Abstract
Pausing during transcription elongation is a fundamental activity in all kingdoms of life. In bacteria, the essential protein NusA modulates transcriptional pausing, but its mechanism of action has remained enigmatic. By combining structural and functional studies we show that a helical rearrangement induced in NusA upon interaction with RNA polymerase is the key to its modulatory function. This conformational change leads to an allosteric re-positioning of conserved basic residues that could enable their interaction with an RNA pause hairpin that forms in the exit channel of the polymerase. This weak interaction would stabilize the paused complex and increases the duration of the transcriptional pause. Allosteric spatial re-positioning of regulatory elements may represent a general approach used across all taxa for modulation of transcription and protein–RNA interactions.
INTRODUCTION
Transcriptional pausing, where RNA polymerase (RNAP) temporarily stalls prior to resuming RNA synthesis, is a fundamental process in all living cells. In eukaryotes, pausing is required for mRNA capping (1), efficient splicing that occurs co-transcriptionally (2,3), and control of transcript levels during cell differentiation (4). Pausing by RNAPII is enhanced by the RNA binding subunit of the transcription factor NELF which binds RNA within the loop of stem–loop structures (5–7). In bacteria, pausing is required for transcription termination, coordination of transcription and translation, formation of RNA secondary structures, recruitment of regulatory factors, protein folding during coupled transcription-translation and DNA repair (8–11). There are two main classes of transcriptional pausing in bacteria. Class I pauses involve the formation of an RNA hairpin in the exit channel of RNAP, whereas class II pauses do not require the formation of RNA secondary structure and are prone to backtracking (12). Class I pause activity is enhanced by the essential elongation factor NusA. Although the requirement for an RNA stem–loop structure is known, the mechanism of action of NusA remains only partially understood after ∼40 years of study (13–17).
The N-terminal domain of NusA (NusAN), which carries the pause activity, binds adjacent to the RNA exit channel on or near a structure called the β-flap tip helix (FTH) (16,18). Transcriptional pausing is a pre-requisite step in termination and NusAN is sufficient for viability in Escherichia coli strains carrying a mutation (E134D) in the gene encoding the transcription terminator protein Rho (13,15,19). Thus, transcriptional pausing is a fundamentally important process in bacteria. Understanding how NusAN is able to mediate transcription pausing requires detailed structural data on the NusAN-β-flap complex, but this has proved recalcitrant to X-ray crystallographic analysis and the FTH is often not resolved in RNAP structures, presumably due to it being mobile or unstructured.
In this study we used nuclear magnetic resonance (NMR)-based approaches to study the structure, dynamics and interaction of NusAN and the RNAP β-flap from the model Gram-positive organism Bacillus subtilis. We show that NusAN is inherently flexible, adopting more than one conformation in solution. Upon interaction with the FTH, NusAN is allosterically induced to adopt a conformation that changes the arrangement of a distal helical motif. The terminal helix in this motif contains a series of highly conserved basic amino acids that we show are critical for pause enhancement. We propose that the helical remodelling of NusAN enhances the interaction with the phosphodiester backbone of RNA pause hairpins, which leads to stabilization of the pause complex, providing a mechanism by which this key biological function is performed.
MATERIALS AND METHODS
DNA manipulation
All cloning was carried out using E. coli DH5α (Gibco BRL; Supplementary Table S1). All plasmids used and constructed in this work were confirmed by DNA sequencing, and are listed in Supplementary Table S1. Primers are listed in Supplementary Table S2.
A fragment of B. subtilis rpoB containing the β-flap (residues 784–923; corresponding to residues 830–1058 of the E. coli β subunit) was inserted into pETMCSIII to produce pNG812 (Supplementary Table S1) for overproduction of protein used in NMR studies.
B. subtilis NusAN (residues 1–124) was inserted into pETMCSIII to produce pNG1072 (Supplementary Table S1) containing an N-terminal hexahistidine tag that could be removed upon digestion with tobacco etch mosaic virus (TEV) protease. The tag-less B. subtilis NusAN was used for NMR studies.
Wild-type and mutant NusAN sequences from B. subtilis and E. coli were polymerase chain reaction (PCR) amplified and cloned into pETMSCIII (Supplementary Table S1) to create N-terminal His-tagged proteins, and into pNG767 (Supplementary Table S1) for GST-tagging. GST-tagged proteins were used in ELISA (below).
Amino acid substitutions were introduced into RNAP and recombinant enzyme was purified using the system described previously (20).
Protein purification
13C/15N- or 15N-labelled B. subtilis β-flap was overproduced using the approach described previously (21). Cell pellets were resuspended in 20 mM KH2PO4 pH 8.0, 500 mM NaCl, 0.1-mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF) along with a trace of DNase I. Following cell disruption and clarification, lysates were loaded onto Ni-NTA resin (Qiagen), washed in lysis buffer containing 15-mM imidazole and eluted with 200 mM imidazole. Samples were dialysed against 20 mM KH2PO4 pH 7.0, 150 mM NaCl, 0.1 mM ethylenediaminetetraacetic acid (EDTA) for 2.5 h, and protein was concentrated using a spin concentrator (Amicon, 3-kDa cut-off membrane) prior to gel filtration (Superdex 75 10/300; GE Healthcare). Fractions containing β-flap were pooled and reconcentrated prior to use in NMR.
15N/13C-labelled NusAN was produced using an approach similar to that described previously (21). Freshly transformed E. coli BL21(DE3) (Supplementary Table S1) cells were used to inoculate 2 l of LB medium supplemented with 100-μg/ml ampicillin. The culture was grown to A600 of 0.7–0.8, the cells harvested by centrifugation and the pellet washed with 500 ml of NMR salts solution (21). The resuspended cells were again harvested by centrifugation and the pellet resuspended in 250 ml NMR labelling medium (21). The resuspended cells were grown at 37°C for 1 h and protein overproduction induced by addition of IPTG to a final concentration of 0.5 mM. The induced culture was then grown overnight at 18°C. Cell pellets were stored at –80°C.
Cell pellets were resuspended in 20 mM KH2PO4 pH 7.8, 500 mM NaCl, 20 mM imidazole, 0.02% (w/v) phenylmethylsulfonyl fluoride (PMSF) and a trace of DNase I. Following lysis and clarification, cleared lysates were loaded onto a 5 ml HisTrapTM HP column, washed and eluted with resuspension buffer containing 500 mM imidazole. Peak fractions were dialysed against 20 mM KH2PO4 pH 7.0, 150 mM NaCl, 0.1-mM EDTA for 2.5 h, concentrated, gel filtered and reconcentrated as for the β-flap fragment (above).
