Abstract
Proneural bHLH activators are expressed in all neuroectodermal regions prefiguring events of central and peripheral neurogenesis. Drosophila Sc is a prototypical proneural activator that heterodimerizes with the E-protein Daughterless (Da) and is antagonized by, among others, the E(spl) repressors. We determined parameters that regulate Sc stability in Drosophila S2 cells. We found that Sc is a very labile phosphoprotein and its turnover takes place via at least three proteasome-dependent mechanisms. (i) When Sc is in excess of Da, its degradation is promoted via its transactivation domain (TAD). (ii) In a DNA-bound Da/Sc heterodimer, Sc degradation is promoted via an SPTSS phosphorylation motif and the AD1 TAD of Da; Da is spared in the process. (iii) When E(spl)m7 is expressed, it complexes with Sc or Da/Sc and promotes their degradation in a manner that requires the corepressor Groucho and the Sc SPTSS motif. Da/Sc reciprocally promotes E(spl)m7 degradation. Since E(spl)m7 is a direct target of Notch, the mutual destabilization of Sc and E(spl) may contribute in part to the highly conserved anti-neural activity of Notch. Sc variants lacking the SPTSS motif are dramatically stabilized and are hyperactive in transgenic flies. Our results propose a novel mechanism of regulation of neurogenesis, involving the stability of key players in the process.
INTRODUCTION
Transcription factors that belong to the bHLH family play fundamental roles in nearly all developmental programs, including neurogenesis, myogenesis, hematopoiesis and sex determination (1). Proneural bHLH proteins are important transcriptional activators that promote transition of neuroepithelial cells to a more differentiated state (2–4). Scute (Sc) and its vertebrate homologue Ascl1 are of immense importance in the development of central and peripheral neurons. It has been known for a long time that overexpression of Sc can induce peripheral sensory organs at ectopic sites in flies (5–7). It has recently been shown that Ascl1 alone can reprogram fibroblasts to neurons with mature morphological and electrophysiological characteristics (8–10). Other mammalian proneural proteins, e.g. Ngn2 (a more distant relative of Sc, more closely related to Drosophila Tap and Atonal), are more effective in promoting neuronal differentiation when expressed in embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) (11,12).
How do proneural proteins implement such dramatic cell fate switches? They act as transcriptional activators heterodimerized via HLH–HLH interactions with E-proteins, whose sole Drosophila representative is Daughterless (13–17). Proneural genes are dynamically expressed in neuroectodermal anlagen in patterns that prefigure neural differentiation, whereas E-proteins are more ubiquitous (1,17–19). Proneural-E heterodimers recognize their target sites, called EA-boxes, even in closed chromatin, acting as pioneer factors to de novo activate silent genes (10).
Given their potent developmental activities, it is not surprising that proneural factors are regulated by a multitude of intercellular signals (20–25). Foremost amongst these is the Notch signal, which acts throughout the animal kingdom to restrict excessive or untimely differentiation of neural cells (26,27). Despite intensive study, many aspects of the mechanism via which Notch restricts proneural activity still remain mysterious. A number of nuclear proteins have also been shown to interface with proneural protein activity (2,4,28–31). Two potent antagonists of proneural factors are the Id proteins (Extramacrochaetae in flies) and the Hes proteins (Enhancer-of-split in flies) (32–41). Both have HLH domains. Id/Emc lack a basic domain and compete with the proneurals and/or E-proteins by sequestering them in DNA binding incompetent heterodimers (42). Hes/E(spl) are bHLH-Orange repressors that bind chromatin, recruit the corepressor Groucho and repress a number of genes that are activated by proneurals (43). One way they achieve this is by binding to the transactivation domains (TADs) of Sc and Da and inhibiting their function (44,45). Importantly, Hes/E(spl) genes are the most common targets of Notch signalling and thus account to a large extent for Notch's inhibitory effect on neural differentiation46–49).
In contrast to the well-studied Id/Emc and Hes/E(spl) inhibitors of proneural factors, less is known about post-translational modifications that affect the latter's activity. Both Ascl1 and Ngn2 are heavily phosphorylated by, among others, GSK3 and Cdks (50–53). Cdk phosphorylation downregulates the biological activity of Ascl1 and Ngn2, consistent with the fact that cell cycle prolongation is needed to promote neuronal differentiation in vertebrates (50,51). GSK3 phosphorylation of Ngn2, on the other hand, is thought to affect the binding specificity to differential subsets of downstream targets (53,54). Drosophila proteins have been less intensely studied. Sc has been shown to be phosphorylated in vitro by Sgg, the GSK3 homologue, and this is thought to decrease its activity (25,55–56).
Proneural protein activity can also be modulated via effects on their stability. A few instances have been reported where mammalian proneural proteins are degraded upon Notch signalling, although all of these are in non-neural tissue contexts (57–59). For example in the pancreas, Ngn3 is degraded via a Notch/Hes1 signal. During lymphocyte differentiation E47 (an E-protein) is degraded by Notch in a MAP-kinase dependent fashion. Transcriptional activators in general are often intrinsically unstable and many TADs act as degrons (60). In some instances, activator ubiquitylation and turnover have been shown to be needed for their full transcriptional activity, e.g. in the case of c-myc and yeast Gal4 (61–64). The stability of Drosophila Sc has not been studied to date, with the exception of one study which showed that degradation of Sc, but not Da, by the ubiquitin ligase complex Phyl/Sina is required before division of sensory organ precursor (SOP) cells can take place (65).
We have examined the stability of Sc in relation to its interacting proteins Da and E(spl)m7 in both cultured cells and Drosophila tissues. We present data that two of the three TADs in the Da/Sc heterodimer act as degrons. The Da AD1 plays a major role in the degradation of Sc, acting in trans, without promoting simultaneous Da turnover. The Sc TAD, on the other hand, is a major degron only when Sc is not complexed with Da. We also show that a major determinant for Sc turnover is a conserved phosphorylation motif, 268-SPTSS. Abolishing this motif makes Sc more stable and more active in promoting sensory organ formation in vivo. E(spl)m7, a known inhibitor of proneural activity, is now shown to also promote Da and Sc degradation; reciprocally, Da/Sc promote E(spl)m7 degradation. These mutual degradation events use two different mechanisms that have similarities and differences. Both of these events require that E(spl)m7 interact with Da/Sc. Sc degradation by E(spl)m7 also requires the Sc SPTSS motif and, as a consequence, phosphomutant Sc is less sensitive to inhibition by Notch/E(spl). Our analysis reveals new mechanisms of regulation at the post-translational level among these three interacting bHLH proteins, Sc, Da and E(spl)m7, which ultimately ensure robust promotion or inhibition of neural differentiation.
MATERIALS AND METHODS
DNA constructs
The EE4-luc reporter and the following expression constructs in the RactHAdh vector (or Ract for short) have been described in previous publications: myc-GFP, Gro, myc-m7, myc-EQRQTKHQ, mγ, mδ, His-Da, Xpress-Ubi (45,66–68). The 3xmyc-pBS vector was used as the basis for myc-tagging all expression constructs and is described in detail in the Supplementary Materials. Each myc-tagged gene was originally constructed by cloning the open reading frame (ORF) of the gene into the 3xmyc-pBS vector, sequence verification and subsequent subcloning into Ract vector for tissue culture expression (list of these plasmids in supplementary data). For Ract-His a sequence derived from pRSET (Invitrogen) ATGCGGGGTTCTCATCATCATCATCATCATGGTATGGCT AGC was inserted to Ract (encoding the peptide MRGSHHHHHHGMASSRPAGMQAI RCTLTFFS). All His tagged genes were constructed by inserting the coding sequence of the gene into Ract-His vector. For HA-m7 in Ract a sequence encoding MYPYDVPDYAGIPLEF was appended to the 5′ end of the m7 variants and inserted into Ract. Luciferase and lacZ were cloned by PCR into Ract vector (Ract-luc and Ract-lacZ plasmids).
For transgenesis 3xmyc-tagged constructs of Sc[RQEQ], Sc[m3p], Sc[1320] and Sc[m5p] were subcloned into pUAST (69).
More detailed description of all new constructs can be found in the Supplementary Materials.
Drosophila stocks and crosses
The new UAS-sc variants were generated by P-element transformation into a yw67c23 strain. Other UAS-E(spl) variants, as well as UAS-sc (and variants) and EE4-lacZ, are described in (45,67–68). Gal4 lines and other stocks were obtained from the Bloomington Drosophila Stock Center.
Histochemistry
Immunofluorescence was performed as in (70). Antibodies used were guinea-pig anti-Sens (a gift from Hugo Bellen), rabbit anti-myc (SantaCruz) and mouse anti β-galactosidase (Promega), mouse-anti-E(spl) (323, a gift from Sarah Bray).
Cell culture assays
S2 cell transfections and luciferase assays were performed as previously described (44). For luciferase assays each transfection experiment was performed in triplicate and repeated at least three times to ensure reproducibility. Data shown represent the average and standard deviation of triplicates for a typical assay. For proteasomal inhibition cells were treated for 3–6 h with 50 μM of MG132 (Sigma, C 2211). For half-life experiments, transfected cells were treated with 50 μg/ml cycloheximide (CHX, Sigma, C7698) to block translation. For Sgg kinase inhibition, cells were incubated with 80 mM LiCl for 4 h. For Cdk8 kinase inhibition, cells were treated with Senexin A (10 μM) (a gift from Igor B. Roninson, (71)).
Cell lysis under non denaturing conditions
Cells were lysed in NP40 lysis buffer (1% Nonidet P-40, 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM PMSF, 1 μg/ml Leupeptin, 1 μg/ml Pepstatin, 1 μg/ml Aprotinin, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF), by using 3x freeze/ thaw cycles. Lysates were cleared by centrifugation (20 min, 13 000 rpm, 4°C) and transfection efficiency was determined by either luciferase or beta-galactosidase activity. For western blots analysis, samples were loaded after adding 3x Laemmli buffer according to their transfection efficiency.
Co-immunoprecipitation
Transfected cells were lysed with NP40 Lysis buffer and were incubated with 4 μg of rabbit anti-myc for 3 h at 4°C with mixing. Protein A -agarose (Millipore) beads were then added and the reaction mixtures were further mixed for 3 h at 4°C. The immunoprecipitates were separated from supernatant by centrifugation and washed three times with NP40 Lysis buffer for 7 min each wash. Proteins were extracted from the agarose beads by boiling in 1x Laemmli loading buffer and resolved on 10% sodium dodecyl sulphate (SDS)–polyacrylamide gels. For the IP of the ubiquitination experiment in the Figure 1E, 50 μM MG132 and 10 mM N-ethylmaleimide was added to the lysis buffer.
Figure 1.
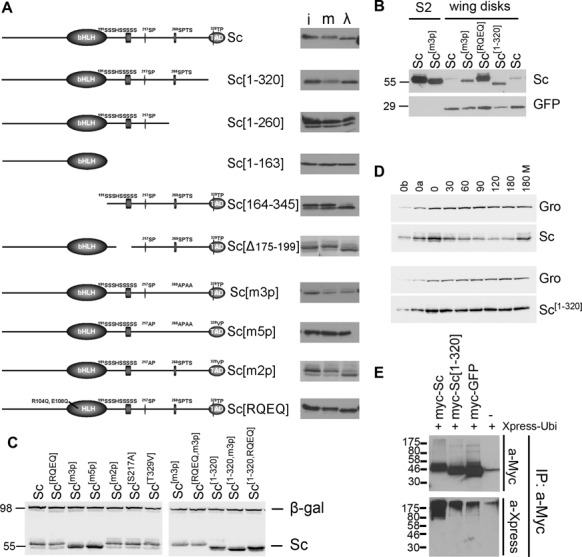
Phosphorylation and stability of Sc. In all panels N-terminally 3xmyc-tagged Sc proteins were detected by anti-Myc tag antibody. (A) The schematic depicts the Sc protein, highlighting its bHLH domain, a Ser-rich stretch and three (S/T)P conserved motifs in the C-terminal half. A series of deletion and point mutations are shown. Next to each the western blot shows (i) the Sc variant expressed in S2 cells (input), (m) the S2 cell extract mock-treated with λ-phosphatase buffer and (λ) the same extract treated with λ-phosphatase. An increase in migration indicates dephosphorylation. (B) Western blot containing extracts from Sc-transfected S2 cells or Sc expressing wing disks (ap-Gal4; UAS-GFP UAS-myc-sc). Note that Sc[m3p] migrates faster than Sc(wt) both in S2 cells and in fly tissue. GFP is shown as a loading control. (C) A comparison of all Sc variants from S2 cells co-transfected with Ract-myc-sc and Ract-lacZ; the β-galactosidase protein is detected as a loading control. Sc[m3p], Sc[m5p] and the versions truncated after residue 320 (lacking the acidic C-terminal TAD) accumulate to higher levels. Also note the faster migration of the phosphomutants (m3p and m5p). (D) Degradation kinetics of transfected myc-Sc and myc-Sc[1–320] after protein synthesis inhibition by cycloheximide. Endogenous Groucho (a stable protein) is used as a loading control. 0, 30,…, 180 refer to minutes after cycloheximide addition. 180M: 180 min after cycloheximide+MG132 addition. 0a, 0b: 1/2 or 1/4, respectively, of input (0 min) for densitometry calibration. One indicative experiment is shown out of four (Sc) or five (Sc[1–320]) repeats. (E) The indicated myc-tagged protein expression constructs were co-transfected with Xpress-tagged ubiquitin. Ubiquitylated species were detected after IP with anti-Myc and western blot (WB) with anti-Xpress (bottom panel). The top panel shows an anti-Myc WB to estimate the amount of Myc-tagged protein in each IP. Cells had been treated with MG132 (for 5 h) before lysis. Note higher level of ubiquitin signal in the Sc lane versus Sc[1–320] and GFP.
