termination sequencing technology
The great success of Human Genome Project significantly energized the development of more cost-effective sequencing technology [2]. Especially since 2005, the development and application of next generation sequencing (NGS) technology, characterized by its low cost and high throughput through massively parallel sequencing, displays an unprecedented technological breakthrough in biology, medicine and many related fields. Due to various advantages, reversible termination sequencing technology has been widely accepted and used in multiple NGS platforms, including the most successful NGS, Illumina/Seloxa sequencer and the first third-generation single molecule sequencer, Helicos sequencer [3,4,12,19]. This technology uses fluorescent labels as sequencing reporter, which not only affords great sensitivity and sequencing accuracy but also markedly decreases the amount of sample required. In addition, the application of reversible termination strategy effectively solves the problem of accurate identifying homopolymeric runs (such as PolyA) which are difficult to achieve using pyrosequencing technology. It was reported that the reversible termination sequencing could accurately read out more than 18 consecutive A [26,27].
Although “sequencing using cyclic reversible terminator” brings about new opportunities to the development of sequencing techniques, some problems still exists, among which, short read length has always been a major weakness. As the most popular NGS platform, the read length of the Illumina/Solexa sequencing platform is only around 100–150 bp (Tables 1 and 2). Drawbacks of short read length in shot-gun sequencing, especially de novo, are obvious.
Table 1.
Comparison of four main second-generation sequencing platforms
| Feature | Roche/454 | Ion-torrent (318) | SOLiD5500 | Hiseq2000 |
|---|---|---|---|---|
| Read size (bp) | 500–1000 | ∼200 | 60 | 100–150 |
| Throughput | 700 Mb | 1 Gb | 180 Gb | 600 Gb |
| Time required/run | 23 h | 2 h | 14 days | 8 days |
| Coverage/dpl h-genome | 10× | 20× | 100× | 60× |
| Run/dpl h-genome | 43 | 60 | 0.5 | 0.33 |
| Days/h-genome | 43 | 5 | 7 | 8 |
| Cost ($USD)/run | 7000 | 4000 | 27,000 | 22,000 |
| Cost ($USD)/h-genome | 300,000 | 240,000 | 13,500 | 2400 |
Note: dpl stands for diploid.
Table 2.
Comparison of several commercial reversible terminators
One of the fundamental reasons for short read length in “sequencing using cyclic reversible terminator” is that reversible terminator nucleotide analogs developed so far leave behind a vestige (“scar”) after cleavage of the linker carrying the fluorophore. Figure 6 illustrates the accumulation of such scars along the major groove of the DNA duplex after two rounds of sequencing extension on Illumina/Solexa platform. Accompanied by primer extension, such accumulation of scars snowballed, impairing the stability of DNA double-helix structure adversely and, thus, hindering substrate recognization and primer extension. Through adding a certain proportion of reversible terminator nucleotide analogs without the fluorophore, some base incorporations (e.g., Illumina/Solexa) leave no vestige, alleviating the impact of scars accumulation to a certain extent and extending read length. However, in order to overcome this deficiency thoroughly, new solutions are still a matter with great urgency. Recently, Metzker and colleagues made some advancement in this respect [20–23]. In their system, the scar left after cleavage of the linker is very small (Figure 2E), suggesting a potential to completely or at least partially overcome the drawback of short read length in “sequencing using cyclic reversible terminator”.
Figure 6.
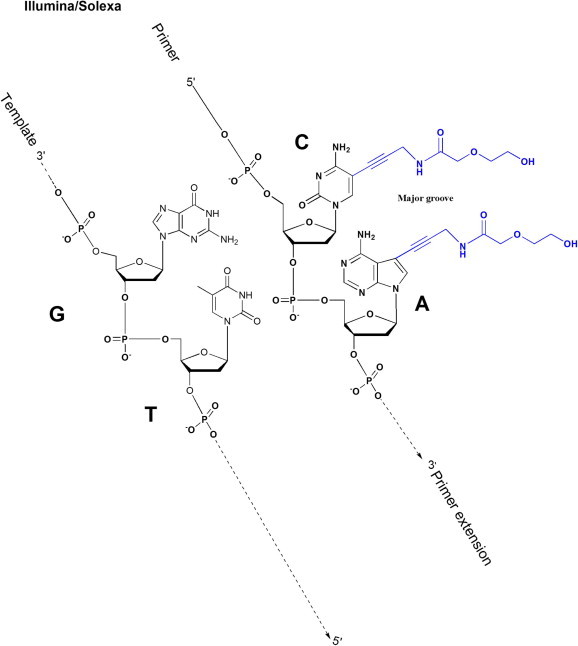
The accumulation of scars in Illumina/Solexa sequencing Schematic shows that two Illumina/Solexa reversible terminators (to cleave the fluorescent group and 3′ blocking group from the nucleotides) are added to the 3′-end of a primer in a template-directed extension reaction. The chemical structures in blue denote the molecular scars left behind.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We are indebted to the National Natural Science Foundation of China (Grant No. 31270846) and the Chinese Academy of Sciences “100-Talent Program” for the support of this work.