Purified NusAN was then dialysed into TEV protease digestion buffer (20 mM Tris-HCl pH 7.8, 150-mM NaCl, 0.5 mM EDTA, 1 mM DTT). The dialysate was centrifuged at 10 000 g at room temperature for 20 min, then TEV protease (5–20 mg/ml) was added to the supernatant. The reaction mixture was incubated at room temperature overnight and digest efficiency assessed using sodium dodecyl sulphate-polyacrylamide gel electrophoresis. The His-tag was removed by passage of the sample through a 5-ml HisTrap HP column (GE Healthcare). The unbound sample was then washed from the column with 5 ml of 10-mM HEPES pH 7.8, 150 mM NaCl. Flow-through and wash fractions containing the target protein were pooled and re-concentrated.
All unlabelled proteins were overproduced in E. coli BL21(DE3) (Supplementary Table S1) by growing cells for 24 h at room temperature in auto-induction medium (22) supplemented with 100-μg/ml ampicillin. Recombinant E. coli RNAP core enzyme and σ70 were purified as described previously (23). All GST-tagged proteins were purified as described previously (24).
NMR data acquisition
13C/15N-labelled β-flap and NusAN samples containing 5% D2O were filtered using a low-protein-binding Ultrafree-MC centrifugal filter (0.22 μm pore size; Millipore, MA, USA), then 300 μl was added to a susceptibility-matched 5 mm outer-diameter microtube (Shigemi Inc., Japan).
NMR data were acquired at 25°C using a 900-MHz AVANCE spectrometer (Bruker BioSpin, Germany) equipped with a cryogenically cooled probe. Data used for resonance assignment were acquired using non-uniform sampling (NUS); sampling schedules that approximated the rate of signal decay along the various indirect dimensions were generated using sched3D (25). The decay rates used were 1 Hz for all constant-time 15N dimensions, 30 Hz for all 13C dimensions and 15 Hz for the semi-constant indirect 1H dimension of the H(CC)(CO)NH/(H)CC(CO)NH-TOCSY experiments. All pulse programs used were modified from the standard BRUKER library for NUS mode and are available from the authors upon request. 13C- and 15N-edited HSQC-NOESY experiments were acquired using linear sampling. Separate experiments were acquired for the aliphatic and aromatic regions of the 13C dimension.
NUS data were processed using the Rowland NMR toolkit (www.rowland.org/rnmrtk/toolkit.html); maximum entropy parameters were selected automatically as described previously (26,27). NMR spectra were analysed and assigned using the program XEASY (28) or CcpNmr (29). 1HN, 15N, 13Cα, 13Cβ and 13C′ resonance assignments were obtained from analysis of amide-proton strips in 3D HNCACB, CBCA(CO)NH and HNCO spectra. Sidechain 1H and 13C chemical shifts were obtained primarily from 3D H(CC)(CO)NH-TOCSY and (H)CC(CO)NH-TOCSY spectra, respectively. The remaining sidechain assignments were derived from 3D H(C)CH-TOCSY and 15N- and 13C-edited NOESY-HSQC spectra.
Structure determination
Distance restraints for structure calculations were derived from 3D 13C- and 15N-edited NOESY-HSQC spectra acquired with a mixing time of 80 ms. NOESY spectra were manually peak picked and integrated. The peak lists were then assigned and an ensemble of structures calculated automatically using the CANDID module of the torsion angle dynamics package CYANA (30,31). The tolerances used for CANDID were 0.04 ppm in the indirect 1H dimension, 0.02 ppm in the direct 1H dimension, 0.2 ppm for the aromatic 13C and 15N dimensions, and 0.4 ppm for the aliphatic 13C data.
Backbone dihedral-angle restraints (120 ϕ and 125 ψ for β-flap and 114 ϕ and 116 for ψ for NusAN) were derived from TALOS+ chemical shift analysis (32); the restraint range was set to twice the estimated standard deviation. All X-Pro peptide bonds were clearly identified as trans on the basis of characteristic NOEs and the Cβ and Cγ chemical shifts for the Pro residues.
CYANA was used to calculate 200 structures from random starting conformations, then the 20 conformers with the lowest CYANA target function were chosen to represent the structural ensemble. During the automated NOESY assignment/structure calculation process the CANDID module of CYANA assigned 91.3% of all NOESY crosspeaks (4470 out of 4891) for the β-flap and 89.7% (3958 out of 4412) for NusAN. The NusA structure appears to exist in two major stable conformations. Residues that could be assigned to the same atom but at different chemical shifts are given in Supplementary Table S3. Normalized 1H/15N chemical shift changes (Δδ) were calculated using the equation 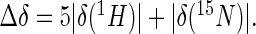
Determination of β-flap tip flexibility
NMR experiments for measuring 1H-15N steady state NOE and 15N R1 and R2 relaxation rates were acquired using single-scan interleaved pulse sequences. A 13C refocussing pulse was used during 15N evolution and a heat compensation cycle was placed in the recovery time of R2 experiments. T1 delays were (in s): 0.01, 0.05, 0.1, 0.3, 0.5, 0.7, 0.9 and 1.1, where delays 0.3 and 0.9 were sampled twice for error estimation. T2 delays (CPMG) were (in s): 0.017, 0.034, 0.068, 0.085, 0.119, 0.170 and 0.204, with delays 0.017, 0.068 and 0.170 sampled twice for error estimation. The NOE saturation time was set to 4 s whilst an additional 5 s of interscan delay was used. The relaxation time in the R1 and R2 experiments was set to 3 s. NMR spectra were analysed using an in-house line-shape fitting tool to extract nuclear frequencies and integrals. These were subsequently analysed using Relax software (33).
Protein interaction studies by NMR
NMR titrations were conducted using 15N HSQC experiments at 900 MHz, where increasing concentrations of an unlabelled protein were added to a fixed concentration of 15N-labelled protein. The concentrations of proteins were optimized by running a series of trial experiments. In the optimized conditions, 15N-labelled protein concentration was kept at 100 or 200 μM (NusAN and β-flap, respectively) whilst unlabelled protein concentrations were varied at ratios of 1:0.1, 1:0.5, 1:1 and 1:2 for β-flap and 1:0.2, 1:0.4 and 1:0.8 for NusAN. Higher ratios resulted in extensive exchange broadening where only the terminal residues were detectable, thus saturation could not be achieved (see Supplementary Figures S2 and S5).