His pull down
107 transfected cells were resuspended and mechanically lysed in 1 ml Urea lysis buffer (8M Urea, 50 mM Tris pH 8.0, 50 mM NaCl, 1% Tween 20, 5 mM mercaptoethanol, 1 mM PMSF) by passing through a 25-gauge needle 7–10 times to shear DNA, and clearing by centrifugation (10 min, 13 000 rpm, RT). Lysates were incubated with 75 μl bed volume of Ni2+-NTA-agarose (Qiagen) for 2–3 h at room temperature in a rotator. The resin was washed three times with Urea Lysis buffer for 10 min each and 2× quick washes with PBS+1 mM PMSF. Beads were resuspended and boiled in 100 μl 1× Lammli buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 5% beta-mercaptoethanol and 0.012% bromphenol blue).
Normalization of sample loading
Fifty nanogram of Ract-luc (or 200 ng of Ract-lacZ) was included in each transfection (106 cells) of a total of 2 μg DNA—transfections were topped up using empty Ract vector. We lysed the cells in 100 μl and 20 μl were kept for luciferase or β-galactosidase assay. In the rest of the lysate we added 3× Laemmli buffer (1× final) and froze them at −80°C, until the electrophoresis of the samples. The linearity of the enzymatic assays was tested using pilot transfections.
Protein extracts from Drosophila tissue
Dissected wing imaginal disks were added directly to 1× Laemmli buffer and frozen at −80°C. Samples were mechanically disrupted, boiled and spun down before loading.
λ phosphatase treatment
Protein phosphatase treatment was conducted by incubating S2 lysate (lysed with NP40 Lysis buffer, without the phosphatase inhibitors Na3VO4/NaF) with 400 U of lambda phosphatase (Sigma, P 9614) or without phosphatase (for mock treatment) for 30 min at 30°C.
Western blotting
Protein lysates were separated on SDS–polyacrylamide gels (10 or 12%) and electrotransferred onto nitrocellulose membranes. The membranes were blocked with PBST buffer (1× PBS, 0.05% Tween20) containing 5% nonfat dry milk and probed overnight using the following primary antibodies: rabbit anti-myc (Santa Cruz), mouse anti-Gro (our lab), mouse anti-RGSHis (Qiagen), mouse anti-HA (Covance), rabbit anti-β-galactosidase (Cappel), mouse -anti-Arm N2 7A1 (DSHB), rabbit anti-SUMO (a gift from Spyros Artavanis-Tsakonas), mouse-anti-myc (9B11, Cell Signaling) or anti-m-Xpress. For ubiquitin detection (Santa Cruz -8017, mouse anti-Ubi, P4D1), before blocking, the blots were boiled for 30–40 min in water to denature Ubi. Membranes were incubated with horseradish peroxidase-linked secondary anti-mouse or anti-Rabbit antibodies (Jackson Immunoresearch) and bound antibodies were visualized using ECL chemiluminescent substrate (Pierce).
Densitometry of western blots
To quantitate protein levels after cycloheximide treatment of cells, we imaged chemiluminescence on a Fujifilm LAS-3000 CCD camera. We took care to ensure that the image was not saturated by using control lanes where 1/2 or 1/4 of the starting sample (heaviest band) was loaded. Densitometry was performed using Image J software. The data were fitted to exponential decay curves using the OriginPro v7 software. Experiments were performed 2–5 times to ensure consistency. Means and standard deviations are presented in the text and figures. As cells started dying by 5 h after cycloheximide application, we ended all our timecourses at ∼4 h. Therefore, half-lives much longer than 4 h could not be reliably estimated.
Electromobility shift assays
Electromobility shift assays were performed using in vitro transcribed-translated proteins (TnT reticulocyte lysate, Promega), as described in (70). The oligonucleotides used as a probe are the EAB oligos described in (68).
RESULTS
Sc is phosphorylated in S2 cells and fly epithelia
As stability of many transcription factors is regulated by their phosphorylation state, we sought to determine whether Sc is phosphorylated, before embarking on studies of its turnover. For this reason we N-terminally tagged it with a 3×Myc epitope and expressed it in Drosophila cultured S2 cells, since there is no available antibody to detect endogenous Sc protein. We confirmed that the epitope tag did not compromise the activity of Sc by various functional assays in both S2 cells and after expression in Drosophila tissues (see Materials and Methods, e.g. Figures 3,6,7). Sc produced in S2 cells migrated as a single band in western blots, whose mobility was markedly decreased upon incubation with λ phosphatase (Figure 1A). This suggests that the bulk of Sc protein is phosphorylated.
Figure 3.
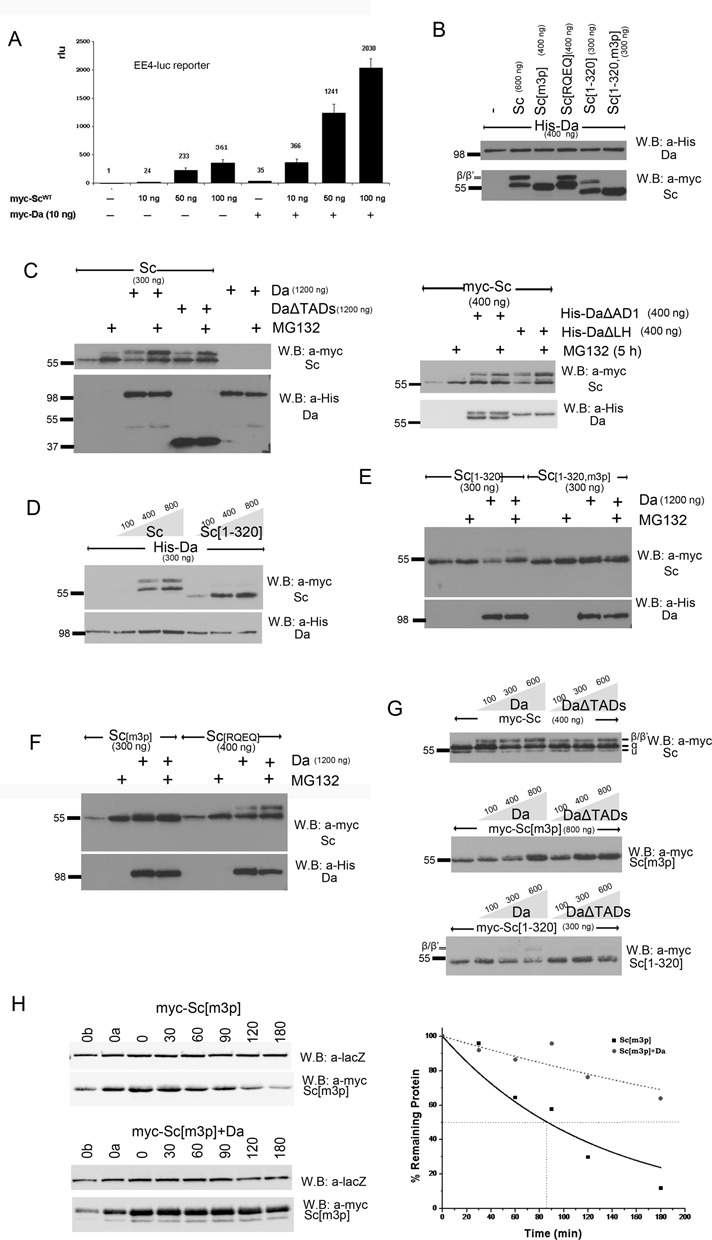
Proteasomal degradation of Sc in the presence of Da depends on phosphorylation of Sc, its DNA binding ability and on the AD1 domain of Da. (A) Luciferase assays on transiently transfected S2 cells with an EE4-luc reporter. Activation levels by increasing amounts of myc-Sc in the presence or absence of co-transfected Da. Relative luciferase units (rlu) are shown and are normalized on the activity of the reporter gene alone, set to 1. Averages/standard deviations of triplicates are shown. In all subsequent panels N-terminally 3xmyc-tagged Sc proteins or His-tagged Da were detected. The amount loaded was adjusted according to the activity of luciferase expressed from control plasmids (Ract-luc) transfected at the same time as the indicated plasmids (to measure transfection efficiency). (B) Whole cell extracts from S2 cells co-transfected with expression constructs as indicated. Note that Da co-expression causes the appearance of new Sc bands that run about 5 kd above the major band. The triple phosphomutant (m3p) abolishes this modification. (C) S2 cells transfected with the indicated constructs were treated (+) or not (-) with MG132 (for ∼5 h) before lysis. Proteasome inhibition increased the levels of Sc when it was expressed alone or together with Da. However, upon co-expression with DaΔTADs, only the β/β’ forms showed significant stabilization. Proteasome inhibition did not seem to increase the levels of Da (lower panel). The same experiment was done using deletion variants DaΔAD1 and DaΔLH (right panel). Proteasome inhibition increased the levels of Sc when it was expressed alone or together with DaΔLH. However, upon co-expression with DaΔAD1, only the β/β’ forms showed significant stabilization. Proteasome inhibition did not seem to increase the levels of Da variants (lower panel). (D) Blots from S2 cell extracts transfected with Da and increasing amounts of Sc or Sc[1–320]. Note that increasing Sc or Sc[1–320] did not diminish the levels of Da; if anything, it caused a small upregulation. (E,F) Cells transfected with Sc[1–320], Sc[1–320, m3p], Sc[m3p] and Sc[RQEQ] in the absence or presence of His-Da were treated or not with MG132. Note that proteasome inhibition stabilized solo Sc[m3p] and Sc[RQEQ], but had no effect when Da is co-expressed (D). Sc[1–320] solo was barely stabilized by MG132 treatment, but when Da was co-expressed MG132 stabilized Sc[1–320]. Finally, MG132 did not seem to increase the levels of Sc[1–320, m3p], whether expressed alone or with Da. (G) Steady-state levels of Sc, Sc[m3p] or Sc[1–320] with increasing amounts of Da variants. Sc[m3p] levels increased with increasing Da or DaΔTADs. Sc[1–320] levels dropped with increasing Da, but were not affected by DaΔTADs. For comparison, Sc levels were not significantly affected by either Da or DaΔTADs; if anything, a weak stabilization at low Da levels was observed. Note that the β form is much more prominent for Sc than for Sc[1–320] and is absent in Sc[m3p]. (H) Degradation kinetics of myc-Sc[m3p] transfected alone or co-transfected with Da after protein synthesis inhibition by cycloheximide. Co-transfected β-galactosidase (lacZ, a stable protein) was used as a loading control. 0, 30,…, 180 refer to minutes after cycloheximide addition. 0a, 0b: 1/2 or 1/4, respectively, of input (0 min) for densitometry calibration. One indicative blot and densitometry plot is shown for each condition. The estimated half-lives are (number of repeats in parenthesis): Sc[m3p] 75±4 min (n = 4), Sc[m3p]/Da 230±130 min (n = 2).
Figure 6.