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Shendure J., Ji H.L. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 4.Metzker M.L. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 5.Guo J., Yu L., Turro N.J., Ju J. An integrated system for DNA sequencing by synthesis using novel nucleotide analogues. Acc Chem Res. 2010;43:551–563. doi: 10.1021/ar900255c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X., Ren L., Meng Q., Li Y., Yu Y., Yu J. The next-generation sequencing technology and application. Protein Cell. 2010;1:520–536. doi: 10.1007/s13238-010-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X., Ren L., Li Y., Zhang M., Yu Y., Yu J. The next-generation sequencing technology: a technology review and future perspective. Sci China Life Sci. 2010;53:44–57. doi: 10.1007/s11427-010-0023-6. [DOI] [PubMed] [Google Scholar]
- 8.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X., Davenport K.W., Gu W., Daligault H.E., Munk A.C., Tashima H. Improving genome assemblies by sequencing PCR products with PacBio. Biotechniques. 2012;53:61–62. doi: 10.2144/0000113891. [DOI] [PubMed] [Google Scholar]
- 10.Liu L., Li Y., Li S., Hu N., He Y., Pong R. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller C.W., Middendorf L.R., Benner S.A., Church G.M., Harris T., Huang X. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27:1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 12.Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z., Bai X., Ruparel H., Kim S., Turro N.J., Ju J. A photocleavable fluorescent nucleotide for DNA sequencing and analysis. Proc Natl Acad Sci U S A. 2003;100:414–419. doi: 10.1073/pnas.242729199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J., Zhang S., Meng Q., Cao H., Li Z., Li X. 3’-O-modified nucleotides as reversible terminators for pyrosequencing. Proc Natl Acad Sci U S A. 2007;104:16462–16467. doi: 10.1073/pnas.0707495104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J., Xu N., Li Z., Zhang S., Wu J., Kim D.H. Four-color DNA sequencing with 3′-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides. Proc Natl Acad Sci U S A. 2008;105:9145–9150. doi: 10.1073/pnas.0804023105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju, J, Kim DH, Guo J, Meng Q, Li Z, Cao H, et al. DNA sequencing with non-fluorescent nucleotide reversible terminators and cleavable label modified nucleotide terminators. PCT Int Appl Publ 2008;WO2009054922.
- 17.Chen F., Gaucher E.A., Leal N.A., Hutter D., Havemann S.A., Govindarajan S. Reconstructed evolutionary adaptive paths give polymerases accepting reversible terminators for sequencing and SNP detection. Proc Natl Acad Sci U S A. 2010;107:1948–1953. doi: 10.1073/pnas.0908463107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutter D., Kim M.J., Karalkar N., Leal N., Chen F., Guggenheim E. Labeled nucleoside triphosphates with reversibly terminating aminoalkoxyl groups. Nucleosides Nucleotides Nucleic Acids. 2010;29:879–895. doi: 10.1080/15257770.2010.536191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowers J., Mitchell J., Beer E., Buzby P.R., Causey M., Efcavitch J.W. Virtual terminator nucleotides for next-generation DNA sequencing. Nat Methods. 2009;6:593–595. doi: 10.1038/nmeth.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu W., Stupi B.P., Litosh V.A., Mansouri D., Farley D., Morris S. Termination of DNA synthesis by N6-alkylated, not 3′-O-alkylated, photocleavable 2′-deoxyadenosine triphosphates. Nucleic Acids Res. 2007;35:6339–6349. doi: 10.1093/nar/gkm689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner A.F., Wang J., Wu W., Karouby J., Li H., Stupi B.P. Rapid incorporation kinetics and improved fidelity of a novel class of 3′-OH unblocked reversible terminators. Nucleic Acids Res. 2012;40:7404–7415. doi: 10.1093/nar/gks330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stupi B.P., Li H., Wang J., Wu W., Morris S.E., Litosh V.A. Stereochemistry of benzylic carbon substitution coupled with ring modification of 2-nitrobenzyl groups as key determinants for fast-cleaving reversible terminators. Angew Chem Int Ed Engl. 2012;51:1724–1727. doi: 10.1002/anie.201106516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litosh V.A., Wu W., Stupi B.P., Wang J., Morris S.E., Hersh M.N. Improved nucleotide selectivity and termination of 3′-OH unblocked reversible terminators by molecular tuning of 2-nitrobenzyl alkylated HOMedU triphosphates. Nucleic Acids Res. 2011;39:e39. doi: 10.1093/nar/gkq1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benner S.A. Understanding nucleic acids using synthetic chemistry. Acc Chem Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 25.Benner S.A., Gerloff D. Patterns of divergence in homologous proteins as indicators of secondary and tertiary structure: a prediction of the structure of the catalytic domain of protein kinases. Adv Enzyme Regul. 1991;31:121–181. doi: 10.1016/0065-2571(91)90012-b. [DOI] [PubMed] [Google Scholar]
- 26.Bentley D.R. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Fields S. Molecular biology. Site-seeing by sequencing. Science. 2007;316:1441–1442. doi: 10.1126/science.1144479. [DOI] [PubMed] [Google Scholar]