Molecular modelling
Molecular modelling was performed with UCSF Chimera (34). The previously published negative-stain electron microscopy structures of B. subtilis RNAP (EMDB: 1577) and RNAP in complex with NusA (EMDB: 1578) (18) were employed to guide the docking. The Thermus thermophilus elemental paused elongation complex (ePEC; PDB: 4GZZ) was docked into the RNAP electron density map by superimposing it onto the previously docked homology model of a B. subtilis elongation complex (18). The position of the FTH was manually rotated around the β-sheet region to mimic the flexibility of the FTH to open up the RNA exit channel. The NMR structure of the human PGY/MDR1 mRNA (PDB: 2GVO) was docked in the RNA exit channel by superimposing the 3′ end onto the 5′ end of the –11 position of the RNA transcript in the B. subtilis elongation complex (18). The hairpin was slightly rotated around its 3′ end to minimize steric clash with the β’ and β subunits. The E. coli/B. subtilis NusAN NMR structures (PDB: 2KWP and 2MT4, respectively) were docked as rigid bodies into the electron density corresponding to NusAN of the RNAP–NusA complex structure (EMDB:1578), by placing residues important for RNAP binding in close proximity to the FTH. Independent experiments using HADDOCK (35) gave almost identical results. Images were created with PyMOL (DeLano Scientific LLC, Palo Alto, CA, USA).
Protein interaction assays
ELISA was performed as described previously (24). To determine binding constants, serial dilutions of GST-tagged NusAN constructs were added to wells coated with β-flap fragment and binding constants calculated through double reciprocal plots.
Far-western blots were performed as described (18) except that NusA was detected using anti-NusA polyclonal antibodies (36) followed by overnight washing in wash buffer (20 mM Tris, 100 mM NaCl, 0.5 mM EDTA, 5% (w/v) glycerol, 0.1% (w/v) TWEEN, 5% (w/v) skim milk pH 7.6). Bound protein was detected using HRP-conjugated goat anti-rabbit antibody as described previously (37).
Single-round transcription assays
Template DNA used for transcription assays comprised a 477-bp fragment amplified from pIA226 (38) using primers 5′-GTGCTCATACGTTAAATCT-3′ (forward) and 5′-CAGTTCCCTACTCTCGCATG-3′ (reverse). The PCR product was purified using a PCR clean-up kit (Promega) and eluted with DEPC-treated water. In vitro single-cycle transcription pausing assays were performed as described previously (16,39): Briefly, 0.5 μl of 2 μM core enzyme (Epicentre) was mixed with 1 μl of 3 μM σ70, followed by 3 μl (0.4 μg) of template DNA at room temperature and incubated at 37°C for 10 min to form an RNAP–promoter open complex. The transcription reaction was then made up to a final volume of 50 μl containing 40 mM Tris HCl (pH 7.9), 160-mM KCl, 10-mM MgCl2, 1-mM DTT, 5% glycerol, 10 μM each UTP and GTP, 0.2 mM ATP and 10 μCi of α-32P UTP (3000 Ci/mmol). The reaction was allowed to proceed at 37°C for 5 min to form the elongation complex stalled at position +26 (EC26). All following reactions were performed at room temperature. Rifampicin was added to 10 μM to prevent transcription reinitiation, 1 μl of 100-μM wild-type or mutant NusAN was then added to the reaction, followed by 2 μl of 5 mM CTP to make the final concentration 0.2 mM. Five microliter of reaction mixture was transferred into 2 μl of RNA gel loading buffer (95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol) at 30 s, 60 s, 90 s, 2 min, 5 min and 10 min. The samples were heated at 90°C for 30 s and run on an 8% denaturing polyacrylamide gel that was pre-run for at least 90 min at 50°C. The gel was dried at 60°C for 1.5 h and imaged using a Typhoon Trio+ imager (GE healthcare). The relative band intensity was determined using ImageJ software (Version 1.46; NIH) and the T1/2 was calculated as described (40).
RESULTS
Structure of NusAN and the RNAP β-flap
Prior to protein interaction and dynamics studies we used NMR to determine the structures of the B. subtilis RNAP β-flap (residues 784–923 of the RpoB subunit, equivalent to residues 830–1058 in E. coli RpoB) and the N-terminal domain of NusA (NusAN; residues 1–124). These proteins were highly amenable to NMR analysis; in particular the lack of the ∼100-residue βi9 insert in the B. subtilis β-flap fragment expedited the NMR analysis (∼16 versus ∼26 kDa).
The β-flap structure was determined using triple-resonance heteronuclear NMR experiments acquired predominantly using NUS and processed using maximum entropy reconstruction; NMR refinement statistics are shown in Table 1, and the final ensemble of 20 structures is shown in Figure 1A. The structure is similar to those previously described for RNAP (RMSD <4.0 Å when compared against 65 β-flap structures) where the FTH is connected to a long antiparallel β-sheet motif via two short β-strands (arms, Figure 1A).
Table 1. Statistical analysis of NMR structuresa.
| β-flap | NusAN | ||
|---|---|---|---|
| Experimental restraintsb | |||
| Interproton distance restraints | |||
| Intraresidue | 587 | 567 | |
| Sequential | 655 | 627 | |
| Medium range (i–j < 5) | 301 | 541 | |
| Long range (i–j > 53) | 607 | 672 | |
| Dihedral-angle restraints (f, y, c1) | 245 | 230 | |
| Total number of restraints per residue | 15.4 | 19.4 | |
| rms deviation from mean coordinate structure (Å) | |||
| Backbone atoms (residues 8–66 and 88–135) | 0.44 ± 0.09 | ||
| All heavy atoms (residues 8–66 and 88–135) | 1.05 ± 0.10 | ||
| Backbone atoms (residues 74–88) | 0.27 ± 0.10 | ||
| All heavy atoms (residues 74–88) | 0.90 ± 0.25 | ||
| rms deviation from mean coordinate structure (Å) | |||
| Backbone atoms (residues 4–118) | 0.51 ± 0.11 | ||
| All heavy atoms (residues 4–118) | 0.92 ± 0.10 | ||
| Stereochemical qualityc | |||
| Residues in most favoured Ramachandran region (%) | 93.9 ± 1.0 | 91.9 ± 1.6 | |
| Ramachandran outliers (%) | 0 ± 0 | 0.3 ± 0.5 | |
| Unfavourable sidechain rotamers (%) | 10.4 ± 1.6 | 11.0 ± 1.9 | |
| Clashscore, all atomsd | 0 ± 0 | 0.07 ± 0.1 | |
| Overall MolProbity score | 1.69 ± 0.08 | 1.77 ± 0.08 | |
| (90th percentile) | (86th percentile) | ||
aAll statistics are given as mean ± SD.
bOnly structurally relevant restraints, as defined by CYANA, are included.
cAs reported by MolProbity (http://molprobity.biochem.duke.edu).
dDefined as the number of steric overlaps >0.4 Å per thousand atoms.
Figure 1.
Structures of B. subtilis β-flap and NusAN. (A) Structure of β-flap shown as a chainbow from blue N-terminus to red C-terminus. FTH, β-flap tip helix. Arms, flexible regions lining the body of the β-flap to the FTH (see text for details). (B) NusAN shown as a chainbow from blue N-terminus to red C-terminus in two orientations. Bottom view is a rotation of the top view 90° into the page. The α-helices and β-strands are numbered in NusAN for reference in text.