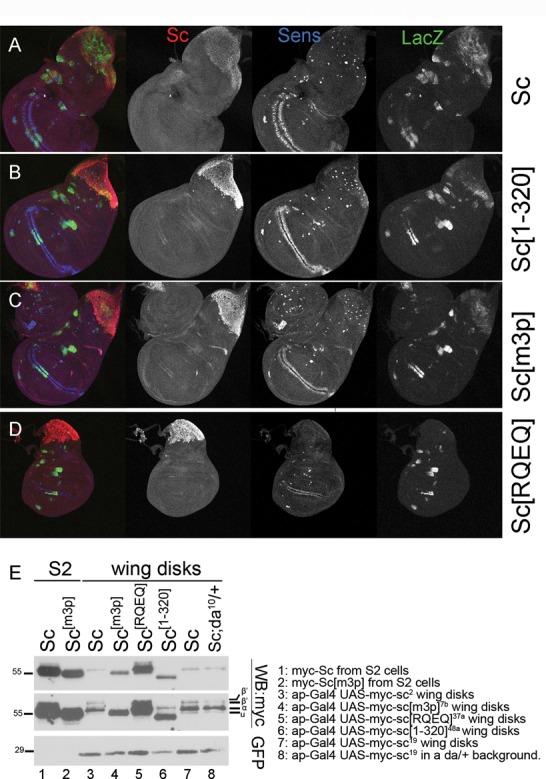
Differential activity and stability of Sc variants in vivo. (A-D) All panels show wing disks carrying one copy of the EE4-lacZ reporter and overexpressing the indicated myc-tagged Sc variants in the pnr domain (pnrGal4 driver). Immunostaining: Sc (anti-myc, red), β-galactosidase (anti-β-gal, green) and Sens, a SOP marker, (anti-Sens, blue). Note that Sc[m3p] and Sc[1–320] produced a higher number of SOPs than wt Sc. The transgenic lines used for Sc[1–320] (line 48a) and Sc[m3p] (line 7b) cause pupal/pharate lethality, whereas expression of UAS-myc-sc (line 2) or UAS-myc-sc[RQEQ] (line 13a) do not affect viability. (E) (Same blot as Figure 1B with a longer exposure added.) Western blot containing extracts from Sc-transfected S2 cells or Sc expressing wing disks (ap-Gal4; UAS-GFP UAS-myc-sc), as indicated. The transgenic lines used are listed next to the blot. UAS-GFP was co-expressed in all wing disk samples and is detected for quantitative comparison. Sc[RQEQ], Sc[1–320] and Sc[m3p] accumulated to higher levels than Sc (wt). Note that the Da-dependent modifications of Sc (the β/β’ bands) were very pronounced in wing disks, suggesting that Da is expressed in relatively high endogenous levels in this tissue. Upon halving the dose of da+ (last lane), we noticed that the α band was enhanced while β/β’ were reduced.
Figure 7.
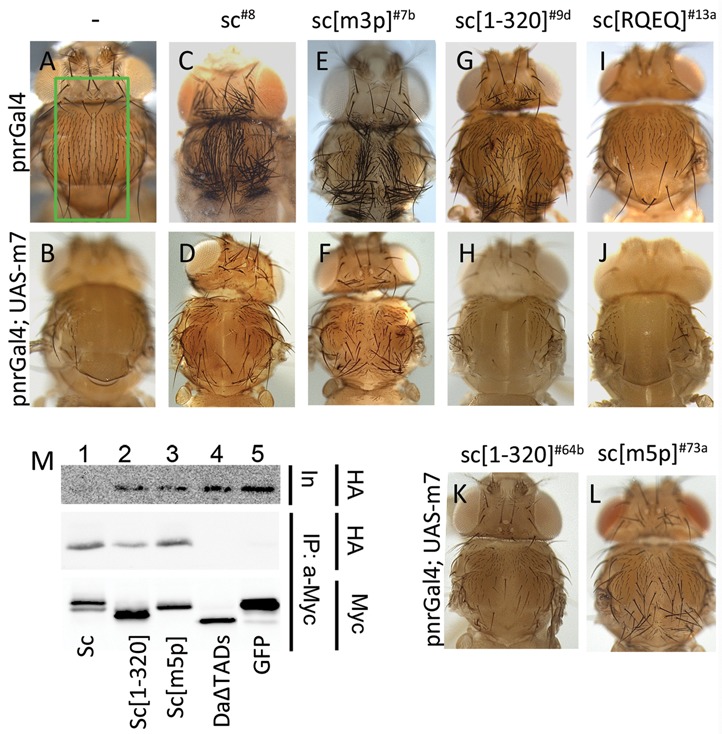
Phosphorylation of Sc in vivo modulates its susceptibility to E(spl) m7 repression. (A, C, E, G, I) Thoraces from pnr-Gal4 flies overexpressing the indicated Sc variants. The pnr expression domain is boxed in panel A. Note the production of ectopic bristles by all Sc variants, except Sc[RQEQ], where mild bristle loss is seen (I). C (UAS-myc-sc8) and E (UAS-myc-sc[m3p]7b) are pharate escapers. G (UAS-myc-sc[1–320]9d) is the only viable line with pnr-Gal4 and I (UAS-myc-sc[RQEQ]13a) is viable, as are all Sc[RQEQ] lines. (B, D, F, H, J) Thoraces from pnr-Gal4 flies overexpressing the indicated Sc variants along with E(spl)m7. m7 strongly inhibits ectopic bristle production, with the exception of panel F, where a large number of bristles persist. (K) pnr-Gal4; UAS-m7. UAS-myc-sc[1–320]64b; line 64b never gives pharate escapers when expressed alone (without m7) and produces many more SOPs in larvae than line 9d (G). Still it is more effectively suppressed by UAS-m7 than Sc[m3p] (F). (L) pnr-Gal4; UAS-m7. UAS-myc-sc[m5p]73a; line 73a never gives pharate escapers when expressed alone (without m7) but is less susceptible than the equally inviable Sc[1–320]64b to m7 overexpression. (M) myc-tagged proteins (as indicated) were expressed and immunoprecipitated from S2 cells. Immunoprecipitation efficiency (Myc, lower panel) and the presence of coprecipitated HA-tagged m7 (HA, middle panel) were assayed. Upper panel shows the levels of the HA-m7 in the cell extracts (input). All Sc variants interact with HA-m7. GFP, the negative control and DaΔTADs (lacks the AD1 and LH domains) do not immunoprecipitate HA-m7.
To map the regions subject to phosphorylation we generated a number of deletions and point mutations and assayed their mobility with or without λ phosphatase treatment. Phosphorylation sites were mapped to the C-teminal half of the protein, beyond residue 260, while a serine-rich stretch from residue 175 to 199 was dispensable for the phosphorylation of Sc. We focused on the (S/T)P sites of the C-terminal half (S217, S268 and T329), as such motifs are often kinase targets, e.g. for Cdk's and MAPK's. S268 is part of a highly conserved SPxxS motif, found in all Sc homologues (data not shown). We altered two, three or five S/T residues to non-phosphorylatable alanines or valines and named the mutants m2p, m3p or m5p, respectively; see Figure 1A and Supplementary Data for description. From these mutants we concluded that phosphorylation of Sc is primarily dependent on this 268-SPTSS motif (Figure 1A). We also tested a basic domain point mutant, Sc[RQEQ]. This carries two amino acid changes, R104Q and E108Q, which abolish DNA binding with consequent loss of activity (Supplementary Figure S1). In agreement with the Sc[164–345] truncation, which lacks the entire bHLH region, the basic domain mutant did not abolish phosphorylation. When isolated from larval wing disks Sc[m3p] migrated faster than wt Sc, consistent with Sc being phosphorylated via its SPTSS motif also in native tissues (Figure 1B).
To get some insight on the possible kinases that modify Sc, we tested two kinase inhibitors. As it has been suggested that the 268-SPTSS motif is a target of Sgg (56), we treated transfected cells with LiCl, an inhibitor of Sgg/GSK3. This did not greatly affect the major bands of Sc or Sc[m3p], but it did cause a significant reduction of minor bands migrating just above the major bands (Supplementary Figure S2A). These minor bands become more prominent upon co-expression with the Sc heterodimerization partner, Da, and depend on residue S217 (Supplementary Figure S2B). We therefore think that Sc can be phosphorylated by Sgg in the S217 region, whereas 268-SPTSS is at best a minor target of Sgg. We noted that co-expression of Sc with Da, besides increasing the slightly more retarded hyperphosphorylated bands, also produced a much more retarded higher molecular weight (MW) form of Sc (which we named β). However, this upper β band is not abolished upon λ-phosphatase treatment (Supplementary Figure S2C), suggesting that it does not represent a phosphorylated form.
Recently an inhibitor specific for Cdk8, a kinase implicated in transcriptional regulation, has been developed (71). Treatment of transfected S2 cells with this inhibitor, Senexin A, did not affect the mobility of the main band of Sc or Sc[1–320], but, surprisingly, it almost completely abolished the high MW β form (Supplementary Figure S2D). Cdk8 is, therefore, not likely responsible for the basal phosphorylation of Sc, but is indirectly needed for the generation of the novel β form. To confirm the effectiveness of the inhibitor, we immunoblotted treated cell extracts for Armadillo. We found that Senexin A causes a reduction in Arm levels (Supplementary Figure S2E), consistent with a role of Cdk8 to inhibit the transcription factor dE2F1, which in turn promotes Arm degradation (72).
The Sc TAD is a degron
In the course of our analysis of the mutant 3xMyc-tagged Sc proteins, we noticed that some variants, most notably the C-terminal truncations, accumulated to significantly higher steady-state levels than wt Sc (Figure 1C), even though all were expressed from the same actin 5C promoter. A similar effect was observed when Sc variants were expressed in fly wing disks (Figure 1B). We measured protein half-lives after protein synthesis inhibition by cycloheximide. Sc showed a very short half-life of 40±10 min (n = 4), whereas Sc[1–320] was stable with a half-life of 127±12 min (n = 5) (Figure 1D, Supplementary Figure S3A and B). Addition of the proteasome inhibitor MG132 stabilized Sc, implicating the proteasome in its turnover. After immunoprecipitating Sc, we detected significantly more poly-Ub adducts on wt Sc than on Sc[1–320], in agreement with the former's higher turnover rate (Figure 1E). This suggests that the Sc TAD (residues 321–345) (45) acts as a degron as do many other TADs (60).
Compared to wt Sc, we found the half-lives of Sc[m3p] and Sc[RQEQ] to be somewhat prolonged, at 75±4 min (n = 4) and 60±20 min (n = 8), respectively (Supplementary Figure S3A and B). Therefore DNA binding and phosphorylation seem to contribute to the rapid turnover of Sc at the proteasome, although the most critical factor is the presence of the TAD.
In an earlier study of the Sc dimerization partner Da, we characterized two TADs on Da, the N-terminal AD1 (aa 1–160) and a centrally located one, named LH (aa 265–417) (68). To ask whether these might also act as degrons, we fused each one to the C-terminus of Sc[1–318] and measured their half-lives in S2 cells after protein synthesis arrest (Supplementary Figure S3C and D). Whereas the Sc[1–318]-LH half-life of >3 h was longer than that of Sc[1–320], Sc[1–318]-AD1 had a significantly shorter half-life (78±11 min), still longer than that of wt Sc. Therefore, Da AD1 also acts as a degron, but is weaker than the Sc TAD.
Da is a very stable phosphoprotein
We wanted to test the ability of the Da TADs to act as destabilizing signals in their native context. We therefore measured the half-life of Da after a cycloheximide block of translation and found it to exceed 4 h (275±90 min, n = 3) (Figure 2A and B). No significant stabilization was detectable for DaΔTADs, an N-terminal truncation up to aa414 (68), which deletes both TADs. Upon incubating cells with the proteasome inhibitor MG132, no significant accumulation of Da was detected. The same was true for deletion variants lacking one or both TADs (Figure 2C). We conclude that Da is a very stable protein and AD1, which has a degron activity when fused to Sc (Figure 1D), does not act as a degron in its native context.
Figure 2.