The NMR-derived structure of NusAN revealed a bipartite fold with a globular domain dominated by three anti-parallel β-sheets and an α-helical bundle comprising helices H0, H1 and H3. The final ensemble of 20 structures is shown in Figure 1B and structural statistics are shown in Table 1. Crucially, detailed analysis of the NMR data revealed that NusAN forms more than one stable conformation, with the dominant form, representing ∼80% of the sample in solution, presented in Figure 1B. It was not possible to determine any alternative structures as they only represented ∼20% of the population, but residues that existed in two stable conformations could be identified. Significantly, the data indicate that the N-terminal domain of NusA is a flexible structure that is in dynamic equilibrium between two different conformations in solution (see below).
NusA binds to RNAP through interaction with the highly mobile β-FTH
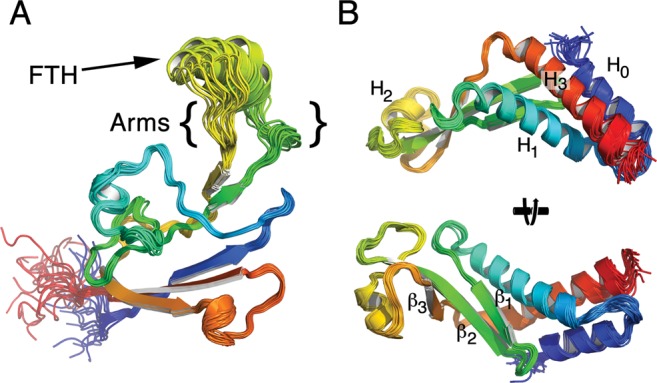
Our NMR-based approach allowed us to examine conformational flexibility in solution (Supplementary Information and Supplementary Figure S1). The FTH region is assumed to be flexible as it is often un-resolved in crystal structures, and indeed in our structure the position of the FTH could not be well defined. Examination of T1/T2 ratios that measure rotational and translational diffusion indicated that the FTH and arms were highly mobile with T1/T2 ratios <12 (Figure 2A and C). The fast rotational correlation times (low T1/T2 ratio) for this region could be an indication that it is unstructured, however, heteronuclear NOEs (Supplementary Figure S1A) and the random coil index (RCI S2; Figure 2B) revealed that most of the β-flap, including the FTH, is well structured. Therefore, the FTH is highly mobile due to flexibility in the arms around G850-E853 and G866-R870 (B. subtilis RpoB (β-subunit) coordinates; Figure 2B and arrows in Figure 2C) connecting it with the main body of the β-flap. This corresponds to E. coli β-subunit residues G891–Q894 and D912–S916. Such flexibility fits well with the known requirement for the RNA exit channel, of which the β-flap forms the outer wall, to accommodate up to three strands of RNA.
Figure 2.
The FTH is structured but highly mobile. (A) T1/T2 ratios that measure the rotational and translational diffusion rates plotted against residue number. (B) Random coil index (RCI)-based order parameters (S2) as predicted by Talos+ plotted against residue number. The disordered arm regions are shaded and the location of the FTH is indicated. (C) The FTH is structured but highly mobile due to flexible arms connecting it to the main body of the β-flap. T1/T2 ratios from panel (A) mapped onto the structure from red (low) to green (high) indicate that the FTH is highly mobile with respect to the rest of the β-flap. The mobility of the well-defined FTH is due to flexibility in the arms (black arrowheads) that connect it to the β-flap.
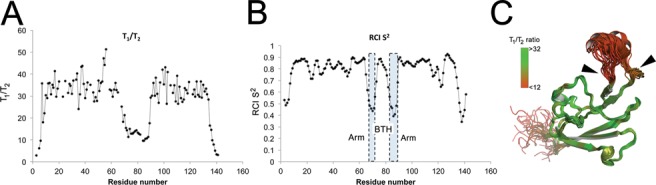
To determine the NusA binding site on RNAP, unlabelled NusAN was titrated against 15N-labelled β-flap (Figure 3A). In these experiments increasing amounts of unlabelled NusAN were added and changes in the position (shift) and intensity (quenching) of the 15N-labelled β-flap peaks were monitored (see solid line boxes in Figure 3A for examples). Shifts and/or quenching of peaks indicates a change in the chemical environment of that residue that can point to its involvement in a protein–protein interaction. The NMR data revealed that a relatively small number of residues were affected, indicative of a highly localized interaction between NusAN and the β-flap. Analysis of the NMR data indicated that the interaction was concentrated at the FTH (Figure 3B), and this information was used to guide in vitro mutagenesis for functional studies using E. coli proteins. ELISA-based protein interaction assays indicated that residues L895, E899, K900, R903 and E908 (E. coli β subunit) are required for interaction with NusAN (Figure 3B). These results were confirmed using transcription pause assays with recombinant RNAP carrying alanine substitutions at E898, E899, R903 and I905 in the FTH. Based on the results of the protein binding assays shown in Figure 3C, changes at E899 and R903 would be expected to reduce/abolish NusAN binding, whereas changes at E898, or I905 which is implicated in binding to initiation factor σ region 4 (41), would not. As shown in Figure 3D, transcription pause activity was only affected by mutations at positions E899 and R903, indicating that NusAN binding is required for transcription pausing and that its binding site does not overlap with that of σ region 4, although binding of the proteins is mutually exclusive due to steric hindrance. Mapping on the structure of E. coli RNAP indicated that the residues required for interaction with NusAN reside on the outside edge of, and the arms adjacent to, the FTH (Figure 3E and F), the most mobile region of the β-flap.
Figure 3.
NusAN binds to the outside edge of the mobile FTH. (A) Overlay of 15N HSQC spectra of β-flap in the presence of increasing concentrations of unlabelled NusAN (red = control, green = 1:0.1, blue = 1:0.5, black = 1:1). Lines indicate glutamine or asparagine sidechain NH2 groups and asterisks indicate arginine sidechain NH groups. (B) Schematic of the structure of β-flap colour coded according to the chemical shift perturbations observed upon titration with NusAN (brown, no perturbation; grey, cyan, yellow and magenta, increasing levels of perturbation). (C, D) Data for binding of NusAN to β-flap with residue changes at the positions indicated along the x-axis (E. coli β subunit numbering) determined by ELISA (C) and transcription pause assay (D). Binding strength or pause activity is relative to wild-type β-flap (WT, 100%). Results are from triplicate samples ± SD. (E, F) NusAN binding data mapped onto E. coli RNAP ((E), modified from PDB 4IGC) and zoomed onto the FTH (F). The location of the FTH is indicated by a dotted circle in (E) and the arrow indicates the position of the view in (F). α, β, β′ subunits are shown in dark, mid and light grey, respectively; ω-subunit, white. Relative contribution of residues to binding is colour coded from most (magenta) to least (cyan) important.
NusAN is a dynamic structure that exists in more than one conformation in solution
Determination of the NMR structure of NusAN was complicated by the presence of line broadening in some spectral regions and the fact that it adopted at least two different conformations. This is shown in Figure 4A where a portion of the 2D 1H/15N spectrum has been enlarged. For some residues peaks can be clearly identified for a major (black) and minor (red) conformation (e.g. I56, see magenta and cyan asterisks, respectively, Figure 4A). Residues most affected by changes in conformation were identified based on the differences in their chemical shifts in the two forms (see Supplementary Table S3) and normalized 1H/15N chemical shift changes (Δδ) are plotted for these residues in Figure 4B. These residues mapped almost exclusively to the α-helical bundle and residues within the hydrophobic core (Figure 4B). There were also three distinct regions at the ends of H0, H1 and H3 that displayed resonance broadening beyond detection (indicative of high disorder), suggesting that the entire helix bundle can undergo movement independent of the rest of the molecule and that the helices themselves are not rigidly held together (magenta arrowheads, Figure 4C).
Figure 4.
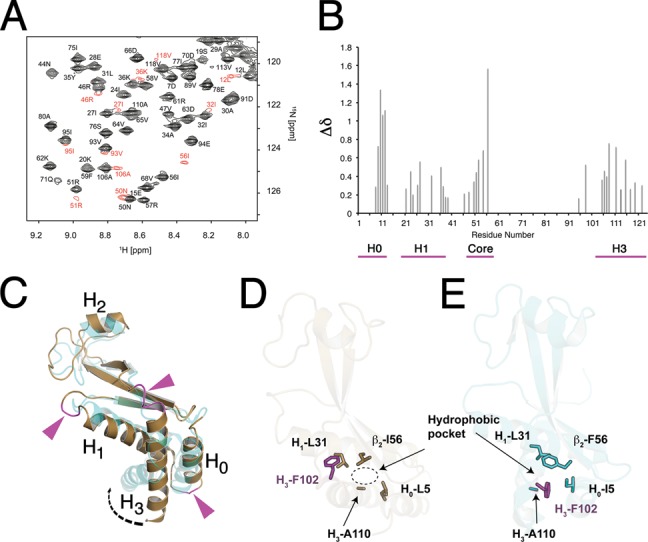
NusAN is in equilibrium between a major and minor structural form. (A) A section of the HSQC spectrum of NusAN is shown with residues that exhibit a large difference in chemical shift in the minor conformation coloured in red. Note the particularly large chemical shift for I56 in the major (magenta asterisk) and minor (cyan asterisk) conformations. (B) Δδ plots based on chemical shift changes highlighting residues present in multiple conformations in B. subtilis NusAN plotted against residue number. Magenta line below indicates residues in the core and helical bundle in multiple conformations. (C) Superposition of the structure of B. subtilis (bronze) and E. coli (PDB 2KWP, cyan) NusAN showing how alternative conformations could form due to flexibility in regions of signal broadening (magenta sections and arrows) connecting the α-helices to the rest of the molecule. α-Helices are labelled from H0 to H3. (D, E) Identification of a hydrophobic pocket and rotation of F102 (magenta) around residue 56 in B. subtilis ((D); bronze) and E. coli ((E); cyan). NusAN structures.
Comparison of all available NusA structures (NusAN and full length NusA) indicates that H3 adopts one of the two positions with respect to the remainder of the N-terminal domain (Supplementary Figure S2) (42,43). Thus, the C-terminal helix of NusAN can adopt two different conformations that presumably exist naturally due to inherent flexibility of the α-helical bundle in NusAN. This is particularly evident when comparing our structure with that from E. coli, which showed a significant translation of H3 and minor translation of H0 and H1 relative to the B. subtilis structure (dashed arrow, Figure 4C). This can be quantified by comparison of the RMSD values in a structural alignment of the two molecules. In the presence of H3 the RMSD between the two structures is relatively large (4.0 Å), but exclusion of this helix improves the RMSD to 2.6 Å, highlighting the large conformational change in H3 (Supplementary Table S4).
The dramatic change in chemical shift seen for residue I56 (a residue that is conserved with respect to having a hydrophobic sidechain) is consistent with proximity to an aromatic side chain, where ring current effects could significantly alter chemical shifts of nearby residues in a distance- and orientation-dependent manner. The close proximity of F102 located at the edge of H3 would suggest a reorientation of F102 with respect to I56, consistent with a repositioning of H3. Examination of the B. subtilis NusAN structure reveals a hydrophobic pocket near the head of H3 (dotted circle, Figure 4D), and the same region is occupied by F102 in E. coli NusAN (Figure 4E). This suggests that rotation of F102 around I56 would: (1) lead to a local unwinding at the head of H3; (2) cause a translation of H3 to a position consistent with that observed for E. coli and P. limnophilus NusAN; and (3) allow F102 to stabilize this conformation through occupation of the hydrophobic pocket and formation of alternative hydrophobic interactions (Figure 4D and E).
Combined, the NMR chemical shift, line broadening and structure comparison data indicate that the α-helical bundle is mobile with respect to the rest of NusAN. To visualize the conformational change a homology model of NusAN from B. subtilis was created based on the reported E. coli NusAN structure. The change in structure required to morph between the two different conformational states provides a model that explains the chemical shift changes observed in the minor state of B. subtilis, and a structural basis for the conformational plasticity of H3 in different NusAN structures (Supplementary Movie 1). We therefore propose that in solution NusAN exists in two alternate conformations, one consistent with our reported structure (conformation I) and the other similar to that reported for NusAN from E. coli (conformation II) (Supplementary Figure S2).
Binding to RNAP induces conformational change in NusA
In stark contrast to the reciprocal experiment to identify the NusAN binding site on the β-flap (above), titration of 15N-labelled NusAN with unlabelled β-flap led to extensive line broadening for ∼65% of residues (Figure 5A, yellow and cyan Figure 5B top). Strikingly, most of the broadened residues correspond to those that adopt different conformations in solution (magenta sticks, Figure 5B top) indicating that NusAN undergoes a conformational rearrangement upon binding the FTH that involves the α-helical bundle and hydrophobic core. Extensive peak broadening in the β-sheet region was also observed exclusively in the titration experiments, suggesting that this region may be directly involved in interaction with the FTH (cyan on NusAN; Figure 5B). We performed extensive mutagenesis of NusAN (Figure 5C), but the only residues found by ELISA to play a major role in the interaction with the β-flap were F59, R61, E94 and Q96 on β-strands 2 and 3 of NusAN (Figure 5D). When mapped onto the NusAN structure, residue changes that resulted in ≤40% binding activity formed a contiguous patch across the edge of the cleft (Figure 5B, bottom). These results were confirmed in transcription pause assays using a 100-fold molar excess of NusAN over RNAP (Figure 5E). The fact that transcription pause activity was ≤20% for the R61A and Q96A mutants suggests that these two NusAN residues are particularly important for interaction with the FTH. Overall, these results show that interaction between NusAN and the FTH is largely driven by charged residues that place NusAN on the outside edge of the RNA exit channel and that this interaction stimulates conformational rearrangement in the NusAN α-helical bundle and hydrophobic core.