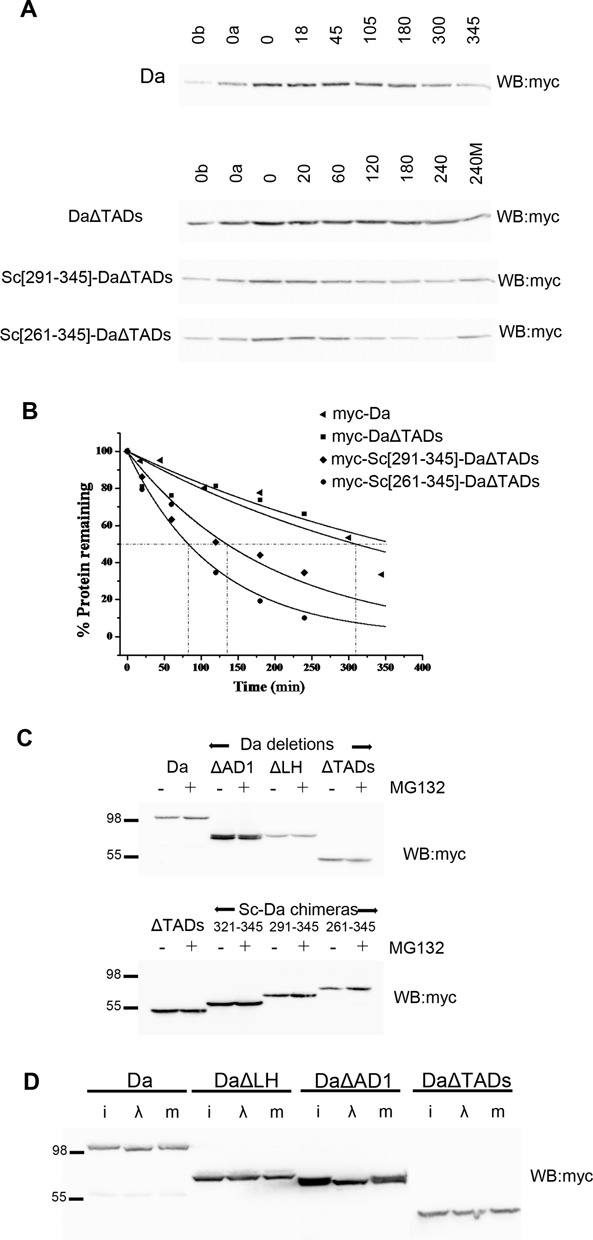
Stability and phosphorylation of Da. In all panels N-terminally 3xmyc-tagged Da proteins were detected. (A) Turnover kinetics of Da and three variants after protein synthesis block. Lanes are labeled as in Figure 1D. (B) The densitometry plots from the blots of panel B. Each measurement was repeated 2–3 times; one indicative experiment is shown. The estimated half-lives (number of repeats in parenthesis) are: Da 275±90 min (n = 3), DaΔTADs > 240 min (n = 3), Sc[291–345]-DaΔTADs 141±43 (n = 2), Sc[261–345]-DaΔTADs 83±10 (n = 2). (C) Da proteins detected from transfected S2 cells treated (+) or not (-) with MG132 for 5 h before lysis. Proteasome inhibition does not seem to increase the levels of Da or versions deleted for one or both of its TADs. Instead, it detectably increased the levels of Da–Sc chimeras bearing the Sc TAD and nearby residues. (D) λ-phosphatase treatment of S2 cell extracts expressing the indicated Da variants. i: input, λ: treated, m: mock-treated. Note the increase in migration for myc-Da (wt) and myc-DaΔAD1, but not myc-DaΔLH or myc-DaΔTADs.
To examine if the Sc TAD (aa 321–345) is a transferable degron we fused different lengths of the Sc C-terminus to the N-terminus of DaΔTADs (Da[415–710]). The constructs Sc[321–345]-DaΔTADs, Sc[291–345]-DaΔTADs and Sc[261–345]-DaΔTADs were all expressed under the same promoter in S2 cells and their half-life was estimated after protein synthesis block. The Sc[261–345] fragment produced a dramatic destabilization of DaΔTADs: from a half-life of >4 h to 83±10 min (n = 2), Sc[291–345] had a milder effect (141±43 min; n = 2) (Figure 2), whereas Sc[321–345] gave no destabilization (data not shown). MG132 treatment stabilized Sc[291–345]-DaΔTADs and Sc[261–345]-DaΔTADs, suggesting that Sc–Da fusions are degraded at the proteasome (Figure 2). Therefore, the Sc TAD acts as a transferable degron only if extended by at least 31aa and is even more effective if extended by 61aa, to encompass the conserved SPTSS motif. It should be noted that in the context of Da, the Sc degron cannot achieve the extremely high turnover rates seen in its native Sc context (40 min half-life), suggesting that other regions of the Sc and Da proteins enhance and suppress (respectively) degradation.
Finally, we asked whether Da is phosphorylated. Upon treatment with λ phosphatase the MW of Da was significantly decreased, whereas that of DaΔTADs was unaltered (Figure 2D). Da phosphorylation was shown to be dependent on the LH region (aa 211–463), as its deletion abolished the MW shift by λ phosphatase.
Da binding alters the turnover parameters of Sc
Since Da and Sc act as a heterodimer, we decided to test whether they affect each other's phosphorylation and stability. Although S2 cells express basal levels of Da (and no detectable Sc; (73)), this is not sufficient to heterodimerize with the high amount of Sc expressed under our transfection conditions. We concluded this by comparing induction levels of the Da/Sc responding EE4-luc reporter: whereas Sc transfection activated this reporter weakly, co-transfection of Da was needed to activate it to higher levels (Figure 3A), suggesting that endogenous Da was limiting. We therefore re-evaluated Sc electrophoretic mobility as well as stability upon co-transfection with Da.
As already mentioned, Da co-expression caused the appearance of a high MW Sc band (form β), which ran about 5 kd above the main Sc form and was not abolished by λ phosphatase treatment (Supplementary Figure S2B and C). Testing our cohort of Sc variants, we found that appearance of the β form was critically dependent on the integrity of the SPTS motif, as neither Sc[m3p] nor Sc[m5p] showed this modification (Figure 3B and data not shown). Sc[RQEQ] and Sc[1–320], on the other hand, did show the higher MW band, albeit at reduced levels. We considered the hypothesis that the β form of Sc might represent a monoubiquitylated form. We tested this by immunoprecipitating Sc and immunoblotting for ubiquitin. No monoubiquitylated form was detectable, although polyubiquitylated adducts were seen both in the presence and absence of co-expressed Da (Figure 1E and data not shown).
Since Da co-expression has dramatic effects on Sc post-translational modification, we asked whether it might also affect its turnover, especially since Da is a very stable protein (Figure 2) compared to unstable Sc (Figure 1). To determine the factors needed for Sc turnover in the presence of Da, we monitored levels of Sc accumulation in the presence or absence of proteasome inhibitor MG132. When expressed alone, Sc was significantly stabilized by MG132 treatment, as expected. This was also the case upon Da co-expression, where both low (α) and high (β) MW forms overaccumulated (Figure 3C), However, upon co-expression with DaΔTADs, only the β form showed significant stabilization. It therefore seems that the presence of the Da TADs is needed to promote proteasome degradation of Sc, with the exception of a small fraction that gets modified to the β form, which is unstable anyway. By co-expressing Sc with the DaΔAD1 and DaΔLH variants, we mapped this destabilizing activity of Da to AD1 (Figure 3C). If Da and Sc are coordinately turned over upon heterodimerization, we would predict that co-expression with Sc would promote Da degradation. However, there was no increase of Da upon MG132 treatment. Also, gradually increasing Sc did not diminish the levels of Da; if anything, it caused a small upregulation (Figure 3D). Therefore, heterodimerization with Sc cannot destabilize Da. Instead, in the Da/Sc heterodimer, Da AD1 acts as a trans-degron, promoting the turnover of the partner protein Sc, while at the same time, Da, which is stable to start with, is further stabilized by Sc.
When we tested our panel of Sc variants with Da co-expression, we found that Sc[1–320] was stabilized by MG132, but Sc[m3p] and Sc[1–320, m3p] were not (Figure 3E and F). Based on our earlier mapping of the major Sc degron to its C-terminal TAD, this was unexpected; Sc[1–320] should not be much stabilized by MG132. Sc[RQEQ]/Da was also quite insensitive to MG132, implicating DNA binding as a further prerequisite for Da/Sc turnover. It therefore appears that Sc has two distinct modes of degradation. In the presence of Da, Sc turnover requires the SPTSS motif and DNA binding, as well as the presence of Da AD1. In the absence of Da, Sc turnover depends critically on the Sc TAD.
The transition from one to the other mode of Sc degradation can be nicely visualized by measuring its steady-state levels upon gradually increasing Da. Sc[1–320] levels dropped with increasing Da (Figure 3G). This is consistent with this originally stable protein (in the absence of Da) being shunted to a different degradation pathway, which does not strongly require the Sc TAD anymore. In contrast, Sc[m3p] (Figure 3G) and Sc[RQEQ] (data not shown) were stabilized upon increasing Da. This is consistent with originally unstable proteins being shunted to the Da-dependent pathway, which now requires the SPTSS motif and DNA binding. Wt Sc on the other hand was not dramatically affected by Da co-expression, consistent with it being unstable both on its own and as a Da heterodimer (Figure 3G). Using a block in protein synthesis, we estimated the half-life of Sc[m3p] and found it to increase from 74±4 min to >3 h (230±130 min), when Da was co-expressed (Figure 3H, cf with Supplementary Figure S3A and B). Green fluorescent protein (GFP) and three other Sc variants, Sc[164–345], Sc[1–163] and Sc[1–320, m3p], were unaffected by increasing Da (Supplementary Figure S4). The latter two lack both degrons, the C-terminal TAD and the SPTSS, so they were very stable under all conditions. GFP and Sc[164–345] cannot interact with Da and so its inability to be affected by increasing Da levels is consistent with a model where the effects of Da on other Sc variants are elicited by direct Da–Sc interaction.
Enhancer of split proteins degrade Scute and Da
Since Sc exhibited dramatic post-translational modification and stability changes when its interactor Da was present, we examined the effects of co-expressing another interactor, an E(spl) protein. We used myc-tagged versions of both sc and E(spl)m7 in order to be able to simultaneously detect their protein products, which separate well in SDS PAGE. When we expressed myc-Sc together with myc-E(spl)m7 (which we will shorten to ‘m7’ from here onward for simplicity) in S2 Drosophila cells we observed that Sc protein is reduced the more m7 we provide (Figure 4A). This was not due to repression of the act5C promoter, as an act5C-mycGFP construct did not show a similar response to myc-Sc (Figure 4B). We therefore conclude that m7 specifically induces the degradation of Sc.
Figure 4.
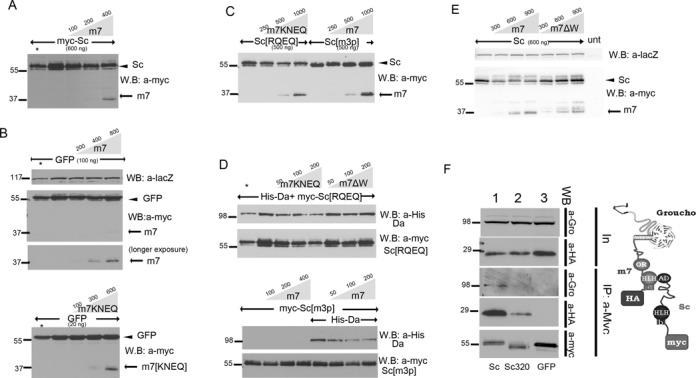
Scute and Da degradation by E(spl)m7 is independent of DNA binding, but depends on m7–Gro interaction and the Sc SPTSS motif. (A–E) Whole cell extracts from S2 cells co-transfected with expression constructs as indicated. In all panels N-terminally 3xmyc-tagged Sc or E(spl)m7 proteins or His-tagged Da were detected. The amount loaded was adjusted according to the activity of luciferase (or β-galactosidase, for blot 4E) expressed from control plasmids (Ract-luc or Ract-lacZ) transfected at the same time as the indicated plasmids (to measure transfection efficiency). In the anti-myc blots, the upper band (arrowhead) is myc-Sc or myc-GFP and the lower (arrow) is myc-m7. (A) Increasing levels of m7 led to degradation of Sc. (B) The same effect was not observed for GFP. (C) Increasing levels of m7KNEQ degraded Sc[RQEQ]; whereas increasing levels of m7 did not degrade Sc[m3p]. (D) Increasing levels of m7KNEQ could degrade Da together with Sc[RQEQ]; whereas m7ΔW had impaired ability to do so. Also Sc[m3p] could not be degraded by m7, even in the presence of Da, which was diminished by increasing levels of m7 (lower panel). Asterisk in A and D: half amount of the adjacent sample is loaded for quantity validation. One hundred nanogram of Gro expressing plasmid was co-transfected in experiments 4E and 4D. (E) myc-tagged proteins (Sc, Sc[1–320] and GFP, as indicated) were immunoprecipitated from S2 cells. Immunoprecipitation efficiency (a-Myc panel) and the presence of coprecipitated HA-tagged m7 (a-HA panel) and endogenous Gro (a-Gro) were assayed. Input for the levels of endogenous Gro and the co-transfected HA-m7 are shown in the two upper panels (In). HA-m7 interacts strongly with myc-Sc and weakly with myc-Sc[1–320]. We had shown earlier that, although the major interaction domain for E(spl)m7 is the Sc C-terminal TAD, a weaker interaction exists with the Sc[1–260] fragment (45). In the case of the strong complex formation (Sc–m7), Gro is also co-immunoprecipitated, showing the existence of a super-complex as it shown in the schematic.