Figure 5.
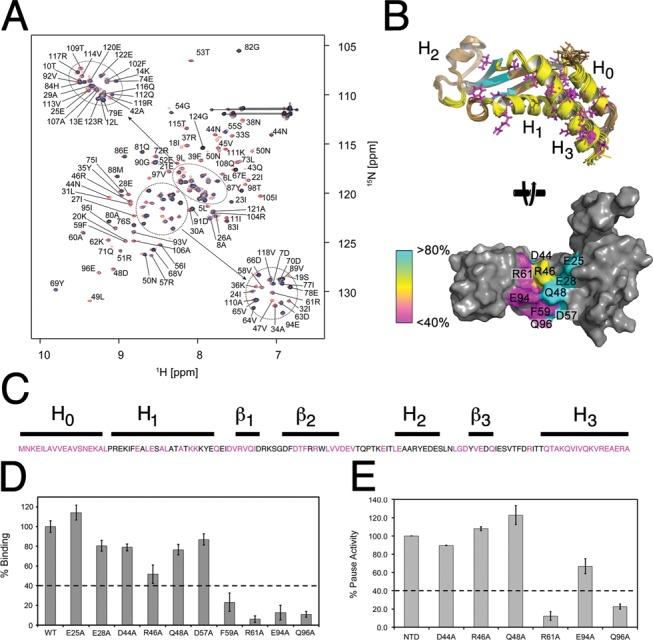
Identification of the FTH binding site on NusAN. (A) Overlay of 15N HSQC spectra of NusAN in the presence of increasing concentrations of unlabelled β-flap (red = control, blue = 1:0.2 and black = 1:0.4). Lines indicate glutamine or asparagine sidechain NH2 groups. (B) Top: B. subtilis NusAN with residues in more than one conformation shown in magenta, and those affected by binding to the β-flap shown in yellow (helical bundle and core) and cyan (β-sheet domain). Bottom: E. coli NusAN rotated 90° from the top view to show residues involved in interaction with the β-flap. Magenta, yellow and cyan colouring denotes importance from most to least important. (C) Sequence of E. coli NusAN with residues altered for binding/transcription pause assays in magenta. Lines above are labelled corresponding to secondary structure. (D, E) Data for binding of β-flap to NusAN with residue changes at the positions indicated along the x-axis (E. coli NusAN numbering) determined by ELISA (D) and transcription pause assay (E). Binding strength or pause activity is relative to wild-type NusAN (WT, 100%). Results are from triplicate samples ± SD.
Highly conserved residues in a conformationally flexible region of NusAN are essential for transcriptional pause activity
Comparison of NusAN sequences from a wide range of clinically and industrially important bacteria showed that sequence conservation is relatively low, even at the locations of residues shown to be required for interaction with the highly conserved FTH (Supplementary Figure S3), consistent with the relatively low binding affinities reported for the NusA–RNAP interaction (16,44). Strikingly, however, sequence conservation is high in the C-terminal portion encompassing H3 (Supplementary Figure S3). This helix contains a series of highly conserved basic residues that, along with K36 and K37 in H1, form part of a basic patch that extends into the C-terminal S1, KH1 and KH2 domains of NusA that have been assigned a potential role in NusA function through interaction with the RNA transcript (45). Using our mutagenesis data that identified the residues involved in interaction between NusAN and the FTH, coupled with previous data obtained from low resolution EM and chemical crosslinking studies, we performed in silico docking onto the structure of RNAP in the elemental pause conformation (46) to produce a model of the complex (16,18). This docking indicated that NusAN must localize over the FTH so that β-strands 2 and 3 are adjacent to the outside edge of the FTH, resulting in the basic patch (magenta) lying proximal to the RNA exit channel in close proximity to the RNA pause hairpin (yellow, Figure 6A). Using this model we then investigated the importance of this basic patch that principally lies within the mobile H3 region of NusAN on transcriptional pausing.
Figure 6.
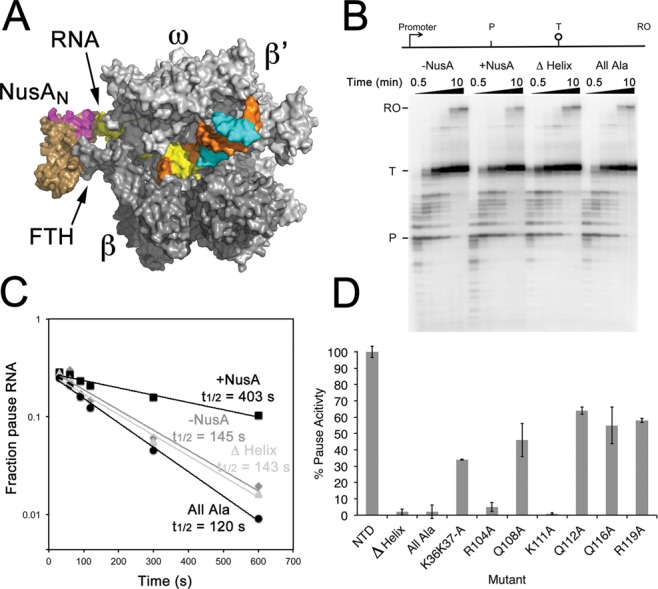
Identification of residues in the mobile H3 region of NusAN required for transcription pause enhancement. (A) Model of NusAN (bronze) bound to RNAP (grey) over the β-FTH (FTH). RNA within the exit channel is shown in yellow and NusAN residues that form the basic patch are shown in magenta. Template DNA strand (orange), non-template strand (cyan). (B–D) Measurement of transcription pause half lives. (B) Transcription pause assays with a schematic of the template shown above the gel. P, his pause site; T, termination site; RO, run-off (when RNAP falls off the end of template DNA). Wedges above gel indicate the relative time points at which samples were taken from 0.5 to 10 min (0.5, 1, 1.5, 2, 5 and 10 min). (C) Determination of transcription pause half-life (t1/2) using data from (B). (D) Half lives for NusAN constructs with altered basic patch residues. Half lives were determined in triplicate and presented as% pause activity relative to wild-type ± SD. Δ Helix, H3 deleted; all Ala, residues K36, K37, R104, Q108, K111, Q112, Q116, R119 altered to alanine. Other amino acid changes are shown below their respective columns.
Since RNA duplex/hairpin formation is important for NusA-stimulated pause activity (16,17) we hypothesized that residues in the basic patch that our modelling shows to be in close proximity to the RNA exit channel may be functionally required for stabilization of pause hairpins. Alteration to alanine of K36 and K37 on H1 along with the basic residues on H3, as well as deletion of H3, showed little effect on the ability of NusAN to bind RNAP (Supplementary Table S5). Therefore, these highly conserved basic residues and H3 have no role in NusAN binding to the β-flap of RNAP. Crucially, however, deletion of H3 or alteration of the NusAN basic patch resulted in total loss of transcription pause activity (Figure 6B–D), indicating that H3 is functionally essential as it confers pause activity on NusAN. Closer examination of the contribution of individual amino acids to transcriptional pausing showed that most had little effect, but alteration of K36 and K37 on H1 resulted in a reduction of pause activity to ∼40% wild-type level indicating that they can contribute to the process (Figure 6D; Supplementary Figure S4). Strikingly, however, changing either R104 or K111 to alanine led to total loss of activity (Figure 6D; Supplementary Figure S4), showing that these highly conserved residues are essential for pause activity and are non-redundant. Thus, the basic patch and, in particular, R104 and K111 are required for RNA hairpin-dependent (class I) transcriptional pause activity.
DISCUSSION
We have shown that formation of a transcription pause complex involves the interaction between two highly flexible components of the transcription machinery, the FTH and NusAN. Furthermore, this interaction places key basic residues in close proximity to the RNA pause hairpin in the exit channel, which provides a mechanism for NusA-stimulated pausing that is outlined below.
The NMR titration experiments showed that the binding surface of the β-flap is localized to the relatively small FTH region. By contrast, reciprocal experiments revealed a significant perturbation of the majority of the NusAN signals, spanning a much larger surface. Line broadening of residues far from the binding site of NusAN shows that the binding event allosterically modulates the chemical environment of these distal residues. The line broadening was observed for both the major and minor conformations and indicated that both conformations of NusA bind the β-flap with the same affinity, which is consistent with the binding site being in a non-flexible region of NusA. However, as the resonances of both conformations experience line broadening it suggests that there is an increase in the rate of interconversion between conformations I and II. This would increase the probability of a particular RNA motif to encounter conformation II, once the NusAN/β-flap complex is formed. The data, therefore, suggest that the β-flap is an allosteric modulator of NusAN, where the primary binding site, which serves to localize NusAN to RNAP, also leads to changes in the distal RNA binding region (H3) of NusAN.
Intriguingly, the transition of NusAN from conformation I to II has a dramatic effect on the conformation of two invariant basic residues R104 and K111, which are essential for pause activity (Figure 6B). In conformation I (consistent with the structure shown in Figure 1B) the distance between the charges at the end of the sidechains is ∼9–15 Å, whereas in the alternative structure (conformation II—consistent with E. coli NusAN) they are 15–23 Å apart, providing a separation range of 9–23 Å between the two states, which could allow for dynamic interaction with the emerging RNA transcript (Figure 7). The allosteric conformational change induced in NusAN upon binding to the FTH causes the separation between R104 and K111 to increase, leading to an enhanced ability to interact with the phosphodiester backbone of the RNA duplex (Figure 7B). This interaction stochastically stabilizes the pause hairpin, increasing ∼3-fold the average time RNAP remains in a pause conformation (Figure 6B–D) (16,46,47).
Figure 7.
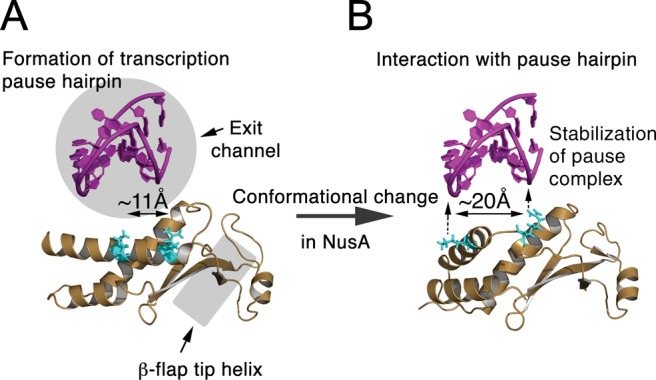
Conformational change in NusAN leads to transcription pause enhancement. (A) NusAN in complex with the β-flap tip helix (grey oblong) adjacent to a transcription pause hairpin (magenta) that has formed in the exit channel of RNAP (grey circle) viewed from a similar position to that shown in Figure 3F. R104 and K111 that are required for transcription pause enhancement are shown in cyan. (B) Following an FTH-induced conformational change, the spacing of R104 and K111 increases to ∼20 Å, thereby permitting interaction with the phosphodiester backbone of the pause hairpin and stabilization of the pause event.
NusA is known to associate with the bulk of transcribing RNAP in both B. subtilis and E. coli, and it is able to bind early in the elongation phase, prior to full dissociation of the initiation factor σ (18,36,48). When RNAP reaches a pause region, it enters the elemental pause conformation that involves opening of the DNA-binding jaws and reduction in the number of contacts made with the DNA (46). In turn, opening of the jaws leads to loss of interaction with the FTH that permits opening of the RNA exit channel (47). The loss of constraints on the FTH facilitates pause hairpin formation within the exit channel. Formation of the pause hairpin would permit interaction of R104 and K111 of NusAN with the phosphodiester backbone of the RNA, increasing the duration of the transcriptional pause. Pausing is a stochastic process and not all RNAP molecules that reach a pause site do pause, even when in complex with full-length NusA or NusAN (16,49) (Figure 6B–D). Allosteric control of the conformational change in NusAN by RNAP appears to be on the same timescale as the rate of transcription, so that upon binding, the rate of NusAN interconversion is increased to the low ms range, similar to the rate of transcription (20 ms/nt at a rate of 50 nt/s), increasing the probability of interaction with the pause hairpin as it forms, accounting for the increase in average pause time for RNAP in complex with NusA. Therefore, regulation of transcription by NusA involves interaction with the emerging transcript and is responsive to allosteric conformational change induced by interaction with the β-flap tip of RNAP, which is itself highly mobile. Such plasticity of the spatial location of these key regulatory elements helps provide the exquisite modulation of transcriptional activity needed to permit cells to respond appropriately to changing circumstances.