In order to gain insight into the mechanism of E(spl)m7-mediated degradation of Scute, we co-expressed different mutant forms of Scute and m7. Sc[RQEQ] and m7KNEQ are basic domain mutants of Sc and m7 (74), which cannot bind DNA. m7ΔW lacks the C-terminal Trp residue and is unable to interact with the corepressor Groucho (43,75–76). Sc[1–320], Sc[1–318]-AD1 and Sc[1–318]-LH have been presented earlier in this paper. The expression levels of all m7 variants were similar (Supplementary Figure S5A).
m7KNEQ was able to degrade Sc[RQEQ] (Figure 4C), despite the inability of either protein to bind DNA, but had no effect on GFP (Figure 4B). The phosphorylation status of Scute was important for its degradation by m7, as the mutants Sc[m3p] and [m5p] could not be degraded by m7 (Figure 4C, Supplementary Figure S5B). When we co-expressed Sc[RQEQ] with Da, m7KNEQ was able to degrade both Sc[RQEQ] and Da (Figure 4D). However, upon co-expression of Sc[m3p] with Da, m7 was able to degrade Da, but Sc[m3p] remained insusceptible to degradation (Figure 4D). We conclude that (i) m7 can promote Sc degradation off DNA (based on the basic domain mutant results) and regardless of the latter's association with Da; (ii) the SPTSS motif is needed for m7-induced degradation of Sc and (iii) Da is also targeted for degradation by m7.
Unlike m7 and m7KNEQ, m7ΔW was unable to degrade Sc (Figure 4E) or Da (Figure 4D), raising the possibility that Gro might be necessary to increase Da/Sc turnover, since the C-terminal Trp residue is necessary for Gro recruitment. We performed co-immunoprecipitation experiments to test the existence of a higher order Sc–m7–Gro complex. We first showed that m7 can interact with the corepressor Gro off DNA (Supplementary Figure S5C)—m7 and m7KNEQ could co-immunoprecipitate Gro with the same efficiency. As expected, m7ΔW or GFP were unable to interact with Groucho. Integrity of the Orange domain was dispensable for Gro interaction, although it was needed for Da interaction (Supplementary Figure S5C and (68))—conversely, the C-terminal W was not needed for Da interaction (Supplementary Figure S5C). When Sc and E(spl)m7 were co-expressed, immunoprecipitation of Sc effectively precipitated Gro, supporting the existence of a Sc–m7–Gro complex (Figure 4F).
We examined the possibility that other E(spl) proteins have the ability to degrade Sc. By overexpressing E(spl)mγ and mδ we noticed that mγ was also able to degrade Sc, but mδ had no obvious effect on the steady-state levels of Sc (Supplementary Figure S5D). As mγ, like m7, is capable of interacting with Sc, whereas mδ is not (45,75), this result is consistent with the hypothesis that interaction between Sc and an E(spl) protein is a prerequisite for the former's destabilization and probably occurs via recruitment of Gro onto the Sc/E(spl) complex.
In support of this hypothesis, m7 displayed minimal destabilization of Sc[1–320] (Supplementary Figure S5E), which lacks the C-terminal Sc TAD, the major interaction surface with m7 (45). We were able to restore degradation of Sc[1–320] by m7 when we fused the AD1 domain of Da in place of the Sc TAD (Supplementary Figure S5F and G), since AD1 also strongly interacts with m7 via the latter's Orange domain (68). Fusion of a non-interacting TAD, the Da LH, did not restore Sc[1–320] degradation (Supplementary Figure S5G). Collectively, our results indicate that E(spl) proteins can induce Sc and Da turnover by associating with them and recruiting Groucho. Sc destabilization requires the SPTSS motif, but is independent of DNA binding.
Da/Sc complex leads to degradation of E(spl)m7
Prompted by the fact that E(spl)m7 and mγ promote Da/Sc degradation we decided to ask whether the reciprocal is also true by studying the effect of the Da/Sc complex on the stability of E(spl)m7. m7 is a very short-lived protein to start with (half-life 26±3 min, Figure 5A). It was further degraded by increasing amounts of Sc (Figure 5B), Sc co-expressed with Da, or even Da alone (Figure 5B and C). Given the high instability of E(spl)m7, we decided to test its ubiquitylation status and its response to proteasome inhibition. Both mono and poly/multi-ubiquitylated forms of m7 were detected from cells that had not been exposed to proteasome inhibitors (Supplementary Figure S6A); in the same lysates SUMOylated species were also detected. Surprisingly, despite the appearance of high MW ubiquitylated species, steady-state levels of m7 were not increased by MG132 treatment of transfected cells, suggesting that m7 is degraded via a proteasome-independent route (Supplementary Figure S6B). In addition, E(spl)m7 is subject to phosphorylation (Supplementary Figure S6C).
Figure 5.
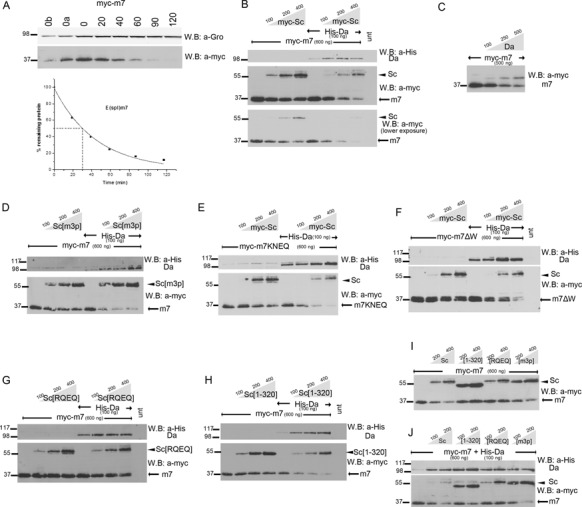
Degradation of E(spl)m7 is enhanced by Scute and Da proteins. (A) Degradation kinetics of transfected myc-m7 after protein synthesis inhibition by cycloheximide. 0, 20,…, 120 refer to minutes after cycloheximide addition. 0a, 0b: 1/2 or 1/4, respectively, of input (0 min) for densitometry calibration. Myc-tagged protein levels were estimated by densitometry and normalized against endogenous Gro. Graph shows the estimated protein levels at each time-point and the half-life of myc-m7. The experiment was repeated three times; only one is shown for clarity. The estimated half-life was 26±2 min. B-J: Whole cell extracts from S2 cells transfected with expression constructs as indicated. The amount loaded was adjusted according to the activity of luciferase (or β-galactosidase, for blot 5C) expressed from control plasmids transfected at the same time as the indicated plasmids (to control for variability in transfection efficiency). In the anti-myc blots, the upper band (arrowhead) is myc-Sc and the lower (arrow) is myc-m7. (B) Increasing Sc expression reduces m7 levels, especially upon Da co-expression. (C) Da alone can reduce m7 levels. Note that it also causes the accumulation of a high MW form of m7. Although m7 is monoubiquitylated (see Supplementary Figure S6A), the monoubiquitylated form did not increase upon Da co-expression (data not shown). (D) The Sc SPTSS motif is dispensable for m7 degradation. (E) m7 DNA binding or (F) Gro binding are dispensable for its degradation. (G) Sc DNA binding is needed for m7 degradation. (H) The Sc TAD is dispensable for m7 degradation. (I,J) Side-by-side comparison of m7 degradation promoted by different Sc variants. Sc[RQEQ] is the least active. Sc[1–320] is also less active than wt Sc, especially in the absence of co-expressed Da; this may be due to inability to get recruited onto m7.
We used our different mutant proteins of Sc and m7 to shed light on the parameters of m7 degradation by Sc. Sc phosphorylation was not needed since Sc[m3p] was able to degrade m7 (Figure 5D). DNA binding of m7 and Gro interaction was not necessary, either, since both m7KNEQ and m7ΔW were degraded by Sc (Figure 5E and F). In all cases Da co-expression increased the degradation of m7 by these Sc variants (Figure 5D–F, I, J). However, Sc[RQEQ] and Sc[RQEQ]/Da were less efficient at degrading m7 (Figure 5G). Therefore, m7 degradation most likely takes place on DNA. This explains the more efficient degradation by Da/Sc versus Sc alone, as the latter binds DNA only weakly using endogenous Da. Da homodimers can bind DNA consistent with the activity of Da to degrade m7 by itself (Figure 5C).
Sc[1–320] was inefficient at m7 degradation, (Figure 5H), but co-expression of Da with Sc[1–320] restored degradation to normal levels. We attribute this to the absence of the Sc TAD, which is the main interaction surface between Sc and m7; in the presence of Da a new interaction surface is supplied in the form of the Da AD1 domain, which interacts with the Orange domain of m7. Consistent with this, when the Orange domain mutant m7EQRQTKHQ was tested, we saw normal degradation by Da/Sc (Sc can still interact with the Orange-mutant m7 (68)), but diminished degradation by Da/Sc[1–320] (Supplementary Figure S6D and E), since in the latter combination both Sc–m7 and Da–m7 interactions have been compromised. These data support the necessity for complex formation of Da/Sc/m7 for degradation of m7 to take place. Besides Da/Sc/m7 complex formation, Da/Sc DNA binding seems to also be important in this process. The requirements for proneural induced m7 degradation are collectively shown in the blots of Figure 5I and J.
It is worth noting that the protein levels of Sc were consistently lower in the Da versus no-Da samples (Figure 5B, E, F, H), except in the case of Sc[m3p] and Sc[RQEQ] (Figure 5D and G). This suggests that high levels of E(spl)m7 somehow stimulate Sc degradation by the Da-dependent mode; remember that in the absence of m7 co-expression wt Sc steady-state levels were approximately the same with or without Da (Figure 3G). This degradation mode needs DNA binding and the SPTSS motif, accounting for the refractoriness of Sc[RQEQ] and [m3p] (Figure 5D and G). Integrity of the m7 Orange domain is needed for this stimulation, since m7EQRQTKHQ was unable to stimulate Sc degradation (Supplementary Figure S6D and E).
Another interesting observation is that when increasing amounts of Sc were provided with constant amounts of Da and m7 proteins, m7 levels dropped, but at the same time Da levels increased (Figure 5B, D–H, J). Two possible explanations for this phenomenon could be that either reduction of m7 levels diminishes Da degradation or that increasing amounts of Sc directly stabilize Da protein (as observed before, Figure 3D). Perhaps a combination of the two mechanisms is true: Da levels are suppressed by m7 co-expression (in the no-Sc samples), but increasing Sc protects Da from degradation while simultaneously reducing m7 levels, thus further favoring Da accumulation.
Levels and activity of Sc are modulated post-translationally also in vivo
We wondered whether the post-translational regulation of Sc stability by Da and E(spl) could also be observed in fly tissues. For this reason we generated flies bearing UAS-driven transgenes of myc-sc, myc-sc[1–320], myc-sc[RQEQ], myc-sc[m3p] and myc-sc[m5p]. Using three different Gal4 drivers (pnr-Gal4, ap-Gal4, act>CD2>Gal4), we analyzed the protein levels as well as the transcriptional activity of these variants, by assaying their ability to promote neurogenesis and to activate lacZ reporters.