ACCESSION NUMBERS
Protein structures and NMR data have been deposited in the PDB/BioMagResBank with accession numbers 2LY7/18712 (β-flap) and 2MT4/25148 (NusAN).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors wish to thank Irina Artsimovitch for the supply plasmids for use in transcription pause assays, and Rick Lewis, Nick Dixon and Bostjan Kobe for their helpful comments on the manuscript.
Footnotes
These authors contributed equally to the paper as first authors.
FUNDING
Australian Research Council [DP110100190 to P.L.]. Funding for open access charge: Australian Research Council [DP110100190 to P.L.]; Australian Research Council Fellowships [FT110100925 to M.M., DP110100190 to X.Y.]; National Health and Medical Research Council Principal Research Fellowship [APP1044414 to G.F.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Sims R.J., III, Belotserkovskaya R., Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 2.Alexander R.D., Innocente S.A., Barrass J.D., Beggs J.D. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrillo Oesterreich F., Preibisch S., Neugebauer K.M. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Saunders A., Core L.J., Sutcliffe C., Lis J.T., Ashe H.L. Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes Dev. 2013;27:1146–1158. doi: 10.1101/gad.215459.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaukowitch K., Joo J.Y., Liu X., Watts J.K., Martinez C., Kim T.K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagano J.M., Kwak H., Waters C.T., Sprouse R.O., White B.S., Ozer A., Szeto K., Shalloway D., Craighead H.G., Lis J.T. Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet. 2014;10:e1004090. doi: 10.1371/journal.pgen.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tome J.M., Ozer A., Pagano J.M., Gheba D., Schroth G.P., Lis J.T. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat. Methods. 2014;11:683–688. doi: 10.1038/nmeth.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gusarov I., Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 9.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 10.Epshtein V., Kamarthapu V., McGary K., Svetlov V., Ueberheide B., Proshkin S., Mironov A., Nudler E. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artsimovitch I., Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnham P.J., Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981;9:563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnham P.J., Greenblatt J., Platt T. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 1982;29:945–951. doi: 10.1016/0092-8674(82)90457-3. [DOI] [PubMed] [Google Scholar]
- 15.Lau L.F., Roberts J.W., Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J. Biol. Chem. 1983;258:9391–9397. [PubMed] [Google Scholar]
- 16.Ha K.S., Toulokhonov I., Vassylyev D.G., Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 2010;401:708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolb K.E., Hein P.P., Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J. Biol. Chem. 2014;289:1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X., Molimau S., Doherty G.P., Johnston E.B., Marles-Wright J., Rothnagel R., Hankamer B., Lewis R.J., Lewis P.J. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 2009;10:997–1002. doi: 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng C., Friedman D.I. Reduced Rho-dependent transcription termination permits NusA-independent growth of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7543–7547. doi: 10.1073/pnas.91.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Ma C., Lewis P. A vector system that allows simple generation of mutant Escherichia coli RNA polymerase. Plasmid. 2014;75:37–41. doi: 10.1016/j.plasmid.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Marley J., Lu M., Bracken C. A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 22.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Yang X., Lewis P.J. Overproduction and purification of recombinant Bacillus subtilis RNA polymerase. Protein. Expr. Purif. 2008;59:86–93. doi: 10.1016/j.pep.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Ma C., Yang X., Kandemir H., Mielczarek M., Johnston E.B., Griffith R., Kumar N., Lewis P.J. Inhibitors of bacterial transcription initiation complex formation. ACS Chem. Biol. 2013;8:1972–1980. doi: 10.1021/cb400231p. [DOI] [PubMed] [Google Scholar]
- 25.Mobli M., Stern A.S., Bermel W., King G.F., Hoch J.C. A non-uniformly sampled 4D HCC(CO)NH-TOCSY experiment processed using maximum entropy for rapid protein sidechain assignment. J. Magn. Reson. 2010;204:160–164. doi: 10.1016/j.jmr.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mobli M., Maciejewski M.W., Gryk M.R., Hoch J.C. An automated tool for maximum entropy reconstruction of biomolecular NMR spectra. Nat. Methods. 2007;4:467–468. doi: 10.1038/nmeth0607-467. [DOI] [PubMed] [Google Scholar]
- 27.Mobli M., Maciejewski M.W., Gryk M.R., Hoch J.C. Automatic maximum entropy spectral reconstruction in NMR. J. Biomol. NMR. 2007;39:133–139. doi: 10.1007/s10858-007-9180-8. [DOI] [PubMed] [Google Scholar]
- 28.Bartels C., Xia T.H., Billeter M., Guntert P., Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 29.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann T., Guntert P., Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 31.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 32.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 33.d'Auvergne E.J., Gooley P.R. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR. 2008;40:107–119. doi: 10.1007/s10858-007-9214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.de Vries S.J., van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 36.Davies K.M., Dedman A.J., van Horck S., Lewis P.J. The NusA:RNA polymerase ratio is increased at sites of rRNA synthesis in Bacillus subtilis. Mol. Microbiol. 2005;57:366–379. doi: 10.1111/j.1365-2958.2005.04469.x. [DOI] [PubMed] [Google Scholar]
- 37.Johnston E.B., Lewis P.J., Griffith R. The interaction of Bacillus subtilis sigmaA with RNA polymerase. Protein Sci. 2009;18:2287–2297. doi: 10.1002/pro.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Artsimovitch I., Svetlov V., Anthony L., Burgess R.R., Landick R. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J. Bacteriol. 2000;182:6027–6035. doi: 10.1128/jb.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artsimovitch I., Henkin T.M. In vitro approaches to analysis of transcription termination. Methods. 2009;47:37–43. doi: 10.1016/j.ymeth.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landick R., Wang D., Chan C.L. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 41.Vassylyev D.G., Sekine S., Laptenko O., Lee J., Vassylyeva M.N., Borukhov S., Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 42.Borukhov S., Lee J., Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 2005;55:1315–1324. doi: 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- 43.Shin D.H., Nguyen H.H., Jancarik J., Yokota H., Kim R., Kim S.H. Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain. Biochemistry. 2003;42:13429–13437. doi: 10.1021/bi035118h. [DOI] [PubMed] [Google Scholar]
- 44.Gill S.C., Weitzel S.E., von Hippel P.H. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J. Mol. Biol. 1991;220:307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 45.Worbs M., Bourenkov G.P., Bartunik H.D., Huber R., Wahl M.C. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- 46.Weixlbaumer A., Leon K., Landick R., Darst S.A. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hein P.P., Kolb K.E., Windgassen T., Bellecourt M.J., Darst S.A., Mooney R.A., Landick R. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat. Struct. Mol. Biol. 2014;21:794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mooney R.A., Davis S.E., Peters J.M., Rowland J.L., Ansari A.Z., Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J., Ha K.S., La Porta A., Landick R., Block S.M. Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA. Mol. Cell. 2011;44:635–646. doi: 10.1016/j.molcel.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