In flies carrying the EE4-lacZ reporter, which consists of a simple tandem array of Da/Sc-binding EA-boxes (26,44), we co-expressed UAS-myc-Sc and UAS-GFP in random clones of cells in wing imaginal disks under the control of the uniform actin5C promoter (act>CD2>Gal4 driver). We noticed that, unlike GFP, Sc did not accumulate to uniform levels inside the clones. Nor was the EE4-lacZ reporter uniformly activated. In addition to cells where Sc protein was detectable (revealed by myc staining) and transcriptionally active (lacZ staining) (Supplementary Figure S7A and B, area 1), we could find cells in which Sc was active, but barely detectable (area 3) and cells in which Sc was highly expressed but was not transcriptionally active (area 2). These results showed that Sc levels and activity are dynamically modulated in vivo, consistent with the complex post-translational regulation of Sc documented in S2 cells. To address whether E(spl) proteins may participate in this post-translational modulation of Sc levels we counterstained such act-FLPout clones with mAb323, which recognizes endogenous E(spl) proteins. E(spl) were ectopically induced in Sc overexpressing clones (Supplementary Figure S7C and D), as expected by the fact that Sc/Da transcriptionally induces E(spl) expression. However on a nucleus-by-nucleus basis, the levels of Sc accumulation were inversely correlated with the levels of E(spl) accumulation. This is consistent with a mechanism of mutual destabilization that we documented for these proteins.
Overexpression of myc-sc using the dorsal body specific pnr-Gal4 driver resulted in flies with greatly increased number of sensory bristles (Figure 7C), indicating enhanced neurogenesis; the same phenotype had been shown earlier for untagged UAS-sc transgenes (44) and shows that the myc tag did not adversely affect Sc protein function. Two out of 5 UAS-myc-sc lines showed a severe effect, resulting in pupal/pharate lethality. Lethality was more pronounced upon expression of myc-Sc[m3p] (9/9 lines), myc-Sc[m5p] (16/16 lines) or Sc[1–320] (11/12 lines). Few rare escapers showed strong induction of neurogenesis (indicative phenotypes shown in Figure 7). To bypass the problem of pupal lethality we scored neurogenesis in late larvae using the SOP marker Sens. Sc[m3p] and Sc[1–320] produced a higher number of SOPs than wt Sc (Figure 6A–D). Detection of the myc-Sc protein itself in these genetic backgrounds showed that all Sc[m3p], Sc[1–320] and Sc[RQEQ] accumulate to much higher levels than wt Sc (Figure 6A–D). Sc[RQEQ] was unable to induce ectopic SOPs, consistent with its inability to bind DNA. Despite their increased proneural activity and increased levels, Sc[m3p], and Sc[1–320] were less active on the EE4-lacZ reporter compared to wt Sc. This is an indication that different conserved domains of Sc may play distinct roles in different target enhancers. We confirmed the differential stability of Sc variants by western blot analysis of wing disk lysates from larvae bearing the UAS transgenes together with an apGal4 driver and UAS-GFP (Figure 6E). We lysed approximately equal numbers of disks and we used GFP as a loading control. All three variants, Sc[RQEQ], Sc[m3p] and Sc[1–320], were more stable than Sc (wt), as expected from the immunofluorescence results.
We wondered how the stabilized Sc variants, [m3p], [m5p] and [1–320], would behave upon co-expression with E(spl)m7. Our earlier work had established that co-expression of UAS-m7—which on its own deletes all sensory bristles, resulting in a bald thorax—is epistatic to UAS-sc (44). We had attributed this to m7/Gro- mediated repression of SOP promoting genes. When m7 was co-expressed with the stabilized Sc variants, an interesting dichotomy was observed. Whereas m7 strongly suppressed the proneural activity of Sc[1–320], it was impressively less effective on Sc[m3p] and Sc[m5p], where a large number of bristles persisted despite m7 expression (Figure 7A–H, K, L). The diminished susceptibility of Sc[m3p] and Sc[m5p] to inhibition by m7 is unlikely to be caused by their inability to interact with m7, since the Sc–m7 interaction domain has been mapped to the C-terminal TAD of Sc and, consistently, Sc[m5p] coIPed m7 more efficiently than Sc[1–320] (Figure 7M). A more likely explanation is that the phosphomutants, unlike Sc (wt) and Sc[1–320], have diminished ability to be degraded by m7 (Figure 4D), whereas they themselves robustly degrade m7 (Figure 5D). This probably ensures that there is residual Sc phosphomutant protein to activate neurogenesis even in the presence of high levels of m7.
DISCUSSION
By generating a large collection of variants, we studied the structural basis of the stability of Sc and E(spl)m7, which together with Da constitute a group of intimately interconnected bHLH proteins that regulate neural differentiation. Our major findings were the following: (i) TADs act as degrons, but they do so in a highly context-dependent manner; (ii) A conserved phosphorylation motif of Sc is crucial for its stability and influences its in vivo activity and (iii) Sc and E(spl)m7 mutually promote their turnover. We also found that Da binding induces post-translational modifications on both Sc and E(spl)m7, seen as retarded bands, but their significance and nature remain unclear.
TADs act as degrons
Sc and Da have extremely different half-lives. Sc is very unstable, with a half-life of <1 h (in S2 cells), whereas the Da half-life is >4 h. Sc contains a C-terminal TAD, whereas Da contains two TADs, the N-terminal AD1 and the central LH (45,68). Within their protein backbones, only Sc TAD behaved as a degron, since its deletion greatly prolonged Sc half-life, something that was also seen in fly tissues. In contrast, AD1 and LH appeared to have no degron activity on Da. An unexpected finding was that Da AD1 behaved as a trans-degron for Sc in the Da/Sc complex. To our knowledge, the only other instance of a TAD acting as a trans-degron is TAD2 of the Mastermind-like (Maml) protein. Upon binding to chromatin in a complex with CBF1 and Nicd, the intracellular fragment of Notch, TAD2 promotes the phosphorylation and turnover of Nicd (77). The fact that Da AD1 was capable of increasing Sc turnover when transferred onto a TAD-less (stabilized) Sc (i.e. acted as a cis-degron) suggests that AD1 can promote degradation of a nearby ‘susceptible’ substrate. Apparently the Da protein does not have such a ‘susceptible’ site or has ‘stabilons’ (e.g. motifs that recruit deubiquitylases), which antagonize the action of degrons (reviewed in (78)). Unlike AD1, LH never behaved as a degron in any of the contexts tested.
In some transcriptional activators, TAD-dependent degradation has been shown to enhance their transcriptional activity ((60,79) reviewed in (64)). In the case of yeast Gal4, its turnover has been shown to enhance RNAPolII CTD phosphorylation and consequent transcript maturation by a yet undefined mechanism (80). In the case of Sc, we have earlier shown that its TAD is not necessary for transactivation of target genes, since the C-terminal truncations Sc[1–260], [1–290] and [1–320] all can activate target genes and are potent inducers of bristles when expressed in Drosophila tissues (68). This proneural activity of truncated (as well as full length) Sc proteins is absolutely dependent on the Da TADs (68). Since Da AD1 is a major degron for the Da/Sc complex, the Da/Sc activator could be envisaged as belonging to the class of activators that have to be turned over in order to achieve high levels of transcription. Two pieces of data seem to disfavor this categorization: (i) Sc or TAD-less truncated versions can promote peripheral neurogenesis in the presence of DaΔAD1, which contains only the LH TAD, which did not appear to be a degron in our assays. (ii) Sc phosphomutants [m3p] and [m5p] are extremely stable in the presence of Da (containing the AD1) and, despite their stability, they strongly activate target genes and sensory organ generation. Therefore, at least as far as Da/Sc is concerned, its turnover does not seem to be necessary for achieving high levels of transcription. Note, however, that the stabilized variants Sc[m3p] and [m5p] were less active on the EE4 artificial enhancer (Figure 6), so the possibility remains that in some enhancer contexts activator turnover is favorable for Da/Sc induced transcription.
Upon closer comparison, we note that one of the prototypical degradation-dependent activators, yeast Gcn4 (79), behaves in a highly similar manner to Sc. wt Gcn4 requires proteasome activity (and consequent degradation) for target gene activation (its activity is inhibited by MG132). In contrast, a phosphomutant of all S/TP sites (Gcn4–3T2S) that is resistant to phosphorylation by Pho85 (Cdk5) and Srb10 (Cdk8) (81) is capable of activating target genes and is even resistant to the inhibitory effect of MG132 (79). This is consistent with a model whereby Gcn4 phosphorylation reduces activator potency and simultaneously recruits a ubiquitin ligase to remove this ‘spent’ activator and replace it with an active unphosphorylated one. When the proteasome is inhibited, inactive phosphorylated Gcn4 accumulates, which decreases target gene expression. Sc[m3p], like Gcn4–3T2S, could retain activity by virtue of not being able to be marked as ‘spent’. Unlike Gcn4, mammalian Smad1 and Smad3 need phosphorylation by Cdk8 and Cdk9 in order to stimulate their transcriptional activity. This is achieved by recognition of p-SP sites in their linker regions by coactivators Yap and Pin1 (82–84). Smad degradation ensues, because p-SP linker motifs prime subsequent GSK3 phosphorylation and the multiply phosphorylated Smads are recognized by Nedd4-family ubiquitin ligases (85). In this case activation and degradation are temporally ordered and degradation simply serves to terminate the activation response. The distinguishing feature between the Gcn4 and Smad mechanisms of activation-coupled degradation seems to be whether the kinases that trigger degradation play a positive or negative role in transcription. Further work in characterizing Sc kinases will allow a genetic approach to this question.
Sc stability and activity depends on a conserved phosphorylation motif
S/TP motifs are often kinase targets, especially for MAPKs and Cdks. Phosphorylation of such motifs on transcriptional activators is often followed by ubiquitylation and degradation, as described above for Gcn4, Smad1 and Smad3. In vertebrates phosphorylation of S/TP motifs has been described for proneural proteins, such as Ascl1, Ngn2, Xath5, XNeuroD (50–52,86–87). We showed that 268-SPTSS of Sc is its major phosphorylation site, whereas minor phospho-species depend on 217-SPLQQ. An SPxxS motif is highly conserved in Sc homologues across species (data not shown). For example, mammalian Ascl1 contains three such motifs in a similar location, between the bHLH domain and the C-terminal acidic TAD (185-SPTIS, 189-SPNYS and 202-SPVSS; mouse numbering). Mutating the Drosophila Sc SPTSS motif to APAAS greatly reduced Sc phosphorylation, although the additional mutation of S217 to alanine was needed to completely eliminate phosphorylation, at least as detected by mobility shift upon treatment by λ-phosphatase. This suggests that the remaining five S/TP motifs, or indeed numerous other S/T residues, in Sc are not heavily phosphorylated in S2 cells. Importantly, the non-phosphorylatable [m3p] and [m5p] mutants were defective in proteasome-dependent turnover when co-expressed with Da, displaying a half-life of >3 h, compared to ∼40 min for wt Sc. Although a different degradation mechanism predominated when Sc was expressed alone, the phosphomutants had a significantly increased half-life (∼75 min) even under this condition. In yet a third context, that of degradation of Sc by high levels of E(spl)m7, the SPTSS motif was also necessary (see Figure 8 for a summary of the different modes of Sc degradation). Phosphorylation-dependent destabilization of Sc is important for its function, since the phosphomutants displayed a reduced response to inhibition by E(spl)m7 (Figure 7; see the next section).
Figure 8.
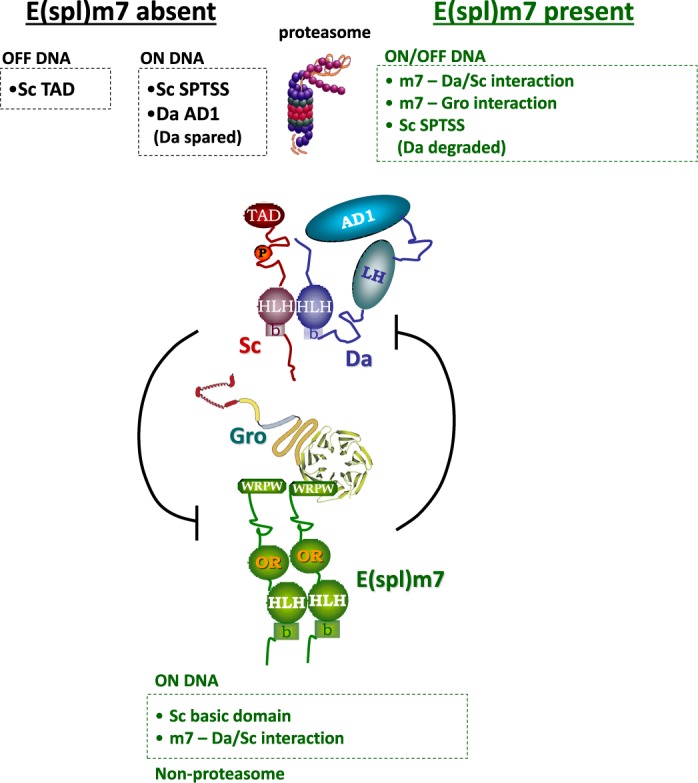
Summary of the main features of Sc and E(spl)m7 degradation. In the absence of E(spl)m7 (black font) Sc can be degraded by two proteasome-dependent mechanisms, one that predominates in the presence of Da (the on-DNA mechanism) and the other that predominates in the absence of Da (the off-DNA mechanism). In the off-DNA mode the most important degron is the C-terminal TAD of Sc. In the on-DNA mechanism, the SPTSS motif becomes more important. Also of great importance is the Da AD1 motif, which acts as a trans-degron for Sc, as Da itself is spared from degradation. In the presence of E(spl)m7 (green font), degradation can take place either on or off DNA and requires the recruitment of Gro onto the Sc–(Da)–m7 complex. The SPTSS motif is also important. In this instance, Da is also degraded. Reciprocally Da/Sc stimulate the turnover of E(spl)m7, which happens on DNA, as it needs the Sc basic domain. Besides the Sc basic domain, only the ability of Da/Sc to recruit E(spl)m7 is needed for m7 degradation, which takes place via a non-proteasomal pathway.
In our preliminary analysis using kinase inhibitors, we were unable to eliminate the major SPTSS-dependent Sc phospho-species (band α). With the reservations inherent to inhibitor studies, it appears that neither Cdk8 nor Sgg/GSK3 is the kinase responsible, although the SPTSS motif conforms to a consensus site for both. Instead, Sgg inhibition by LiCl eliminated the same minor phospho-species as those eliminated by the S217A mutation (α’, β’ bands, Figure S2A-B). S217 lies within a motif 213-TISVSPLQQQQ. It could therefore conceivably act as a priming phospho-Ser for the subsequent Sgg phosphorylation of T213 (GSK3 consensus site is SxxxpS, where pS is a priming phospho-serine). We speculate that, like mammalian Ascl1 and Ngn2, the most likely kinase for the SPTSS motif is Cdk1 and/or Cdk2; alternatively the SPTSS motif may be phosphorylated by a combination of kinases (e.g. Cdk8 or Sgg) or by another Cdk or MAPK. The difference between Sc and murine Ascl1 is that for the latter all six SP motifs had to be converted to AP in order to eliminate phosphorylation (50), whereas in Sc the remaining five S/TP motifs seem to play a minor role, if any. Another difference is that mammalian S>A mutants (all SP sites) of both Ngn2 and Ascl1 showed only moderately increased stability (∼2-fold) (50,51), whereas the Sc phosphomutants were dramatically stabilized both in S2 cells (at least in the presence of Da) and in imaginal disks.
Multiple mechanisms of neural bHLH protein turnover
Sc carries two major destabilizing motifs, its TAD (aa 321–345) and the SPTSS motif (aa 268–272). Interestingly, the former is more important when Sc is expressed in excess of Da, whereas the latter predominates when Sc is complexed with Da, although both play a role in both contexts. The Da/Sc complex is known to bind DNA much better than Sc homodimers, which may not bind at all. In the Da-dependent degradation mode the integrity of the Sc basic domain is important (Sc[RQEQ] is much more stable than wt), suggesting that Sc degradation takes place on chromatin. We presume that when Sc is present in excess of Da, its degradation takes place off chromatin. A major finding of this work was that in the on-chromatin mechanism (Da/Sc complex) Da AD1 acts in trans to destabilize Sc; it seems to act as a composite phosphodegron, as it absolutely requires the Sc SPTSS motif. In the same context, AD1 does not destabilize Da. In fact, providing increasing amounts of Sc seems to stabilize Da, which is already a very stable protein. Although we have not addressed the mechanism of this effect, we speculate that it may be due to the displacement of a negative cofactor from Da upon Da/Sc dimerization. This could be the Id-homologue of Drosophila, Extramacrochaetae (Emc), a ubiquitously expressed protein. Indeed in emc− tissues (mosaics) Da is massively upregulated (38). Although this was attributed to transcriptional effects, additional post-translational effects have not been ruled out. Interactions among E47 (Da homologue), Ascl1 and Id1 can affect their stability in mammalian cells (88), although this study did not address the role of TADs or SP motifs.
Another major finding of our work was the mutual destabilization of Sc/Da and E(spl) proteins. Both of these destabilizations require Da/Sc/E(spl) complex formation, as they get progressively weaker when interaction domains are compromised. Whereas m7 degradation by Sc seems to happen on chromatin (it requires the Sc basic domain, but not the m7 basic domain), Sc degradation by m7 can happen also off DNA, since neither of the basic domains is required. Interestingly, Sc degradation in this context requires the Gro co-repressor, which is recruited onto the Da/Sc complex by m7. This suggests a novel function for this multifunctional corepressor, which is not necessarily carried out on chromatin. This function may be conserved, since the WRPW peptide, the Gro interaction motif found at the C-terminus of all E(spl) homologues, acts as a proteasome-dependent degradation signal in the case of mammalian Hes6 (89). The reciprocal degradation of E(spl)m7 by Da/Sc only requires DNA binding and m7-Da/Sc complex formation. Gro and the Sc SPTSS motif are dispensable in this case. Interestingly m7 degradation does not seem to be mediated by the proteasome, as it is not inhibited by MG132. Sc degradation, on the other hand, is proteasome dependent in all cases (high or low Da; high or low E(spl)). Figure 8 summarizes the major requirements for each case of bHLH protein turnover.
E(spl)/Hes genes are often induced by Notch signaling (51,57–59,87) and their products are known antagonists of proneural activators in many contexts, such as the fly and mammalian neuroectoderm, fly retina, mammalian pancreas and fly intestine, to name a few (35,40,44,49,59,90–93). In many of these tissues E(spl)/Hes and proneural expression are complementary (e.g. 59,91,94–95). Ever since the discovery of the Hes/ E(spl) proteins, the reigning paradigm to explain the Notch/Hes-proneural antagonism has been the repression of proneural gene expression by Hes/E(spl). Here we show that a post-translational component also exists (Supplementary Figure S7D), based on their mutual degradation. Our results might also explain the observation that Hes1 can downregulate Ngn3 post-translationally in the mammalian pancreas (59). In that context, the Gro-interacting C-terminus of Hes1 was required, reminiscent of the destabilization of Sc by E(spl)m7. However, it was not addressed whether interaction between Hes1 and Ngn3 was responsible for the latter's degradation or whether a transcriptional target of Hes1 was involved.
Our results propose a novel role for Notch, namely the induction of Hes/E(spl) proteins as degradation adaptors for proneural proteins. In early Xenopus embryos, overexpression of either Ascl1 or Ngn2 induces ectopic neurogenesis and this effect is inhibited by co-expression of active Notch (50–52). Although in this system it was not tested whether Hes proteins recapitulate the effect of Notch, Ascl1 or Ngn2 mutants that eliminate all putative SP phosphorylation sites were less susceptible to inhibition by Notch, reminiscent of the decreased response of Sc[m3p] and [m5p] to E(spl)m7 (Figure 7). It is therefore conceivable that SP motif phosphorylation may play a conserved role to sensitize proneural proteins to inhibition by their Hes antagonists. It is possible that, similarly to the Smads (85), Sc is kept unstable during recruitment to its target genes by phosphorylation by a transcription-coupled kinase, such that it is poised to respond rapidly to inhibitory signals, like E(spl) proteins.
The present work has shed light to some new mechanisms of bHLH interactions, but also poses many questions for future research, such as: Which enzymes mediate phosphorylation and degradation of Sc? What is the role of ubiquitylation and SUMOylation of E(spl)m7? Which enzymes are involved in the reciprocal degradations of m7 and Da/Sc? Extrapolation of the present work to mammalian systems should reveal whether similar mechanisms regulate the activity of the Sc orthologue Ascl1, a pioneer factor in cellular reprogramming toward the neural fate. Our study reconfirms that life has devised a plethora of interlinked mechanisms to safeguard its existence.
SUPPLEMENTARY DATA
Supplementary Data available at NAR Online.
Acknowledgments
We are grateful to Konstantinos Koumbanakis and Nikolaos Giagtzoglou for their help in cell culture and Drosophila protocols. We thank Giannis Livadaras for Drosophila transgenesis and Marina Theodosiou, Ioanna Koltsaki for their support. We are thankful to Spyros Artavanis-Tsakonas for the SUMO antibody and to Hippokrates Kiaris, Elena Farmaki and Igor Roninson for the Senexin reagent. We thank Nikolaos Giagtzoglou, Nicholas Baker and Iannis Talianidis for their comments on the manuscript.
Footnotes
Present address: Marianthi Kiparaki, Department of Genetics, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA.
FUNDING
Funding for work in the Delidakis lab and for open access charge: Grants from the Greek General Secretariat of Research & Technology (Aristeia II/ grant no. 4436 and PENED).
Conflict of interest statement. None declared.
REFERENCES
- 1.Massari M.E., Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villares R., Cabrera C.V. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- 3.Guillemot F., Lo L.C., Johnson J.E., Auerbach A., Anderson D.J., Joyner A.L. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand N., Castro D.S., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 5.Campuzano S., Balcells L., Villares R., Carramolino L., Garcia-Alonso L., Modolell J. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell. 1986;44:303–312. doi: 10.1016/0092-8674(86)90764-6. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez M., Campuzano S. asense, a member of the Drosophila achaete-scute complex, is a proneural and neural differentiation gene. EMBO J. 1993;12:2049–2060. doi: 10.1002/j.1460-2075.1993.tb05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez I., Hernandez R., Modolell J., Ruiz-Gomez M. Competence to develop sensory organs is temporally and spatially regulated in Drosophila epidermal primordia. EMBO J. 1990;9:3583–3592. doi: 10.1002/j.1460-2075.1990.tb07569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanda S., Ang C.E., Davila J., Pak C., Mall M., Lee Q.Y., Ahlenius H., Jung S.W., Sudhof T.C., Wernig M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wapinski O.L., Vierbuchen T., Qu K., Lee Q.Y., Chanda S., Fuentes D.R., Giresi P.G., Ng Y.H., Marro S., Neff N.F., et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thoma E.C., Wischmeyer E., Offen N., Maurus K., Siren A.L., Schartl M., Wagner T.U. Ectopic expression of neurogenin 2 alone is sufficient to induce differentiation of embryonic stem cells into mature neurons. PLoS One. 2012;7:e38651. doi: 10.1371/journal.pone.0038651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caudy M., Vassin H., Brand M., Tuma R., Jan L.Y., Jan Y.N. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988;55:1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- 14.Murre C., McCaw P.S., Vaessin H., Caudy M., Jan L.Y., Jan Y.N., Cabrera C.V., Buskin J.N., Hauschka S.D., Lassar A.B., et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 15.Cabrera C.V., Alonso M.C. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campuzano S., Modolell J. Patterning of the Drosophila nervous system: the achaete-scute gene complex. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- 17.Cronmiller C., Cummings C.A. The daughterless gene product in Drosophila is a nuclear protein that is broadly expressed throughout the organism during development. Mech. Dev. 1993;42:159–169. doi: 10.1016/0925-4773(93)90005-i. [DOI] [PubMed] [Google Scholar]
- 18.Skeath J.B., Carroll S.B. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes Dev. 1991;5:984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- 19.Cubas P., de Celis J.F., Campuzano S., Modolell J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- 20.zur Lage P., Jan Y.N., Jarman A.P. Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Curr. Biol. 1997;7:166–175. doi: 10.1016/s0960-9822(97)70087-3. [DOI] [PubMed] [Google Scholar]
- 21.Okabe M., Okano H. Two-step induction of chordotonal organ precursors in Drosophila embryogenesis. Development. 1997;124:1045–1053. doi: 10.1242/dev.124.5.1045. [DOI] [PubMed] [Google Scholar]
- 22.zur Lage P.I., Powell L.M., Prentice D.R., McLaughlin P., Jarman A.P. EGF receptor signaling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev. Cell. 2004;7:687–696. doi: 10.1016/j.devcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Doroquez D.B., Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit. Rev. Biochem. Mol. Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- 24.Culi J., Martin-Blanco E., Modolell J. The EGF receptor and N signalling pathways act antagonistically in Drosophila mesothorax bristle patterning. Development. 2001;128:299–308. doi: 10.1242/dev.128.2.299. [DOI] [PubMed] [Google Scholar]
- 25.Simpson P., Carteret C. A study of shaggy reveals spatial domains of expression of achaete-scute alleles on the thorax of Drosophila. Development. 1989;106:57–66. doi: 10.1242/dev.106.1.57. [DOI] [PubMed] [Google Scholar]
- 26.Culi J., Modolell J. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 1998;12:2036–2047. doi: 10.1101/gad.12.13.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 28.Campuzano S., Carramolino L., Cabrera C.V., Ruiz-Gomez M., Villares R., Boronat A., Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985;40:327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- 29.Chien C.T., Hsiao C.D., Jan L.Y., Jan Y.N. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc. Natl Acad. Sci. U.S.A. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Bellido A. Genetic analysis of the achaete-scute system of Drosophila melanogaster. Genetics. 1979;91:491–520. doi: 10.1093/genetics/91.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghysen A., Richelle J. Determination of sensory bristles and pattern formation in Drosophila. II. The achaete-scute locus. Dev. Biol. 1979;70:438–452. doi: 10.1016/0012-1606(79)90037-x. [DOI] [PubMed] [Google Scholar]
- 32.Delidakis C., Preiss A., Hartley D.A., Artavanis-Tsakonas S. Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer of split locus of Drosophila melanogaster. Genetics. 1991;129:803–823. doi: 10.1093/genetics/129.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kageyama R., Nakanishi S. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr. Opin. Genet. Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- 34.Fisher A., Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Singson A., Leviten M.W., Bang A.G., Hua X.H., Posakony J.W. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- 36.Van Doren M., Powell P.A., Pasternak D., Singson A., Posakony J.W. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- 37.Ruzinova M.B., Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharya A., Baker N.E. A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell. 2011;147:881–892. doi: 10.1016/j.cell.2011.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delidakis C., Artavanis-Tsakonas S. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc. Natl Acad. Sci. U.S.A. 1992;89:8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramatschek B., Campos-Ortega J.A. Neuroectodermal transcription of the Drosophila neurogenic genes E(spl) and HLH-m5 is regulated by proneural genes. Development. 1994;120:815–826. doi: 10.1242/dev.120.4.815. [DOI] [PubMed] [Google Scholar]
- 41.Garrell J., Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- 42.Van Doren M., Ellis H.M., Posakony J.W. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- 43.Paroush Z., Finley R.L., Jr, Kidd T., Wainwright S.M., Ingham P.W., Brent R., Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 44.Giagtzoglou N., Alifragis P., Koumbanakis K.A., Delidakis C. Two modes of recruitment of E(spl) repressors onto target genes. Development. 2003;130:259–270. doi: 10.1242/dev.00206. [DOI] [PubMed] [Google Scholar]
- 45.Giagtzoglou N., Koumbanakis K.A., Fullard J., Zarifi I., Delidakis C. Role of the Sc C terminus in transcriptional activation and E(spl) repressor recruitment. J. Biol. Chem. 2005;280:1299–1305. doi: 10.1074/jbc.M408949200. [DOI] [PubMed] [Google Scholar]
- 46.Delidakis C., Monastirioti M., Magadi S.S. E(spl): genetic, developmental, and evolutionary aspects of a group of invertebrate Hes proteins with close ties to Notch signaling. Curr. Top Dev. Biol. 2014;110:217–262. doi: 10.1016/B978-0-12-405943-6.00006-3. [DOI] [PubMed] [Google Scholar]
- 47.Bailey A.M., Posakony J.W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 48.Lecourtois M., Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 49.Kageyama R., Ohtsuka T., Kobayashi T. Roles of Hes genes in neural development. Dev. Growth Differ. 2008;50(Suppl. 1):S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 50.Ali F., Hindley C., McDowell G., Deibler R., Jones A., Kirschner M., Guillemot F., Philpott A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138:4267–4277. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali F.R., Cheng K., Kirwan P., Metcalfe S., Livesey F.J., Barker R.A., Philpott A. The phosphorylation status of Ascl1 is a key determinant of neuronal differentiation and maturation in vivo and in vitro. Development. 2014;141:2216–2224. doi: 10.1242/dev.106377. [DOI] [PubMed] [Google Scholar]
- 52.Hindley C., Ali F., McDowell G., Cheng K., Jones A., Guillemot F., Philpott A. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development. 2012;139:1718–1723. doi: 10.1242/dev.077552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Y.C., Song M.R., Park J.P., Henry Ho H.Y., Hu L., Kurtev M.V., Zieg J., Ma Q., Pfaff S.L., Greenberg M.E. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron. 2008;58:65–77. doi: 10.1016/j.neuron.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li S., Mattar P., Zinyk D., Singh K., Chaturvedi C.P., Kovach C., Dixit R., Kurrasch D.M., Ma Y.C., Chan J.A., et al. GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 2012;32:7791–7805. doi: 10.1523/JNEUROSCI.1309-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neumann C.J., Cohen S.M. Sternopleural is a regulatory mutation of wingless with both dominant and recessive effects on larval development of Drosophila melanogaster. Genetics. 1996;142:1147–1155. doi: 10.1093/genetics/142.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang M., Hatton-Ellis E., Simpson P. The kinase Sgg modulates temporal development of macrochaetes in Drosophila by phosphorylation of Scute and Pannier. Development. 2012;139:325–334. doi: 10.1242/dev.074260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sriuranpong V., Borges M.W., Strock C.L., Nakakura E.K., Watkins D.N., Blaumueller C.M., Nelkin B.D., Ball D.W. Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol. Cell. Biol. 2002;22:3129–3139. doi: 10.1128/MCB.22.9.3129-3139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nie L., Xu M., Vladimirova A., Sun X.H. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 2003;22:5780–5792. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qu X., Afelik S., Jensen J.N., Bukys M.A., Kobberup S., Schmerr M., Xiao F., Nyeng P., Veronica Albertoni M., Grapin-Botton A., et al. Notch-mediated post-translational control of Ngn3 protein stability regulates pancreatic patterning and cell fate commitment. Dev Biol. 2013;376:1–12. doi: 10.1016/j.ydbio.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 60.Salghetti S.E., Muratani M., Wijnen H., Futcher B., Tansey W.P. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl Acad. Sci. U.S.A. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salghetti S.E., Kim S.Y., Tansey W.P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molinari E., Gilman M., Natesan S. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 1999;18:6439–6447. doi: 10.1093/emboj/18.22.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salghetti S.E., Caudy A.A., Chenoweth J.G., Tansey W.P. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 64.Geng F., Wenzel S., Tansey W.P. Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang P.J., Hsiao Y.L., Tien A.C., Li Y.C., Pi H. Negative-feedback regulation of proneural proteins controls the timing of neural precursor division. Development. 2008;135:3021–3030. doi: 10.1242/dev.021923. [DOI] [PubMed] [Google Scholar]
- 66.Daskalaki A., Shalaby N.A., Kux K., Tsoumpekos G., Tsibidis G.D., Muskavitch M.A., Delidakis C. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. J. Cell Biol. 2011;195:1017–1031. doi: 10.1083/jcb.201105166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kux K., Kiparaki M., Delidakis C. The two Tribolium E(spl) genes show evolutionarily conserved expression and function during embryonic neurogenesis. Mech. Dev. 2013;130:207–225. doi: 10.1016/j.mod.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 68.Zarifi I., Kiparaki M., Koumbanakis K.A., Giagtzoglou N., Zacharioudaki E., Alexiadis A., Livadaras I., Delidakis C. Essential roles of Da transactivation domains in neurogenesis and in E(spl)-mediated repression. Mol. Cell. Biol. 2012;32:4534–4548. doi: 10.1128/MCB.00827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 70.Zacharioudaki E., Magadi S.S., Delidakis C. bHLH-O proteins are crucial for Drosophila neuroblast self-renewal and mediate Notch-induced overproliferation. Development. 2012;139:1258–1269. doi: 10.1242/dev.071779. [DOI] [PubMed] [Google Scholar]
- 71.Porter D.C., Farmaki E., Altilia S., Schools G.P., West D.K., Chen M., Chang B.D., Puzyrev A.T., Lim C.U., Rokow-Kittell R., et al. Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities. Proc. Natl Acad. Sci. U.S.A. 2012;109:13799–13804. doi: 10.1073/pnas.1206906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morris E.J., Ji J.Y., Yang F., Di Stefano L., Herr A., Moon N.S., Kwon E.J., Haigis K.M., Naar A.M., Dyson N.J. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Celniker S.E., Dillon L.A., Gerstein M.B., Gunsalus K.C., Henikoff S., Karpen G.H., Kellis M., Lai E.C., Lieb J.D., MacAlpine D.M., et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jimenez G., Ish-Horowicz D. A chimeric enhancer-of-split transcriptional activator drives neural development and achaete-scute expression. Mol. Cell. Biol. 1997;17:4355–4362. doi: 10.1128/mcb.17.8.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alifragis P., Poortinga G., Parkhurst S.M., Delidakis C. A network of interacting transcriptional regulators involved in Drosophila neural fate specification revealed by the yeast two-hybrid system. Proc. Natl Acad. Sci. U.S.A. 1997;94:13099–13104. doi: 10.1073/pnas.94.24.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fisher A.L., Ohsako S., Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fryer C.J., White J.B., Jones K.A. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 78.Dantuma N.P., Masucci M.G. Stabilization signals: a novel regulatory mechanism in the ubiquitin/proteasome system. FEBS Lett. 2002;529:22–26. doi: 10.1016/s0014-5793(02)03252-0. [DOI] [PubMed] [Google Scholar]
- 79.Lipford J.R., Smith G.T., Chi Y., Deshaies R.J. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 80.Muratani M., Kung C., Shokat K.M., Tansey W.P. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 81.Chi Y., Huddleston M.J., Zhang X., Young R.A., Annan R.S., Carr S.A., Deshaies R.J. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alarcon C., Zaromytidou A.I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A.N., Manova-Todorova K., Macias M.J., et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao S., Alarcon C., Sapkota G., Rahman S., Chen P.Y., Goerner N., Macias M.J., Erdjument-Bromage H., Tempst P., Massague J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol. Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matsuura I., Chiang K.N., Lai C.Y., He D., Wang G., Ramkumar R., Uchida T., Ryo A., Lu K., Liu F. Pin1 promotes transforming growth factor-beta-induced migration and invasion. J. Biol. Chem. 2010;285:1754–1764. doi: 10.1074/jbc.M109.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aragon E., Goerner N., Zaromytidou A.I., Xi Q., Escobedo A., Massague J., Macias M.J. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hand R., Bortone D., Mattar P., Nguyen L., Heng J.I., Guerrier S., Boutt E., Peters E., Barnes A.P., Parras C., et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 87.Moore K.B., Schneider M.L., Vetter M.L. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/s0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- 88.Vinals F., Reiriz J., Ambrosio S., Bartrons R., Rosa J.L., Ventura F. BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J. 2004;23:3527–3537. doi: 10.1038/sj.emboj.7600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kang S.A., Seol J.H., Kim J. The conserved WRPW motif of Hes6 mediates proteasomal degradation. Biochem. Biophys. Res. Commun. 2005;332:33–36. doi: 10.1016/j.bbrc.2005.04.089. [DOI] [PubMed] [Google Scholar]
- 90.Giebel B., Campos-Ortega J.A. Functional dissection of the Drosophila enhancer of split protein, a suppressor of neurogenesis. Proc. Natl Acad. Sci. U.S.A. 1997;94:6250–6254. doi: 10.1073/pnas.94.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jennings B., Preiss A., Delidakis C., Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- 92.Bardin A.J., Perdigoto C.N., Southall T.D., Brand A.H., Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baker N.E., Yu S., Han D. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr. Biol. 1996;6:1290–1301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- 94.Ligoxygakis P., Yu S.Y., Delidakis C., Baker N.E. A subset of Notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development. 1998;125:2893–2900. doi: 10.1242/dev.125.15.2893. [DOI] [PubMed] [Google Scholar]
- 95.Imayoshi I., Isomura A., Harima Y., Kawaguchi K., Kori H., Miyachi H., Fujiwara T., Ishidate F., Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


